Abstract
It has been postulated that multiple-marker methods may have added ability, over single-marker methods, to detect genetic variants associated with disease. The Wellcome Trust Case Control Consortium (WTCCC) provided the first successful large genome-wide association studies (GWAS) which included single-marker association analyses for seven common complex diseases. Of those signals detected, only one was associated with coronary artery disease (CAD), and none were identified for hypertension (HTN). Our objective was to find additional genetic associations and pathways for cardiovascular disease by examining the WTCCC data for variants associated with CAD and HTN using two-marker testing methods. We applied two-marker association testing to the WTCCC dataset, which includes ~2,000 affected individuals with each disorder, and a shared pool of ~3,000 controls, all genotyped using Affymetrix GeneChip 500 K arrays. For CAD, we detected single nucleotide polymorphisms (SNP) pairs in three genes showing genome-wide significance: HFE2, STK32B, and DIPC2. The most notable SNP pairs in a non-protein-coding region were at 9p21, a known major CAD-associated region. For HTN, we detected SNP pairs in five genes: GPR39, XRCC4, MYO6, ZFAT, and MACROD2. Four further associated SNP pair regions were at least 70 kb from any known gene. We have shown that novel, multiple-marker, statistical methods can be of use in finding variants in GWAS. We describe many new, associated variants for both CAD and HTN and describe their known genetic mechanisms.
Introduction
In spite of the recent success of genome-wide association studies (GWAS) in searching for common or rare variants of complex diseases, it has been observed that the genetic variants identified through GWAS have accounted for only a small proportion of the presumed genotypic variation and hence, many variants remain to be discovered (McCarthy et al. 2008).
The SNP chips used in GWAS provide a good coverage of the human genome by genotyping hundreds of thousands of SNPs at a time. Although the coverage provided for common variants is good, it is still unclear exactly how many SNPs are in the genome. Therefore, it is more likely that the variants of interest will not be directly observed and it is important to develop the methods of searching for variants without typing the variant directly.
Two categories of tests have been applied to search for the disease association in GWAS: single-marker association tests, which examine a single SNP at a time and multiple-marker tests, which examine multiple SNPs simultaneously (Figs. 1, 2). The Wellcome Trust Case Control Consortium (WTCCC) provided the first successful large comprehensive GWAS, performing single-marker association data analysis for seven complex diseases: bipolar disease (BD), Crohn’s disease (CD), rheumatoid arthritis (RA), type 1 diabetes (T1D), type 2 diabetes (T2D), CAD, and HTN—with 2,000 cases for each disease and 3,000 shared common controls (Wellcome Trust Case Control Consortium 2007). Twenty-four independent association signals had a genome-wide significance of P <5 × 10−7, many of which have been replicated in the later independent studies (Wellcome Trust Case Control Consortium 2007, Zhu et al. 2010, Browning and Browning 2008). Of those signals detected in the original WTCCC, only one was associated with CAD, none were identified for HTN (Wellcome Trust Case Control Consortium 2007).
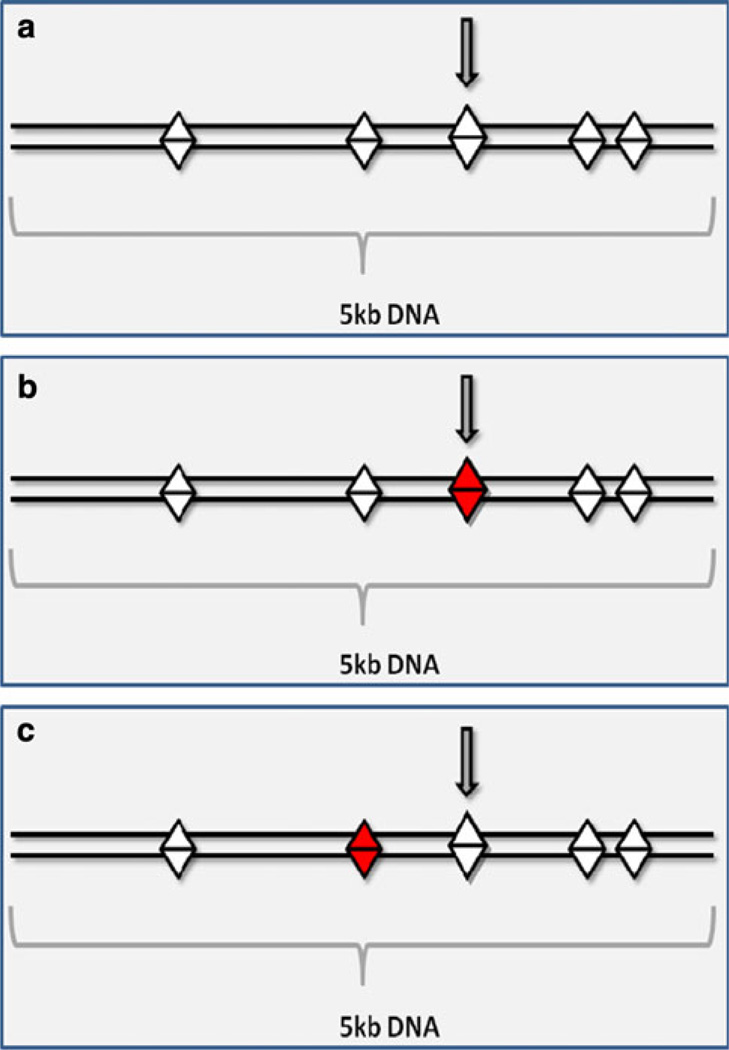
Fig. 1.
Hypothetical SNP chip typing for single-marker method. Straight lines depict a pair of chromosomes. SNP positions are represented by the diamonds. a shows a normal tagged SNP (pointed at by arrow), b and c show the disease scenarios. Ideally, the disease variant of interest (shaded diamond) would be directly typed, as in b; however, as current platforms likely only type a small portion of possible SNPs, it is more likely that the disease causing SNP will not be tagged directly, as in c
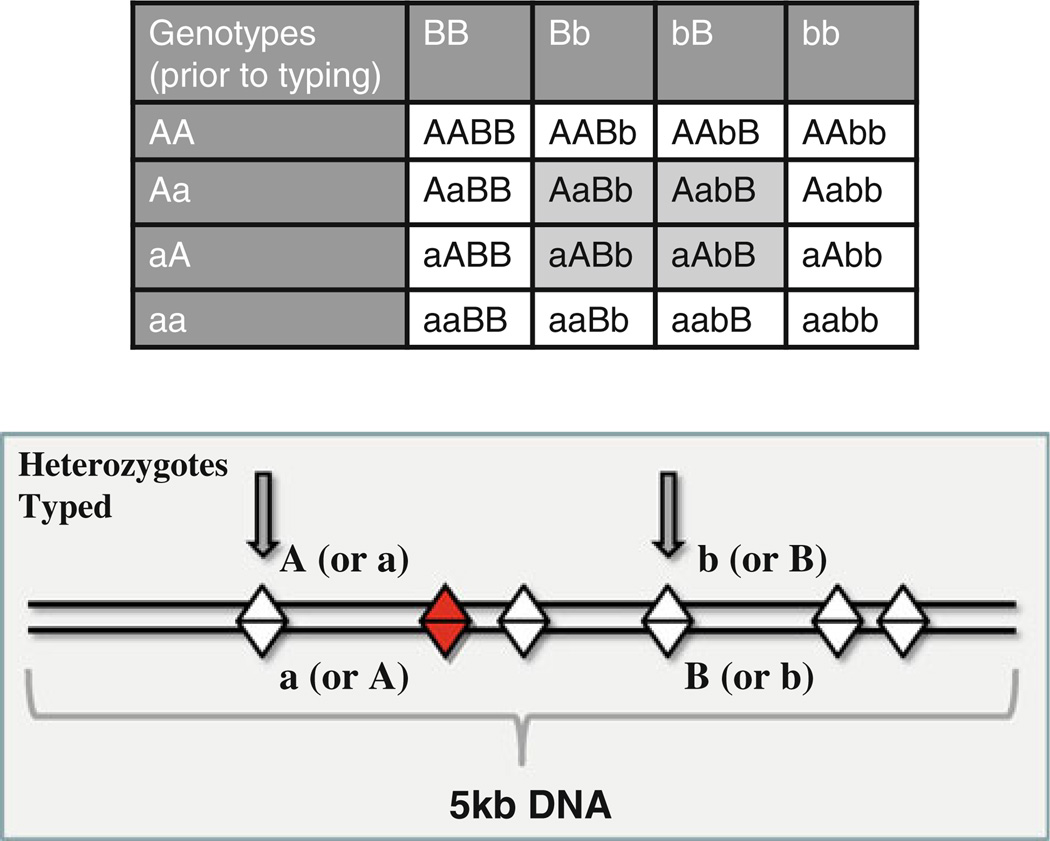
Fig. 2.
Hypothetical SNP chip typing using a two-marker method. Prior to typing, phased diplotypes for the AA, Aa, or aa genotypes and BB, Bb, or bb genotypes could include sixteen possibilities. However, only the four heterozygous combinations (light gray shading) are informative about the phase of the diplotype. The reduction in possible diplotypes when the flanking markers are both heterozygous makes the information gained more powerful. This concept is also depicted in the bottom figure, where straight lines depict a pair of chromosomes, diamonds indicate hypothetical SNP positions along 5 kb of DNA (horizontal lines), the shaded diamond indicates the disease SNP of interest, and arrows indicate the SNPs typed by the chip platform. Thus, when the markers are close enough together so that they are in linkage disequilibrium, two-marker association testing methods have increased ability to find non-typed variants: both SNPs near the disease causing variant combine to reach genome-wide significance
One possible explanation for the limited results for CAD and HTN may have been due to lack of statistical power provided by single-marker analysis to detect the disease-susceptibility variants. The purpose of this study is to find additional genetic associations and pathways for cardiovascular disease by examining the WTCCC data for variants associated with CAD and HTN using two-marker testing methods as described byKim et al. (2010). We show that these novel statistical methods can be of use in finding variants of genome-wide significance for complex diseases. We describe multiple, new, disease associations for both CAD and HTN and describe the known genetic mechanisms that may be relevant.
Methods
Detailed description of the study samples can be found in the original WTCCC GWAS paper (Wellcome Trust Case Control Consortium 2007). In brief, the WTCCC dataset includes seven major complex diseases: BD, CD, RA, T1D, T2D, CAD, and HTN; each has ~2,000 affected individuals and a shared pool of ~3,000 controls (which consist of a 1958 Birth Cohort and a recently recruited UK Blood Service sample). The majority of subjects were of European ancestry. All the individuals were genotyped using Affymetrix GeneChip 500K arrays. We downloaded the genotype data called with the algorithm CHIAMO only for the CAD and HTN disease cases and the shared controls from the WTCCC website.
CAD phenotype description
CAD cases had a validated history of myocardial infarction, coronary artery bypass surgery, and/or percutaneous coronary angioplasty prior to their 66th birthday. Verification was required either from hospital records or from a primary care physician. More details about the sample collection and ascertainment can be found in the original WTCCC GWAS paper (Wellcome Trust Case Control Consortium 2007).
HT phenotype description
Cases of HTN had a history of HTN diagnosed before 60 years of age, with blood pressure (BP) levels of 150/100 mmHg (if based on a single reading), or if the mean of three readings was >145/95 mmHg. Hypertensive individuals were excluded if they: self-reportedly consumed >21 units of alcohol per week, had diabetes, had intrinsic renal disease, or had a history of secondary hypertension or other co-morbid condition. Cases were not screened for rare, known monogenic causes of HTN. Recruitment was focused on individuals with a body mass index (BMI) under 30 kg/m2. More details in the sample collection and ascertainment can be found in the original WTCCC GWAS paper (Wellcome Trust Case Control Consortium 2007).
Quality controls
The majority of subjects had at least one sibling also affected, whether by CAD or HTN, but only one subject from each family was included in the ~2,000 cases. Individuals were further dropped in the WTCCC study because of evidence of non-European ancestry or poor call rates (Wellcome Trust Case Control Consortium 2007). We applied the following criteria to call SNPs: (1) CHIAMO probability >0.99; (2) Hardy–Weinberg Equilibrium exact test P value >5.7 × 10−7 in controls; (3) allele frequency difference test based on 1 degree of freedom (df) trend test P value >5.7 × 10−7, or genotype frequency difference based on 2 df general test >5.7 × 10−7, between the two control groups (the 1958) Birth Cohort and the recently recruited UK Blood Service sample); (4) missing genotype proportions <1% and (5) minor allele frequencies >1%. We dropped SNPs with bad genotype calling, as suggested in the original WTCCC analysis (Wellcome Trust Case Control Consortium 2007). This resulted in a total of 407,576 SNPs that were used in this genome-wide scan for CAD and 405,022 SNPs for HTN.
Association test methods
As the WTCCC used the Cochran–Armitage trend test (and not a logistic regression model) for single-marker analysis, we first performed a single-marker logistic regression test on the entire dataset as inKim et al. (2010) on the CAD and HTN WTCCC data (supplemental material, Table 1). The model is log (μ/1 − μ) = βo + βxXi, where μ/1 − μ is the odds of being affected, βo is the intercept, βx is the additive effect of SNP X on the logit scale and Xi is the value of the i-th person’s SNP coded as the number of minor alleles in person i’s genotype. In large enough samples, this test should be identical to the trend test used by the WTCCC, and indeed gave essentially similar results in this sample.
We then applied two-marker analysis with the 2 df and 3 df models as described inKim et al. (2010). Test 2–2 (2 df) jointly tests allelic contrasts for two neighboring SNPs (SNP X and SNP Y), and test 2–3 (3 df) additionally tests the product of the two additively coded SNPs (i.e. the term XiYi). Thus, the model for test 2–2 is log (μ/1 − μ) = βo + βxXi + βyYi and that for test 2–3 is log (μ/1 − μ) = βo + βxXi + βyYi + βxyXiYi. Under this latter model, we can test the coefficient βxy, and hence determine whether including this product term significantly helps us to detect associations. Note that, since we only included consecutive adjacent SNP pairs [(SNP1, SNP2), (SNP2, SNP3), (SNP3, SNP4), etc.] in the model, in each case we performed slightly fewer tests in the two-marker analyses than in the single-marker analysis. We further included as covariates in the model the first ten principal components of all the SNPs, cases and controls combined, which can control for any population structure (Zhu et al. 2008, 2002).
Using the SNP map annotations provided by Affymetrix 5.0 GeneChip as a reference (https://www.affymetrix.com/support/technical/annotationfilesmain.affx), we evaluated all SNPs with a P value <5 × 10−7, the same as used by the WTCCC. As seen in the results, tests 2–2 and 2–3 are highly correlated; therefore, performing both tests does not double the number of independent tests performed. Furthermore, since the tests, whether test 2–2 or tests 2–3, are no longer independent, but positively correlated [e.g., the tests for (SNP1, SNP2), and (SNP2, SNP3) both include testing SNP2], the effective number of independent tests was smaller. If the SNP was in a known gene, the gene was evaluated through published cardiovascular literature for a possible pathogenic link. If SNP pairs were located between two neighboring genes, we mapped them to the nearest gene and also evaluated the locus for possible published cardiovascular pathogenic links.
Results
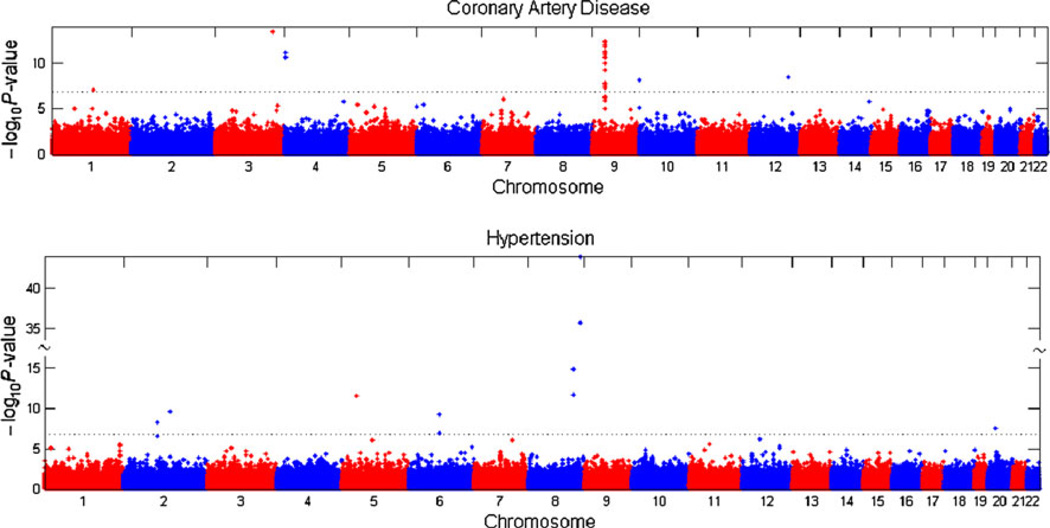
Results for the two-marker association analysis for CAD and HTN using the 2 and 3 df models are shown in Tables 1 and 2. The 2 df model yielded the exact same loci with genome-wide significance as the 3 df model for both CAD and HTN, with the exception of one additional SNP pair locus for CAD (Table 1, asterisk). The term representing the extra df in the 3 df model was calculated and statistically significant for only the CAD SNP pair on chromosome 10 (supplemental material, Table 2) and therefore, we choose to display only the 2 df test Manhattan plot results for CAD and HTN (Fig. 3).
Table 1.
Two-marker logistic model association test results for coronary artery disease
| Ch | SNP1 | MAF1 | SNP2 | MAF2 | 2 df P value | 3 df P value | Hap | R2 | D′ | Pairs | Gene | CAD Link? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs12091564 | 0.037 | rs10218795 | 0.034 | 1.75 × 10−7 | 1.75 × 10−7 | CC (R) | 0.92 | 1 | 1 | HFE2 | Yes |
| 3 | rs11924705 | 0.423 | rs6789378 | 0.416 | 4.04 × 10−14 | 7.34 × 10−14 | CA (P) | 0.96 | 0.98 | 1 | >1 Mb from any gene |
No |
| 4 | rs7697839 | 0.026 | rs7673097 | 0.023 | 2.19 × 10−11 | 2.19 × 10−11 | GG (R) | 0.89 | 1 | 2 | STK32B | No |
| 9 | rs1333048 | 0.477 | rs1333049 | 0.494 | 7.08 × 10−14 | 2.55 × 10−13 | GG (P) | 0.92 | 0.99 | 18 | 115 kb from CDKN2B/2A |
Yes |
| 10 | rs2066314 | 0.016 | rs2066315 | 0.034 | 8.6 × 10−2* | 1.18 × 10−8 | GT (R) | 0.36 | 0.87 | 1 | DIP2C | No |
| 12 | rs1165668 | 0.260 | rs1165669 | 0.261 | 3.05 × 10−9 | 5.32 × 10−9 | GC (R) | 0.98 | 0.99 | 1 | 10 kb from HSP90B1 |
Yes |
Two and three degree of freedom (df) logistic model results for CAD. If multiple SNP pairs showed significant association with a P value <5 × 10−7, only the lowest P value pair is shown. The asterisk for the SNP pair on chromosome (Ch) 10 denotes a SNP pair only reaching genome-wide significance with 3 df model testing. Mean allele frequency (MAF) refers to the MAF of SNP 1 and SNP 2, respectively. The associated risk (R) or protective (P) haplotypes (Hap) are given. Correlation (R 2) and linkage disequilibrium (D′) values for the individual SNP pairs are listed. The “Pairs” column displays the number of SNP pairs in the area with a P value <5 × 10−7. Please refer to the text for specific information on the CAD linking literature. Italics indicate genes
Table 2.
Two-marker logistic model association test results for hypertension
| Ch | SNP1 | MAF1 | SNP2 | MAF2 | 2 df P value | 3 df P value | Hap | R2 | D′ | Pairs | Gene | HTN Link? |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | rs10496288 | 0.104 | rs10496289 | 0.098 | 2.08 × 10−9 | 7.32 × 10−9 | GC (P) | 0.96 | 1 | 1 | H: > 1 Mb from any gene |
No |
| 2 | rs13420028 | 0.200 | rs10188442 | 0.204 | 1.03 × 10−10 | 4.98 × 10−10 | AA (P) | 0.97 | 0.99 | 1 | GPR39 | No |
| 5 | rs7735940 | 0.240 | rs12522034 | 0.250 | 5.47 × 10−13 | 3.04 × 10−12 | AC (R) | 0.95 | 0.99 | 1 | >200 kb from RANBP3L |
No |
| 5 | rs6452524 | 0.442 | rs6887846 | 0.448 | 1.69 × 10−7 | 7.12 × 10−7 | GA (P) | 0.98 | 0.99 | 1 | XRCC4 | No |
| 6 | rs3798440 | 0.157 | rs9350602 | 0.151 | 3.38 × 10−10 | 7.43 × 10−10 | AC (P) | 0.96 | 1 | 2 | MYO6 | No |
| 8 | rs2469997 | 0.186 | rs6469823 | 0.181 | 3.25 × 10−16 | 2.22 × 10−15 | GC (R) | 0.97 | 1 | 2 | 70 kb from NOV | No (?) |
| 8 | rs7827545 | 0.321 | rs1372662 | 0.333 | 1.65 × 10−44 | 4.11 × 10−44 | CG (R) | 0.95 | 1 | 2 | ZFAT | Yes |
| 12 | rs7960483 | 0.331 | rs10785581 | 0.338 | 1.43 × 10−7 | 6.27 × 10−7 | TC (P) | 0.97 | 0.99 | 1 | ~100 kb from AN06 |
No |
| 20 | rs200752 | 0.115 | rs200759 | 0.113 | 6.65 × 10−09 | 3.34 × 10−08 | TA (R) | 0.92 | 0.96 | 1 | MACROD2 | Yes |
Two degree of freedom (df) logistic model results for HTN. If multiple SNP pairs showed significant association with a P value <5 × 10−7, only the lowest P value pair is shown. Mean allele frequency (MAF) refers to the MAF of SNP 1 and SNP 2, respectively. The associated risk (R) or protective (P) haplotypes (Hap) are given. Correlation (R 2) and linkage disequilibrium (D′) values for the individual SNP pairs are listed by their respective chromosome (Ch). The “Pairs” column displays the number of SNP pairs in the area with a P value <5 × 10−7. Please refer to the text for specific information on the HTN linking literature. Italics indicate genes. The (?) symbol is used for the SNP pair on chromosome (ch) 8, as NOV, which is 70 kb away from the associated SNP pair and has HTN linking literature
Fig. 3.
Manhattan plots of 2 df test results for CAD and HTN. Two df Manhattan plots for CAD and HTN are as marked. Three df testing methods yielded the same loci with the exception of one additional chromosome 10 SNP pair for CAD marked by the asterisk in Table 1
Coronary artery disease results
Results of the single-marker logistic regression (test 1–1) indicated multiple SNPs for CAD in the 9p21 region. The 9p21 region was the same area associated with CAD in the original WTCCC GWAS paper (Wellcome Trust Case Control Consortium 2007). This was to be expected as in large samples, the trend test is equivalent to logistic regression; however, we report here the P values after including the first ten principal components in the model, so there are some minor discrepancies when comparing with the P values reported by the WTCCC (supplemental material, Table 1).
Using the two-marker methods, we detected SNP pairs in three genes, and three non-protein coding regions showing genome-wide significant association evidence for CAD (Table 1). An additional SNP pair on chromosome 10 was found to attain genome-wide significance with the 3 df model, two-marker analysis (Table 1, asterisk). If more than one SNP pair in a particular region showed significant association, only the lowest P value pair is shown. The three associated genes include: hemochromatosis type 2 (HFE2), serine/threonine kinase 32B (STK32B), and disco-interacting protein 2 homolog C (DIP2C).
Of the other non-protein-coding regions shown in Table 1, the chromosome 9p21 region was the only area reaching genome-wide significance in the original WTCCC study. Additionally, the associated non-protein-coding region on chromosome 12 is only 10 kb from heat shock protein 90 kDa beta (HSP90B1).
Hypertension results
There was no SNP reaching genome-wide significance for HTN in the original WTCCC report. Similarly, no SNPs in our single-marker logistic regression (test 1–1), with or without inclusion of the principal components in the model to control for population stratification, had a P value <5 × 10−7 (supplemental material, Table 1).
Using the two-marker methods, we detected SNP pairs with significant association for HTN in five genes, and four non-protein-coding regions (Table 2). If more than one SNP pair showed significant association, only the lowest P value pair is shown. The five associated genes include: G protein-coupled receptor 39 (GPR39), X-ray repair, complementing defective, in Chinese Hamster 4 (XRCC4), myosin VI (MYO6), zinc finger and AT hook domain (ZFAT), and MACRO domain containing 2 (MACROD2). The remaining four associated non-protein-coding regions were at least 70 kb from any known gene.
Discussion
Since multiple forms of statistical testing methods are used in GWAS, it is important to use a model that can provide dependable, replicable results without inflation of significance. We have shown our experience using two, two-marker association tests on the WTCCC CAD and HTN data. We specifically chose tests 2–2 and 2–3, because they were previously predicted to provide reliable power for disease models inKim et al. (2010).
Our results show new SNPs associated with both CAD and HTN. Although some associations have been previously described, others may include new variants found by using two-marker methods. Our results did not find any overlapping SNP pairs between HTN and CAD.
For CAD, the study identified six separate genome regions with SNP pairs showing a strong association, three of which were located in genes: HFE2, STK32B, and DIP2C. HFE2 is the cause of hemochromatosis type 2A, a form of juvenile hemochromatosis (JH), a disorder that can cause cardiomyopathy from iron overload (Rivard et al. 2003). HFE2 was also detected for CAD with haplotype analysis using roughly 30 SNPs at a time byZhu et al. (2010). As proper iron metabolism is necessary for good heart function, disruptions in this pathway may be deleterious to cardiac function. HFE2 is also in a region associated with copy number variation (CNV), and CNV, as it relates to disease, is not well understood. Additionally, HFE2 is expressed in the heart.
No cardiovascular link was determined for the following: STK32B, which belongs to the serine threonine protein kinase superfamily, and DIP2C, which has an unknown genetic function but moderate expression in most fetal and adult tissues (Nagase et al. 1999). DIP2C is in a region associated with CNV.
The most notable SNP pairs that were in non-protein-coding regions include those on chromosome 9, near the coding sequences of two cyclin dependent kinase inhibitors, CDKN2A and CDKN2B. We found 20 SNP pairs with P values <5 × 10−7 in the area. This was also the only area of genome-wide significance in both the WTCCC and our single-marker logistic regression analysis. As the 9p21 locus has been replicated in multiple studies (Wellcome Trust Case Control Consortium 2007; Horne et al. 2008; Samani and Schunkert 2008), there is little doubt that it is a major CAD locus.
The associated SNP pair on chromosome 12 is only 10 kb from HSP90B1. HSP90B1 is a highly conserved molecular chaperone protein. HSP90B1 has been shown to form a chaperone complex that interacts with p38, an important stress activated protein kinase involved in gene regulation, proliferation, differentiation, and cell death regulation in the cardiomyocyte (Ota et al. 2010). It also has been shown to associate with nitric oxide synthase (NOS3) to aid rats in myocardial ischemia–reperfusion injury resistance (An et al. 2009). The other reported associated SNP pair on chromosome 3 for CAD was far removed from any known gene.
For HTN, we detected SNP pairs in five genes and four non-protein-coding regions reaching genome-wide significance using the two-marker methods. The five associated genes are: ZFAT, MACROD2, GPR39, XRCC4, and MYO6. Of note, ZFAT conferred the lowest P value in our entire study. It is a newly described immune regulatory gene and has previously been linked to HTN (Zhu et al. 2010), although the mechanism is unclear. Furthermore, the 8q22–23 locus has shown significant linkage (i.e., genomic location) evidence to essential hypertension (Ciullo et al. 2006). MACROD2, a newly described gene with unclear function, was also found to be associated with HTN (Zhu et al. 2010). In addition, it has been associated with extreme obesity (Cotsapas et al. 2009).
Genes without literature evidence of cardiovascular involvement include GPR39, XRCC4, and MYO6. GPR39 has an unknown protein function, but has been shown to have expression in human atrial tissue (Iglesias et al. 2007); it is also in an area of CNV. XRCC4 repairs double-strand DNA breaks through nonhomologous end-joining recombination (Mari et al. 2006). It has no known link to HTN. MYO6 encodes an actin-based molecular motor involved in intracellular vesicle and organelle transport. It also has no known link to HTN.Avraham et al. (1995) found that the MYO6 gene is defective in certain types of deafness.
Of the remaining associated SNP pairs for HTN, two on chromosome 8 were 70 kb from NOV, a member of the CCN family of regulatory proteins. CCN regulatory proteins are extracellular matrix-associated proteins that play crucial roles in cardiovascular and skeletal development, injury repair, cancer, and fibrotic diseases (Chen and Lau. 2009). The remaining four HTN associated SNP pairs were at least 100 kb from any known gene.
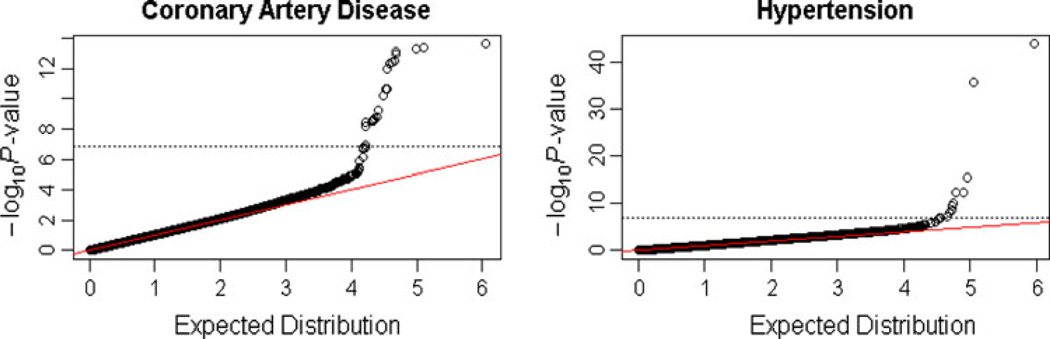
Typically, genome-wide association studies include a quantile–quantile (Q–Q) plot of the results, i.e., a plot of the observed results against what would be expected under the null hypothesis, in order to check the correctness of the found P values. Since our tests are correlated, we do not know the large-sample theoretical null distribution of our −log10 P values, so we estimated it from the permutation distribution. We permuted the cases and controls, repeated the analysis and ranked the −log10 P values. We similarly ranked the −log10 P values for the observed data and then formed the Q–Q plot by plotting the observed values against the permutation values (Fig. 4). The Q–Q plot appears as a straight diagonal line except at the tail, suggesting that our significant findings cannot be attributed to population structure or cryptic relatedness. When we plotted the observed −log10 P values against the −log10 P values of another independent permutation, the result was essentially the same (data not shown).
Fig. 4.
Q–Q plots of −log10 P for test 2–2: left, coronary artery disease; right, hypertension. See text for how the expected distribution was computed
However, one of our main concerns was that the correlations between some of the SNP pairs in Tables 1 and 2 are close to 1, suggesting that the pair should have also been found on single-marker analysis. We attribute the fact that these SNPs were not found on single-marker analysis, and yet significant by two-marker analysis, to the extra power two-marker analysis affords to provide haplotype information when the markers are both heterozygous and in high enough linkage disequilibrium. However, it is known that multicollinearity of the predictors can inflate significance levels in regression analysis. Therefore, to check our results, for each SNP pair given in Tables 1 and 2, we used the software PHASE, version 2.1 (Stephens et al. 2001; Stephens and Scheet 2005) to infer the haplotypes and found that, with few exceptions, only three of the four possible haplotypes were present; in all but one of these exceptions the fourth haplotype was rare in both cases and controls. We excluded those rare haplotypes (the largest excluded total count being 12, five cases and seven controls), and then the smallest expected value (on the assumption of no association between haplotypes and disease status) was 11.47 haplotypes. Furthermore, in all but two of the fifteen SNP pair tables, the expected haplotype count was always >15, so for these there is no reason to doubt the validity of the P value based on asymptotic assumptions. For the two tables for which the smallest expected value was <15, we calculated P values by Fisher’s exact test, two-sided because the association could have been with risk or protection, then multiplied by four because the associated haplotype could have been any one of the four haplotypes. The P values for these extreme cases were within 1–2 orders of magnitude of those based on asymptotic assumptions; therefore, we believe the increased evidence for association to be valid and due to real haplotype effects rather than an artifact as a result of either multicollinearity or small sample numbers (actual haplotype counts and odds ratios are presented in supplemental material, Table 3).
Furthermore, for both CAD and HTN, the 3 df two-marker model, containing the addition of a cross-product term, yielded roughly the same SNP pairs with a P value of <5 × 10−7 as the 2 df purely additive model. The only exception is noted by the asterisk in Table 1. Both models gave similar P values, even though we were expecting the 3 df test to yield more significant association because of the additional cross-product term. In all but the above case, the extra term in the 3 df test proved to not be statistically significant (supplemental material, Table 2). The reason for this appears to be that in the situation where two disease flanking heterozygote genotypes are called, only four diplotypes are possible instead of the prior 16 possible diplotypes (Fig. 2), yielding two-locus phased genotype (diplotype) information. Therefore, the SNP pair becomes more informative and hence the association with disease would be less likely to be due to chance alone—regardless of any additional effect of the cross-product term. Whenever one of the SNPs in the pair is called as homozygous, the two-marker testing would be expected to perform as a single- marker analysis, though with slightly less power owing to the extra df.
Ideally, all information from GWAS should be replicated using a separate dataset. Replication is challenging, especially for rare variants, as the necessary large available datasets are frequently not available and, if available, may involve variable population demographics, quality control, and phenotypic disease descriptors. We reviewed some additional associations that some recent GWAS for CAD and HTN report that are not listed in this study but, since CAD and HTN are disorders with complex inheritance, the list of genes that could be involved with disease would be lengthy and vary significantly from population to population (Newton-Cheh et al. 2009; Reilly et al. 2011; Levy et al. 2009). Of the eight genes cited by this study, three have been previously found in GWAS; HFE2, ZFAT, and MACROD2 (Zhu et al. 2010). The only non-protein-coding locus with previous association was 9p21, which was the only area reaching genome-wide significance in the WTCCC (Wellcome Trust Case Control Consortium 2007).
In conclusion, using two-marker methods, we found many interesting SNP pairs and associated genes not previously reported to be linked to CAD or HTN. Of note, CAD associated genes HFE2, CDKN2A/CDKN2B, and HSP90B1, and HTN associated genes ZFAT, MACROD2 and NOV, have known cardiovascular literature and/or known genetic mechanisms that may be relevant to disease.
We believe that multiple-marker association models may be better suited to detect variants because they are more informative of phased diplotypes than single-marker tests. Additional associated variants could also be found by using less stringent quality control filters. As many methods are available for multiple-marker association tests, further analysis using different techniques will need to be compared to determine the best fitting model for the detection of causal and protective variants for common, complex diseases such as HTN and CAD.
Supplementary Material
Acknowledgments
This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from http://www.wtccc.org.uk/info/participants.shtml. The work was supported by the National Institutes of Health, grant numbers: HL074166 and HL086718 from the National Heart, Lung, Blood Institute, HG003054 from the National Human Genome Research Institute, and, RR03655 from the National Center for Research Resources. Funding for the original WTCCC project was provided by the Wellcome Trust under award 076113.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00439-011-1009-6) contains supplementary material, which is available to authorized users.
Conflict of interest None.
Contributor Information
Thomas P. Slavin, Department of Genetics, Center for Human Genetics, University Hospitals of Cleveland and Case Western Reserve University, 10524 Euclid Avenue, Cleveland, OH, USA Hawai’i Community Genetics, 1441 Kapi‘olani Blvd., Ste 1800, Honolulu, HI 96814, USA, thomas.slavin@kapiolani.org.
Tao Feng, Department of Epidemiology and Biostatistics, Case Western Reserve University, Cleveland, OH 44106, USA; Department of Mathematics, Heilongjiang University, Harbin 150086, China.
Audrey Schnell, Department of Epidemiology and Biostatistics, Case Western Reserve University, Cleveland, OH 44106, USA.
Xiaofeng Zhu, Department of Epidemiology and Biostatistics, Case Western Reserve University, Cleveland, OH 44106, USA.
Robert C. Elston, Department of Epidemiology and Biostatistics, Case Western Reserve University, Cleveland, OH 44106, USA
References
- An J, Du J, Wei N, Xu H, Pritchard KA, Jr, Shi Y. Role of tetrahydrobiopterin in resistance to myocardial ischemia in Brown Norway and Dahl S rats. Am J Physiol Heart Circ Physiol. 2009;297:H1783–H1791. doi: 10.1152/ajpheart.00364.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham KB, Hasson T, Steel KP, Kingsley DM, Russell LB, Mooseker MS, Copeland NG, Jenkins NA. The mouse Snell’s waltzer deafness gene encodes an unconventional myosin required for structural integrity of inner ear hair cells. Nat Genet. 1995;11:369–375. doi: 10.1038/ng1295-369. [DOI] [PubMed] [Google Scholar]
- Browning BL, Browning SR. Haplotypic analysis of Wellcome Trust Case Control Consortium data. Hum Genet. 2008;123:273–280. doi: 10.1007/s00439-008-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771–783. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciullo M, Bellenguez C, Colonna V, Nutile T, Calabria A, Pacente R, Iovino G, Trimarco B, Bourgain C, Persico MG. New susceptibility locus for hypertension on chromosome 8q by efficient pedigree-breaking in an Italian isolate. Hum Mol Genet. 2006;15:1735–1743. doi: 10.1093/hmg/ddl097. [DOI] [PubMed] [Google Scholar]
- Cotsapas C, Speliotes EK, Hatoum IJ, Greenawalt DM, Dobrin R, Lum PY, Suver C, Chudin E, Kemp D, Reitman M, Voight BF, Neale BM, Schadt EE, Hirschhorn JN, Kaplan LM, Daly MJ GIANT Consortium. Common body mass index-associated variants confer risk of extreme obesity. Hum Mol Genet. 2009;18:3502–3507. doi: 10.1093/hmg/ddp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne BD, Carlquist JF, Muhlestein JB, Bair TL, Anderson JL. Association of variation in the chromosome 9p21 locus with myocardial infarction versus chronic coronary artery disease. Circ Cardiovasc Genet. 2008;1:85–92. doi: 10.1161/CIRCGENETICS.108.793158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias MJ, Salgado A, Pineiro R, Rodino BK, Otero MF, Grigorian L, Gallego R, Dieguez C, Gualillo O, Gonzalez-Juanatey JR, Lago F. Lack of effect of the ghrelin gene-derived peptide obestatin on cardiomyocyte viability and metabolism. J Endocrinol Invest. 2007;30:470–476. doi: 10.1007/BF03346330. [DOI] [PubMed] [Google Scholar]
- Kim S, Morris NJ, Won S, Elston RC. Single-marker and twomarker association tests for unphased case-control genotype data, with a power comparison. Genet Epidemiol. 2010;34:67–77. doi: 10.1002/gepi.20436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Kottgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, Smith AV, Taylor K, Scharpf RB, Hwang SJ, Sijbrands EJ, Bis J, Harris TB, Ganesh SK, O’Donnell CJ, Hofman A, Rotter JI, Coresh J, Benjamin EJ, Uitterlinden AG, Heiss G, Fox CS, Witteman JC, Boerwinkle E, Wang TJ, Gudnason V, Larson MG, Chakravarti A, Psaty BM, van Duijn CM. Genomewide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari PO, Florea BI, Persengiev SP, Verkaik NS, Bruggenwirth HT, Modesti M, Giglia-Mari G, Bezstarosti K, Demmers JA, Luider TM, Houtsmuller AB, van Gent DC. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc Natl Acad Sci USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:63–70. doi: 10.1093/dnares/6.1.63. [DOI] [PubMed] [Google Scholar]
- Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S, Papadakis K, Voight BF, Scott LJ, Zhang F, Farrall M, Tanaka T, Wallace C, Chambers JC, Khaw KT, Nilsson P, van der Harst P, Polidoro S, Grobbee DE, Onland-Moret NC, Bots ML, Wain LV, Elliott KS, Teumer A, Luan J, Lucas G, Kuusisto J, Burton PR, Hadley D, McArdle WL, Wellcome Trust Case Control Consortium, Brown M, Dominiczak A, Newhouse SJ, Samani NJ, Webster J, Zeggini E, Beckmann JS, Bergmann S, Lim N, Song K, Vollenweider P, Waeber G, Waterworth DM, Yuan X, Groop L, Orho-Melander M, Allione A, Di Gregorio A, Guarrera S, Panico S, Ricceri F, Romanazzi V, Sacerdote C, Vineis P, Barroso I, Sandhu MS, Luben RN, Crawford GJ, Jousilahti P, Perola M, Boehnke M, Bonnycastle LL, Collins FS, Jackson AU, Mohlke KL, Stringham HM, Valle TT, Willer CJ, Bergman RN, Morken MA, Doring A, Gieger C, Illig T, Meitinger T, Org E, Pfeufer A, Wichmann HE, Kathiresan S, Marrugat J, O’Donnell CJ, Schwartz SM, Siscovick DS, Subirana I, Freimer NB, Hartikainen AL, McCarthy MI, O’Reilly PF, Peltonen L, Pouta A, de Jong PE, Snieder H, van Gilst WH, Clarke R, Goel A, Hamsten A, Peden JF, Seedorf U, Syvanen AC, Tognoni G, Lakatta EG, Sanna S, Scheet P, Schlessinger D, Scuteri A, Dorr M, Ernst F, Felix SB, Homuth G, Lorbeer R, Reffelmann T, Rettig R, Volker U, Galan P, Gut IG, Hercberg S, Lathrop GM, Zelenika D, Deloukas P, Soranzo N, Williams FM, Zhai G, Salomaa V, Laakso M, Elosua R, Forouhi NG, Volzke H, Uiterwaal CS, van der Schouw YT, Numans ME, Matullo G, Navis G, Berglund G, Bingham SA, Kooner JS, Connell JM, Bandinelli S, Ferrucci L, Watkins H, Spector TD, Tuomilehto J, Altshuler D, Strachan DP, Laan M, Meneton P, Wareham NJ, Uda M, Jarvelin MR, Mooser V, Melander O, Loos RJ, Elliott P, Abecasis GR, Caulfield M, Munroe PB. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–676. doi: 10.1038/ng.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota A, Zhang J, Ping P, Han J, Wang Y. Specific regulation of noncanonical p38alpha activation by Hsp90-Cdc37 chaperone complex in cardiomyocyte. Circ Res. 2010;106:1404–1412. doi: 10.1161/CIRCRESAHA.109.213769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly MP, Li M, He J, Ferguson JF, Stylianou IM, Mehta NN, Burnett MS, Devaney JM, Knouff CW, Thompson JR, Horne BD, Stewart AF, Assimes TL, Wild PS, Allayee H, Nitschke PL, Patel RS, Myocardial Infarction Genetics Consortium, Wellcome Trust Case Control Consortium, Martinelli N, Girelli D, Quyyumi AA, Anderson JL, Erdmann J, Hall AS, Schunkert H, Quertermous T, Blankenberg S, Hazen SL, Roberts R, Kathiresan S, Samani NJ, Epstein SE, Rader DJ, Qasim AN, DerOhannessian SL, Qu L, Cappola TP, Chen Z, Matthai W, Hakonarson HH, Wilensky R, Kent KM, Lindsay JM, Pichard AD, Satler L, Waksman R. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet. 2011;377:383–392. doi: 10.1016/S0140-6736(10)61996-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivard SR, Lanzara C, Grimard D, Carella M, Simard H, Ficarella R, Simard R, D’Adamo AP, Ferec C, Camaschella C, Mura C, Roetto A, De Braekeleer M, Bechner L, Gasparini P. Juvenile hemochromatosis locus maps to chromosome 1q in a French Canadian population. Eur J Hum Genet. 2003;11:585–589. doi: 10.1038/sj.ejhg.5201009. [DOI] [PubMed] [Google Scholar]
- Samani NJ, Schunkert H. Chromosome 9p21 and cardiovascular disease: the story unfolds. Circ Cardiovasc Genet. 2008;1:81–84. doi: 10.1161/CIRCGENETICS.108.832527. [DOI] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–462. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Zhang S, Zhao H, Cooper RS. Association mapping, using a mixture model for complex traits. Genet Epidemiol. 2002;23:181–196. doi: 10.1002/gepi.210. [DOI] [PubMed] [Google Scholar]
- Zhu X, Li S, Cooper RS, Elston RC. A unified association analysis approach for family and unrelated samples correcting for stratification. Am J Hum Genet. 2008;82:352–365. doi: 10.1016/j.ajhg.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Feng T, Li Y, Lu Q, Elston RC. Detecting rare variants for complex traits using family and unrelated data. Genet Epidemiol. 2010;34:171–187. doi: 10.1002/gepi.20449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.