Abstract
Natural Regulatory T (Treg) cells are a subset of CD4+ T cells characterized by expression of the transcription factor Foxp3 and the ability to suppress immune responses. Treg cells develop in the thymus in response to highly specific interactions between the T cell receptor (TCR) and self-antigens. These processes can be recapitulated in antigen-specific systems using transgenic mice that co-express a TCR with its cognate peptide as a neo-self antigen. Here we describe a method for using such a system to establish a flow cytometric profile of phenotype markers expressed by developing and mature Treg cells in the thymus. Our approach is to compare antigen-specific thymocytes developing in the presence or absence of Treg cell-selecting ligands to identify phenotypic changes that characterize thymocytes undergoing selection into the Treg cell lineage.
Keywords: Thymocyte, Foxp3, Immune regulation, Treg progenitor cell, immunophenotyping
1. Introduction
T cell development in the thymus can be broadly categorized into four stages based on expression of the co-receptors CD4 and CD8 (1). The most immature thymocytes express neither co-receptor and are termed double negative (DN). Double positive (DP) cells have passed the β-selection checkpoint and express both CD4 and CD8. Thymocytes that have been selected on class II major histocompatibility complex (MHC) downregulate CD8 and are termed CD4 single positive (CD4SP); their class I MHC-selected counterparts become CD8SP. Mature SP thymocytes exit the thymus and join the pool of naïve CD4+ and CD8+ T cells that circulate between the blood and peripheral lymphoid organs.
Natural regulatory T (Treg) cells are a distinct subset of CD4+ T cells that develop in the thymus and are required for the maintenance of immune tolerance in the periphery (reviewed in reference 2). Briefly, Treg cells express a number of cell-surface markers associated with activated T cells including CD25, GITR and CTLA-4, and require the γc-chain cytokine IL-2 for their development in the thymus and survival in the periphery. Maintenance of Treg cell phenotype and function requires expression of the lineage-specific transcription factor Foxp3, and mutation of the Foxp3 gene leads to Treg cell deficiency and autoimmunity in both mouse and man.
Thymic selection of Treg cells is thought to occur by a two step process requiring both T cell receptor (TCR)-dependent and -independent signals (2). Evidence from our lab and others indicates that Treg cell selection occurs by a TCR-instructive process requiring highly specific interactions between the TCR and self-antigens (3, 4). Maturation of committed Treg cell precursors into Foxp3+ cells, however, may be TCR independent and instead requires γc-chain-dependent signals downstream of IL-2 (5).
While there is now convincing evidence that Foxp3+ CD4SP thymocytes represent mature cells that have acquired regulatory function, the identification of progenitors that will give rise to Foxp3+ CD4SP cells is less well advanced. Here we describe a method for using flow cytometry to analyze subsets of thymocytes that are undergoing Treg cell selection in order to identify phenotypic markers that characterize mature and developing Treg cells. Our approach is to use an antigen-specific transgenic mouse system to identify phenotypic changes that characterize thymocytes developing in the presence or absence of Treg cell-selecting ligands. These characteristics are then used as a basis for the identification of mature and developing Treg cells in a non-transgenic system.
2. Materials
2.1. Isolation of thymocytes
Mice. The experiments outlined in this protocol make use of single transgenic (ST) TS1 mice and double transgenic (DT) TS1xHA28 mice; however, any suitable transgenic mouse model1 can be used. For simplicity we will refer to TS1 as “ST mice” and TS1xHA28 as “DT mice”. A non-transgenic BALB/c mouse (Charles River, Wilmington, MA) will be used for analysis in the last section of this protocol.
Dissection bed with restraining pins. The lid to a Styrofoam shipping container and 27 G needles can be used for this purpose.
Dissection scissors and fine-point forceps (Fisher Scientific, Pittsburg, PA)
Phosphate buffered saline (PBS): 1.9 mM NaH2PO4, 8.1 mM Na2HPO4, 154 mM NaCl; prepared in dd-H2O.
Stainless steel mesh; 316 stainless steel, 200×200 mesh (W.S. Tyler, Mentor, OH). The stainless steel mesh should be cut into 1 inch squares, washed by submersion in 100% ethanol, rinsed with double distilled water and autoclaved prior to use.
1cc syringes (BD, San Jose, CA)
2.2. Immunostaining antigen-specific thymocytes for flow cytometry
96-well microtiter plate with v-shaped wells (Costar, Corning, NY)
Antibodies for the Treg cell profiling panel shown in Table 1. With the exception of 6.5, all of the antibodies used in this procedure can be obtained from eBioscience (San Diego, CA), Biolegend (San Diego, CA) or BD (San Jose, CA). The antibodies should be titrated prior to use to determine the optimal dilution for staining. The 6.5 antibody is produced and biotinylated in-house following standard procedures.
A fluorescent conjugate of streptavidin for detection of biotin-6.5 in the secondary detection step.
PBS supplemented with 2% heat inactivated FBS (Tissue Culture Biologicals, Tulare, CA) and 5 mM EDTA (FACSwash)
1% paraformaldehyde (PFA, USB, Cleveland, OH) in PBS
FACSwash supplemented with 0.1% Triton X-100 (Intracellular staining wash, ICSwash)
A flow cytometer capable of at least 5-channel fluorescence.
Table 1.
| Antibody staining panels
| |||||
|---|---|---|---|---|---|
| Treg cell profiling panel
|
BALB/c analysis panel
|
||||
| Developmental markers
|
Profiling markersd
|
antigen | recommended fluorochrome | ||
| antigen | clone | antigen | clone | ||
|
|
|
|
|
|
|
| Foxp3b | FJK-16s | CD25 | PC61.5 | Foxp3b | efluor450 |
| CD4 | GK1.5 | CD62L | MEL-14 | CD4 | APC-efluor780 |
| CD8 | 53–6.7 | CD69 | H1.2F3 | CD8 | efluor650 |
| TS1-TCR | 6.5a | CTLA-4b | UC10-4B9 | TS1-TCR | Sav-APC |
| TNFRII | TR75-89 | TNFRII | PE | ||
| GITR | DTA-1 | GITR | PE-Cy7 | ||
| Isotypec | N/A | CD25 | PerCP-Cy5.5 | ||
| CD69 | FITC | ||||
The 6.5 antibody used in this procedure is biotinylated and is detected with a fluorescent conjugate of Streptavidin in the secondary detection step.
CTLA-4 and/or Foxp3 should be stained during the intracellular staining step
The isotype control for these stains will be a single sample that is stained with the phenotype marker panel plus rat IgG1, IgG2a, IgG2b and Armenian hamster IgG
All of these antibodies including the isotype controls should be on the same fluorochrome. Seven staining panels should be prepared for this experiment. Each staining panel consists of all of the phenotype markers plus one of the comparison markers.
2.3. Data analysis: gating strategies and comparisons for phenotypic profiling transgenic thymocytes
FlowJo (Tree Star, Ashland, OR) or similar software2 for analysis of flow cytometric data.
2.4. Analysis of non-transgenic thymocytes by flow cytometry
96-well microtiter plates, FACSwash, PFA, and ICSwash, as in section 2.2.
Antibodies for the BALB/c analysis panel shown in Table 1. See note for suppliers in the previous section.
A flow cytometer capable of at least 8-channel fluorescence.
3. Methods
In transgenic mice that co-express a defined TCR with its cognate peptide as a neo self-antigen, thymocytes expressing the transgenic TCR can undergo enhanced selection to become Treg cells (3, 4, 6, 8, 9). The TS1 transgene encodes a MHCII-restricted TCR recognizing the site 1 (S1) determinant of PR8 influenza hemagglutinin, and can be identified by the clonotypic antibody 6.5 (10). HA28-transgenic mice constitutively express low levels of the S1 peptide, and in DT TS1xHA28 mice a significant fraction of thymocytes expressing the TS1-TCR are selected to become Treg cells (7). Using this system we can track populations of antigen-specific thymocytes from Treg cell-selecting (DT) or non-selecting environments (ST). In the first section of this protocol we make direct comparisons between these two populations of cells in order to determine the phenotypic profile of thymocytes undergoing selection into the Treg cell lineage. In the final section of this procedure we apply this profile to a BALB/c mouse to show that an equivalent population can be identified in a non-transgenic system.
3.1. Isolation of thymocytes
The thymus3 is a bi-lobed organ located in the thoracic cavity resting on top of the heart. Due to its proximity to the cardiac vasculature, it is essential to make a clean dissection to avoid contamination of the thymocytes with peripheral blood leukocytes.
All dissection tools should be washed and autoclaved prior to use. One stainless steel mesh and one 1cc syringe will be needed per thymus. For each thymus prepare a Petri dish containing 5 ml PBS.
Euthanize the mouse according to animal welfare regulations
Immobilize the mouse for dissection by pinning each paw to the dissection bed.
Using surgical scissors make sub-dermal cuts from groin to jaw and from the groin to each hind paw as illustrated in Figure 1A. Using forceps to grasp the skin on either side of the abdominal incision pull the skin away from the body of the mouse and pin it to the dissection bed to expose the ribcage and peritoneal membrane.
Expose the thymus by cutting through the ribcage as shown in Figure 1B. First, make a centerline cut through the peritoneal membrane and into the sternum. This cut will puncture diaphragm and the heart and lungs should be just visible through the incision. Second make two lateral cuts running between the ribs and diaphragm. Third, using forceps to push the lungs aside, cut through the ribs as near to the base of the thoracic cavity as possible. Fourth, trim the pectoral muscles away from the ribcage so that the ribcage is cantilevered from the sternum and can be lifted upwards to expose the thoracic cavity as shown in Figure 1C.
The thymus will be pulled up and away from the heart along with the ribcage4. Using scissors cut the ribcage and thymus away from the thoracic cavity. This cut should be made in one stroke to minimize contact between the thymus and cardiac blood.
Using forceps tease the thymus away from the ribcage and place in a Petri dish containing PBS until further processing.
Place a stainless steel mesh over the mouth of a 15 ml centrifuge tube and seat in place using the butt of a 1cc syringe to indent the mesh into the mouth of the tube.
Transfer the thymus onto the mesh and use the plunger from a 1cc syringe to gently mash the organ through the mesh. Wash the mesh and plunger with 2 ml of PBS and repeat the process until the thymus is completely disrupted and only white connective tissue remains on the mesh.
Pellet the cells by centrifugation at 400×g for 4 min.
Decant the supernatant and resuspend the pellet in 10 ml of FACSwash. Repeat this step once more for a total of two washes. During the second wash count the cells using a hemocytometer.
Following the last wash resuspend the cells at 20×106/ml in FACSwash.
Figure 1. Isolation of the mouse thymus.

The procedure illustrated here allows removal of the thymus with minimal exposure to peripheral blood. A. Euthanize and immobilize the mouse. Make three sub-dermal incisions as illustrated by the dashed lines and peel the skin away from the abdominal cavity and ribcage. B. Cut through the ribs and pectoral muscles as shown and using forceps pull the ribcage up and away from the thoracic cavity to reveal the heart and thymus. C. The thymus will be pulled away from the heart by the ribcage. Using scissors, cut the ribcage and thymus away from the thoracic cavity, and subsequently remove the thymus from the ribcage.
3.2. Immunostaining of antigen-specific thymocytes for analysis by flow cytometry
Experimental setup. Purify thymocytes as described above from a single- and a double-transgenic mouse. Transfer 200 ul/well (4×106 cells) of each cell suspension into a 96-well plate for staining. Plate 7 replicate wells from each cell suspension. In this experiment, cells will be stained with seven different antibody panels for analysis by 5-color flow cytometry (Table 1). Each panel will contain the same four antibodies for determining developmental stage (developmental markers), but will vary in the final antibody (profiling marker) that will be used in comparisons. One of these panels will contain a mixture of isotype control antibodies and will be used as the negative control for staining. To simplify analysis and cytometer setup, the profiling antibodies should all be conjugated to the same fluorochrome. This protocol assumes readers are familiar with the requirements for setting up and compensating a flow cytometer and the preparation of single-color compensation control samples will not be explicitly addressed here.
This is a three-step staining protocol. Cells are first stained for surface markers. The 6.5 antibody used here is biotinylated, and the second stain is with streptavidin-conjugated APC. In the final step the cells are fixed, permeabilized and stained for the intracellular markers Foxp3 and CTLA-4.
All reagents used in this procedure should be ice-cold, and all incubations are carried out on ice and protected from light. Centrifugation steps should be at 400×g for 4 min (surface staining) or at 800×g for 5 min (intracellular staining) in a 4 °C centrifuge.
Prepare the antibody panels for surface staining. Make sufficient cocktail for 200 ul/sample plus 5% excess. The antibody panels should be prepared in FACSwash.
Pellet the cells in the 96-well plate by centrifugation and discard the supernatant.
Resuspend the cells in 200 ul of the appropriate antibody panel and incubate for 30 min on ice. Note that the cells plated for CTLA-4 staining should only be stained with the developmental panel during this step.
Pellet the cells by centrifugation, discard the supernatant and resuspend the pellet in 200 ul of FACSwash. Repeat three times.
Perform steps 8 and 9 only if using biotinylated or unconjugated antibodies that require secondary detection, otherwise skip to step 10. Following the last wash resuspend the cells in 200 ul of FACSwash + a florescent-conjugate of streptavidin. Incubate for 30 min on ice.
Pellet the cells by centrifugation and wash thrice as in step 7.
Resuspend the cells in 200 ul of 1% PFA and incubate for at least 30 min on ice5.
Pellet the cells by centrifugation, discard the supernatant and wash twice with ICSwash. Remember that all post-fixation centrifugation steps should be performed for 5 min at 800×g.
Incubate the cells 10 min in 200 ul of ICSwash.
Prepare the intracellular staining antibody panels. One panel should contain both anti-CTLA-4 and anti-Foxp3, and will only be applied to the two samples plated for profiling CTLA-4 expression. The second panel should contain both anti-Foxp3 and hamster IgG, and will only be applied to the isotype control sample. The final panel will contain only anti-Foxp3 and will be applied to all the remaining samples. Make sufficient volume for 200 ul/sample plus 5% excess. The antibody panels used in this step should be prepared in ICSwash.
Pellet the cells by centrifugation, resuspend in 200 ul of the appropriate ICS antibody panel and incubate for 30 min on ice.
Pellet the cells by centrifugation and wash twice with ICSwash and once with FACSwash.
Following the last wash resuspend the cells in 200 ul of FACSwash. The cells are now ready for analysis by flow cytometry.
3. 3. Data analysis: gating strategies and comparisons for phenotypic profiling of transgenic thymocytes
Acquisition. Some of the populations that will be analyzed here are present at very low frequencies in the thymus. It is essential that enough events are collected to allow statistically valid comparisons. Be sure to collect a sufficient number of events so that there are at least 100 cells (and preferably more) of the lowest frequency population to be analyzed6. Also, set the flow cytometer to collect forward scatter height (FSC-H) as well as area (FSC-A) to allow exclusion of doublets.
Analysis. Import the data into an analysis program such as FlowJo. The following gating strategy should be used to make comparisons between samples. Be sure to apply these gates uniformly to all of the samples being analyzed.
Stringency gates (Figure 2A). For accurate results it is important to exclude false positives that arise from clumps of cells being acquired by the cytometer as a single event (doublets). This can be accomplished using a plot of FSC-A vs. FSC-H to exclude doublets from further analysis. Single cells exhibit a 1:1 relationship between these two parameters and fall along a 45° angle from the origin. When two or more cells are acquired simultaneously the FSC-A is increased disproportionately to the FSC-H, and these cells will stray significantly from 45°. Establish a gate that includes only single cells (“singlets”) and plot the FSC-A of the included events against their side scatter area (SSC-A). When plotted in this manner, singlet thymocytes will form a distinct population of cells that can be distinguished from cellular debris and many types of accessory cells. Use this plot to set a “thymocyte” gate. Only cells that fall within this gate should be included in the analyses described below.
Developmental gates (Figure 2B). Establish gates to segregate the cells into developmental stages by plotting the singlet thymocyte population from the ST mouse on a graph of CD4 vs. CD8. Define four regions on this plot7: CD4−CD8− (DN), CD4+CD8+ (DP), CD4−CD8+ (CD8SP), CD4+CD8−(CD4SP). At this point, the cells that fall within these gates will not be analyzed. Instead, the gates established here will be used to identify specific populations for analysis in the steps that follow.
Identification of a non-Treg cell forming control population. Clonotype+ thymocytes from a ST mouse do not form Treg cells in vivo and are therefore used as a control population that is devoid of Treg cells or their progenitors. Plot singlet thymocytes from the ST mouse with clonotype and Foxp3 on the axes and gate clonotype+Foxp3− cells as shown in the left panel of Figure 2C.
Identification of mature Treg cells. Here we define clonotype+Foxp3+ thymocytes from a DT mouse as mature Treg cells. Identify these cells by plotting singlet thymocytes as Foxp3 vs. clonotype and gating the Foxp3+clonotype+ cells as indicated in the right panel of Figure 2C.
Analysis of mature Treg cell development. The first step in the analysis of mature Treg cells is to determine the ontogeny of Foxp3 expression during thymic development (Figure 2D). Plot the DT clonotype+Foxp3+ cells gated in the previous step with CD4 and CD8 on the axes and apply the developmental gates established in step 4. The results of this analysis clearly show that mature Foxp3+ cells predominantly fall within the CD4SP subset of thymocytes, and we will therefore limit our investigation of the profiling markers to cells at this stage of development.
Analysis of mature Treg cell phenotype. Next, use the profiling markers to determine the phenotypic profile of mature Treg cells (Figure 2E). The appropriate comparison to be made here is the expression of each of these markers by the CD4SP, mature Treg cells identified in steps 6 and 7, to CD4SP cells from the ST control population identified in step 5. Plot these two populations of cells as histograms of the fluorescence intensity of staining for each profiling marker. Based on this analysis we conclude that mature Treg cells in the thymus express high levels of GITR, CTLA-4, TNFRII, and CD25 relative to Foxp3− CD4SP thymocytes from a ST mouse. They also express marginally higher levels of CD62L and equal to marginally lower levels of CD69 than Foxp3− CD4SP thymocytes from ST mice.
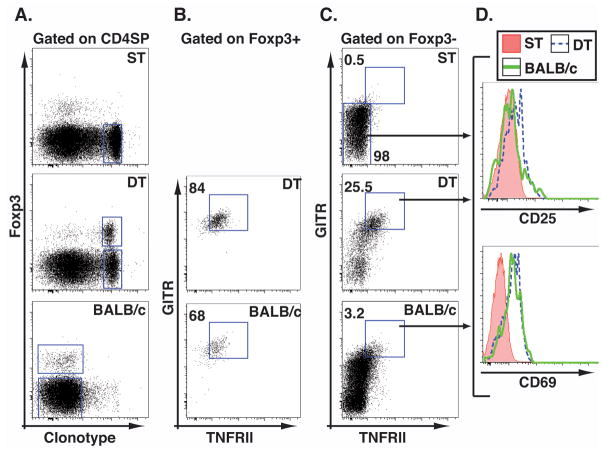
Analysis of developing Treg cells. Here, we will define developing Treg cells as being enriched within the clonotype+Foxp3− subset of thymocytes in a DT mouse, but absent within the same subset of thymocytes from a ST mouse. The latter was gated in step 5 of this procedure. Gate the former population by plotting singlet thymocytes from the DT mouse as clonotype vs. Foxp3 and gate on clontoype+Foxp3− cells as shown in Figure 3A. Plot the developing Treg cells with CD4 and CD8 on the axes and apply the developmental gates that were established in step 4. This analysis shows that Foxp3− cells expressing the clonotypic TCR can be found at all four stages of thymic development (Figure 3B) in thymocytes from both ST and DT mice. The analysis of the profiling markers will be limited to DN, DP, and CD4SP cells, however, since 6.5 is a MHCII-restricted TCR, and the significance of CD8+Foxp3+ cells remains to be established.
Analysis of developing Treg cell phenotype. To establish a phenotypic profile for developing Treg cells, compare the expression of the profiling markers by each developmental subset within the DT clonotype+Foxp3− cells with the corresponding population of ST control cells. Make this comparison by plotting a histogram of the fluorescence intensity of staining for each marker (Figure 3C). Based on these plots we conclude that upregulation of GITR, TNFRII and CD69 are most strongly associated with a population of cells containing putative Treg cell progenitors that can be found at the CD4SP stage, and to a lesser degree at the DP stage, in Foxp3− thymocytes from DT but not ST mice.
Figure 2. Flow cytometric profiling of mature Treg cells in the thymus.
Clonotype+Foxp3+ thymocytes from a DT mouse were used to develop a profile of mature Treg cell-associated phenotype markers. A. Stringency gates. Left panel shows the gate used to exclude doublets from analysis. Right panel shows the thymocyte gate. B. Developmental gates based upon CD4 and CD8 expression by singlet thymocytes from a ST mouse. C. Left panel shows the gate used to identify clonotype+Foxp3− control cells from a ST mouse. Right panel shows the gate used to identify clonotype+Foxp3+ mature Treg cells from a DT mouse. D. Developmental gates applied to clonotype+Foxp3+ Treg cells. E. histograms show the fluorescence intensity of staining by the indicated subsets for each of the profiling markers.
Figure 3. Flow cytometric profiling of developing Treg cells in the thymus.
Clonotype+Foxp3− thymocytes from a DT mouse were used to establish a profile of phenotype markers enriched on thymocytes developing in the presence of Treg cell-selecting ligands. A. Plots show the gates used to limit analysis to clonotype+Foxp3− cells from ST and DT mice. B. Gates used to segregate clonotype+Foxp3− thymocytes from ST and DT mice by developmental stage. C. histograms show the fluorescence intensity of staining by the indicated subsets for each of the profiling markers. Shaded histograms represent staining by an isotype control antibody except for the CD25 data, which shows an unstained control sample.
3.4. Analysis of non-transgenic thymocytes by flow cytometry
The phenotypic profiles generated in section 3.3 indicate that GITR, TNFRII, CD69, and CD25 may be useful surface markers for both mature and developing Treg cells. In this section of the protocol we will determine whether or not equivalent populations of cells can be identified in a non-transgenic BALB/c mouse.
Experimental setup. Purify thymocytes as described in section 3.1 from a BALB/c mouse, and also from ST and DT mice for comparison. Plate 4×106 cells from each mouse into a 96-well plate for immunostaining.
Stain the cells as described in section 3.2, but replace the phenotype and profiling markers with the BALB/c analysis panel listed in Table 1. Stain thymocytes from all three mice with this panel.
Collect the data on a flow cytometer, taking care to record enough events for analysis.
Using FlowJo or equivalent analysis software apply stringency gates as described in section 3.3.
Define the thymocyte populations for analysis. Plot the singlet thymocytes gated in the previous step with CD4 and CD8 on the axes. Set developmental gates to segregate the DN, DP, CD8SP and CD4SP subsets. Based on the profiles generated in the previous section, analysis will be limited to CD4SP cells. Plot the CD4SP subset from each mouse on a graph of clonotype vs. Foxp3. Gate the clonotype+Foxp3− cells from the ST mouse, clonotype+Foxp3− and clonotype+Foxp3+ cells from the DT mouse, and clonotype−Foxp3− and clonotype−Foxp3+ cells from the BALB/c mouse for analysis (Figure 4A). Plot these populations as GITR vs. TNFRII (Figure 4B and C).
Establishment of the GITR+TNFRII+ gate. Using the clonotype+Foxp3− cells from the ST mouse as a negative control and the clonotype+Foxp3+ cells from the DT mouse as a positive control establish a gate for GITR+TNFRII+ cells.
Identification of mature Treg cells. Apply the GITR+TNFRII+ gate to the Foxp3+ BALB/c thymocytes gated in step 6. Gating the Foxp3+ cells in this manner clearly shows that a majority of mature Treg cells from BALB/c mice express high levels of both TNFRII and GITR (Figure 4B).
Identification of developing Treg cells. Apply the GITR+TNFRII+ gate to the Foxp3− cells identified in step 6. When the Foxp3− cells are viewed in this manner the GITR+TNFRII+ population of cells that is enriched in the DT but absent in ST mice can also be found in a non-transgenic BALB/c mouse (Figure 4C). Plot the fluorescence intensity of staining by these cells for CD25 and CD69 to verify that these two markers are upregulated along with GITR and TNFRII (Figure 4D). We can conclude based on these histograms that GITR and TNFRII mark a population of cells in the BALB/c thymus that also express CD25 and CD69 at similar levels to the presumptive Foxp3− Treg cell precursors identified in DT mice (see section 3.3.10).
Figure 4. GITR and TNFRII expression by developing and mature Treg cells in the thymus of a BALB/c mouse.
The flow cytometric profiles of mature and developing Treg cells from transgenic mice were validated by assessing their expression on BALB/c thymocytes. A. Expression of clonotypic TCR and Foxp3 by the indicated mice. The gated populations were used for analysis. B. GITR and TNFRII expression by Foxp3+ thymocytes from DT and BALB/c mice. C. GITR and TNFRII expression by Foxp3− cells from ST, DT and BALB/c mice. D. Histograms of the fluorescence intensity of CD25 and CD69 staining by the indicated populations.
Acknowledgments
The authors would like to thank Malinda Aitken, Christina Mergenthaler, Abigail Liebow, Alissa Basehoar, and Lori Mroz for their invaluable help in maintaining the transgenic mouse lineages described here. This work was supported by R01-AI59166 and by the Commonwealth Universal Research Enhancement Program, Pennsylvania department of Health. DMS is supported by T32 CA09171.
Footnotes
Treg cell formation using the TS1xHA system has been reported with HA expression driven by the β-globin locus control region, the β-myoglobin heavy chain promoter, and by SV40, AIRE and Igκ promoters (4, 6). The DO11xOVA system can also be used to track the development of antigen specific Treg cells using the KJ-126 clonotypic antibody. Thymic Treg cell formation has been reported in this system using both the insulin promoter to drive OVA expression and also when OVA is targeted to the nucleus (7–9).
All data displayed in this protocol was generated using FlowJo. Equivalent analyses can be performed using a number of alternative software suites including FCS Express by De Novo Software, Venturi One by Applied Cytometry, Cyflogic, and Weasel (developed by the Walter and Eliza Hall Institute of Medical Research).
The size and cellularity of the thymus can vary greatly depending on the age of the mouse being dissected. Thymic involution occurs between 8 and 10 weeks of age in mice resulting in a 50% to 75% reduction in thymic cellularity. DT mice will also have reduced thymic cellularity due to the presence of deleting antigen in the thymus. The reduction in thymus size will be dependent upon the amount of antigen in the thymus and must be determined empirically for each transgenic strain.
If the thymus is not pulled away from the heart with the ribcage, use fine-point forceps to gently tease away the connective tissue connecting the two organs. Be careful not to puncture the heart.
The sensitivity of an antibody to fixation time must be determined empirically for each clone. We have left cells in fixative for as long as overnight without significant loss of Foxp3 staining using the FJK-16s clone.
We typically only collect events that fall within the stringency gates described in section 3.3. Using these gating criteria the number of events required for valid analyses is typically between 300,000 and 500,000.
The relative proportions of the developmental subsets can be substantially skewed by the transgenic expression of TCRs and/or antigen. For example, expression of an MCH class II-restricted TCR by the TS1 mouse results in an enrichment of CD4SP cells compared to a BALB/c thymus. Although it is still relatively straight-forward to distinguish between the developmental subsets in a transgenic mouse, it may be useful to include a single well of BALB/c thymocytes in your surface stains to establish these gates.
References
- 1.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30(5):616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordan MS, Boesteanu A, Reed AJ, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol. 2001;2(4):301–306. doi: 10.1038/86302. [DOI] [PubMed] [Google Scholar]
- 4.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol. 2002;3(8):756–763. doi: 10.1038/ni816. [DOI] [PubMed] [Google Scholar]
- 5.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28(1):100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aschenbrenner K, D’Cruz LM, Vollmann EH, et al. Selection of Foxp3(+) regulatory T cells specific for self antigen expressed and presented by Aire(+) medullary thymic epithelial cells. Nat Immunol. 2007;8(4):351–358. doi: 10.1038/ni1444. [DOI] [PubMed] [Google Scholar]
- 7.Cozzo PC, Oh S, Panarey L, Aitken M, Basehoar A, Caton AJ. Thymocyte deletion can bias Treg formation toward low-abundance self-peptide. Eur J Immunol. 2009 doi: 10.1002/eji.200939709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker LS, Chodos A, Eggena M, Dooms H, Abbas AK. Antigen-dependent proliferation of CD4+ CD25+ regulatory T cells in vivo. J Exp Med. 2003;198(2):249–258. doi: 10.1084/jem.20030315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawahata K, Misaki Y, Yamauchi M, et al. Generation of CD4(+)CD25(+) regulatory T cells from autoreactive T cells simultaneously with their negative selection in the thymus and from nonautoreactive T cells by endogenous TCR expression. J Immunol. 2002;168(9):4399–4405. doi: 10.4049/jimmunol.168.9.4399. [DOI] [PubMed] [Google Scholar]
- 10.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J Exp Med. 1994;180(1):25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]