Abstract
Validation of a recent finding linking a rare variant in TP53 to the risk of glioma, the most common primary brain tumour, is reported here. This study genotyped the single nucleotide polymorphism (SNP) rs78378222 in 566 glioma cases and 603 controls. The variant ‘C’ allele (with an allelic frequency of 1.1% in controls) was associated with a 3.5-fold excess in glioma risk (odds ratio 3.54; p=0.0001). Variant carriers had significantly improved survival (hazard ratio 0.52; p=0.009) when compared to non-carriers. The rs78378222 SNP is the first confirmed rare susceptibility variant in glioma. Results may shed light on the aetiology and progression of these tumours.
Keywords: glioma, glioblastoma, risk, prognosis, single nucleotide polymorphism
Rare (minor allele frequency <5%) variants have been proposed to contribute to the “missing heritability” in cancer (1) and may contribute to recently identified signals reported in genome-wide association studies (2). Rare variants are more likely than common variants to have a functional impact, and tend to have a stronger effect size than do common variants (3). For these reasons, rare variants are likely to be a crucial element of the genetic architecture of common human diseases. However, few rare variants have yet been definitively linked to cancer risk.
A rare variant conferring glioma risk was recently reported in the Icelandic population and provisionally validated in two separate US case-control studies (4). The implicated single nucleotide polymorphism (SNP), rs78378222, occurs in the sole polyadenylation signal of TP53 and is predicted to disrupt the signal sequence. In functional assays, the rs78378222[C] variant was shown to impair proper termination and polyadenylation of the TP53 transcript (4). The SNP was linked to risk of a spectrum of diverse cancers including glioma. This is the first rare variant linked to glioma risk.
We genotyped the rs78378222 SNP in a large, ongoing, clinic based, case-control study and considered associations according to glioma histological subtype. We also examined for the first time the prognostic impact of this new candidate susceptibility allele in glioma and other primary tumours.
A description of the study population has been published (5). Briefly, glioma cases were individuals aged 18 years and older with a recent diagnosis of glioma identified in neurosurgery and neuro-oncology clinics at medical centres in the southeastern USA. Controls were persons sampled from communities giving rise to the cases with no personal history of brain tumour supplemented with friends and non-blood relatives of the cases. Glioma cases were enrolled a median of 1.0 month following glioma diagnosis (interquartile range 2 weeks to 1.7 months). Oral genomic DNA was available for all subjects. Study protocols were approved by the institutional review committees at each participating centre and all study participants provided written informed consent.
A total of 566 cases and 603 controls, all Caucasian, were genotyped for the rs78378222 variant using TaqMan. Laboratory personnel were masked to the case-control status of the samples. A single homozygous carrier of the variant ‘C’ allele, a case with glioblastoma, was identified in the series of 1169 genotyped subjects. All remaining variant allele carriers were heterozygous at this locus. The minor allele frequency was 0.037 in the cases and 0.011 in the controls. Genotype frequencies were in Hardy-Weinberg equilibrium (p=0.789).
Results for the examination of risk associations are shown in the table 1. Odds of glioma were increased 3.5-fold among variant allele carriers (odds ratio (OR) 3.54, 95% confidence interval (CI) 1.87 to 6.71; p=0.0001) compared to non-carriers. An increased risk associated with the C allele was observed regardless of glioma histologic subtype (test for heterogeneity in multinomial regression: p=0.779).
Table 1.
Risk associations for rs78378222 overall and according to glioma histologic subtype
| Glioma subtype* | Cases (AA/AC or CC) | Controls (AA/AC or CC) | OR (95% CI)† | P value |
|---|---|---|---|---|
| All Gliomas | 525/41 | 590/13 | 3.54 (1.87, 6.71) | 0.0001 |
| Glioblastoma | 292/24 | 590/13 | 3.51 (1.75, 7.04) | 0.0006 |
| Astrocytic tumors | 130/11 | 590/13 | 4.04 (1.72, 9.50) | 0.0016 |
| Oligodendrogliomas | 85/5 | 590/13 | 2.83 (0.95, 8.40) | 0.0610 |
Histology was unknown in 19 of 566 glioma cases.
Odds ratio (OR) and 95% confidence interval (CI) under a dominant model adjusting for age and sex.
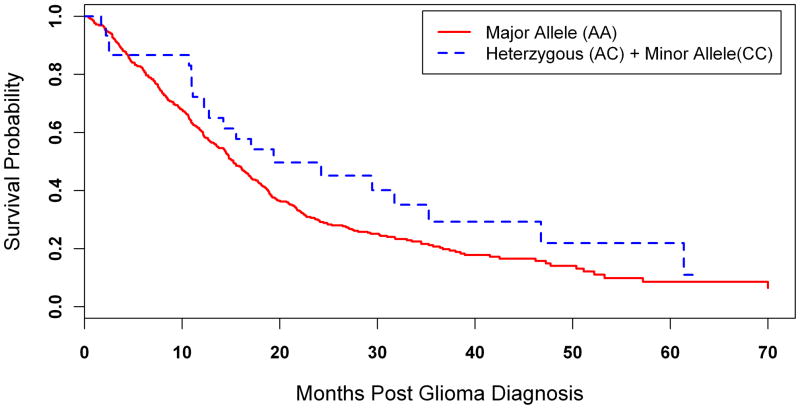
We further examined the impact of the rs78378222 variant allele on survival among the 413 patients with high grade gliomas (316 glioblastomas and 97 grade III astrocytomas or oligodendrogliomas) in whom 328 glioma related deaths were documented (figure 1). Carriers of the variant ‘C’ allele had longer survival times when compared to persons homozygous for the wild type ‘A’ allele (median Kaplan-Meier survival probability: 19.4 months and 15.2 months, respectively), with an approximately 50% reduction in death rates observed among variant allele carriers (hazard ratio adjusted for patient age and gender: 0.52, 95% CI 0.32 to 0.85; p=0.0094).
Figure 1.
Kaplan-Meier curves depicting probability of survival following diagnosis of high grade glioma according to rs78378222 genotype.
The present study confirms the reported association of the rare TP53 variant with glioma risk and demonstrates that the association is consistent across glioma subtypes. Odds ratios in the present study (OR 3.54 for all gliomas combined) are more prominent than those from the Icelandic population (OR 2.36) and the two validation studies from the USA in the study by Stacey et al (combined OR 2.34) (4). Similar to the previous analyses, all of the subjects in the present study were Caucasian with the great majority reporting European ancestry.
We also report an apparent survival advantage in carriers of the rare allele. Prolongation of survival among carriers of a genetic variant that increases the incidence of the tumour is an unexpected finding. (We attempted to validate this finding in The Cancer Genome Atlas (6) which includes mortality information and results of a genome-wide scan on approximately 300 patients with glioblastoma; however, neither rs78378222 nor any suitable proxy SNP was included in the The Cancer Genome Atlas array.). We note that in the report of Stacey et al (4), the rs78378222 variant was associated with the risk of colorectal adenoma but not colorectal cancer, suggesting the SNP may select for neoplasms with more indolent behaviour. If not due to chance, the observed association may offer some insights into the mechanistic aspects of cancer development and progression. Since the risk allele affects a regulatory sequence in the untranslated region (3′UTR), the likely impact of this variant is a reduction in TP53 gene dosage and not in the production of a mutated protein. It is tempting to speculate that the presence of the risk allele could direct tumour development into a less aggressive path by not requiring loss of the wild-type TP53 allele for tumour development. Alternatively, in tumours that undergo loss of the wild-type TP53 allele, the reduced levels of wild-type TP53 protein produced by the variant allele could provide residual TP53 activity sufficient to maintain a less aggressive phenotype.
Glioma remains a poorly understood neoplasm with a high morbidity and devastating outcome. The discovery of the association of the TP53 variant rs78378222 with glioma offers new insights into these tumours and the prospects for better delineating populations at risk.
Acknowledgments
Funding Source
Financial support was received from Public Health Service Grants R01CA116174 from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services and institutional funding from the Moffitt Cancer Center, Tampa, FL and the Vanderbilt-Ingram Comprehensive Cancer Center, Nashville, TN.
Footnotes
Competing Interests
The authors declare that they have no competing interests.
Contributorship Statement
K. Egan participated in designing the study, generated and gathered data for the study, analyzed the data, and wrote the majority of the original draft of the paper. She is the corresponding author and guarantor. L.B. Nabors, J. Olson and R. Thompson participated in gathering data for the study and in writing the paper. A Monteiro participated in designing the study, in writing the paper and in reviewed the pertinent raw data on which the study was based. J. Browning analyzed the data and participated in writing the paper. M. Madden gathered data for the study, participated in writing the paper and reviewed the pertinent raw data on which the study was based. All authors reviewed and approved the final version.
References
- 1.Bodmer W, Tomlinson I. Rare genetic variants and the risk of cancer. Curr Opin Genet Dev. 2010 Jun;20(3):262–7. doi: 10.1016/j.gde.2010.04.016. Review. [DOI] [PubMed] [Google Scholar]
- 2.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010 Jan 26;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorlov IP, Gorlova OY, Frazier ML, Spitz MR, Amos CI. Evolutionary evidence of the effect of rare variants on disease etiology. Clin Genet. 2011 Mar;79(3):199–206. doi: 10.1111/j.1399 0004.2010.01535.x. Epub 2010 Sep 10. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey SN, Sulem P, Jonasdottir A, Masson G, Gudmundsson J, Gudbjartsson DF, Magnusson OT, Gudjonsson SA, Sigurgeirsson B, Thorisdottir K, Ragnarsson R, Benediktsdottir KR, Nexø BA, Tjønneland A, Overvad K, Rudnai P, Gurzau E, Koppova K, Hemminki K, Corredera C, Fuentelsaz V, Grasa P, Navarrete S, Fuertes F, García-Prats MD, Sanambrosio E, Panadero A, De Juan A, Garcia A, Rivera F, Planelles D, Soriano V, Requena C, Aben KK, van Rossum MM, Cremers RG, van Oort IM, van Spronsen DJ, Schalken JA, Peters WH, Helfand BT, Donovan JL, Hamdy FC, Badescu D, Codreanu O, Jinga M, Csiki IE, Constantinescu V, Badea P, Mates IN, Dinu DE, Constantin A, Mates D, Kristjansdottir S, Agnarsson BA, Jonsson E, Barkardottir RB, Einarsson GV, Sigurdsson F, Moller PH, Stefansson T, Valdimarsson T, Johannsson OT, Sigurdsson H, Jonsson T, Jonasson JG, Tryggvadottir L, Rice T, Hansen HM, Xiao Y, Lachance DH, O Neill BP, Kosel ML, Decker PA, Thorleifsson G, Johannsdottir H, Helgadottir HT, Sigurdsson A, Steinthorsdottir V, Lindblom A, Sandler RS, Keku TO, Banasik K, Jørgensen T, Witte DR, Hansen T, Pedersen O, Jinga V, Neal DE, Catalona WJ, Wrensch M, Wiencke J, Jenkins RB, Nagore E, Vogel U, Kiemeney LA, Kumar R, Mayordomo JI, Olafsson JH, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K Swedish Low-risk Colorectal Cancer Study Group. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011 Sep 25;43(11):1098–103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egan KM, Thompson RC, Nabors LB, Olson JJ, Brat DJ, Larocca RV, Brem S, Moots PL, Madden MH, Browning JE, Ann Chen Y. Cancer susceptibility variants and the risk of adult glioma in a US case-control study. J Neurooncol. 2011 Sep;104(2):535–42. doi: 10.1007/s11060-010-0506-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–8. doi: 10.1038/nature07385. Epub 2008 Sep 4. [DOI] [PMC free article] [PubMed] [Google Scholar]