Abstract
The association between promoter methylation status and survival was investigated in a large cohort of women with breast cancer, participants in the Long Island Breast Cancer Study Project. Archived tumor tissues (n=839) were collected from women diagnosed with a first primary invasive or in situ breast cancer in 1996-1997. Vital status was followed through the end of 2005 with a mean follow up time of 8 years. Promoter methylation of 8 breast cancer-related genes was assessed by MethyLight. The frequencies of methylation for HIN1, RASSF1A, DAPK1, GSTP1, CyclinD2, TWIST, CDH1 and RARβ were 62.9%, 85.2%, 14.1%, 27.8%, 19.6%, 15.3%, 5.8% and 27.6%, respectively. Since survival rates of in situ and invasive breast cancers are substantially different, survival analyses were conducted within 670 invasive cases with complete data on all genes. Age-adjusted Cox-proportional hazards models revealed that GSTP1, TWIST and RARβ methylation was significantly associated with higher breast cancer-specific mortality. Methylation of GSTP1 and RARβ were significantly associated with higher all-cause mortality. To investigate the relationship between the number of methylated genes and breast cancer-specific mortality, we included previously published MethyLight data on p16 and APC methylation status. Breast cancer-specific mortality increased in a dose-dependent manner with increasing number of methylated genes (Ptrend = 0.002), although confidence intervals were wide. Our results suggest that promoter methylation, particularly for a panel of genes, has the potential to be used as a biomarker for predicting prognosis in breast cancer.
Keywords: Promoter methylation, Tumor suppressor gene, Breast cancer, Mortality
INTRODUCTION
Breast cancer is the most common cancer and second cause of cancer-death among females in most Western countries [1]. Although early detection and novel treatments have improved outcomes and survival rates for women with breast cancer, disease progression is still poorly understood. Recently, gene expression status [1] is being used to predict cancer outcomes, but its prognostic relevance is still undergoing evaluation. Therefore, research efforts continue to focus on identifying more sensitive and specific biomarkers that can reliably predict clinical outcomes and enhance treatment options.
Epigenetic alterations are one of the most common molecular alterations in human neoplasia [2]. In particular, aberrant promoter methylation occurs in numerous genes in cancer development and progression [3]. Among these genes, APC [4], p16 [5], RASSF1A [6], DAPK1 [7], CDH1 [8], RARβ [9], GSTP1 [10], CyclinD2 [11], HIN1 [12] and TWIST [13] are frequently methylated in breast cancer. Different studies have confirmed the hypothesis that aberrant methylation at specific genes contributes to the malignant phenotype and survival rates of breast cancer. Nimmrich et al. [14] reported that hypermethylation of PITX2 was negatively associated with PITX2 gene expression in cell lines and positively associated with breast cancer disease progression in lymph node negative and steroid hormone receptor positive breast cancer patients. Several studies by Kioulafe et al. [15-17] showed that promoter hypermethylation in RASSF1A, Kallikrein 10 (KLK10) and cystatin M (CST6) in tumor tissue are independent prognostic marker in early stage breast cancer patients. More recently methylation of 773 genes was examined in breast tumors and 8 methylation classes were identified; associations between methylation class with age, tumor size, alcohol intake and total dietary folate were observed [18].
We previously reported positive associations between survival and promoter methylation status of BRCA1 using methylation-specific PCR [19] and APC and p16 using MethyLight [20] in a large cohort of women with a first primary breast cancer who were participants in the Long Island Breast Cancer Study Project. In the present study, we expanded tumor DNA methylation analysis to an additional eight tumor suppressor genes (HIN1, RASSF1A, DAPK1, GSTP1, CyclinD2, TWIST, CDH1 and RARβ) to investigate the association of aberrant methylation with clinicopathologic characteristics and all-cause and breast cancer-specific mortality in women with breast cancer.
MATERIALS AND METHODS
This project draws upon resources of the Long Island Breast Cancer Study Project (LIBCSP), a population-based study of English-speaking residents of Nassau and Suffolk counties of Long Island. Details of the study participants and study design for the parent case-control and follow-up studies have been described previously [21-23]. The study protocol was approved by the Institutional Review Boards of the collaborating institutions.
Study population
Briefly, eligible participants were adult women who were newly diagnosed with a first primary in situ or invasive breast cancer between August 1, 1996, and July 31, 1997, and were residents of Nassau and Suffolk counties on Long Island NY. A total of 1,508 women with breast cancer participated in the baseline interview. At diagnosis, the participants ranged in age from 20 to 98 years; 94% self-reported their race as white, 4% as black, and 2% as other, which reflects the underlying population of these two counties [21-23]. As part of the follow-up study, the case participants, or their next of kin, were re-interviewed approximately five years after diagnosis.
Data collection
Information used in this study was obtained as part of the: (1) baseline in-home interview; (2) follow-up telephone interview; (3) medical record abstraction (complete course of treatment and tumor characteristics for the primary breast cancer), which occurred at baseline and again at follow-up; (4) the National Death Index (NDI); and (5) retrieved archived tumor tissue, as described previously [21-24].
Subject interviews. Structured questionnaires were interviewer-administered to assess demographic characteristics and breast cancer-related factors.
Medical records. First course of treatment for the primary breast cancer, and characteristics of estrogen and progesterone receptor (ER and PR) status were abstracted from medical records. Among cases included in the present project, 337 (59.8%) were ER+/PR+; 91 (16.1%) were ER+/PR-; 21 (3.7%) were ER-/PR+; and 115 (20.4%) were ER-/PR-.
Vital status at the follow-up. The NDI was used to ascertain all-cause and breast cancer-specific mortality among case participants, as previously described [23]. Participants diagnosed with breast cancer in 1996–1997 were followed until December 31, 2005, for a mean of 8.0 yrs (range, 0.3-9.4). Among the entire cohort of 1508 women, 308 (20.4%) deaths occurred, of which 164 (53.2%) were due to breast cancer. In the present study, a total of 179 all-cause deaths and 99 (55.3%) breast cancer-specific deaths were observed among the participant subset with available tumor DNA (n=859).
Tumor block retrieval, microdissection and DNA extraction. REMARK criteria for reporting tumor studies are used throughout this report [25].
Archived pathology blocks for the first primary breast cancer from 962 (63.8%) women were successfully retrieved from the 33 hospitals in the Long Island study area [24]. After review by the study pathologist (HH), 859 tumor tissue blocks (89.3%) were appropriate for laboratory analyses. There were no differences in demographic and clinicopathological features between cases with and without tumor blocks available for methylation analysis in our study, except for age, post-menopausal status, and percent invasive tumors. At diagnosis, compared to cases without tumor samples, cases with available tumor samples were: older (mean age, 59.6 yrs vs 57.9 yrs; P=0.005); were more frequently post-menopausal (70.7% vs 64.6%; P=0.01); and a greater percent had invasive tumors (87.8% vs 80.1%; P<0.001).
Tumor tissues were isolated from 2 × 10 micron thick slides of paraffin sections by microdissection; at least 90% of the cells were tumor cells. Tumor DNA was isolated by adding 30 μl of proteinase K-digestion buffer (50mM Tris, pH 8.1, 1 mM EDTA, 0.5% Tween 20, 10 μg/ml proteinase K) and incubating overnight at 37°C. Proteinase K was inactivated by heating at 95°C for 10 min and centrifugation.
Laboratory analysis of gene promoter methylation
MethyLight methods for p16 and APC were reported previously [20]. Among 859 cases with tumor DNA, 20 subjects were excluded because of insufficient DNA. Thus, a total of 839 tumor DNAs were available for this project, however, only 765 (95 in situ and 670 invasive breast cancer cases) had complete data on all 10 genes. Tumor DNAs first underwent bisulfite modification to convert unmethylated cytosine residues to uracil using the CpGnome DNA Modification Kit (Chemicon International, Purchase, NY) following the protocol from the manufacturer. Sodium bisulfite treated DNA was analyzed by the MethyLight technique as described previously [26]. The primers and probes for the selected genes and β-actin (ACTB) were previously described [12;26-28]. Specificity of the reactions for methylated DNA was confirmed separately using CpGenome™ Universal methylated DNA (Chemicon, MA, USA) and unmethylated human sperm DNA. TaqMan PCR reactions with primers specific for the bisulfite-converted methylated sequence for a particular locus and with the ACTB reference primers were performed separately. The values obtained in these two TaqMan analyses were used as a measure of the degree of methylation at that locus. Ct value for ACTB ranged from 25.7 to 39.5 with a median of 29.9. Relative quantification was determined based on the threshold cycles of the gene of interest and of the internal reference gene. The percentage of methylation at a specific locus was calculated by the 2-ΔΔCT method, where ΔΔCT = (CT,Target-CT,Reference)sample - (CT,Target-CT,Reference)fully methylated DNA [29] and multiplying by 100. Percentage methylation for selected genes was dichotomized to methylated or unmethylated using a cut-off of 4%, as used in a number of prior publications (e.g.,[30]). All samples were assayed in duplicate and the MethyLight assay was further validated by using mixtures of fully methylated and unmethylated DNA to give 0, 1, 5, 10, 25, 50 or 100% methylation. Intra- and interassay coefficients of variation (CVs) were 0.5 and 1.9, respectively.
To validate the 2-ΔΔCT method, the amplification efficiencies of the test genes and reference gene, ACTB, were examined using serial dilutions of DNA with a 100-fold range and gene-specific primers of each gene and ACTB. The ΔCT (CT,Target gene - CT,Reference) was calculated for each DNA dilution and a plot of the log DNA dilution vs ΔCT was made. All amplification efficiencies were similar (data not shown).
Statistical analysis
DNA methylation status was dichotomized using 4% as a cut-off point (≥4% as methylated vs. <4% as unmethylated) based on previous validated data [31;32]. While in agreement with a prior study [33], promoter methylation was evident in <10% of the tumor samples for CDH1; thus, this gene was omitted from the analyses focused on the relationship between risk factors and patient characteristics (Table 1), due to small cell sizes and unstable results. For the remaining markers, the Chi-square test was used to examine the association between each gene promoter marker and selected patient factors, which were considered as categorical variables including age at diagnosis (<50 years, >50 years), race (white, other), menopausal status (pre-, post-), body mass index (BMI = weight in kilograms (kg) divided by height in meters squared (m2) (<25, >25)), family history of breast cancer (no, yes in mother or sister), history of benign breast disease (yes, no), cancer type (in situ, invasive) and ER/PR status (both positive, either positive, both negative).
Table 1.
Association between gene promoter methylation and general patient characteristics in a population-based cohort of women diagnosed with a first primary breast cancer in 1996-1997, Long Island Breast Cancer Study Project
| No.* | HIN1 | RASSF1A | DAPK1 | GSTP1 | CyclinD2 | TWIST1 | RARβ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. positive (%) | P | No. positive (%) | P | No. positive (%) | P | No. positive (%) | P | No. positive (%) | P | No. positive (%) | P | No. positive (%) | P | ||
| Total | 481/765 (62.9) | 652/765 (85.2) | 108/765 (14.1) | 213/765 (27.8) | 150/765 (19.6) | 117/765 (15.3) | 211/765 (27.6) | ||||||||
| Age at diagnosis (y) | |||||||||||||||
| < 50 | 194 | 126 (65.0) | 165 (85.1) | 16 (8.3) | 54 (27.8) | 27 (13.9) | 26 (13.4) | 46 (23.7) | |||||||
| ≥ 50 | 571 | 355 (62.2) | 0.49 | 487 (85.3) | 0.94 | 92 (16.1) | 0.007 | 159 (27.9) | 0.99 | 123 (21.54) | 0.02 | 91 (15.9) | 0.40 | 165 (28.9) | 0.16 |
| Menopausal status* | |||||||||||||||
| Pre- | 217 | 144 (66.4) | 190 (87.6) | 24 (11.1) | 65 (30.0) | 31 (14.3) | 22 (10.1) | 59 (27.1) | |||||||
| Post- | 532 | 331 (62.2) | 0.29 | 450 (84.6) | 0.30 | 83 (15.6) | 0.11 | 144 (27.1) | 0.42 | 117 (22.0) | 0.02 | 91 (17.1) | 0.02 | 150 (28.2) | 0.78 |
| Cancer type | |||||||||||||||
| In situ | 95 | 70 (73.7) | 82 (86.3) | 11 (11.6) | 32 (33.7) | 17 (17.9) | 13 (13.7) | 34 (35.8) | |||||||
| Invasive | 670 | 411 (61.3) | 0.02 | 570 (85.1) | 0.75 | 97 (14.5) | 0.45 | 181 (27.0) | 0.17 | 133 (19.9) | 0.65 | 104 (15.5) | 0.64 | 177 (26.4) | 0.06 |
| BMI | |||||||||||||||
| < 25 | 345 | 202 (58.6) | 293 (84.9) | 42 (12.2) | 92 (26.7) | 63 (18.3) | 50 (14.5) | 96 (27.8) | |||||||
| ≥ 25 | 420 | 279 (66.4) | 0.07 | 359 (85.5) | 0.83 | 66 (15.7) | 0.16 | 121 (28.8) | 0.51 | 87 (20.7) | 0.40 | 67 (16.0) | 0.58 | 115 (27.4) | 0.89 |
| Family history of breast cancer* | |||||||||||||||
| No | 604 | 375 (62.1) | 511 (84.6) | 83 (13.7) | 171 (28.3) | 123 (20.4) | 94 (15.6) | 165 (27.3) | |||||||
| Yes | 136 | 93 (68.4) | 0.17 | 119 (87.5) | 0.39 | 21 (15.4) | 0.61 | 35 (25.7) | 0.54 | 22 (16.2) | 0.27 | 21 (15.4) | 0.97 | 36 (26.5) | 0.84 |
| ER status* | |||||||||||||||
| Negative | 136 | 61 (44.9) | 101 (74.3) | 14 (10.3) | 37 (27.2) | 21 (15.4) | 22 (16.2) | 41 (30.1) | |||||||
| Positive | 428 | 282 (65.9) | <0.01 | 383 (89.5) | <0.01 | 73 (17.1) | 0.06 | 122 (28.5) | 0.77 | 96 (22.4) | 0.08 | 70 (16.4) | 0.96 | 117 (27.3) | 0.52 |
| PR status* | |||||||||||||||
| Negative | 206 | 106 (51.5) | 167 (81.1) | 28 (13.6) | 65 (31.6) | 47 (22.8) | 41 (19.9) | 69 (33.5) | |||||||
| Positive | 358 | 237 (66.2) | <0.01 | 317(88.5) | 0.01 | 59 (16.5) | 0.36 | 94 (26.3) | 0.18 | 70 (19.6) | 0.36 | 51 (14.3) | 0.08 | 89 (24.9) | 0.03 |
When < 765 data unknown or missing
Since survival rates of in situ and invasive breast cancers are substantially different, survival analyses were only conducted within 670 invasive cases. Cox proportional hazard regression [34] was used to estimate the hazard ratios (HR) and 95% confidence intervals (95% CI) for the association between gene promoter methylation status and breast cancer-specific or all-cause mortality. We first considered each gene separately in relation to mortality, adjusting only for age at diagnosis. Then we used a backward elimination strategy to test for potential confounding by the following covariates (race, BMI, family history of breast cancer, and history of benign breast disease) on the associations between methylation markers and breast cancer-specific or all-cause mortality. None of these covariates changed the effect estimate for any of the methylation markers by 10% or more [35], and thus were not included in the final models. We did not consider as potential confounders, clinical characteristics (including tumor size, nodal involvement or first course of treatment), because under our hypothesis, these factors can be considered to be on the causal pathway between the tumor marker and the study outcome (mortality). As epidemiologic methodologists have long argued, inclusion of such mediators in our models would result in biased estimates of effect [36]. Thus, our underlying hypothesis is that these tumor markers of methylation are not influenced by clinical characteristics such as tumor size and nodal involvement, but instead occur prior to disease progression. All statistical analyses were completed using Statistical Analysis System 9.0 (SAS Institute, Cary, NC).
RESULTS
We examined promoter methylation status for a panel of 8 genes in a population-based cohort of 839 women with breast cancer. Among the 839 cases with tumor tissue DNA, we obtained methylation data on all 8 genes for 765 tumors; failures were due to insufficient amount or poor quality DNA. Promoter hypermethylation of HIN1, RASSF1A, DAPK1, GSTP1, CyclinD2, TWIST, CDH1 and RARβ was observed in 62.9%, 85.2%, 14.1%, 27.8%, 19.6%, 15.3%, 5.8% and 27.6% of the samples, respectively.
For those 7 genes for which the prevalence of methylation was greater than 10%, the relationships with selected clinical/pathological parameters are summarized in Table 1. Methylation in DAPK1 and CyclinD2 was more frequent in women with breast cancer over 50 years of age at diagnosis (P=0.007 and P=0.02, respectively), and CyclinD2 and TWIST methylation was more frequent among women who were postmenopausal at the time of diagnosis (both, P=0.02). In situ case tumors had more frequent methylation of HIN1 than invasive cancers (P=0.02). Hormone receptor status of the case tumor was also positively associated with a high frequency of HIN1 and RASSF1A methylation (P=0.02 and P=0.009, respectively). Specifically, patients with ER+ or PR+ tumors had more frequent methylation of HIN1 and RASSF1A than those with ER- or PR- tumors.
Among the 308 (20.4%) participants who died within the follow-up period, tumor DNA methylation data were available for 161 women with invasive breast cancer. As shown in Table 2, methylation status of GSTP1, TWIST and RARβ in the case tumor was associated with higher breast cancer-specific mortality, after adjustment for age at diagnosis. Compared to cases with an unmethylated promoter in tumor tissue, those with a methylated promoter had a 71%, 67% and 78% increased risk of dying from breast cancer at the end of follow up for GSTP1 (HR: 1.71; 95% CI: 1.10-2.65), TWIST (HR: 1.67; 95% CI: 1.01-2.79) and RARβ (HR: 1.78; 95% CI: 1.15-2.76), respectively. Similar but somewhat weaker associations between methylation status in these genes and all-cause mortality were also observed; the associations with GSTP1 and RARβ were significant (HR: 1.49; 95% CI: 1.08-2.07 and HR: 1.45; 95% CI: 1.05-2.02, respectively).
Table 2.
Age-adjusted hazard ratios (HRs) and 95% confidence intervals (CI) for methylation status of selected tumor markers and mortality after 8 years of follow up among a population-based cohort of women diagnosed with a first primary breast cancer in 1996-1997, Long Island Breast Cancer Study Project
| Genes | No. of cases | All-cause mortality | Breast cancer-specific mortality | ||
|---|---|---|---|---|---|
| No. of death | Age-adjusted HR (95% CI) | No. of death | Age-adjusted HR (95% CI) | ||
| HIN1 | |||||
| Unmethylated | 284 | 62 | 1.00 (Ref.) | 31 | 1.00 (Ref.) |
| Methylated | 481 | 110 | 1.05 (0.77-1.44) | 59 | 1.12 (0.72-1.73) |
| RASSF1A | |||||
| Unmethylated | 113 | 21 | 1.00 (Ref.) | 9 | 1.00 (Ref.) |
| Methylated | 652 | 151 | 1.24 (0.78-1.95) | 81 | 1.61 (0.81-3.21) |
| DAPK1 | |||||
| Unmethylated | 657 | 143 | 1.00 (Ref.) | 74 | 1.00 (Ref.) |
| Methylated | 108 | 29 | 1.12 (0.75-1.67) | 16 | 1.33 (0.77-2.29) |
| GSTP1 | |||||
| Unmethylated | 552 | 113 | 1.00 (Ref.) | 56 | 1.00 (Ref.) |
| Methylated | 213 | 59 | 1.43 (1.05-1.97) | 34 | 1.66 (1.09-2.54) |
| CyclinD2 | |||||
| Unmethylated | 615 | 128 | 1.00 (Ref.) | 69 | 1.00 (Ref.) |
| Methylated | 150 | 44 | 1.23 (0.87-1.74) | 21 | 1.27 (0.77-2.08) |
| TWIST1 | |||||
| Unmethylated | 648 | 138 | 1.00 (Ref.) | 70 | 1.00 (Ref.) |
| Methylated | 117 | 34 | 1.28 (0.88-1.87) | 20 | 1.69 (1.02-2.78) |
| RARβ | |||||
| Unmethylated | 554 | 114 | 1.00 (Ref.) | 56 | 1.00 (Ref.) |
| Methylated | 211 | 58 | 1.37 (1.00-1.89) | 34 | 1.69 (1.10-2.59) |
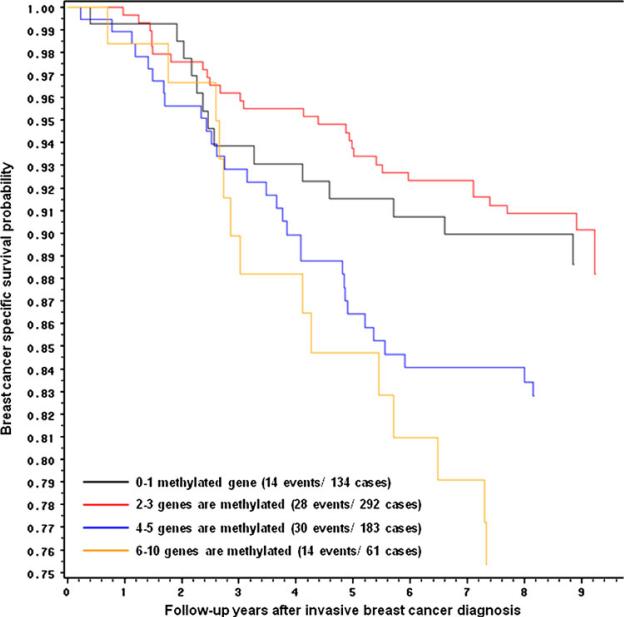
We also considered joint gene effects, combining cases into 4 groups according to the number of genes methylated. For this analysis we also included MethyLight data on p16 and APC methylation status from our prior studies [20]. As shown in Table 3, a dose-response association emerged; as the number of methylated genes increased, the magnitude of the associations with all-cause and breast cancer-specific mortality increased (Ptrend=0.02, Ptrend=0.002, respectively). With 6-10 genes methylated, the breast cancer-specific mortality HR was 2.45 (95%CI: 1.16-5.17). Similar but weaker associations were observed for all-cause mortality (HR: 1.49; 95%CI: 0.87-2.57). Figure 1 presents Kaplan-Meier breast cancer-specific survival curves, which illustrates the decreasing survival associated with increasing number of genes methylated.
Table 3.
Number of methylated genes in relation to all-cause or breast cancer-specific mortality after 8 years of follow-up among a population-based cohort of women diagnosed with breast cancer in 1996-1997, Long Island Breast Cancer Study Project
| No of genes Methylated* | No. of cases | All-cause mortality | Breast cancer-specific mortality | ||
|---|---|---|---|---|---|
| No. of Death | HR** (95% CI) | No. of death | HR** (95% CI) | ||
| 0-1 | 149 | 32 | 1.00 | 14 | 1.00 |
| 2-3 | 329 | 59 | 0.76 (0.49-1.16) | 30 | 0.95 (0.50-1.79) |
| 4-5 | 215 | 57 | 1.24 (0.80-1.91) | 31 | 1.61 (0.85-3.02) |
| 6-10 | 72 | 24 | 1.41 (0.83-2.40) | 15 | 2.38 (1.14-4.96) |
Data were combined with previously published data (20, 21) on APC and p16.
Adjusted for age at diagnosis as continuous, Ptrend = 0.03, HR = 1.21 (95%CI: 1.02-1.43) for all-cause mortality; Ptrend = 0.004, HR = 1.41 (95%CI: 1.12-1.78) for breast cancer-specific mortality
Fig. 1.
Kaplan–Meier breast cancer survival curves for number of methylated genes in tumor tissue among a population-based cohort of women diagnosed with a first primary invasive breast cancer in 1996–1997 and followed for 8 years, Long Island Breast Cancer Study Project. Black carrying 0–1, red 2–3, blue 4–5 or yellow 6–10 methylated genes
DISCUSSION
Patient age, axillary lymph node status, tumor size, histologic grade, hormone receptor status and HER2 status have been used historically to determine prognosis in women with breast cancer [37]. More recently, gene expression data have been used with two commercial laboratory tests available to assist in treatment decisions [1]. However these assays have limitations suggesting that other approaches maybe of use. We began conducting studies to understand the relationship between gene-specific methylation and prognosis using data on the population-based cohort of women diagnosed with a first primary breast cancer in the LIBCSP. We previously reported positive associations between promoter methylation status of BRCA1, APC and p16 and poor disease outcome among women with breast cancer [19;20]. In the present study, we evaluated methylation in 8 additional genes in the same cohort of breast cancer cases. Our univariate results showed that promoter hypermethylation of three tumor suppressor genes, GSTP1, TWIST1 and RARβ was associated with breast cancer-specific mortality, while hypermethylation in GSTP1 and RARβ showed significant associations with all-cause mortality. We also found significant inverse associations between decreasing breast cancer survival and increasing number of genes methylated.
Few prior studies have investigated tumor methylation status and prognosis. While we observed an association with TWIST1 methylation and prognosis, a prior smaller study (N=151) [38] did not find an association with sentinel lymph node tumor status, a marker of metastasis. In contrast, our positive results for methylation in GSTP1 and poorer prognosis had been reported previously in a study of 100 breast cancer cases [39]. Another study reported that RASSF1A methylation was associated with poorer prognosis [16]; and although we also found a positive association with RASSF1A methylation and breast cancer mortality (HR: 1.77; 95%CI: 0.86-3.67), the effect estimate was not significant possibly due to the high frequency of positive tumors. In a study of 99 tumors from carriers of BRCA1 or 2 mutations, methylation of six genes; ERα, TWIST, Cyclin D2, CDH1, APC and RASSF1 was elevated for at least one of the histopathological variables tested (ER, HER2, lymph node status, tumor stage and recurrent or metastatic disease) [40]. One previous study [41] that examined 7 genes, along with our own prior study with 3 genes [20], observed that a panel of hypermethylated genes has higher predictive power than single genes in determining breast cancer survival. In the current study, data on 10 genes were analyzed, and a trend was observed in which increasing number of methylated genes was associated with poorer survival. With 6-10 genes methylated, the HR was 2.45 (1.16-5.17). These findings suggest that hypermethylation-mediated silencing of multiple tumor-suppressor genes influences prognosis of breast cancer.
We found few correlations of tumor methylation status with clinical/pathological factors. While older women had more frequent DAPK1 and CyclinD2 methylation then younger women, there were fewer women <50 years of age at diagnosis in our study cohort. Similarly, associations were found for CyclinD2 and TWIST1 methylation and menopausal status and for methylation in HIN1 and RASSF1A and ER/PR status, but cell sizes were limited. Although the higher frequency of methylation for RASSF1A among women with ER/PR positive tumors has been observed previously [38], most of these associations await confirmation by others. Thus, these results should be interpreted with caution.
Since we expanded our study with additional breast cancer-related tumor suppressor genes on the same study population and used the same tumor tissue blocks as our previous study [20], the limitations in the present study are as previously described. First, since the blocks were collected from multiple hospitals with non-standardized protocols, some clinicopathologic parameters which are known as independent breast cancer prognostic factors were not completed on the medical records for all cases. This limited our more detailed investigation of the predictive effect of gene methylation status and prognosis. However, among women with this information available, we explored the association between methylation status and these clinical covariates, according to cancer type, and found no substantial differences. Second, the frequency of hypermethylation of CDH1 and the number of deaths in some of the subgroups are relatively small. Thus, some subgroup analyses are based on small number of patients, which are reflected in the wide confidence intervals. These results should be interpreted carefully. Third, the overwhelming proportion of our study participants are white women with breast cancer (which reflects the underlying racial distribution of the geographic area of the LIBCSP), and thus our results are not generalizable to other race/ethnicities. Another limitation of our study is the possibility of misclassification on the cause of death. Uncertainty regarding cause of death on death certificates is an ongoing issue in most studies of cause-specific mortality, including our own. In the current study, we used the National Death Index, which has been validated and is considered the gold standard source of mortality data for epidemiologic studies [42].
Advantages of our study approach include a large population-based cohort of women diagnosed with a first primary breast cancer, with both biospecimens and a comprehensive assessment of factors suspected of influencing prognosis. This approach allows for greater generalizability and higher sensitivity, since multiple factors were taken into consideration in examining the associations with survival. Moreover, there are few epidemiologic studies that have reported on the prognostic value of multiple gene promoter methylation status in breast cancer. In summary, we examined promoter methylation status of 8 additional breast cancer-related genes and explored their relationship with clinical/pathological factors as well as survival among a cohort of women diagnosed with breast cancer. Our results suggest that promoter methylation, particularly for a panel of multiple genes, could offer additional value to predict prognosis of breast cancer.
Acknowledgment
This work was supported by a grant from the National Institutes of Health (CA109753 to JC) and in part by grants from Department of Defense (BC031746), National Cancer Institute and the National Institutes of Environmental Health and Sciences (UO1CA/ES66572, UO1CA66572, P30CA013696, P30ES009089, and P30ES10126); Xu, X. is a recipient of the Predoctoral Traineeship Award (W81XWH-06-1-0298) of Department of Defense Breast Cancer Research Program.
Reference List
- 1.Kim C, Paik S. Gene-expression-based prognostic assays for breast cancer. Nat.Rev.Clin.Oncol. 2010;7:340–347. doi: 10.1038/nrclinonc.2010.61. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat.Rev.Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 3.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 4.Jin Z, Tamura G, Tsuchiya T, Sakata K, Kashiwaba M, Osakabe M, Motoyama T. Adenomatous polyposis coli (APC) gene promoter hypermethylation in primary breast cancers. Br J Cancer. 2001;85:69–73. doi: 10.1054/bjoc.2001.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva JM, Dominguez G, Villanueva MJ, Gonzalez R, Garcia JM, Corbacho C, Provencio M, Espana P, Bonilla F. Aberrant DNA methylation of the p16INK4a gene in plasma DNA of breast cancer patients. British Journal of Cancer. 1999;80:1262–1264. doi: 10.1038/sj.bjc.6690495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agathanggelou A, Honorio S, Macartney DP, Martinez A, Dallol A, Rader J, Fullwood P, Chauhan A, Walker R, Shaw JA, Hosoe S, Lerman MI, Minna JD, Maher ER, Latif F. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001;20:1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- 7.Dulaimi E, Hillinck J, Ibanez dC, I, Al Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin.Cancer Res. 2004;10:6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- 8.Nass SJ, Herman J, Gabrielson E, Iversen P, Parl F, Davidson N, Graff J. Aberrant Methylation of the Estrogen Receptor and E-Cadherin 5' CpG Islands Increases with Malignant Progression in Human Breast Cancer. Cancer Research. 2000;60:4346–4348. [PubMed] [Google Scholar]
- 9.Sirchia SM, Ferguson AT, Sironi E, Subramanyan S, Orlandi R, Sukumar S, Sacchi N. Evidence of epigenetic changes affecting the chromatin state of the retinoic acid receptor beta2 promoter in breast cancer cells. Oncogene. 2000;19:1556–1563. doi: 10.1038/sj.onc.1203456. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M, Corn PG, Urena JM, Gabrielson E, Baylin SB, Herman JG. Inactivation of glutathione S-transferase P1 gene by promoter hypermethylation in human neoplasia. Cancer Res. 1998;58:4515–4518. [PubMed] [Google Scholar]
- 11.Evron E, Umbricht C, Korz D, Raman V, Loeb D, Niranjan B, Buluwela L, Weitzman S, Marks J, Sukumar S. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Research. 2001;61:2782–2787. [PubMed] [Google Scholar]
- 12.Fackler M, McVeigh M, Mehrotra J, Blum M, Lange J, Lapides A, Garrett E, Argani P, Sukumar S. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Research. 2004;64:4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 13.Gort EH, Suijkerbuijk KP, Roothaan SM, Raman V, Vooijs M, van der WE, van Diest PJ. Methylation of the TWIST1 promoter, TWIST1 mRNA levels, and immunohistochemical expression of TWIST1 in breast cancer. Cancer Epidemiol.Biomarkers Prev. 2008;17:3325–3330. doi: 10.1158/1055-9965.EPI-08-0472. [DOI] [PubMed] [Google Scholar]
- 14.Nimmrich I, Sieuwerts AM, Meijer-van Gelder ME, Schwope I, Bolt-de VJ, Harbeck N, Koenig T, Hartmann O, Kluth A, Dietrich D, Magdolen V, Portengen H, Look MP, Klijn JG, Lesche R, Schmitt M, Maier S, Foekens JA, Martens JW. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Res.Treat. 2008;111:429–437. doi: 10.1007/s10549-007-9800-8. [DOI] [PubMed] [Google Scholar]
- 15.Kioulafa M, Balkouranidou I, Sotiropoulou G, Kaklamanis L, Mavroudis D, Georgoulias V, Lianidou ES. Methylation of cystatin M promoter is associated with unfavorable prognosis in operable breast cancer. Int.J.Cancer. 2009;125:2887–2892. doi: 10.1002/ijc.24686. [DOI] [PubMed] [Google Scholar]
- 16.Kioulafa M, Kaklamanis L, Mavroudis D, Georgoulias V, Lianidou ES. Prognostic significance of RASSF1A promoter methylation in operable breast cancer. Clin.Biochem. 2009;42:970–975. doi: 10.1016/j.clinbiochem.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Kioulafa M, Kaklamanis L, Stathopoulos E, Mavroudis D, Georgoulias V, Lianidou ES. Kallikrein 10 (KLK10) methylation as a novel prognostic biomarker in early breast cancer. Ann.Oncol. 2009;20:1020–1025. doi: 10.1093/annonc/mdn733. [DOI] [PubMed] [Google Scholar]
- 18.Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Kushi LH, Kwan ML, Wiencke JK. Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS.Genet. 2010;6:e1001043. doi: 10.1371/journal.pgen.1001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, Gammon MD, Zhang Y, Bestor TH, Zeisel SH, Wetmur JG, Wallenstein S, Bradshaw PT, Garbowski G, Teitelbaum SL, Neugut AI, Santella RM, Chen J. BRCA1 promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res.Treat. 2009;115:397–404. doi: 10.1007/s10549-008-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Gammon MD, Zhang Y, Cho YH, Wetmur JG, Bradshaw PT, Garbowski G, Hibshoosh H, Teitelbaum SL, Neugut AI, Santella RM, Chen J. Gene promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res.Treat. 2010;121:685–692. doi: 10.1007/s10549-009-0628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gammon MD, Neugut AI, Santella RM, Teitelbaum SL, Britton JA, Terry MB, Eng SM, Wolff MS, Stellman SD, Kabat GC, Levin B, Bradlow HL, Hatch M, Beyea J, Camann D, Trent M, Senie RT, Garbowski G, Maffeo C, Montalvan P, Berkowitz GS, Kemeny M, Citron C, Schnabel F, Schuss A, Hajdu S, Vinceguerra V, Collman GW, Obrams G. The Long Island Breast Cancer Study Project: description of a multi-institutional collaboration to indentify environmental risk factors for breast cancer. Breast Cancer Res.Treat. 2002;74:235–254. doi: 10.1023/a:1016387020854. [DOI] [PubMed] [Google Scholar]
- 22.Gammon MD, Santella RM, Neugut AI, Eng SM, Teitelbaum SL, Paykin A, Levin B, Terry MB, Young TL, Wang LW, Wang Q, Britton JA, Wolff MS, Stellman SD, Hatch M, Kabat GC, Senie R, Garbowski G, Maffeo C, Montalvan P, Berkowitz GS, Kemeny M, Citron M, Schnabel F, Schuss A, Hajdu S, Vinceguerra V. Environemntal toxins and breast cancer on Long Island. I. Polycyclic aromatic hydrocarbon DNA adducts. Cancer Epidemiol.Biomarkers & Prev. 2002;11:677–685. [PubMed] [Google Scholar]
- 23.Cleveland RJ, Eng SM, Abrahamson PE, Britton JA, Teitelbaum SL, Neugut AI, Gammon MD. Weight gain prior to diagnosis and survival from breast cancer. Cancer Epidemiol.Biomarkers Prev. 2007;16:1803–1811. doi: 10.1158/1055-9965.EPI-06-0889. [DOI] [PubMed] [Google Scholar]
- 24.Rossner P, Jr., Gammon MD, Zhang YJ, Terry MB, Hibshoosh H, Memeo L, Mansukhani M, Long CM, Garbowski G, Agrawal M, Kalra TS, Gaudet MM, Teitelbaum SL, Neugut AI, Santella RM. Mutations in p53, p53 protein overexpression and breast cancer survival. J.Cell Mol.Med. 2009;13:3847–3857. doi: 10.1111/j.1582-4934.2008.00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res.Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 26.Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA, Laird PW. Epigenetic Patterns in the Progression of Esophageal Adenocarcinoma. Cancer Research. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 27.Widschwendter M, Siegmund KD, Muller HM, Fiegl H, Marth C, Muller-Holzner E, Jones PA, Laird PW. Association of Breast Cancer DNA Methylation Profiles with Hormone Receptor Status and Response to Tamoxifen. Cancer Research. 2004;64:3807–3813. doi: 10.1158/0008-5472.CAN-03-3852. [DOI] [PubMed] [Google Scholar]
- 28.Muller HM, Widschwendter A, Fiegl H, Goebel G, Wiedemair A, Muller-Holzner E, Marth C, Widschwendter M. A DNA methylation pattern similar to normal tissue is associated with better prognosis in human cervical cancer. Cancer Lett. 2004;209:231–236. doi: 10.1016/j.canlet.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, Laird PW, Skinner KA. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- 31.Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ, Laird PW, Loda M, Fuchs CS. Precision and performance characteristics of bisulfite conversion and real-time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn. 2006;8:209–217. doi: 10.2353/jmoldx.2006.050135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SY, Kwon HJ, Lee HE, Ryu HS, Kim SW, Kim JH, Kim IA, Jung N, Cho NY, Kang GH. Promoter CpG island hypermethylation during breast cancer progression. Virchows Arch. 2011;458:73–84. doi: 10.1007/s00428-010-1013-6. [DOI] [PubMed] [Google Scholar]
- 33.Sproul D, Nestor C, Culley J, Dickson JH, Dixon JM, Harrison DJ, Meehan RR, Sims AH, Ramsahoye BH. Transcriptionally repressed genes become aberrantly methylated and distinguish tumors of different lineages in breast cancer. Proc.Natl.Acad.Sci.U.S.A. 2011;108:4364–4369. doi: 10.1073/pnas.1013224108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosmer DW, Lemeshow S. Applied Survival Analysis: Regression Modeling of Time to Event Data. John Wiley & Sons, Inc; New York: 1999. [Google Scholar]
- 35.Hosmer DW, Lemenshow S. Applied Logistic Regression. John Wiley & Sons; New York: 1989. [Google Scholar]
- 36.Rothman KJ, Greenland S. Modern Epidmeiology. Lippcott-Raven; New York: 1998. [Google Scholar]
- 37.Schnitt SJ. Classification and prognosis of invasive breast cancer: from morphology to molecular taxonomy. Mod.Pathol. 2010;23(Suppl 2):S60–S64. doi: 10.1038/modpathol.2010.33. [DOI] [PubMed] [Google Scholar]
- 38.Shinozaki M, Hoon DS, Giuliano AE, Hansen NM, Wang HJ, Turner R, Taback B. Distinct hypermethylation profile of primary breast cancer is associated with sentinel lymph node metastasis. Clin.Cancer Res. 2005;11:2156–2162. doi: 10.1158/1078-0432.CCR-04-1810. [DOI] [PubMed] [Google Scholar]
- 39.Sharma G, Mirza S, Parshad R, Srivastava A, Gupta SD, Pandya P, Ralhan R. Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci. 2010;87:83–91. doi: 10.1016/j.lfs.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Swift-Scanlan T, Vang R, Blackford A, Fackler MJ, Sukumar S. Methylated genes in breast cancer: Associations with clinical and histopathological features in a familial breast cancer cohort. Cancer Biol.Ther. 11:2011. doi: 10.4161/cbt.11.10.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma G, Mirza S, Yang YH, Parshad R, Hazrah P, Datta GS, Ralhan R. Prognostic relevance of promoter hypermethylation of multiple genes in breast cancer patients. Cell Oncol. 2009;31:487–500. doi: 10.3233/CLO-2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rich-Edwards JW, Corsano KA, Stampfer MJ. Test of the National Death Index and Equifax Nationwide Death Search. Am.J.Epidemiol. 1994;140:1016–1019. doi: 10.1093/oxfordjournals.aje.a117191. [DOI] [PubMed] [Google Scholar]