INTRODUCTION
One of the major barriers for clinical pharmacogenomics has been the efficient integration of rapid turnaround time genetic testing into routine clinical practice. To address this specific challenge, both point-of-care and pre-emptive pharmacogenomic testing programs, initially centered on cardiovascular pharmacogenomics, have recently been deployed. Early results indicate that these strategies are both feasible and likely beneficial clinically; however, despite these exciting advances towards implementing clinical pharmacogenomics, challenges for widespread adoption still remain.
The field of pharmacogenomics has grown dramatically since the initial scientific discoveries in the 1950’s that identified interindividual drug response variability within unique medical contexts. Subsequent work established important enzymes implicated in drug metabolism and pharmacodynamic pathways, whose genes have since been characterized at the nucleotide level by the identification of functionally relevant variant alleles. Consequently, pharmacogenomics is often considered one of the most actionable areas of the personalized medicine paradigm, as evidenced by the increased availability of clinical testing for selected genes involved in drug response among Clinical Laboratory Improvement Amendments (CLIA)-certified laboratories. Moreover, multiplexed genotyping and sequencing panels enriched for important pharmacogenes have been commercially developed for both research and clinical use, prompting the possibility of providing patients with a predicted drug metabolism phenotype profile. However, despite continued discoveries and the advances in molecular technologies, effective clinical implementation remains a challenge for pharmacogenomics. Some reasons for this include professional education, reimbursement issues, continued debate over clinical utility, and the feasibility of incorporating rapid turnaround time genetic testing into routine clinical practice. Although addressing all of these barriers are beyond the scope of this Practice, recent developments regarding point-of-care and pre-emptive testing for constitutional pharmacogenomic variants are reviewed.
Clinical DNA-based testing traditionally has been limited to constitutional mutations implicated in rare Mendelian disorders and acquired somatic mutations involved in cancer pathology. As such, CLIA-certified laboratories and relevant professional societies have successfully developed molecular testing programs for carrier-screening of autosomal recessive disorders, postnatal molecular diagnosis, prenatal mutation analysis, and preimplantation genetic diagnosis. Although rapid testing can be performed in selected scenarios, typical turnaround times for genetic testing in a clinical laboratory are usually days to weeks depending on the testing technology [e.g., genotyping (shorter turnaround time) versus sequencing (longer turnaround time)] (Figure 1). Unfortunately, for the majority of currently actionable pharmacogenomic variants, these turnaround times are unacceptable for efficient implementation into routine clinical care. For example, in the cardiovascular pharmacogenomics field, the often-cited examples of warfarin and clopidogrel require knowledge of CYP2C9/VKORC1 and CYP2C19 genotypes, respectively, at the time of drug initiation for their most effective use (1). Moreover, given the demanding environments common to most anticoagulation clinics and cardiac catheterization laboratories, disruption of routine care by interfacing with an external clinical laboratory that has additional genetic testing logistics and unique laboratory information management systems can present further complexities for effective testing of pharmacogenetic variants.
Figure 1.
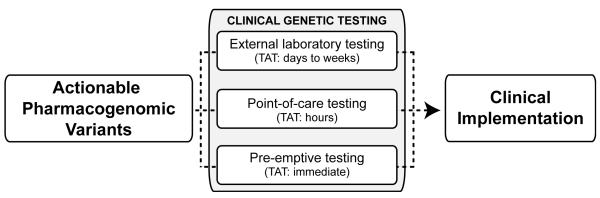
Illustrated is a conceptual diagram highlighting the different mechanisms of testing for actionable pharmacogenomic variants and their implementation into routine clinical practice. Notable are the turnaround times for each testing scenario and the need for clinical decision support when deploying pre-emptive genetic testing. TAT: turnaround time.
To address this issue, many commercial companies have been developing genotyping platforms that offer rapid sample-to-result assays that will be highly beneficial for integrating pharmacogenomics at the point-of-care (2) (Figure 1). A notable example is the recently reported implementation of CYP2C19*2 (c.681G>A) point-of-care genetic testing for cardiac patients initiating clopidogrel therapy following percutaneous coronary intervention (PCI) in the Reassessment of Anti-Platelet Therapy Using An Individualized Strategy Based on Genetic Evaluation (RAPID GENE) trial (3). As a proof-of-concept study, patients were randomly assigned to rapid point-of-care genotyping or to standard treatment and those in the rapid genotyping group were tested for CYP2C19*2 using a buccal swab genetic testing device that reported results within 60 minutes. Carriers were treated with prasugrel, and non-carriers and patients in the standard treatment group were treated with clopidogrel. Notably, no carriers in the rapid genotyping group had high on-treatment platelet reactivity (HTPR) at day seven (the primary endpoint), compared with 30% of patients given standard treatment (p=0.0092). This important study shows that point-of-care genetic testing following PCI can be performed effectively by nursing staff and that personalized antiplatelet therapy can reduce HTPR in this patient population. Related studies measuring clinical outcomes following point-of-care CYP2C19*2 testing with the same device are currently ongoing [e.g., ClinicalTrials.gov Identifier: NCT01452139 (RAPID STEMI); NCT01477775 (GENE-MATRIX)].
Despite the enthusiasm for rapid turnaround time pharmacogenomic testing and the successes of the RAPID GENE trial, issues remain when one considers transferring this type of genetic testing program from research study to routine clinical care. For example, the regulatory landscape of point-of-care testing, particularly that involving genetic testing, can be complicated and no point-of-care genetic tests are currently approved for in vitro diagnostic use by the U.S. Food and Drug Administration (FDA). Point-of-care testing is, by definition, clinical laboratory testing performed at or near the site of clinical care delivery by personnel (or patients) whose primary training is not in the clinical laboratory sciences. The pathway for FDA approval of point-of-care devices includes 510(k) clearance, premarket approval applications or CLIA waivers when a device has a negligible likelihood of erroneous results and has no risk of harm if performed incorrectly. Point-of-care pharmacogenomic testing may not be amenable to a CLIA waiver, which highlights a potential challenge when performing clinical genetic testing at the point-of-care in the absence of personnel with certified genetics expertise. Depending on regional regulations, it is possible that a local CLIA-certified clinical genetics laboratory may be required to oversee the point-of-care testing by remotely managing interpretation, performance, quality control/assurance, and participation in relevant proficiency testing programs. This could increase the overhead costs of point-of-care testing and add potential difficulties when defining the relationship between point-of-care clinical staff and CLIA-certified genetic laboratories.
A technical challenge for point-of-care pharmacogenomic testing involves the content of the genotyping assays themselves. The RAPID GENE trial was centered on a single polymorphic allele, which for future pharmacogenomic assays will not be adequate. This will become relevant when deploying point-of-care pharmacogenomic testing across more diverse patient populations, as the frequencies of relevant variant alleles differ between racial groups and ethnicities. For example, the CYP2C19*3 (c.636G>A; p.W212X) loss-of-function allele has a frequency of ~5% among Asians but is generally not found in other racial and ethnic groups. In addition, more genes and functional variants are necessary for some currently actionable pharmacogenomic examples (e.g., warfarin) and ongoing whole-genome sequencing studies are likely to identify less common variants with large effect sizes that will justify inclusion in future point-of-care testing panels. Moreover, some of the more robust pharmacogenomic associations at the present time involve specific human leukocyte antigen (HLA) alleles of the major histocompatibility complex (e.g., HLA-B*5701 and flucloxacillin-induced liver injury and abacavir-induced hypersensitivity; HLA-B*1502/HLA-A*3101 and carbamazepine-induced hypersensitivity). Unfortunately, HLA genotyping is one of the more challenging molecular assays, requiring combinatorial multiplexing that is beyond the technical capacity of current point-of-care platforms. Despite these content concerns, it is highly likely that future point-of-care platforms will overcome the technical needs for multiplexed sample-to-answer genotyping given the ongoing rapid advances in DNA-based technologies.
Another recently actualized strategy for clinical pharmacogenomics that can circumvent some of these issues surrounding both traditional laboratory and point-of-care testing is pre-emptive clinical genotyping (Figure 1). Although this model has its own set of obstacles for effective clinical implementation, pre-emptive pharmacogenomic testing has recently been deployed at selected academic medical centers (4-5). This approach deposits clinical genotype data in patient records pre-emptively, usually through coordinated biobanking, CLIA-certified testing and informatics, and alerts prescribers at the point-of-care through sophisticated electronic clinical decision support (CDS) systems when a drug is ordered for a patient with an at-risk genotype. The immediate knowledge of personalized and relevant pharmacogenomic variation, with interpretation and possible/recommended actions, without any disruption of routine clinical care is the clear advantage to this strategy. Additionally, pre-emptive genotyping can still utilize CLIA-certified laboratory testing, but without the usual concerns regarding turnaround time. However, the necessary institutional investments in informatics, CDS, health-care provider participation and education, and infrastructure for biobanking and testing in a CLIA-certified environment suggest that this exciting mode of clinical pharmacogenomic delivery will be limited to large academic medical centers.
In conclusion, clinical pharmacogenomics has been greatly reinforced by the recent advances in point-of-care and pre-emptive genetic testing. Both strategies provide the opportunity to integrate pharmacogenomic test results into routine care by addressing the important issue of turnaround time, which previously has been one of the major barriers towards effective clinical implementation. Of note, the specific intricacies of laboratory, point-of-care, and pre-emptive genetic testing suggest that no one mechanism will be sufficient for widespread adoption of all actionable pharmacogenomic examples throughout the health care system. Rather, one can envision a near future where rapid multiplexed devices are commonplace among primary care providers and clinics, with pre-emptive genotyping programs prevalent across most academic medical centers. For the more distant future, the increasing accessibility of whole-genome sequencing programs and direct-to-consumer predisposition testing suggest the interesting possibility of a health care landscape that no longer needs point-of-care pharmacogenomic testing based on that information already being available for all patients. Of utmost importance to that future direction, however, is the continued and much-needed effort toward health care practitioner education in pharmacogenomics, accessible and intuitive CDS, and continued rigorous assessment of clinical utility by the pharmacogenomics communities.
ACKNOWLEDGEMENTS
This work was supported in part by the National Center for Research Resources (NCRR) and the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH), through Grant KL2TR000069.
Footnotes
CONFLICT OF INTEREST
The author has consulted for USDS, Inc.
REFERENCES
- (1).Collet JP, et al. High doses of clopidogrel to overcome genetic resistance: the randomized crossover CLOVIS-2 (Clopidogrel and Response Variability Investigation Study 2) JACC Cardiovasc Interv. 2011;4:392–402. doi: 10.1016/j.jcin.2011.03.002. [DOI] [PubMed] [Google Scholar]
- (2).Dobson MG, Galvin P, Barton DE. Emerging technologies for point-of-care genetic testing. Expert Rev Mol Diagn. 2007;7:359–70. doi: 10.1586/14737159.7.4.359. [DOI] [PubMed] [Google Scholar]
- (3).Roberts JD, et al. Point-of-care genetic testing for personalisation of antiplatelet treatment (RAPID GENE): a prospective, randomised, proof-of-concept trial. Lancet. 2012;379:1705–11. doi: 10.1016/S0140-6736(12)60161-5. [DOI] [PubMed] [Google Scholar]
- (4).Pulley JM, et al. Operational Implementation of Prospective Genotyping for Personalized Medicine: The Design of the Vanderbilt PREDICT Project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Overby LC, et al. Attitudes toward adoption as a potential predictor for the translation of genome-informed interventions: a pharmacogenomic study protocol. Submitted. 2012 [Google Scholar]


