Abstract
The extent to which RNA stability differs between individuals and its contribution to the interindividual expression variation remain unknown. We conducted a genome-wide analysis of RNA stability in seven human HapMap lymphoblastoid cell lines (LCLs) and analyzed the effect of DNA sequence variation on RNA half-life differences. Twenty-six percent of the expressed genes exhibited RNA half-life differences between LCLs at a false discovery rate (FDR) < 0.05, which accounted for ~ 37% of the gene expression differences between individuals. Nonsense polymorphisms were associated with reduced RNA half-lives. In genes presenting interindividual RNA half-life differences, higher coding GC3 contents (G and C percentages at the third-codon positions) were correlated with increased RNA half-life. Consistently, G and C alleles of single nucleotide polymorphisms (SNPs) in protein coding sequences were associated with enhanced RNA stability. These results suggest widespread interindividual differences in RNA stability related to DNA sequence and composition variation.
Population variation of RNA abundances is widespread1,2, influenced by DNA sequence variation3,4,5,6,7,8,9,10,11, and has a heritability of > 30%4,7,9,12,13,14. RNA abundance is determined by the RNA transcription and RNA degradation (referred to here as RNA stability). Of the two, RNA transcription has been shown to have a larger effect than RNA stability on the cellular variance of total RNA abundances between genes15,16. However, at a population level, the extent of RNA stability differences between individuals and how much the interindividual RNA stability differences contribute to the overall RNA abundances remain unknown.
RNA stability is regulated through interactions between cis-regulatory sequences and various RNA-binding proteins and microRNAs (miRNAs)17. DNA sequence variants (or polymorphisms) can modify the regulation of RNA stability and thereby affect interindividual RNA stability differences. The effects of nonsense mutations and 3′-UTR sequence variants have received the most attention18; transcripts containing premature stop codons (i.e., nonsense mutations) undergo accelerated RNA decay, known as nonsense-mediated RNA decay (NMD)18. Within the 3′-UTR, AU-rich elements (AREs) and miRNA target sites mediate RNA decay and block RNA translation19. Cis-regulatory sequences can also be present in the 5′-UTRs, protein coding regions18,20,21, and intron/exon junctions22,23. Synonymous sequence variants are often thought to be functionally silent, but we and others have shown that they can affect RNA stability24,25,26,27,28,29, where, as predicted, G and C alleles were associated with more stable transcripts. In humans, RNA stability differences have been implicated in inflammation30,31,32,33,34, cardiovascular disease35,36, cancer37,38,39, glaucoma27, age-related macular degeneration40, and neuropsychiatric disorders such as mental retardation41,42, Alzheimer's disease43, and some forms of muscular atrophy44,45.
Using LCLs as a cellular model, we explored the extent of interindividual RNA stability differences, the contribution of RAN stability differences to variation of total RNA abundances between genes and between individuals, and the effects of gene sequence features and sequence variants on RNA stability differences in a genome-wide scale (outlined in Table 1). We employed a non-invasive approach to measure genome-wide RNA half-lives through assaying the ratio of 4sU-labeled nascent (newly transcribed) RNAs and total46,47,48. Seven HapMap LCLs derived from genetically unrelated HapMap individuals (hapmap.ncbi.nlm.nih.gov)3,4,5,6,7,10,11 were analyzed. Three of the analyzed LCLs were part of the 1,000 Genomes Project dataset, thus allowing the examination on a genome-wide scale of the effect of specific types of genetic variants on RNA half-life. We found that ~ 25.7% of the expressed genes exhibited interindividual RNA stability differences, and ~ 37% of genes presenting interindividual expression variation showed detectable interindividual RNA stability differences that could explain the direction of gene expression differences. At a genome-wide scale, we found that nonsense mutations correlated with reduced RNA half-life (as expected) while G or C alleles of synonymous or non-synonymous SNPs weakly correlated with increased RNA half-life.
Table 1. Summary of analyses.
| Motivation | Analysis | Result | Conclusion |
|---|---|---|---|
| Interindividual differences of total RNA abundances are prevalent, but extent of interindividual RNA stability differences is unknown. | Use ANOVA to test for RNA stability variation among 3 LCLs (with 3 biological replicates). | ~ 25.7% (n = 2884) genes with FDR < 0.05, of which 452 with > 2-fold differences. | Interindividual RNA stability differences are common. |
| Relative contribution of de novo transcription and of RNA decay to the total RNA abundance variation between genes in LCLs is unknown. | Examine the Pearson correlation of total RNA abundance with RNA half-life and with nascent RNA abundance across all the genes. | Nascent RNA differences explained ~ 80%, and RNA half-life differences explained ~ 6.4% of the variance in total RNA abundances between genes. | Transcriptional differences are a major determinant of the variance in transcript abundances between genes. |
| At population level, how much the interindividual RNA stability differences contribute to the interindividual total RNA abundance differences is unknown. | Calculate the percentages of genes that show inter-individual differences of nascent RNA abundances and/or RNA half-lives among those with inter-individual total RNA abundance differences. | Out of the 6,536 genes with interindividual total RNA abundance differences, 46% showed only nascent RNA differences, 9% showed only RNA half-life differences, and 28% showed differences of both. | Transcriptional differences are a major determinant of the variance in transcript abundances between individuals. |
| RNA stability differences can be influenced by cis-regulatory sequences and genetic variants, trans-factors (e.g., miRNA or RNA-binding proteins) and environmental factors (e.g., cell line growth condition). We investigate whether the effects of sequence features and genetic variants are detectable in LCLs. | Examine the Spearman's rank correlation between RNA half-life and different gene features across all genes. | RNA half-lives were weakly correlated with a number of gene features (transcript length, 3′-UTR length, miRNA target sites, GC and GC3 contents, etc.). | Multiple gene features contribute to RNA half-life differences between genes but with weak effect. |
| Use linear regression to analyze the association of inter-individual RNA half-life differences with specific type of DNA sequence variants. | Nonsense associated with reduced RNA half-lives, and for genes showing interindividual RNA half-life differences, G and C alleles of coding SNPs weakly associated with enhanced RNA stability. | DNA sequence variants contribute to RNA half-life differences between individuals but with weak effect. |
Results
RNA stability in LCLs and interindividual RNA half-life differences
We examined RNA stability by measuring the ratio of nascent RNAs and total RNAs employing two hour (h) 4sU labeling46,47,48,49 in seven LCLs. Three of them (GM07029, GM10835, and GM12813) had biological replicates (i.e., independent cell cultures for the same LCL) and each had technical duplicates (i.e., separate RNA aliquots from the same cell culture) which allowed us to test the reproducibility of the RNA measurements.
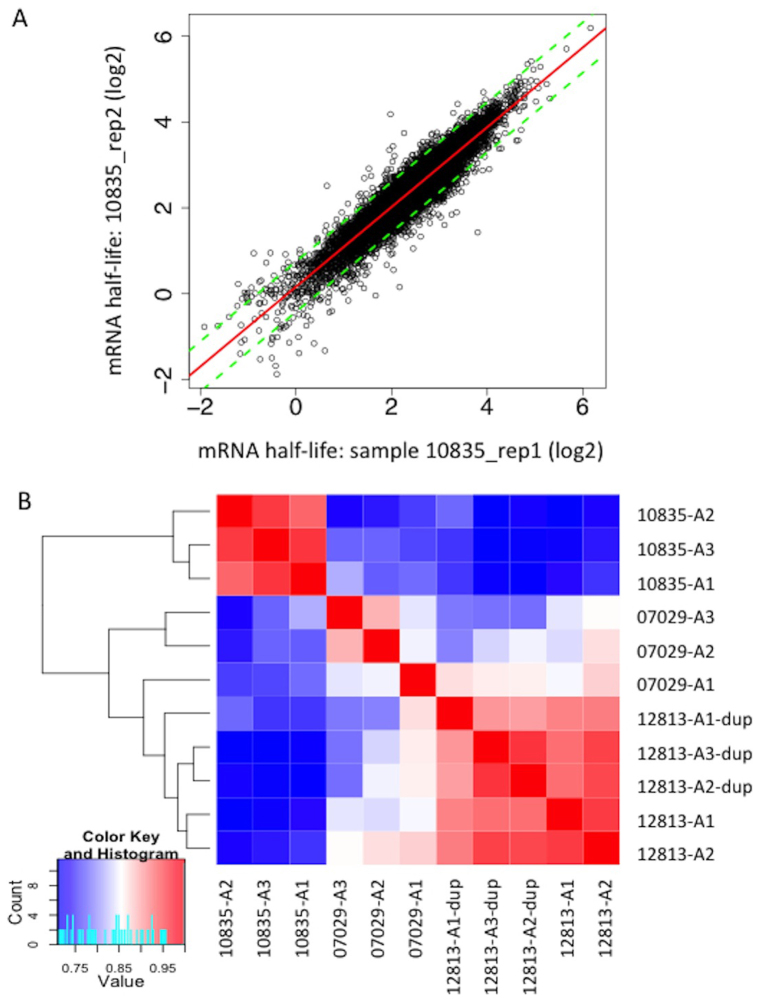
The purified 4sU-labeled nascent RNA accounted for ~ 3% of total RNA. 11,301 RefSeq genes (hg18) passed array quality control metrics and showed detectable expression in both nascent and total RNA samples (see Methods). RNA abundances showed high correlation (Pearson R = 0.91 ~ 0.93; See Supplementary Fig. S1 online) with the measurements on the same specimens that were also used in a previous study50. RNA half-lives were determined for 11,132 genes, and correlated well (Pearson R > 0.5) with those previously reported for a human B cell line48 (See Supplementary Fig. S2 online). While 2 h of 4sU-labeling increases the precision of RNA half-lives for medium and long-lived genes (compared to 1 h of 4sU-labeling), measurements of short RNA half-lives (< 1 h) becomes less precise due to the relatively large nascent/total RNA ratios (> 75%)51. As expected, genes with very short RNA half-life (< 1 h) thus showed rather poor reproducibility of RNA half-life measurements (Figure 1A). However, these genes only accounted for ~ 0.5% of all the expressed genes, and 2 h 4sU-labeling generated reproducible data for the more stable transcripts (Figure 1A). The correlation of RNA half-life (Figure 1B) followed the expected descending order from technical replicates (Pearson R = 0.92 − 0.95) to biological replicates (Pearson R = 0.85 − 0.96), and to unrelated LCLs (Pearson R = 0.72 − 0.82).
Figure 1. Correlation of RNA half-life measurements between replicates and between cell lines.
(A) A representative scatter plot of RNA half-life (log2 scale) from two biological replicates of GM10835. RNA half-life for each gene was calculated from the ratio of total RNA abundances and nascent RNA abundances after 2 h of 4sU-labeling. Green dashed lines indicate the 1.5-fold deviation from the trend line (red) at either side. Pearson R between biological replicates for the three LCLs ranged from 0.85 to 0.96. (B) Heatmap of RNA half-life correlation for all 3 cell lines (GM07029, GM10835, and GM12813) with biological replicates (denoted as A1, A2, and A3) and technical duplicates (denoted as “dup”). As shown in the color key, blue color represents lower correlation (Pearson R) and red color indicates higher correlation.
Gene ontology (GO) analysis of genes with different RNA stability in LCLs using the DAVID tool52 showed consistency with previous reports in other cell types51,53. Rapidly decaying RNAs (n = 396, half-life < 2 h) were found enriched for GO-terms related to transcription (Benjamini family-wide FDR < 0.05; See Supplementary Table S1 online), while slowly decaying RNAs (n = 199, half-life > 15 h) were enriched for GO-terms related to protein synthesis, modification, and trafficking (FDR < 0.05; See Supplementary Table S2 online). For rapidly decaying genes, there was a trend towards enrichment of the GO-term of cell cycle (FDR = 0.058)53.
We then analyzed RNA stability variation in the three LCLs with biological replicates of RNA half-life measurements. Out of the 11,132 genes with measured RNA half-life, analysis of variance (ANOVA) detected 2,884 genes (~ 25.7%) with interindividual RNA half-life differences at a Benjamini & Hochberg54 FDR < 0.05 (corresponding nominal p = 0.013). 452 genes showed ≥ 2-fold interindividual RNA half-life differences (Figure 2A and See Supplementary Table S3). To investigate whether the ANOVA p-value for each transcript is robust to the normality assumption, we performed 1,000 random permutations to approximate the p-values. We found that the p-values based on standard ANOVA and permutations agreed very well (see Supplementary Fig. S3 online), and 21.7% (vs. ~ 25.7% in initial ANOVA) of expressed genes show interindividual RNA half-life differences at FDR < 0.05 (corresponding nominal p = 0.011) in permutation test. Adding the four additional LCLs (GM07019, GM12812, GM12814, and GM12815) for which there were no biological/technical replicates, we found more genes (total n = 3,589) showing interindividual half-life differences at FDR < 0.05 (See Supplementary Table S3), and the statistical significances (ANOVA p-values) of the result from all 7 LCLs correlated well (Pearson R = 0.81; Figure 2B) with that of the three LCLs with biological replicates.
Figure 2. Interindividual differences of RNA half-life.
(A) ANOVA result from using three LCLs (GM07029, GM10835 and GM12813) with 3 biological replicates. The ANOVA p-values (in -log10 scale) are plotted against the between-subject differences of RNA half-life (expressed as the maximum fold of changes between the three LCLs). Red and blue dots represent genes (n = 2,884) showing RNA half-life differences at FDR < 0.05 (p = 0.013), of which red dots indicate genes (n = 452) with 2-fold or higher between-subject RNA half-life differences. (B) ANOVA p-values from using 3 LCLs (GM07029, GM10835 and GM12813) and from using all 7 LCLs (GM07019, GM07029, GM10835, GM12812, GM12813, GM12814, and GM12815) correlate very well (Pearson R = 0.81).
For an independent validation of interindividual differences of RNA half-life, we re-grew the three LCLs (GM07029, GM10835 and GM12813) and performed quantitative PCR (qPCR) on nascent and total RNAs for 15 selected genes. These 15 genes were randomly selected from those exhibiting interindividual RNA half-life differences > 2-fold at FDR < 0.05. We considered a gene as validated if the gene showed significant RNA stability differences (by ANOVA) across the three tested LCLs in qPCR with the same directional changes as in array experiment (based on the positive correlation of RNA stability between qPCR and array experiments). For 12 out of the 15 tested genes, qPCR yielded results consistent with those observed in our array experiment (Table 2). Among the 12 validated genes, 10 showed strong correlations of R > 0.70 (except for LANCL22 with R = 0.49 and C20orf43 with R = 0.59) between qPCR and microarray results, which we considered as a typical validation between qPCR and microarray55,56. One possible explanation for the weaker correlations of those two genes (LANCL22 and C20orf43) could be that multiple probes for each gene were used in exon array experiment while only single probe was used in the qPCR.
Table 2. Confirmation of the interindividual RNA half-life differences of 15 selected genes by qPCR.
| Gene | Method | t1/2-07029 (h) | SD | t1/2-10835 (h) | SD | t1/2-12813 (h) | SD | Fold-diff | ANOVA p | Pearson R (qPCR vs. array) |
|---|---|---|---|---|---|---|---|---|---|---|
| ARPC5L | array | 2.96 | 0.50 | 4.16 | 0.31 | 2.03 | 0.36 | 2.05 | 6.9E-04 | 0.996 |
| qPCR | 2.29 | 0.32 | 3.16 | 0.15 | 1.37 | 0.17 | 2.31 | 2.3E-04 | ||
| MESDC2 | array | 2.61 | 0.25 | 3.87 | 0.45 | 1.81 | 0.44 | 2.14 | 1.4E-03 | 0.964 |
| qPCR | 1.36 | 0.23 | 1.78 | 0.20 | 0.67 | 0.11 | 2.67 | 9.7E-04 | ||
| NFE2L3 | array | 5.64 | 0.99 | 3.07 | 1.07 | 7.81 | 1.81 | 2.54 | 4.4E-03 | −0.998 |
| qPCR | 1.76 | 0.22 | 3.50 | 0.33 | 0.59 | 0.19 | 5.96 | 2.5E-05 | ||
| AMICA1 | array | 4.35 | 0.91 | 9.73 | 2.20 | 5.18 | 0.45 | 2.24 | 8.8E-04 | 0.999 |
| qPCR | 1.70 | 0.11 | 3.67 | 0.34 | 2.10 | 0.41 | 2.16 | 6.0E-04 | ||
| HBXIP | array | 5.54 | 1.13 | 7.95 | 1.08 | 3.84 | 0.78 | 2.07 | 1.7E-03 | 0.695 |
| qPCR | 5.74 | 0.98 | 5.34 | 0.53 | 3.62 | 0.29 | 1.58 | 1.7E-02 | ||
| LANCL2 | array | 2.32 | 0.42 | 2.83 | 0.29 | 1.34 | 0.38 | 2.10 | 2.5E-03 | 0.493 |
| qPCR | 2.44 | 0.10 | 1.70 | 0.18 | 1.33 | 0.08 | 1.83 | 1.3E-04 | ||
| GPC4 | array | 1.45 | 0.30 | 5.70 | 0.56 | 1.19 | 0.21 | 4.80 | 4.3E-06 | 0.837 |
| qPCR | 3.11 | 0.21 | 4.10 | 0.26 | 1.68 | 0.75 | 2.43 | 2.3E-03 | ||
| C20orf43 | array | 3.64 | 0.48 | 6.93 | 0.25 | 2.43 | 0.43 | 2.86 | 2.5E-05 | 0.590 |
| qPCR | 5.38 | 0.41 | 4.97 | 0.40 | 2.93 | 0.33 | 1.83 | 4.9E-04 | ||
| RGS1 | array | 8.41 | 1.60 | 4.18 | 0.57 | 5.45 | 0.46 | 2.01 | 6.6E-04 | −0.156 |
| qPCR | 1.84 | 0.07 | 2.59 | 0.23 | 0.60 | 0.12 | 4.28 | 1.3E-05 | ||
| PDCD4 | array | 4.43 | 0.76 | 5.16 | 0.37 | 2.01 | 0.25 | 2.57 | 9.4E-06 | 0.719 |
| qPCR | 3.27 | 0.22 | 6.39 | 0.20 | 3.06 | 0.11 | 2.09 | 9.7E-07 | ||
| RBMX2 | array | 4.44 | 0.34 | 6.98 | 0.35 | 2.58 | 0.55 | 2.70 | 1.3E-04 | 0.945 |
| qPCR | 2.98 | 0.67 | 3.62 | 0.27 | 1.36 | 0.19 | 2.67 | 1.7E-03 | ||
| DNAJA2 | array | 2.60 | 0.60 | 2.80 | 0.24 | 1.21 | 0.09 | 2.32 | 3.6E-05 | 0.984 |
| qPCR | 1.50 | 0.12 | 1.90 | 0.11 | 0.56 | 0.15 | 3.39 | 2.2E-04 | ||
| RPL14 | array | 7.72 | 1.43 | 5.18 | 0.55 | 3.07 | 0.62 | 2.51 | 6.4E-04 | 0.844 |
| qPCR | 26.60 | 1.31 | 26.50 | 0.84 | 17.49 | 0.38 | 1.52 | 2.8E-05 | ||
| SFXN2 | array | 5.57 | 0.55 | 7.05 | 0.55 | 11.32 | 0.54 | 2.04 | 2.1E-06 | −0.259 |
| qPCR | 3.19 | 0.31 | 7.55 | 0.68 | 3.24 | 0.47 | 2.37 | 6.3E-05 | ||
| IGKC | array | 1.98 | 0.19 | 4.44 | 0.41 | 3.72 | 0.25 | 2.24 | 3.7E-06 | 0.985 |
| qPCR | 0.77 | 0.17 | 13.76 | 0.65 | 12.18 | 0.39 | 17.77 | 6.3E-08 |
Note: RNA half-life (t1/2) measurements from array and qPCR methods are listed. The RNA samples used in array and qPCR experiments were from independent cell cultures for each of the 3 LCLs (GM07029, GM10835, and GM12813). There are 3 biological replicates (i.e., from independent cell cultures) for each measurement and the average RNA half-life and standard deviation (SD) are listed. Fold-diff = the ratio between the largest RNA half-life value and the smallest RNA half-life value among three LCLs. ANOVA is used to test the interindividual differences of RNA half-life. Pearson correlation (R) of RNA half-life measurements between array and qPCR experiments is calculated. Only genes showing both significant ANOVA p value (<0.05) in qPCR validation and positive Pearson R (qPCR vs. array) are considered as independently confirmed (Pearson R in bold).
De novo transcription, RNA half-life, and RNA abundance
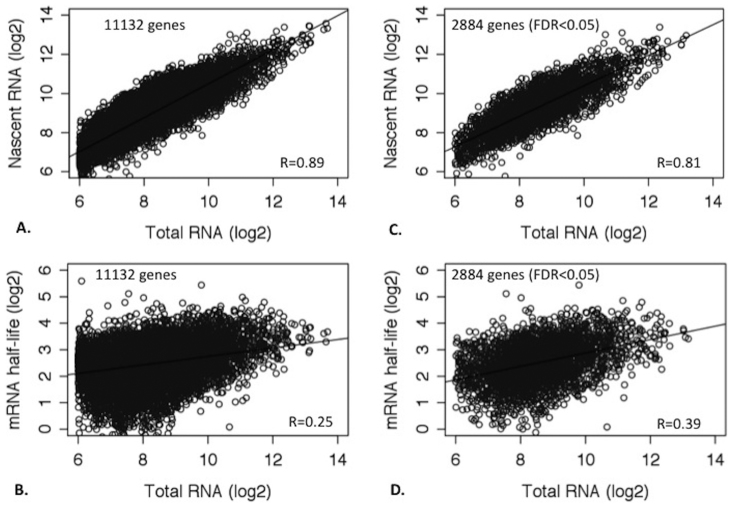
Total RNA abundances are a result of de novo transcription and RNA decay. We estimated the relative contribution of de novo transcription (represented by nascent RNA abundance) and of RNA decay (indexed as RNA half-life) on RNA abundance by examining the coefficient of determination (R2) of the correlations between total RNA, RNA half-life, and nascent RNA abundance. We initially analyzed the 3 LCLs with biological replicates. Total RNA abundances showed a Pearson correlation coefficient (R) of 0.89 (p < 2.2 × 10−16) with nascent RNA abundances, and an R of 0.25 (p < 2.2 × 10−16) with RNA half-lives (Figures 3A and 3B). Hence, based on the R2, nascent RNA abundance differences explained ~ 80%, and RNA half-life differences independently explained ~ 6.4% of the variance in total RNA abundances between genes. For the subset of genes showing interindividual RNA half-life differences (FDR < 5%), the proportion of the variance of total RNA abundances between genes explained by RNA half-life differences between genes increased to ~ 16% as estimated from the correlation between RNA half-life and total RNA abundance (R = 0.39, p < 2.2 × 10−16; Table 3 and Figure 3C and 3D). Analyzing the other 4 LCLs without replicates of measurements gave similar results (See Supplementary Fig. S4 online), with the proportion of variance in RNA abundances explained by RNA half-life differences were at ~ 14.4% (Pearson R = 0.38) for all the expressed genes and ~ 21.2% for the subset of genes showing interindividual RNA half-life differences (FDR < 5%). Including nascent RNA abundance and RNA half-life in a single linear model, we found that de-novo transcription and decay together explained 96.8% of the variance of variance of total RNA.
Figure 3. Correlations of total RNA abundances with nascent RNA abundances and RNA half-lives in LCLs.
Total RNA abundance, nascent RNA abundance and RNA half-life for each of the 11,132 genes (with RNA half-life calculated) were averaged from the 3 LCLs with biological and technical replicates. (A) shows a strong Pearson correlation (R = 0.89) between total RNA and nascent RNA abundances (log2 scale) for all 11,132 genes, indicating that variance of total RNA abundances between genes was predominately determined by transcription. (B) Pearson correlation (R = 0.25) of total RNA abundances with RNA half-life for all 11,132 genes is shown. (C) and (D) show the Pearson correlations of total RNA abundances with nascent RNA abundances and RNA half-life (R = 0.81 and R = 0.39, respectively), for a subset of genes showing interindividual RNA half-life differences at FDR < 0.05 (n = 2,884).
Table 3. Spearman rank correlation of RNA stability with total RNA abundance and gene features.
| All genes (n = 11,132) | Genes with FDR < 0.05 (n = 2,884) | Genes with FDR < 0.01 (n = 1,077) | ||||
|---|---|---|---|---|---|---|
| R | p-value | R | p-value | R | p-value | |
| Total RNA level | 0.253 | < 2.2E-16 | 0.389 | < 2.2E-16 | 0.389 | 2.2E-16 |
| Transcript length | −0.222 | < 2.2E-16 | −0.274 | < 2.2E-16 | -0.242 | 8.5E-16 |
| 3′-UTR length | −0.142 | < 2.2E-16 | −0.171 | < 2.2E-16 | −0.167 | 5.2E-08 |
| # of miRNA targets | −0.146 | < 2.2E-16 | −0.174 | < 2.2E-16 | −0.143 | 4.7E-07 |
| # of miRNA targets/kb 3′-UTR | −0.133 | < 2.2E-16 | −0.151 | 1.7E-15 | −0.120 | 1.0E-04 |
| 3′-UTR folding energy (G) | 0.129 | < 2.2E-16 | 0.142 | 1.1E-13 | 0.115 | 2.4E-04 |
| 5′-UTR length | −0.087 | < 2.2E-16 | −0.094 | 8.6E-10 | −0.143 | 3.6E-06 |
| 5′-UTR folding energy (G) | 0.094 | < 2.2E-16 | 0.111 | 6.9E-09 | 0.131 | 3.0E-05 |
| Coding region length | −0.189 | < 2.2E-16 | −0.209 | < 2.2E-16 | −0.217 | 1.9E-12 |
| Coding GC content | 0.004 | 7.3E-01 | 0.070 | 3.5E-04 | 0.141 | 1.3E-05 |
| Coding GC3 content | 0.029 | 3.7E-03 | 0.113 | 7.9E-09 | 0.197 | 1.0E-09 |
| Coding GC3/GC content | 0.044 | 1.2E-05 | 0.136 | 4.2E-12 | 0.224 | 2.9E-12 |
| # of Intron | −0.014 | 1.4E-01 | −0.038 | 4.6E-02 | −0.064 | 3.8E-02 |
| Intron length | −0.035 | 2.9E-04 | −0.087 | 3.8E-06 | −0.139 | 6.0E-06 |
| Exon junctions/kb transcript | 0.191 | < 2.2E-16 | 0.217 | < 2.2E-16 | 0.176 | 9.2E-09 |
Note: Spearman rank correlation (R) between RNA half-life and each gene feature and the corresponding p-value are listed for all genes (n = 11,132) and genes showing interindividual differences of RNA half-life at FDR < 0.05 (n = 2,884) or FDR < 0.01 (n = 1,077). RNA half-life data are the averages from the 7 LCLs. Gene features for each gene are downloaded from UCSC hg18 (RefSeq genes). RNA folding energies (pre-computed in UCSC hg18) are predicted by RNAfold of Vienna RNA Package81. Bolded are correlations specific to or stronger in genes with interindividual differences of RNA half-life.
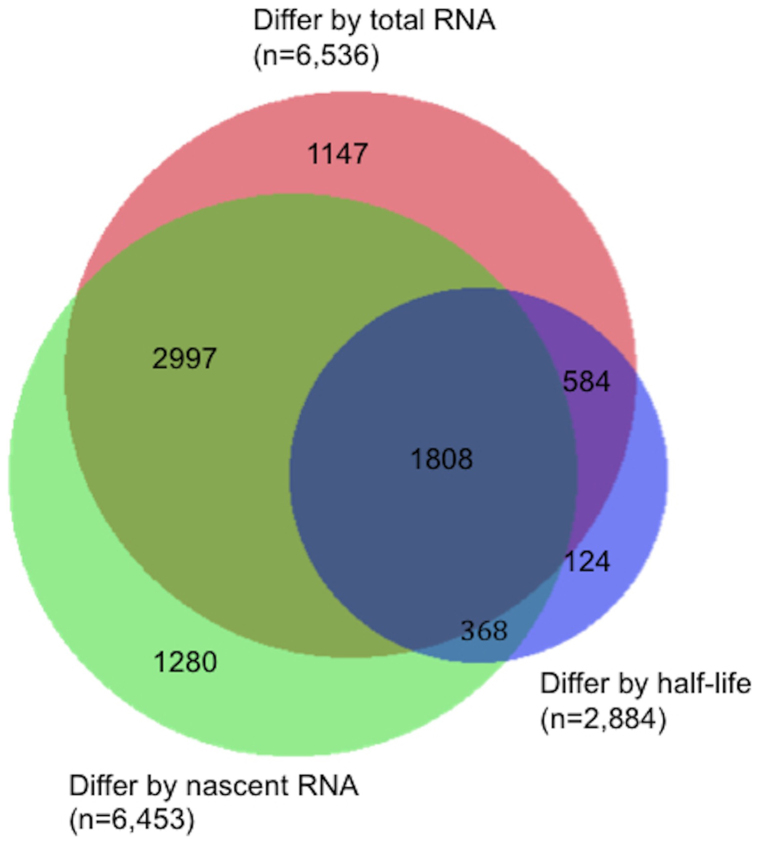
We next estimated the contributions of RNA decay and de novo transcription to the interindividual variation of total RNA abundance. At FDR < 0.05, between the same three LCLs for which we detected 2,884 genes with interindividual RNA half-life differences, 6,536 genes showed differences of total RNA abundances and 6,453 genes showed differences of nascent RNA abundances between LCLs. About 83% of the genes showing interindividual differences of total RNA abundances also showed differences of nascent RNA abundances and/or RNA half-lives (Figure 4). We further determined whether the interindividual differences of total RNA abundances were in the same directions as the interindividual differences of nascent RNA abundances and RNA half-lives by examining the directions of the correlations of total RNA abundances with nascent RNA abundances and with RNA half-lives across the three tested LCLs (See Supplementary Table S4). Out of the 6,536 genes with interindividual total RNA abundance differences, 2,997 (46%) showed differences only at the transcription level and 584 (9%) showed differences only at the RNA decay level, with > 99% of the differences in the same direction as gene expression changes; 1,808 (28%) showed differences at both transcriptional and decay levels, with > 74% of the differences in the same direction as gene expression changes (See Supplementary Table S4 online). These results indicate that transcriptional differences between individuals play a larger role than RNA stability differences in explaining the overall interindividual variation of gene expression. Nevertheless, ~ 37% of all genes presenting significant gene expression variation between individuals also showed RNA half-life differences (28% showing both transcription and RNA half-life differences while 9% showing only RNA_half-life differences), where most (~ 88%) showed the same direction of the differences in RNA half-lives and in total RNA abundances (See Supplementary Table S4).
Figure 4. Venn diagrams showing the number of genes with interindividual differences of total RNA abundances, nascent RNA abundances, RNA half-lives and both.
A total of 11,132 genes were tested by ANOVA for interindividual differences (FDR < 0.05) in total RNA abundances, nascent RNA abundances and RNA half-lives between 3 LCLs. 6,536 genes showed interindividual total RNA abundance differences, of which 2,997 (46%) showed differences only at the transcription level and 584 (9%) showed differences only at the RNA decay level, and 1,808 (28%) showed differences at both transcriptional and RNA decay levels, indicating that transcriptional differences between individuals play a larger role than RNA stability differences in explaining the overall interindividual variation of gene expression. However, genes showing interindividual RNA half-life differences contributed to ~ 37% of all genes showing significant gene expression variation.
Gene features correlated with genome-wide RNA stability
RNA decay is a process that involves both cis-sequence features and trans-protein factors (e.g., RNA-binding proteins and miRNAs). We investigated here whether the effects of cis-sequence features on mRNA stability variation are detectable. As shown in Table 3, using the data from the 7 LCLs, we found a genome-wide negative correlation of RNA half-life with both transcript length (Spearman rank correlation R = − 0.222; p < 2.2 × 10−16) and 3′-UTR length (R = − 0.142; p < 2.2 × 10−16), which is consistent with previous reports22,51,53,57,58,59,60. There was also a positive correlation (R = 0.191; p < 2.2 × 10−16) of RNA half-life with the number of exon/intron junctions per kb transcript (i.e., adjusted for transcript length) as previously reported22,60, which has been suggested to be a result of a stabilizing effect of the exon/intron junction complex (EJC) on spliced transcripts22,61. In addition, we observed a negative correlation of RNA half-life and the number of 3′-UTR miRNA targets (R = − 0.146; p < 2.2 × 10−16), an observation previously reported in plants59 and consistent with the role of miRNAs in destabilizing its target mRNAs in mammalian cells62. For the subset of genes showing interindividual differences of RNA stability, we found a position correlation of RNA half-life with coding GC content (R = 0.141, p = 9.0 × 10−6) and with coding GC3 content (R = 0.197, p = 8.4 × 10−10) (Table 3). The correlation of RNA stability with coding GC3 content was independent from overall coding GC content (R = 0.224, after correction for GC content, p = 2.8 × 10−12). As GC3 content represents the nucleotide sequence composition at the third codon positions (which are mostly synonymous sites), our result suggests a role of synonymous SNPs on affecting RNA stability but with weak effect. The magnitudes of the correlations of RNA half-life with various genomic features in LCLs were low in general (R = 0.1 ~ 0.3), which were consistent to previous reports in other cell types or species22,51,53,57,58,59,60.
As some of these tested genomic features are not fully independent (e.g., 3′-UTR length and the number of miRNA targets), we further examined the overall contribution of these genomic features to the variance of RNA half-life by multiple linear regression analysis. For all the expressed genes, we found that genomic features listed in Table 3 explained ~ 9.5% of the RNA half-life variance (R2 = 0.095). For the subset of genes showing interindividual RNA half-life differences, the fraction of the RNA half-life variance explained by these genomic features increased to ~ 13.4% (R2 = 0.134).
Effect of DNA sequence variant type on RNA stability
Out of the seven LCLs for which we have RNA half-life data, three (GM12812, GM12814 and GM12815) have been sequenced by the 1000 Genomes Project (July 2012 release). Using the resultant genotype data for these three subjects, we examined whether interindividual RNA half-life differences were correlated with specific sequence changes, namely, nonsense SNPs (45 in 41 genes), SNPs disrupting miRNA-target sites (115 in 111 genes), and coding synonymous or non-synonymous SNPs with allele changes between “stable alleles” G or C and “unstable alleles” A or T (14,137 in 5,671 genes) (See Supplementary Table S5 online).
Nonsense mutations represent the most extreme protein coding sequence changes, some of which are known to cause RNA to undergo NMD18. Consistently, nonsense SNPs showed the strongest correlation with RNA half-life differences (See Supplementary Table S6), with premature stop-codons associated with reduced RNA half-lives (regression coefficient β = 0.138, P = 0.009; Figure 5A and See Supplementary Table S5), supporting the validity of our RNA half-life measurements. For SNPs in miRNA target sequences, alleles disrupting the sequence pairing between miRNA and its target sequence are expected to block the miRNA-mediated RNA degradation, thereby increasing RNA half-lives. However, we did not observe a significant correlation between RNA half-life differences and different SNP alleles in miRNA target sites (β = 0.022, p = 0.312; Figure 5B), although the SNP effect appeared to be in the expected direction, which may be due to our underpowered sample size.
Figure 5. Correlation of RNA half-life with sequence variants with 1,000 Genomes genotype data.
For genes with SNPs polymorphic between the three individuals (GM12812, GM12814, and GM12815), RNA half-life (in log10 scale) and genotype for each individual were fitted in a regression analysis to estimate the regression coefficient β and p value for the correlation. (A) For 45 nonsense SNPs in 41 genes, mutant (stop-codon) allele of a nonsense SNP significantly correlates with decreased RNA half-life (homozygous mutant allele < heterozygous < homozygous reference allele) (β = 0.138, p = 0.009). This is consistent with expected NMD effect. One nonsense SNP, rs16982743, in SIGLEC12 was not included in the analysis because it is in exon 1 of the gene and is expected to escape from NMD83,84. (B) For 115 SNPs in miRNA sites of 111 genes, miRNA target-disrupting allele dose not show significant correlation with RNA half-life (β = 0.022, p = 0.312). (C) For 1,080 SNPs in 505 genes showing interindividual RNA half-life differences at FDR < 0.01, G or C allele does not show a correlation with RNA half-life (β = 0.023, p = 0.075). (D) For a subset of genes (n = 261) containing single coding SNPs, the G or C allele shows significant correlation with increased mRNA half-life (homozygous G or C alleles > heterozygous > heterozygous A or T alleles) (β = 0.071, p = 0.021). Single-side p-values are presented, because either a positive or negative correlation is expected between different genotypes and RNA half-life for each type of tested sequence variants.
For coding synonymous and non-synonymous SNPs, our hypothesis was that G or C allele were “stable allele” and thus associated with increased RNA half-life. Our initial analysis of all the available 14,137 synonymous and non-synonymous SNPs in 5,671 genes did not reveal the expected correlation between G or C alleles and increased RNA half-life (See Supplementary Table S5). Given our underpowered sample size, we restricted the analysis to a total of 1,080 SNPs in 505 genes that had interindividual RNA half-life differences at the FDR < 0.01 level to increase the power to detect possible weak effect from G or C alleles. Such analysis showed a trend towards positive correlation between G or C alleles and RNA half-life (β = 0.023, p = 0.075 for untransformed RNA half-life; Figure 5C). Because the correlation analysis for genes with multiple coding SNPs can be confounded by non-independent tests (i.e., multiple SNPs linked to the same RNA half-life measurement) and complicated by possible balancing effects from multiple functional SNPs (e.g., the “stable” allele G or C allele of one SNP may cancel out the effect from an A or T allele of another SNP), we further restricted the analysis to only 261 genes with a single coding SNP (See Supplementary Table S7). We found that G or C allele was nominally associated with increased RNA half-life (β = 0.071, p = 0.021; Figure 5D and see Supplementary Table S5 online), but with very weak effect.
Discussion
About 80% of genes show interindividual transcript abundance differences1,2, resulting from a balance between transcription and RNA decay. This pilot study represents the first attempt to characterize genome-wide interindividual RNA stability differences and the effects from specific types of genetic variants. We found that ~ 25.7% of all the expressed genes presented interindividual RNA stability differences (FDR < 0.05). Factoring in our qPCR validation rate of 80%, ~ 21% of all expressed genes in LCLs showed RNA half-life differences between individuals. Although transcriptional differences remain the largest known factor influencing the variance of total RNA abundances15,16, RNA stability differences explained a larger fraction of the total RNA abundance variance in the subset of genes showing interindividual RNA half-life differences than in all the expressed genes (16% vs. 6%). For genes with interindividual total RNA abundance differences, 37% also showed interindividual RNA half-life differences with most (~ 88%) in the same direction as the gene expression differences. On a genome-wide scale, we have replicated the effect of nonsense SNPs in accelerating RNA decay, and detected weak effect of allele substitutions from A or T to G or C of synonymous and nonsynonmous SNPs in stablizing RNA transcripts.
While mRNA stability analysis in LCLs might be confounded by differences of sex or age of the donor, EBV transformation, and other technical factors (as would be total RNA abundance analysis), these confounding factors are unlikely to explain the observed extensive RNA stability differences between our LCLs. For the three LCLs used to compare RNA half-life differences, the donors' ages at the time of phlebotomy were similar (~ 40 s). Although the donors were not of the same sex, over-representation of sex-chromosome genes among those showing interindividual differences of RNA half-life was not observed (85/301 X chromosome genes, Fisher's exact test p = 0.46). In addition, there was no enrichment of EBV-affected genes63 among those showing RNA half-life differences (32/103 EBV-affected genes, Fisher's exact test p = 0.39), suggesting that the EBV-transformation condition of different LCLs had no major effect on our results. Furthermore, the observed interindividual RNA half-life differences were unlikely to merely be the result of measurement problems and technical artifacts since: (1) The approach of 4sU-labeling of newly synthesized RNAs is less disruptive to cellular function compared to the traditional method of blocking transcription with actinomycin D, thus minimizing the confounding effects on global gene expression inherent to actinomycin D usage48,51. (2) The measurements of RNA half-life were highly reproducible (Pearson correlation R = 0.92 − 0.95 between technical replicates), and the degree of correlation of the RNA half-life measurements was in the expected order of technical replicates > biological replicates > different LCLs, indicating the reliability of the RNA half-life measurements. (3) The array-measured interindividual RNA half-life differences for a random set of genes were validated by qPCR using independent cell cultures, suggesting that systematic bias is unlikely. Because trans-acting factors stabilize/destabilize RNA transcripts in a tissue/cell-specific manner64,65, other cell/tissue types may show different sets of genes with interindividual RNA half-life differences. Nonetheless, our results indicated that interindividual RNA half-life differences are measurable and widespread on a genome-wide scale.
Although the effects of gene sequence features on RNA stability differences between genes and between individuals are detectable, such effects were very weak as indicated by the weak correlations between gene/sequence features and RNA stability (R = 0.1 ~ 0.3) (Table 3). On the other hand, the observed weak correlations are consistent with previous reports22,51,53,57,58,59,60, and have similar magnitudes to that of reported correlation between expression level and gene features, where the maximum R is −0.26 with 3′-UTR length out of ~ 200 gene/sequence features examined66. Some explanations for the observed weak correlations may include: (1) Other unexamined gene/DNA sequence features may contribute to the RNA stability variance, each with a small effect similar to what have been demonstrated for total RNA abundances66. (2) Gene/sequence features in some subset of genes may show stronger effect on RNA half-life. (3) More importantly, as RNA decay is a complicated process that involves not only cis-sequence features but also various trans-factors (e.g., RNA-binding proteins or miRNAs)17, expression variation of such trans-factors may also contribute to the RNA stability differences between cell line. In our dataset, although we did not find over-representation of genes related to RNA decay or RNA binding among (by DAVID tool; data not shown) those showing interindividual differences of total RNA abundances, we did find significant interindividual differences of total RNA abundance for a number of critical RNA decay-related genes17 (17/26 expressed genes with inter-individual differences at FDR < 5%) (Supplementary Table S8).
For genes showing RNA stability differences, we have found a weak correlation between RNA half-life and coding GC and GC3 contents (Table 3). We also found that G or C alleles of coding SNPs were also weakly correlated with more stable RNAs for a subset of genes showing interindividual RNA half-life differences and with a single SNP (Figure 4D). Our results are thus consistent with that GC base pairings are more stable than an AU pairings (3 vs. 2 hydrogen bonds), suggesting a role of thermodynamic stability of RNA molecules in determining RNA stability. On a genome-wide scale, our results are in line with previous observations in individual genes where coding SNPs (synonymous or nonsynonymous SNPs) affecting RNA stability all had allele changes between G or C and A or T24,25,26,67,68.
It is important to acknowledge a number of limitations of this pilot study. Since both transcription and RNA decay are dynamic processes subject to extensive temporal and spatial regulation15,69,70, our analytical model is a simplified representation of a very complex system (e.g, alternative RNA splicing was not considered here). Furthermore, while our results suggest a relatively weaker contribution of RNA decay (compared to de novo transcription) to the overall gene expression variation, RNA stability may have a larger contribution to the plasticity of RNA levels, including the kinetics and the overall response to cellular perturbations. Lastly, due to the small sample size, we have restricted our analyses to examining the collective effects of different types of DNA sequence variants on mRNA stability, rather than identifying specific DNA sequence variants that influence mRNA stability by a genome-wide association study. Nonetheless, our study suggested that interindividual RNA stability differences are pervasive and influenced by specific gene features and sequence variants. These results thus warrant further genome-wide mapping of individual sequence variants associated with interindividual RNA stability differences in larger cohorts of samples. Given the substantial role of gene expression regulation in common disease pathogenesis29,71,72, delineating the regulatory effect of genetic variants on RNA transcription and RNA decay will help elucidate novel causal mechanisms underlying genetic associations with common diseases.
Methods
Cell culture
We obtained LCLs from Coriell Cell Repositories (ccr.coriell.org). These LCLs were derived from 7 unrelated HapMap CEU individuals, GM07019 (female), GM07029 (male), GM10835 (male), GM12812 (male), GM12813 (female), GM12814 (male), and GM12815 (female). All donors were genetically unrelated Caucasians, of which only GM07019, GM07029, and GM10835 had age information (45, 47, and 39 years old, respectively). To minimize the effect of non-genetic factors on gene expression, we kept cell culture conditions consistent across different cell lines, including (1) using the same batch of media and drug (4sU; Sigma), (2) seeding cells at the same density before treatment of 4sU, (3) harvesting the 4sU-treated cells at the same time under similar conditions across cell flasks, and (4) assuring that different LCLs and biological replicates have the same growth rates (i.e., cell number doubling time). In brief, we grew the cells in 15 ml RPMI 1640 media (from ATCC) supplemented with 15% fetal bovine serum (FBS; from ATCC) in T-25 cell culture flasks. We passaged the cells at an initial cell density of 5 × 105 cells/ml and change media every other day. Upon cell growth reaching log phase of growth (usually ~ 5 days after the first passaging), we seeded the cells at a density of 2.5 × 105 cells/ml and counted the viable cell number the next day to make sure the cell growth was in log phase for all the cell lines. To start the 4sU-labeling experiment, we seeded the log-phase growing cells at a density of 2.5 × 105 cells/ml and 24 h later, we treated cells with 200 μM 4sU for 2 h and harvested cells (cell density reached to 5–6 × 105 cells/ml). The concentration of 200 μM of 4sU has been shown to allow efficient labeling of nascent RNAs without affecting global gene expression47,48. The experiments were approved by Northshore Healthsystem Research Institute Institutional Review Board.
Total RNA and nascent RNA isolation
We used TRIzol reagent (Invitrogen) to isolate total RNAs from ~ 7.5 × 106 LCL cells and further purified the total RNAs by the RNeasy Mini Kit (Qiagen). To separate the 4sU-labeled nascent RNAs from total RNAs, we followed the protocol developed by Dolken et al.48. Briefly, ~ 60 μg of total RNAs were subjected to biotinylation in the presence of 1 mg/ml of Biotin-HPDP (Pierce) for 3 h at room temperature, where only 4sU-labeled RNAs can be biotinylated, and then precipitated by 2-propanol. The precipitated RNAs were resuspended and incubated with 100 μL of μMACS streptavidin-coated beads (Miltenyi Biotec) to allow the biotinylated nascent RNAs to bind to μMACS beads. We then applied the mixture onto the top of a column placed in the magnetic field of a μMACs separator (Miltenyi Biotec), where only the nascent RNAs bind with streptavidin-coated beads in high affinity and were retained in the column by the strong magnetic field. After three washes (100 mM Tris-HCl, pH 7.4, 10 mM EDTA, 1 M NaCl, 0.1% Tween20) at 65°C and then another three washes at room temperature, the nascent RNAs were eluted from the column by the addition of 100 mM dithiothreitol (DTT). We found that the binding of nascent RNAs to the streptavidin-coated microbeads was specific and time/dosage-dependent (See Supplementary Fig. S5 online). We were able to recover 2 μg of nascent RNAs, which was sufficient for subsequent exon array profiling, from a total of ~ 60 μg total RNAs derived from ~ 7.5 × 106 LCLs treated with 200 μM of 4sU for 2 h. The recovered nascent RNAs were further purified using RNeasy MinElute Spin columns (Qiagen). Before sending the RNA samples to the array facility, we assessed RNA quality for both total and nascent RNAs by measuring the A260/A280 ratio (all > 1.9) by Biophotometer (Eppendorf). We also examined RNA integrity on agarose gel by visually checking bands of 28S and 18S rRNAs.
Affymetrix Human Exon 1.0 ST array processing
The abundance of total and nascent RNAs was assayed in parallel with Affymetrix Human Exon arrays at the University of Chicago functional genomics core. The array facility rechecked the total and nascent RNAs for concentration, A260/A280 ratio, and RNA integrity by Nanodrop (Thermo Scientific) and an Agilent 2100 Bioanalyzer. As a routine procedure for the exon array assay at the array facility, ribosomal RNA (rRNA) reduction was performed with the RiboMinus Transcriptome Isolation Kit (Invitrogen) before cDNA synthesis to minimize the background hybridization signal. 100 ng of total RNAs was used in cDNA synthesis with the GeneChip WT cDNA Synthesis Kit (Affymetrix), and the synthesized cDNAs were fragmented and end-labeled with the GeneChip WT terminal Labeling Kit (Affymetrix). 5.5 μg fragmented/labeled cDNAs were used to hybridize with probes on the exon array at 45°C for 16–18 h. The hybridized chips were washed and stained on a GeneChip Fluidics Station 450, and scanned on a GCS3000 Scanner (Affymetrix).
Exon array data analyses
We used the Affymetrix Power Tools (APT) software package to normalize the probe intensities and summarize the expression values at the probe set level (representing exon expression) and at the meta-probe set level (representing gene expression). See Supplementary Table S9 summarizes key quality control (QC) metrics. ~ 597 K SNP-containing probes (based on dbSNP build 129)73 were removed before probe intensity normalization, since such a SNP may affect the probe's binding affinity in the array hybridization and thus lead to biased estimates of gene expression level1,74,75. Hybridization intensities of the remaining probes on an array were log2-transformed and then quantile normalized76,77. We carried out the normalization process for the “C” series of arrays with total RNAs and the “N” series of arrays with nascent RNAs separately, because N and C arrays represent different biology and therefore may show different gene expression patterns. To determine the reliability of the signal intensity, we generated a detection P-value of each probe set (core exon) or each meta-probe set (transcript cluster or gene) using the “Detection Above Background” (DABG) algorithm implemented in APT. A detection P ≤ 0.01 (recommended by Affymetrix) was used as a cut-off for determining whether a probe was expressed (i.e., detectable). Furthermore, only genes expressed in > 95% samples and with log-scale expression values > 6 in the “C” series of arrays were considered for further expression analyses. The number of the expressed probe sets (< average plus two standard deviations) on each array was used as one of the key QC metrics to detect array outliers. We identified a “C” array, 12813_rep3 (A12813A3 in Supplementary Fig. S6 online), as an outlier and excluded it from further analysis for the following reasons: (1) it had ~ 120 K expressed probe sets, which is > 2 standard deviations lower than the average number of expressed probe sets; (2) the correlation coefficient of gene expression values in 12813_rep3 with other biological or technical replicates (R = 0.94 ~ 0.98) appeared lower than that between other pairs of replicates of sample 12813 (R = 0.98 ~ 0.99); and (3) a multidimensional scaling (MDS) analysis also indicated that A12813A3 was not clustered together with other replicates.
Estimation of RNA half-life from microarray data
The steady-state level of RNA transcripts measured from “C” arrays reflects the kinetic balance between transcription and RNA degradation. At steady state level, the RNA decay process in the period of 4sU-labeling can be expressed as48,49,78:
where kd is the decay constant, N is the amount of nascent RNAs measured from “N” arrays and C is the amount of the total RNAs measured from “C” arrays. A commonly used term for describing RNA stability, RNA half-life (T1/2), is dependent on the kd and can be calculated by49:
Therefore, to determine the RNA decay constant kd and subsequently to estimate the RNA half-life in each cell line, we needed to determine the ratio N/C from the microarray data with t = 2 h (i.e., the 4sU-labeling time). Because the N and C arrays for each cell line were processed independently, we normalized the scale of N/C arrays using the global median RNA half-life method48. Briefly, based on the median global RNA half-life in human B-cells (~ 5.3 h48, we determined a correction factor for the expression values between a pair of N and C arrays for each LCL, and then calculated the ratio of N/C from which the decay constant kd and T1/2 were calculated49. Because RNA half-life can be calculated only when N/C < 1 (see the formula above), we were able to estimate RNA half-life for 11,132 out of 11,301 expressed genes. Applying such a correction factor based on predefined B-cell median half-life to calculate RNA half-life for each sample should not have distorted the global distribution of RNA half-life between individuals, because the median N/C ratios before applying the correction factor were very similar across different samples (1.66 to 1.69), implying that the median degradation rates and RNA half-lives were very similar between samples.
qPCR
Total and 4sU-labeled nascent RNAs were extracted from LCLs cultured independently from those used in the exon array experiment. We performed reverse transcription from RNAs to cDNAs with random hexamer primers using the Applied Biosystem (ABI) Reverse-transcription kit. With cDNAs as templates, we used one ABI TaqMan gene expression assay per gene in real-time PCR to quantify the expression of a target gene in total and nascent RNAs, respectively. Real-time PCR was performed in 384-well plates with 3 replicates on an ABI 7900 with standard cycling parameters. For quantification of gene expression, we used the comparative delta-Ct method with glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a constitutively expressed gene, as an endogenous control to normalize the quantity of target transcripts (i.e., the relative expression value of a target gene = 2 (Ct_target - Ct_GAPDH)). RNA half-life was then estimated from the ratio of nascent/total RNAs as described above in the microarray experiment.
Bioinformatics
We downloaded RNA length, exon and intron lengths, miRNA target sites, 5′- and 3′-UTR sequences and folding energies from UCSC hg18 (RefSeq genes). For genes with multiple transcripts, the longest transcript was chosen in the analyses. We calculated coding GC content and GC3 content using locally installed CodonW version 1.4.2 (codonw.sourceforge.net//culong.html). Whole genome sequencing data for three individuals (GM12812, GM12814, and GM12815) were from the 1,000 Genomes Project (June 2011 release). The downloaded genotype file from the 1,000 Genomes Project (in VCF 4.0 format) was further processed in VCFtools (vcftools.sourceforge.net) and PLINK79 to keep only genotypes for coding SNPs (synonymous, non-synonymous, and nonsense) and 3′-UTR SNPs disrupting miRNA target sites. We used ANOVAR80 to systematically perform functional SNP annotation and to extract RNA sequences carrying different alleles of a SNP. The minimum free energy of RNA folding structures was predicted by RNAfold of Vienna RNA Package (www.tbi.univie.ac.at/~ivo/RNA/)81 using the default setting. DAVID tools were used for enrichment analyses of GO terms and pathways52. For clustering genes with co-regulated RNA stability, we used the R package for WGCNA, which identifies modules of densely interconnected genes based on a correlation matrix containing all pair-wise Pearson correlations between all genes across all subjects82. We carried out here hierarchical clustering across all 7 LCLs (to boost the power) for RNA half-life of the 4,533 genes showing interindividual RNA half-life differences with a p < 0.05.
Statistical analyses
Statistical analyses were performed using R (www.r-project.org). For each gene, we performed ANOVA to test the null hypothesis that the measurements (total RNA abundances, nascent RNA abundances, and RNA half-life) had no interindividual differences. A P-value was calculated for each gene and each measurement. Genes showing significant interindividual differences in each measurement were identified by controlling the Benjamini & Hochberg FDR < 0.0554, corresponding to the ANOVA nominal P < 0.013. To investigate whether the ANOVA p-value for each transcript is robust to the normality assumption, we first calculated the F statistic using ANOVA, which measured how well the replicates were separated among different cell lines. Then, we performed 1,000 permutations and approximated the p-value as the proportion of permutation with F statistic greater than the observed one. We then examined whether the p-values of standard ANOVA analysis based on normality assumption and the permutations agree well. For the Spearman rank correlation of RNA half-life with gene features, we averaged RNA half-life for each gene from the 3 LCLs with biological replicates to reduce the noise. To evaluate the overall contribution of various genomic features to the RNA half-life variance between genes, we performed multivariate linear regression using log10-transformed RNA half-life as the dependent variable and log10-transformed genomic features (except 5′- and 3′-UTR folding energies were not log10-transformed as they are negative values) as predictors (Table 3).
Furthermore, we investigated whether the RNA half-life was associated with sequence variants from different functional categories, e.g. nonsense SNPs, miRNA target-disrupting SNPs in 3′-UTRs, and coding synonymous and non-synonymous SNPs. Ideally, a linear regression analysis could be done for each SNP with a sufficient sample size. In our study, however, we had no power to detect any association based on only three LCLs with whole genome sequence data available. Therefore, we aggregated the SNPs for each category and tested the overall association for each category. Our test involves the following major steps: (1) We extracted the SNPs polymorphic in the three samples sequenced by the 1000 Genomes Project and classified the extracted SNPs into functional categories. (2) We coded homozygotes of alleles putatively decreasing RNA half-life (i.e., nonsense allele of a nonsense mutation, reference allele of a miRNA target-disrupting SNP, and “unstable” allele A or T of a coding synonymous or non-synonymous SNP) as 0, heterozygotes as 1, and homozygotes of the other allele (i.e., the reference allele of a nonsense SNP, miRNA target-disrupting allele, and “stable” allele G or C of a coding SNP) as 2. (3) A linear regression analysis was performed to test the overall correlation between the RNA half-life and the genotypes across subjects and qualified SNPs from each functional category. We computed single-sided P-values because only positive correlations were expected based on our SNP coding scheme.
Author Contributions
J.D. and P.V.G. conceived and designed the experiments. D.H., S.S. and J.D. performed the experiments. J.S., X.G., L.D., W.M. and J.D. analyzed the data. A.R.S. and J.R. helped to interpret the results. J.D. wrote the paper. All authors reviewed the manuscript.
Supplementary Material
Supplementary Figures
Supplementary Tables
Acknowledgments
This work was supported by NorthShore University HealthSystem (formally Evanston Northwestern Healthcare) Research Career Development Award to J.D.; MRC Fellowship grant G1002523 to L.D.' and grants from National Institute of Health (RC2MH090030 to A.R.S. and R01MH94091 to P.V.G.). We thank Dr. Kai Wang (Zilkha Neurogenetic Institute, CA) for bioinformatic assistance on gene annotation by ANOVAR.
References
- Storey J. D. et al. Gene-expression variation within and among human populations. Am J Hum Genet 80, 502–509 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardson A. S. et al. The effect of food intake on gene expression in human peripheral blood. Hum Mol Genet 19, 159–169 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley M. et al. Genetic analysis of genome-wide variation in human gene expression. Nature 430, 743–747 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks S. A. et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet 75, 1094–1105 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V. G. et al. Mapping determinants of human gene expression by regional and genome-wide association. Nature 437, 1365–1369 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B. E. et al. Genome-wide associations of gene expression variation in humans. PLoS Genet 1, e78 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B. E. et al. Population genomics of human gene expression. Nat Genet 39, 1217–1224 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring H. H. et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet 39, 1208–1216 (2007). [DOI] [PubMed] [Google Scholar]
- Dixon A. L. et al. A genome-wide association study of global gene expression. Nat Genet 39, 1202–1207 (2007). [DOI] [PubMed] [Google Scholar]
- Montgomery S. B. et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature 464, 773–777 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K. et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L. et al. Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet 7, e1001317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae A. F. et al. Replicated effects of sex and genotype on gene expression in human lymphoblastoid cell lines. Hum Mol Genet 16, 364–373 (2007). [DOI] [PubMed] [Google Scholar]
- Emilsson V. et al. Genetics of gene expression and its effect on disease. Nature 452, 423–428 (2008). [DOI] [PubMed] [Google Scholar]
- Rabani M. et al. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29, 436–442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011). [DOI] [PubMed] [Google Scholar]
- Schoenberg D. R. & Maquat L. E. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13, 246–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau N. L., Wilusz J. & Wilusz C. J. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8, 113–126 (2007). [DOI] [PubMed] [Google Scholar]
- Saunders M. A., Liang H. & Li W. H. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A 104, 3300–3305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Marguerat S. & Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci 30, 506–514 (2005). [DOI] [PubMed] [Google Scholar]
- Montgomery S. B., Lappalainen T., Gutierrez-Arcelus M. & Dermitzakis E. T. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genet 7, e1002144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharova L. V. et al. Database for mRNA half-life of 19 977 genes obtained by DNA microarray analysis of pluripotent and differentiating mouse embryonic stem cells. DNA Res 16, 45–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. F., Feng L. & Niu D. K. Relationship between mRNA stability and intron presence. Biochem Biophys Res Commun 354, 203–208 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J. et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet 12, 205–216 (2003). [DOI] [PubMed] [Google Scholar]
- Capon F. et al. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum Mol Genet 13, 2361–2368 (2004). [DOI] [PubMed] [Google Scholar]
- Nackley A. G. et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science 314, 1930–1933 (2006). [DOI] [PubMed] [Google Scholar]
- Thorleifsson G. et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science 317, 1397–1400 (2007). [DOI] [PubMed] [Google Scholar]
- Plotkin J. B. & Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12, 32–42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna Z. E. & Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat Rev Genet 12, 683–691 (2011). [DOI] [PubMed] [Google Scholar]
- Hao S. & Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10, 281–288 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D., Pasparakis M., Pizarro T. T., Cominelli F. & Kollias G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10, 387–398 (1999). [DOI] [PubMed] [Google Scholar]
- Gardner J. et al. Potential mechanisms for astrocyte-TIMP-1 downregulation in chronic inflammatory diseases. J Neurosci Res 83, 1281–1292 (2006). [DOI] [PubMed] [Google Scholar]
- Fan J., Heller N. M., Gorospe M., Atasoy U. & Stellato C. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur Respir J 26, 933–947 (2005). [DOI] [PubMed] [Google Scholar]
- Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol 10, 24–35 (2009). [DOI] [PubMed] [Google Scholar]
- Tiret L. et al. Genetic analysis of the interleukin-18 system highlights the role of the interleukin-18 gene in cardiovascular disease. Circulation 112, 643–650 (2005). [DOI] [PubMed] [Google Scholar]
- Douthwaite J. A., Lees D. M. & Corder R. A role for increased mRNA stability in the induction of endothelin-1 synthesis by lipopolysaccharide. Biochem Pharmacol 66, 589–594 (2003). [DOI] [PubMed] [Google Scholar]
- Denkert C. et al. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol 19, 1261–1269 (2006). [DOI] [PubMed] [Google Scholar]
- Benjamin D. & Moroni C. mRNA stability and cancer: an emerging link? Expert Opin Biol Ther 7, 1515–1529 (2007). [DOI] [PubMed] [Google Scholar]
- Mayr C. & Bartel D. P. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138, 673–684 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche L. G. et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet 40, 892–896 (2008). [DOI] [PubMed] [Google Scholar]
- Tarpey P. S. et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet 39, 1127–1133 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X., Wang Z. & Kiledjian M. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol Cell 24, 713–722 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14, 723–730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers R. J. et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science 329, 1650–1653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brichta L. et al. Nonsense-mediated messenger RNA decay of survival motor neuron 1 causes spinal muscular atrophy. Hum Genet 123, 141–153 (2008). [DOI] [PubMed] [Google Scholar]
- Cleary M. D., Meiering C. D., Jan E., Guymon R. & Boothroyd J. C. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol 23, 232–237 (2005). [DOI] [PubMed] [Google Scholar]
- Kenzelmann M. et al. Microarray analysis of newly synthesized RNA in cells and animals. Proc Natl Acad Sci U S A 104, 6164–6169 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolken L. et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 14, 1959–1972 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. mRNA stability in mammalian cells. Microbiol Rev 59, 423–450 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S. et al. Genetic architecture of transcript-level variation in humans. Am J Hum Genet 82, 1101–1113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel C. C., Dolken L., Ruzsics Z., Koszinowski U. H. & Zimmer R. Conserved principles of mammalian transcriptional regulation revealed by RNA half-life. Nucleic Acids Res 37, e115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T. & Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- Yang E. et al. Decay rates of human mRNAs: correlation with functional characteristics and sequence attributes. Genome Res 13, 1863–1872 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society,Series B 57, 289–300 (1995). [Google Scholar]
- Mittal P. et al. A molecular signature of an arrest of descent in human parturition. American journal of obstetrics and gynecology, 177, e115–133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisha S. Z., Serre D., Schauer P., Kashyap S. R. & Smith J. D. Changes in whole blood gene expression in obese subjects with type 2 diabetes following bariatric surgery: a pilot study. PLoS ONE 6, e16729 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. E., Lee J. Y., Wilusz J., Tian B. & Wilusz C. J. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS ONE 5, e11201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steglich C. et al. Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol 11, R54 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsai R. et al. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell 19, 3418–3436 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L. & Niu D. K. Relationship between mRNA stability and length: an old question with a new twist. Biochem Genet 45, 131–137 (2007). [DOI] [PubMed] [Google Scholar]
- Zhao C. & Hamilton T. Introns regulate the rate of unstable mRNA decay. J Biol Chem 282, 20230–20237 (2007). [DOI] [PubMed] [Google Scholar]
- Baek D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan M., Cusanovich D. A., Ober C. & Gilad Y. The effects of EBV transformation on gene expression levels and methylation profiles. Hum Mol Genet 20, 1643–1652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F., Contente-Cuomo T. & Perrone-Bizzozero N. I. Novel recognition motifs and biological functions of the RNA-binding protein HuD revealed by genome-wide identification of its targets. Nucleic Acids Res 38, 117–130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiruchinapalli D. M., Ehlers M. D. & Keene J. D. Activity-dependent expression of RNA binding protein HuD and its association with mRNAs in neurons. RNA Biol 5, 157–168 (2008). [DOI] [PubMed] [Google Scholar]
- Vogel C. et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Molecular systems biology 6, 400 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J. & Antezana M. A. Mammalian mutation pressure, synonymous codon choice, and mRNA degradation. J Mol Evol 57, 694–701 (2003). [DOI] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C. et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 (2007). [DOI] [PubMed] [Google Scholar]
- Yosef N. & Regev A. Impulse control: temporal dynamics in gene transcription. Cell 144, 886–896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck A. C. & Tollervey D. RNA in pieces. Trends Genet 27, 422–432 (2011). [DOI] [PubMed] [Google Scholar]
- Deutsch S. et al. Gene expression variation and expression quantitative trait mapping of human chromosome 21 genes. Hum Mol Genet 14, 3741–3749 (2005). [DOI] [PubMed] [Google Scholar]
- Hindorff L. A. et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106, 9362–9367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S., Zhang W., Bleibel W. K., Cox N. J. & Dolan M. E. SNPinProbe_1.0: A database for filtering out probes in the Affymetrix GeneChip(R) Human Exon 1.0 ST array potentially affected by SNPs. Bioinformation 2, 469–470 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y., Rifkin S. A., Bertone P., Gerstein M. & White K. P. Multi-species microarrays reveal the effect of sequence divergence on gene expression profiles. Genome Res 15, 674–680 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliwerska E. et al. SNPs on chips: the hidden genetic code in expression arrays. Biol Psychiatry 61, 13–16 (2007). [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Astrand M. & Speed T. P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 (2003). [DOI] [PubMed] [Google Scholar]
- Barbacioru C. C. et al. Effect of various normalization methods on Applied Biosystems expression array system data. BMC Bioinformatics 7, 533 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T. R., Rudin S. D., Blossey B. K. & Ilan J. Newly synthesized RNA: simultaneous measurement in intact cells of transcription rates and RNA stability of insulin-like growth factor I, actin, and albumin in growth hormone-stimulated hepatocytes. Proc Natl Acad Sci U S A 88, 5287–5291 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber A. R., Lorenz R., Bernhart S. H., Neubock R. & Hofacker I. L. The Vienna RNA websuite. Nucleic Acids Res 36, W70–74 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P. & Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu-Yilik G. et al. Mechanism of escape from nonsense-mediated mRNA decay of human beta-globin transcripts with nonsense mutations in the first exon. RNA 17, 843–854 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G. & Maquat L. E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell 106, 607–617 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables