Abstract
Transforming growth factor β isoforms (TGF-β) are among the most recently evolved members of a signaling superfamily with more than 30 members. TGF-β play vital roles in regulating cellular growth and differentiation, and they signal through a highly restricted subset of receptors known as TGF-β type I receptor (TβR-I) and TGF-β type II receptor (TβR-II). TGF-β's specificity for TβR-I has been proposed to arise from its pre-helix extension, a five-residue loop that binds in the cleft between TGF-β and TβR-II. The structure and backbone dynamics of the unbound form of the TβR-I extracellular domain were determined using NMR to investigate the extension's role in binding. This showed that the unbound form is highly similar to the bound form in terms of both the β-strand framework that defines the three-finger toxin fold and the extension and its characteristic cis-Ile54-Pro55 peptide bond. The NMR data further showed that the extension and two flanking 310 helices are rigid on the nanosecond-to-picosecond timescale. The functional significance of several residues within the extension was investigated by binding studies and reporter gene assays in cultured epithelial cells. These demonstrated that the pre-helix extension is essential for binding, with Pro55 and Pro59 each playing a major role. These findings suggest that the pre-helix extension and its flanking prolines evolved to endow the TGF-β signaling complex with its unique specificity, departing from the ancestral promiscuity of the bone morphogenetic protein subfamily, where the binding interface of the type I receptor is highly flexible.
Keywords: TGF-β, type I receptor, cis-proline, specificity, cooperative binding
Introduction
Transforming growth factor β isoforms (TGF-β) are secreted signal ligands that play vital roles in coordinating wound healing, modulating immune cell function, maintaining the extracellular matrix, and regulating epithelial and endothelial cell growth and differentiation.1 The importance of TGF-β is underscored by their conservation among vertebrates and their demonstrated roles in a variety of human diseases, including tissue fibrosis2 and cancer.3 TGF-β are members of an extended signaling superfamily that arose in early metazoans.4 The superfamily has greatly diversified, with more than 30 known members in vertebrates. This includes three TGF-β (TGF-β1, TGF-β2, and TGF-β3); activins and inhibins, which regulate the release of pituitary hormones; bone morphogenetic proteins (BMPs), which play fundamental roles in regulating embryonic patterning; and the closely related growth and differentiation factors (GDFs), which regulate cartilage and skeletal development.
TGF-β transduce their signals by binding and bringing together two structurally related single-pass transmembrane receptor kinases, known as TGF-β type I receptor (TβR-I) and TGF-β type II receptor (TβR-II).5 This triggers a transphosphorylation cascade that begins with TβR-II-mediated activation of TβR-I kinase, and it propagates to intracellular effectors, including both canonical receptor-mediated Smad proteins (R-Smads)1 and non-Smads.6 This manner of signaling is shared by all ligands of the superfamily, although TGF-β and activins bind and signal through a highly restricted subset of type I and type II receptors, namely TβR-I (Alk5)/TβR-II and ActR-Ib (Alk4)/ActR-IIa/b, respectively, whereas the more numerous and varied BMPs/GDFs promiscuously bind and signal through multiple type I and type II receptors, including BMPR-Ia (Alk3), BMPR-Ib (Alk6), Alk1, and Alk2, and ActR-IIa, ActR-IIb, and BMPR-II. TGF-β and activins are further distinguished from BMPs and GDFs in that their type I receptors activate R-Smads 2 and 3, while the BMP and GDF type I receptors activate R-Smads 1, 5, and 8.7 These two subclasses of Smads, upon association with Smad 4, assemble distinct transcriptional complexes and thus activate distinct subsets of genes.8
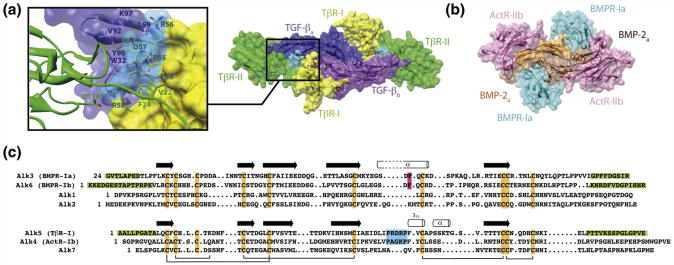
Structural studies have shown that TGF-β and BMPs bind and assemble their receptors in a distinct manner.9–14 TGF-β bind their receptors, TβR-I and TβR-II, on the underside of the “fingers” and “fingertips,” respectively, while the BMPs bind their type I and type II receptors on the “wrist” and “knuckles,” respectively (Fig. 1a and b). This places the type I and type II receptors in direct contact within the TGF-β receptor complex, but not with the BMP. The direct receptor–receptor contact has been shown to be responsible for the pronounced stepwise manner with which TGF β bind TβR-II and recruit TβR-I,11 and is further thought to underlie TGF-β's high specificity for binding and recruiting TβR-I.11,15
Fig. 1.
Distinct modes of receptor binding for TGF-β and BMPs. Surface representations of the ligand/type I receptor/type II receptor ternary complexes with TGF-β3 (a) and BMP-2 (b) (PDB codes: 2PJY and 2H64, respectively). The TβR-I pre-helix extension, Pro55-Arg56-Asp57-Arg58-Pro59 (shaded cyan), fills the cavity between TβR-II and the TGF-β monomer to which TβR-II is bound and completes a hydrophobic pocket into which Val22 and Phe24 from the TβR-II N-terminal tail bind. (c) Sequence alignment of the seven known type I receptors in humans reveals the conserved secondary structural features and disulfides that define the receptor three-finger toxin fold. Secondary structural elements shown above the Alk1, Alk2, Alk3, and Alk6 and Alk4, Alk5, and Alk7 sequences correspond to those present in the bound form of Alk3 and (PDB code: 1REW) and Alk5 (PDB code: 2PJY), respectively. Structural elements that are important in enabling the distinct mode of BMP and TβR-I binding, the phenylalanine knob, and the pre-helix extension are highlighted in magenta and cyan, respectively. Structurally disordered segments in the BMPR-Ia, BMPR-Ib, and TβR-I complex structures (PDB codes: 1REW, 3EVS, and 2PJY) are shaded green.
TβR-I's distinctive manner of binding, where it principally contacts TβR-II and the TGF-β monomer to which TβR-II is bound,11,13 is thought to be driven by its pre-helix extension, a five-residue segment preceding a short solvent-exposed 310 helix (Fig. 1c). The pre-helix extension, which is also present in ActR-Ib but is absent in other type I receptors of the superfamily (Fig. 1c), adopts a tight turn that wedges between TβR-II and the underside of the TGF-β fingers 11,13 (Fig. 1a). The key structural features of the extension include Pro55 at the N-terminal end, which adopts a cis peptide bond; Asp57 and Arg58, which ion pair with TGF-β Lys97 and TβR-II Asp118, respectively; and Pro59, whose pyrrolidine ring forms the edge of a hydrophobic pocket on the surface of TβR-I (Fig. 1a). This pocket accommodates Val22 and Phe24 from the N-terminal tail of TβR-II and represents one of the key receptor–receptor interactions in the complex, as shown through functional analyses of TβR-II variants bearing substitutions in the tail.11
BMPR-I not only lacks the pre-helix extension but also binds in a distinct manner at the “wrist,” where it has extensive contacts with both ligand monomers, but not with the type II receptor. Structural and functional studies have shown that one of the key interaction elements that it employs is the short helix homologous to the short helix of the TβR-I pre-helix extension.12,16 Recent NMR studies of the BMPR-Ia extracellular domain (BMPR-Ia ED) have shown that this helix undergoes a disorder-to-order transition upon binding, suggesting a mechanism by which it promiscuously binds multiple BMPs.17
The solution structure and backbone dynamics of the unbound form of the TβR-I extracellular domain (TβR-I ED) are presented here. TβR-I's principal interaction element, the pre-helix extension, is shown to be structurally ordered and to adopt a configuration highly similar to that of the bound form, including the Ile54-Pro55 cis-prolyl peptide bond. Pro55, Pro59, and, to a lesser extent, Arg58 are further shown to be essential in enabling TβR-I's recruitment into the TGF-β receptor complex. The significance of these findings is discussed in light of TGF-β's reported high specificity for its signaling receptors and recent reports suggesting that TGF-β might recruit and activate, albeit weakly, type I receptors that lack a pre-helix extension.18–20
Results
Resonance assignments
The structural elucidation of the unbound form of BMPR-Ia ED by NMR shed light as to the structural and dynamic changes that occur upon ligand binding.12,17 The objective of this study was to perform a similar assessment for TβR-I ED, with a particular focus on its pre-helix extension. Towards this goal, we took advantage of the previously reported bacterial expression and refolding method21 to generate structurally homogeneous preparations of human TβR-I ED. This expression construct, as well as that used in the crystallization of the TβR-I/TβR-II/TGF-β3 complex,11 included the entire region between the predicted signal peptide cleavage site and the transmembrane domain (101 residues and 10 cysteines).22 This protein, termed TβR-I 1–101 (residues 1–101 of TβR-I ED), yielded a well-dispersed 1H–15N shift correlation spectrum (Supplementary Material, Fig. 1) but slowly precipitated at 25 °C when the concentration was higher than about 0.1 mM, hindering our ability to collect NMR spectra of sufficient signal-to-noise ratio for assignment and structure determination.
Solubility was improved, with stable samples at 0.2–0.3 mM, by eliminating the first 6 residues (residues 1–6) and the last 10 residues (residues 92– 101). Residues 1–6 were structurally disordered in the crystal structure of the TβR-I/TβR-II/TGF-β3 complex and may be responsible for the limited solubility of TβR-I 1–101, as the SignalP algorithm23 indicates that these correspond to the C-terminal portion of the signal peptide, not to the N-terminal region of the mature extracellular domain.11 Residues 88–101 were also structurally disordered in the crystal structure of the TβR-I/TβR-II/TGF-β3 complex; thus, truncation of the N-terminal and C-terminal regions, while serving to improve solubility, would not be expected to affect either the folding properties or the binding properties.
The 1H–15N heteronuclear single-quantum coherence (HSQC) spectrum of the shortened construct, TβR-I 7–91, exhibited a pattern nearly identical with that of TβR-I 1–101, except that it lacked several intense backbone amide resonances in the random-coil region (7.9–8.5 ppm 1H) (Supplementary Material, Fig. 1). The truncation had no detectable effect on its affinity for the TβR-II/TGF-β3 binary complex, as shown through surface plasmon resonance (SPR)-based binding studies in which variable concentrations of TβR-I 7–91 and TβR-I 1– 101 were injected over a TGF-β3 surface in the presence of a near-saturating concentration of the TβR-II extracellular domain (TβR-II ED) (Supplementary Material, Fig. 2), confirming that truncation of the N-terminal and C-terminal regions had no detectable effect on either the folding properties or the binding properties of TβR-I ED.
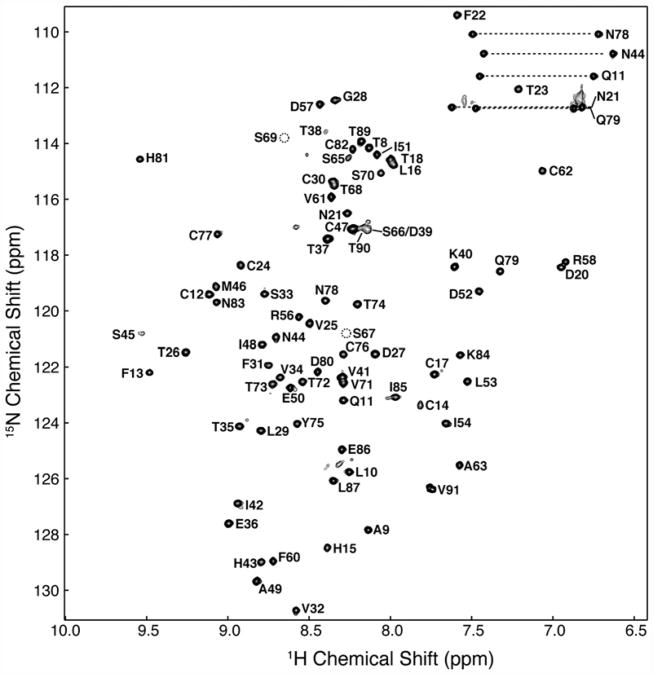
Fig. 2.
Two-dimensional 1H–15N HSQC spectrum of 0.2 mM 15N TβR-I 7–91 in 25 mM sodium phosphate, 0.02% sodium azide, and 5% 2H2O (pH 6.6) recorded at 300 K at a magnetic field strength of 14.1 T (600 MHz 1H). Peaks are labeled according to their resonance assignments (residues are numbered as in Fig. 1c). Broken circles indicate the location of backbone amides of Ser67 and Ser69, which do not appear at the contour level plotted. Horizontal broken bars designate the side-chain –NH2 groups of asparagine and glutamine.
The backbone resonances of TβR-I 7–91 were assigned by uniformly labeling it with 13C and 15N and by acquiring sensitivity-enhanced triple-resonance data sets with 0.2–0.3 mM samples in 25 mM sodium phosphate (pH 7.2) (Materials and Methods). These spectra allowed for the sequence-specific assignment of all the expected backbone amide signals of TβR-I 7–91, except for Lys19 (Fig. 2). The side-chain 1H and 13C assignments, including stereospecific assignments of the side-chain methyl groups of valine and leucine, were obtained by extending from the backbone using established methods (Materials and Methods).
Secondary structure and configuration of the Ile54-Pro55 peptide bond
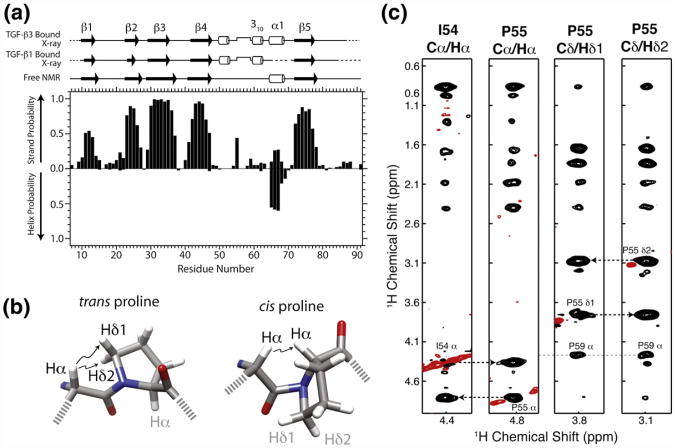
The secondary shifts of TβR-I 7-91 were analyzed using the program PECAN, which provides secondary structure probabilities on a residue-by-residue basis24 (Fig. 3a). This analysis showed that the secondary structure of the uncomplexed form of TβR-I 7–91 is composed of five β-strands: β1 (residues 10–14), β2 (residues 23–27), β3 (residues 29–37), β4 (residues 41–47), and β5 (residues 72–78). PECAN analysis also identified one α-helix (residues 65–68), although this was with reduced probability compared to the regions of β-strand. This framework is in close accord with that from the bound structures, although it lacked the two 310 helices flanking the pre-helix extension: one from residues 50–52 and the other from residues 60–62 (Fig. 3a).
Fig. 3.
TβR-I alone adopts a similar overall secondary structure and cis-prolyl peptide bond compared to the bound form. (a) Secondary structural probabilities for the unbound form of TβR-I, deduced on the basis of secondary shifts using the program PECAN,24 correlate closely with secondary structures for the TGF-β1-bound and TGF-β3-bound forms of TβR-I (PDB codes: 3KFD and 2PJY, respectively). Secondary structures were calculated from the structures of the bound forms using the program DSSP.25 (b) cis-Xaa-Pro and trans-Xaa-Pro peptide bonds are characterized by close interproton distances between Xaa Hα and either Pro Hα or Pro Hδ1/Hδ2, respectively.26 (c) Strips from a 3D 13C-edited NOESY spectrum from the Cα/Hα positions of Ile54 Hα, Pro55 Hα, Pro55 Hδ1, and Pro55 H δ2. NOEs between Ile54 Hα and Pro55 Hα indicative of a cis peptide bond are identified by broken lines. Positive and negative signals are drawn with black and red contours, respectively.
The Ile54-Pro55 peptide bond adopts a near-cis configuration in its bound form (ω equal to 2 ° and −12° in the TGF-β1 and TGF-β3 complex structures, respectively); thus, it was of interest to determine whether this peptide bond was also in the cis configuration in the unbound form. This was initially assessed by comparing the chemical shifts for the Pro Cβ and Cγ resonances relative to the database values for the cis and trans forms.27,28 This showed that the Cβ and Cγ chemical shifts for Pro55 (33.9 and 25.6 ppm, respectively) closely matched the reported database values for the cis configuration29 (33.8±1.2 and 24.4±0.7 ppm), whereas those for Pro59, Pro64, and Pro88 (32.8 and 28.5, 32.1 and 27.4, and 32.0 and 27.3 ppm, respectively) matched the database values for the trans configuration29 (31.8 ±1.0 and 27.4±0.9 ppm).
A three-dimensional (3D) 13C-edited nuclear Overhauser enhancement spectroscopy (NOESY) spectrum of TβR-I 7–91 was recorded and evaluated for nuclear Overhauser enhancements (NOEs) involving Ile54 and Pro55 to directly determine whether the Ile54-Pro55 peptide bond was in cis configuration. The spectrum exhibited intense NOEs between the Hα of Pro55 and the Hα of its preceding residue, Ile54, with the concomitant absence of NOEs between Pro55 Hδ1 and and Hδ2 and Ile54 Hδ (Fig. 3c). The former NOEs are diagnostic of a cis peptide bond, while the latter NOEs are diagnostic of a trans peptide bond (Fig. 3b).26 This supports the conclusions of the indirect analysis and shows that the Pro55 of the unbound form of TβR-I 7–91 adopts the cis configuration.
TβR-I 7–91 solution structure
Chemical shift analysis suggests that the overall structure of the uncomplexed form of TβR-I 7–91 is not significantly different from that of the bound form, although the extent of this similarity, particularly the pre-helix extension and its flanking 310 helices, remains unknown. To investigate this, we determined the solution structure of TβR-I 7–91 using simulated annealing (SA) with torsion-angle dynamics, as implemented in the program ARIA 1.2.30 The input data for the calculations consisted of 1017 experimental restraints, including 856 NOE distance restraints, 106 TALOS-predicted φ and ψ restraints, 24 3JHNHa restraints, and 31 1H–15N residual dipolar couplings (RDCs) (Table 1).
Table 1. Structural statistics for TβR-I 7-91.
| Total restraints | 1017 |
|---|---|
| NOE distance restraints | |
| Sequential restraints (|i–j| = 1) | 355 |
| Short range (2≤ |i–j| ≤ 5) | 147 |
| Long range (|i–j|> 5) | 354 |
| Dihedral restraints | |
| φ | 52 |
| ψ | 54 |
| RDC restraints | |
| 1DNH | 31 |
| Coupling restraints | |
| 3JHNHa | 24 |
| Deviation among ensemble | |
| Bonds (Å) | 0.002±0.001 |
| Angles (°) | 0.41 ±0.04 |
| Impropers (°) | 0.38 ±0.04 |
| Dihedral restraints (°) | 0.45 ±0.10 |
| RDC | |
| 1DNH (Hz) | 0.38 ±0.06 |
| JHNα restraints (Hz) | 0.59 ±0.0.05 |
| Ramachandran plota | |
| Most favored (%) | 65.6 |
| Additionally allowed (%) | 28.7 |
| Generously allowed (%) | 3.1 |
| Disallowed (%) | 2.6 |
| Overall precision | |
| Secondary structure | |
| Backboneb | 0.45 |
| Heavyb | 1.04 |
| Ordered residuesc | |
| Backboneb | 1.14 |
| Heavyb | 1.25 |
Structural statistics are calculated for the ensemble of the 10 lowest-energy structures.
Calculated using the program PROCHECK.25
Backbone atoms include NH, Cα, and CO; heavy includes all non-hydrogen atoms.
Ordered residues correspond to residues 9–37,41–63, and 71–84; secondary structure corresponds to residues 10–13, 23–26, 29–36, 41–48, 50–52, 60–62, and 71–77.
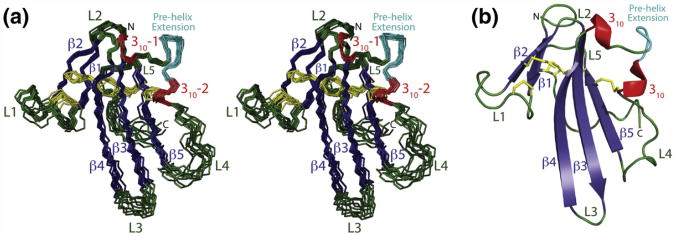
A superposition of the ten lowest-energy structures, consistent with NOE, chemical-shift-derived dihedral, 3JHNHa coupling, and RDC restraints, is shown in Fig. 4a. The regions of regular secondary structure— β1 (residues 10–13), β2 (residues 22–25), β3 (residues 29–36), β4 (residues 41–49), 310-1 (residues 50–52), 310-2 (residues 60–62), and β5 (residues 71–79)—were well-defined, with a backbone root-mean-square deviation (RMSD) of 0.45 Å, while the structurally ordered core, which extends from residue 10 to residue 88 and includes several loops, had a backbone RMSD of 1.14 Å (Table 1). The terminal regions (residues 7–9 and 89–91) yielded very few long-range NOEs and were disordered in the final structures. The stereochemical quality of the core, as assessed by the program PROCHECK, was typical of a well-refined structure, with 94.2% of the residues in the most favored or additionally allowed regions of the Ramachandran plot (Table 1). The residues in the disallowed region of the Ramachandran plot were nearly all positioned in the terminal regions or loops.
Fig. 4.
Ensemble of the 10 lowest-energy NMR structures of the unbound form of TβR-I 7–91. (a) Stereo view of the superimposition of the backbone of the 10 lowest-energy structures of the unbound form of TβR-I 7–91 after refinement (RMSD for backbone atoms in regular secondary structures: 0.49 Å). β-strands, dark blue; 310 helices, red; loops, dark green; disulfide bonds, yellow; pre-helix extension (P55-R56-D57-R58-P59), cyan. Secondary structural elements and other key structural features, including loops, the N-terminus, and the C-terminus, are indicated. (b) Ribbon diagram of a representative low-energy structure highlighting its secondary structural elements and overall fold.
The pre-helix extension resides in an extended segment from residue 49 to residue 71 that connects the C-terminal end of β-strand 4 with the N-terminal end of β-strand 5. This segment is solvent exposed and protrudes significantly from the structured core, yet the N-terminal half (residues 49–62), which includes the pre-helix extension, is surprisingly well-ordered (Fig. 4a). Three structural features appear to contribute to this ordering. These include the Cys24-Cys47 and Cys62-Cys76 disulfide bonds, which serve as rigid anchors on the N-terminal and C-terminal ends, respectively; the two flanking 310 helices, which serve as rigid adaptors; and the pre-helix extension, which adopts a tight turn with the Ile54-Pro55 peptide bond in the cis configuration.
Internal dynamics of TβR-I 7–91
The internal flexibility of TβR-I 7–91 was investigated by measuring 15N T1, 15N T2, and {1H}–15N NOE relaxation parameters at a 15N frequency of 60.8 MHz. The raw relaxation data were first analyzed to determine the extent of diffusional anisotropy (D║/D⊥) by fitting the T1/T2 data to a model with axial symmetry.31 This yielded a D║/D⊥ of 1.32 and an averaged rotational correlation time, τavg, of 7.35 ns. The normalized error for the fit (0.56) was significantly lower than that assuming isotropic diffusion (1.9) or that assuming anisotropic diffusion but with a randomized relaxation data set (1.8), justifying the additional parameters associated with the anisotropic model.
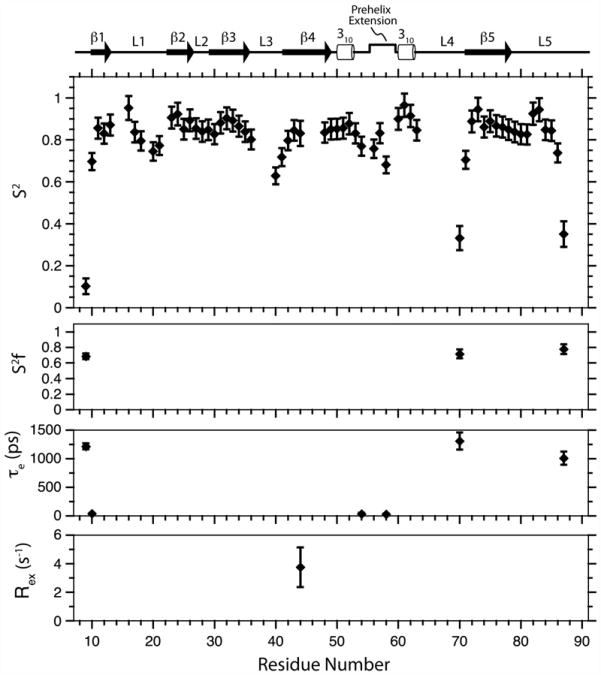
Model-free formalism was used and anisotropic tumbling was assumed, with the parameters for overall diffusion derived by the analysis above (τavg = 7.35 ns, D║/D⊥ =1.32, θ=114 °, φ=160°), to analyze the internal dynamics of TβR-I 7–91. The model-free fits were carried out using the program ModelFree4, and the procedure of Mandel et al. was used for model selection.32 This yielded statistically significant fits for all residues. The derived parameters show that the N-terminal and C-terminal regions are highly flexible on the nanosecond-to-picosecond timescale, while the regions of regular secondary structure are rigid, with a mean S2 of 0.82 ±0.03 (Fig. 5). The boundaries that demarcate the terminal segments from the structured core correspond closely to the boundaries between the structurally ordered regions and the disordered regions in the bound crystal structures.11,13 The internal loops exhibit varying degrees of disorder, with loop 2 exhibiting negligible disorder (minimum S2 =0.8); with loop 1, loop 3, and the pre-helix extension exhibiting moderate disorder (minimum S2 =0.6); and with loop 4 exhibiting significant disorder (minimum S2 =0.3).
Fig. 5.
Model-free parameters for TβR-I backbone amides derived by the fitting of 15N T1, 15N T2, and 15N–{1H} NOE data recorded at a magnetic field strength of 14.1 T. Lipari–Szabo S2, , τe, and Rex parameters are shown from top to bottom, respectively. Missing , τe, and Rex data points indicate that this parameter was not included in the motional model for that residue. Schematic representation of the TβR-I secondary structure shown along the top was derived by DSSP analysis25 of the 10 lowest-energy structures.
The relaxation data further highlight the significant difference in flexibility between the N-terminal half and the C-terminal half of the segment bridging β-strands 4 and 5. The N-terminal half (residues 49– 62), which includes the pre-helix extension and the two flanking 310 helices, is largely rigid, with both 310 helices being highly rigid (S2 =0.85 and higher) and with the intervening pre-helix extension being only moderately flexible, with the most dynamic residue being Arg58 (S2 =0.68). The C-terminal half (residues 63–71), designated as loop 4, is, in contrast, highly flexible, with residue 70 at its tip exhibiting an S2 value comparable to that of the terminal regions (S2 =0.33).
Comparison of the free and bound conformations of TβR-I
The unbound form of TβR-I determined by NMR superimposes well with the bound form of the TβR-I/TβR-II/TGF-β3 and TβR-I/TβR-II/TGF-β1 crystal structures,11,13 with a backbone RMSD of 1.4– 1.5 Å over the regions of regular secondary structure and with an overall RMSD of 3.1–3.2 Å. The high level of similarity of the β-strand framework is shown by the overlay of the unbound and bound forms presented in Fig. 6a (leftmost subpanel). This overlay also highlights the high level of similarity of the pre-helix extension and the two flanking 310 helices, 310-1 and 310-2, which superimpose nearly as well as the β-strand regions. The fact that the two 310 helices are present in the unbound form, even though they were not predicted based on their secondary shifts (Fig. 3a), is likely due to their short length and factors other than backbone dihedral angles that influence their shifts.
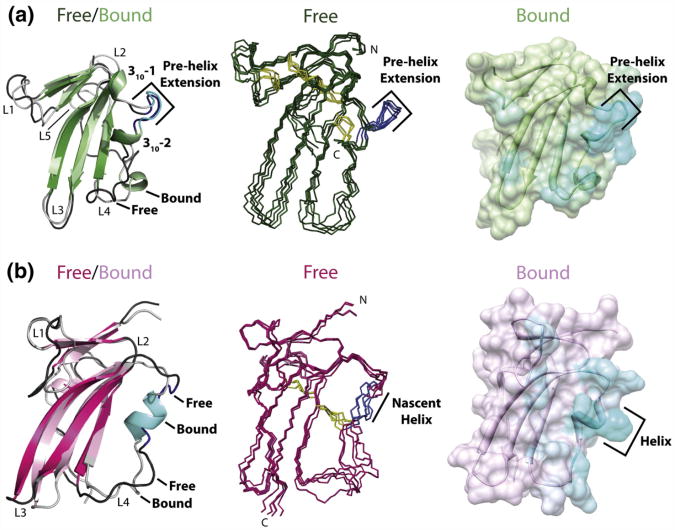
Fig. 6.
The key interaction element of TβR-I, the pre-helix extension, is structurally ordered prior to binding, while that of BMPR-Ia, the nascent helix harboring the “knob,” is not. (a) Left: Superimposition of the cartoon representations of the lowest-energy structures for free TβR-I 7–91 (dark green) and TGF-β3-bound TβR-I (light green). Center: Stick representation of the ensemble of the five lowest-energy solution structures for free TβR-I 7–91. Right: Surface and cartoon representation of the TGF-β3-bound form of TβR-I, with the extent of cyan coloring corresponding to the fraction of the total surface area buried in the TGF-β3/TβR-II/TβR-I crystal structure (PDB code: 2PJY). The pre-helix extension in the unbound form is shaded dark blue in the left and middle panels. (b) Left: Superimposition of the cartoon representations of the lowest-energy structures for free BMPR-Ia ED (magenta) and BMP-2-bound BMPR-Ia ED (pink). Center: Stick representation of the ensemble of BMPR-Ia solution structures (PDB code: 2K3G). Right: Surface and cartoon representation of the BMP-2-bound form of BMPR-Ia ED, with the extent of cyan coloring corresponding to the fraction of the total surface area buried in the BMP-2/ActR-IIb/BMPR-Ia crystal structure (PDB code: 2H64). The nascent helix of BMPR-Ia is shaded dark blue in the left and middle panels.
The region that deviated most from the bound form was loop 4, the extended segment from residues 63–71 (Fig. 6a, left). The difference in structure in loop 4 is likely a consequence of its intrinsic flexibility in both the unbound form and the bound form. The flexibility in the unbound form was directly demonstrated by an analysis of the backbone relaxation parameters, where the order parameter, S2, was as low as 0.33 (Fig. 5). The flexibility in the bound form is suggested by the absence of interpretable electron density in the crystal structure of the TβR-I/TβR-II/TGF-β1 complex from residues 64–71 (in one of the molecules in the asymmetric unit and from residues 67–70 in the other)13 and the reported weak density and elevated B-factors in this region in the crystal structure of the TβR-I/TβR-II/TGF-β3 complex.11 Although flexible, this region also appears to have an intrinsic propensity to form an α-helix, with residues 64–67 having about a 50% probability of forming an α-helix based on the secondary shifts of the unbound form (Fig. 3a). This propensity is also evident in the bound form, where residues 64–68 of TβR-I were modeled as an α-helix in the crystal structure of the TβR-I/TβR-II/TGF-β3 complex. The presence of this short helix in the crystal structure of the TβR-I/TβR-II/TGF-β3 complex, but not in TGF-β1, is likely due to slight differences in the way that TβR-I is positioned in the two complexes, with loop 4 making a slight contact with the C-terminal end of TGF-β α-helix 3 in the TGF- β3 complex, but not in TGF- β1.13 Thus, this loop appears to undergo a transition between a random coil and a α-helix in the unbound state, and while this helix is partially stabilized in the TGF- β3 receptor complex, it is evidently not stabilized in TGF- β1.
Comparison of the unbound and bound forms of TβR-I and BMPR-Ia
The unbound form of TβR-I differs significantly from the unbound form of BMPR-Ia in the extended segment between the C-terminal end of β-strand 4 and the N-terminal end of β-strand 5 (Fig. 6a and b). The N-terminal half up to the second 310 helix (310-2) is highly structured in the unbound form of TβR-I but is disordered in the unbound form of BMPR-Ia (Fig. 6a and b).17 These differences are significant, as TβR-I's primary interaction element, the pre-helix extension, is structurally ordered and conformationally similar to the bound form (Fig. 6a), whereas BMPR-I's primary interaction element, the short helix positionally conserved with respect to TβR-I's 310-2, is structurally disordered and undergoes a disorder-to-order transition upon binding12,17 (Fig. 6b).
Role of pre-helical residues in TβR-I recruitment and signaling
The TβR-I pre-helix extension lies at the center of the interface with TGF-β and TβR-II (Fig. 6a, right) and therefore likely plays a critical role in enabling TβR-I's recruitment by the TGF-β/TβR-II binary complex. To investigate this, we substituted several residues within the extension and evaluated them for their effects on recruitment and signaling. The substituted residues included Pro55, Arg58, and Pro59, all of which fall within the extension and appear to be important in either determining the overall conformation of the extension (cis-Ile54-Pro55) or enabling interactions with TβR-II (Arg58 and Pro59). Pro64, which is outside the extension and contacts neither TGF-β nor TβR-II in the complex, was also substituted to control for possible indirect effects on binding.
TβR-I ED folds poorly, with native species representing only a small fraction of the total pool of folded monomers. The folding mixture is sequentially fractionated on high-resolution cation-exchange and reverse-phase columns to isolate the native species. This procedure is normally implemented in conjunction with a native gel binding activity assay21 that allows native species to be detected. The native gel binding assay is easily applied, but its drawback is that it fails to detect native TβR-I when the Kd value for binding and recruitment by the TβR-II/TGF-β complex is diminished by about 15-fold or more.11
There was detectable native gel activity in the initial ion-exchange eluate for the Pro64-Ala variant (P64A), but not for the Pro55-Gly, Arg58-Ala, and Pro59-Gly variants (P55G, R58A, and P59G, respectively). To work around this, we divided the broad peak from the ion-exchange eluates for the P55G, R58A, and P59G variants into three parts and fractionated them using reverse-phase chromatography. Each of the major peaks from the reverse-phase eluates was exchanged into NMR buffer [25 mM sodium phosphate and 5% 2H2O (pH 7.2)] and examined using one-dimensional 1H NMR to identify the native species. The spectra obtained were examined for the dispersion of methyl and amide signals beyond the random-coil limits and for the correspondence of the overall pattern compared to wild type (WT). This identified one predominant species in the reverse-phase chromatograms of each of the variants, with signals beyond the random-coil limits, downfield of 8.5 ppm for the amides, and upfield of 0.8 ppm for the methyl groups. The predominant native-like species varied though in the similarity of its spectral pattern to WT, with P64A and R58A having the highest similarity, with P59G having intermediate similarity, and with P55G having the least similarity (Supplementary Material, Fig. 3).
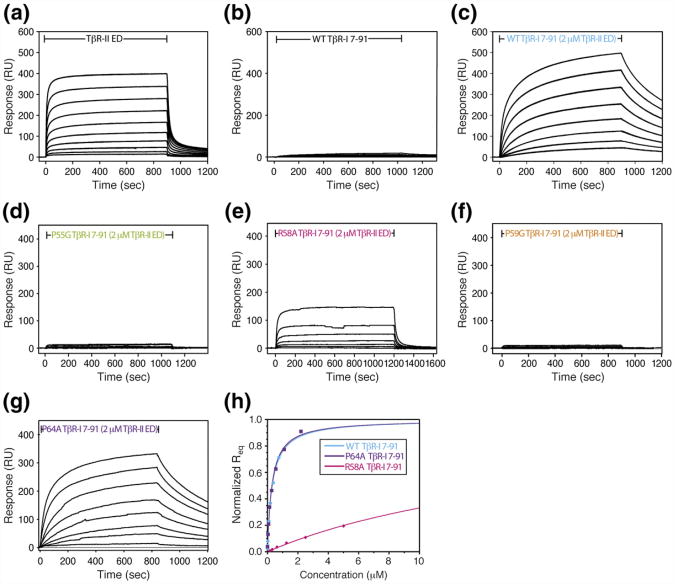
The binding affinity of the TβR-I variants for the TβR-II/TGF-β binary complex was assessed using SPR. This was accomplished by immobilizing TGF-β3 on the sensor surface and by injecting increasing concentrations of WT or variant TβR-I in the presence of 2 μM TβR-II. The assay is demonstrated in Fig. 7a–c, where TβR-II is shown to bind TGF-β3 with high affinity, potentiating the binding of TβR-I several hundred fold. The TβR-II concentration for the recruitment experiments, while only marginally saturating (roughly four times the Kd), proved to be sufficient for the purpose of these experiments, as experiments repeated with WT TβR-I and twice the concentration of TβR-II in the buffer (8 μM instead of 4 μM) led to only minor changes in the measured Kd for TβR-I recruitment. The data for the four TβR-I variants are presented in Fig. 7d–g. As shown, P64A produced a robust concentration-dependent response, R58A produced an intermediate response, and P55G and P59G produced detectable but very low responses. The equilibrium response, Req, as a function of concentration, could be reliably fitted to derive the Kd and maximal response, Rmax, for WT and P64A TβR-I. The response for R58A TβR-I could also be fitted, but only by constraining the maximal response, Rmax, to the same value obtained for TβR-II (which is similar in size to TβR-I). The responses for P55G and P59G TβR-I were so weak that they could not be reliably fitted even by constraining the maximal response, Rmax. The fits for WT, R58A, and P64A TβR-I are shown in Fig. 7h, and the derived values are listed in Table 2. The data show that WT and P64A Tβ-I are indistinguishable (with Kd values of 0.31±0.02 and 0.30±0.03 μM, respectively) and that R58A TβR-I is reduced roughly 65-fold relative to WT (with a Kd of 20.2±2.2 μM). These results show that residues within the extension play critical roles in enabling the recruitment of TβR-I, with Pro55 and Pro59 being absolutely essential and with Arg58 contributing, although to a lesser extent.
Fig. 7.
SPR binding profiles of TβR-I 7–91 variants bearing substitutions within the pre-helix extension. Control experiments in which either TβR-II ED (a) or TβR-I 7–91 (b) alone was injected over an amine-coupled TGF-β3 surface. The sensorgrams shown were obtained with serial 2-fold dilutions of the injected receptor (8.0–0.016 and 0.5–0.002 μM for TβR-II and TβR-I, respectively). (c–g) Recruitment experiments where WT TβR-I 7–91 or P55G, R58A, P59G, and P64A TβR-I 7–91 variants were injected over a TGF-β3 surface in the presence of a near-saturating concentration of TβR-II (2.0 μM). The inclusion of TβR-II was achieved by adding it both to the injected samples and to the SPR running buffer. The sensorgrams shown were obtained with serial 2-fold dilutions of the injected receptor (2.0–0.0156 μM for WT, 10.0– 0.078 μM for P55G, 5.0–0.312 μM for R58A, 32.0–0.063 μM for P59G, and 2.2–0.043 μM for P64A). (h) Plots of the normalized equilibrium response as a function of injected receptor concentration for the recruitment of WT TβR-I and P55G, R58A, P59G, and P64A variants by the TβR-II/TGF-β3 complex. Continuous line corresponds to fits of the experimental data to Req=(Req×concentration)/(Kd+concentration).
Table 2. Dissociation constants for the binding of TβR-I 7– 91 variants to TGF-β3 in the presence of a near-saturating concentration of TβR-II (2 μM).
| Analyte | Saturating receptor | Kd (μM) | Rmax (RU) |
|---|---|---|---|
| TβR-II ED | None | 0.52 ±0.04 | 445 ± 19 |
| TβR-I 7–91 | None | ND | ND |
| TβR-I 7–91 | 2 μM TβR-II ED | 0.31 ±0.02 | 536 ± 28 |
| P55G TβR-I 7–91 | 2 μM TβR-II ED | ND | ND |
| R58A TβR-I 7–91 | 2 μM TβR-II ED | 20.2±2.2 | 750 ±38 |
| P59G TβR-I 7–91 | 2 μM TβR-II ED | ND | ND |
| P64A TβR-I 7–91 | 2 μM TβR-II ED | 0.30 ±0.03 | 362 ± 23 |
All Kd values, except that for R58A TβR-I, were determined by fitting the observed concentration-dependent maximal response to derive both Kd and Rmax; for R58A TβR-I, Kd was fitted, but Rmax was fixed at the same value obtained for TβR-II binding over the same surface.
ND, not determined due to the minimal response observed.
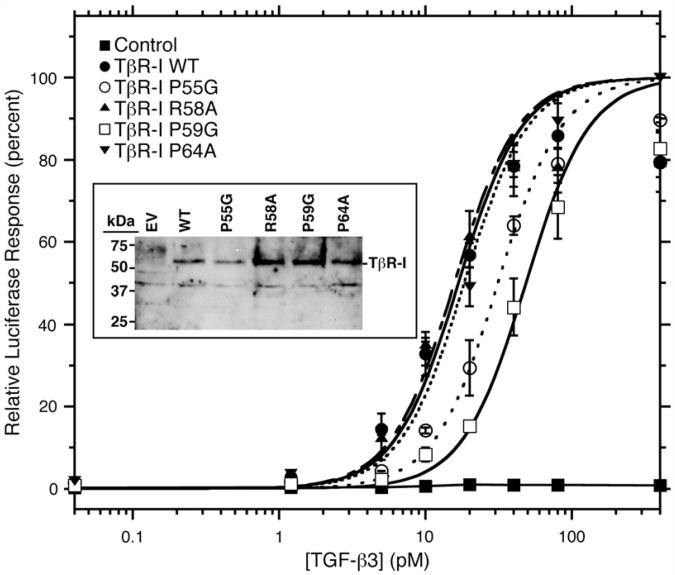
The TβR-I variants were also studied in the context of the full-length receptor in cultured cells. This was accomplished by transiently transfecting a vector expressing WT or variant TβR-I, along with a TGF-β luciferase reporter, into L17-R1b mink lung epithelial cells, a mutagenized cell line that lacks endogenous TβR-I and is not TGF-β responsive.33 The cells were also transfected with a β-galactosidase reporter to normalize for differences in transfection efficiencies. The results showed that there was a robust concentration-dependent luciferase response when the cells were transfected with WT TβR-I, but not with an empty vector control (Fig. 8). The three TβR-I variants, P55G, R58A, and P59G, also induced a robust concentration-dependent luciferase response, but the apparent potency was reduced for the P55G and P59G variants. The differences were quantitated by fitting the observed response as a function of concentration to a standard dose–response curve (Fig. 8, Table 3). The results show that WT, R58A, and P64A TβR-I were essentially indistinguishable, with EC50 values of 16.7±2.3, 15.7±2.5, 18.4±1.5 pM, respectively, whereas P55G and P59G TβR-I were diminished in their potency, with EC50 values of 31.3±2.4 and 48.5±4.7 pM, respectively (Table 3). The differences in activity among the variants could not be attributed to differences in the levels at which the receptors were expressed, as Western blot analysis for TβR-I revealed roughly equal levels of expressed TβR-I in lysates prepared from cells transfected with WT TβR-I and variants (Fig. 8, inset). There was no detectable TβR-I in the cells transfected with the empty vector, demonstrating the specificity of the antibody used in the Western blot analysis and further demonstrating that the activity must arise from the transfected plasmid DNA (not from endogenous WT TβR-I).
Fig. 8.
Reporter gene assay for variant receptor function. Reporter gene activity was assayed by measuring luciferase activity in L17-R1b mink lung epithelial cells transiently transfected with a fixed amount of plasmid expressing WT TβR-I and variants, together with CAGA12-Luc and β-galactosidase reporters, as a function of increasing concentrations of added TGF-β3. Luciferase values reported are normalized by β-gal activity and expressed as a percentage of the maximum value attained by the WT receptor. The Western blot analysis of protein lysates prepared from the transiently transfected cells using a TβR-I polyclonal is shown in the inset. EV, empty-vector-transfected cells.
Table 3. Reporter gene activity of TβR-I variants.
| TβR-I variant | EC50 (pM) |
|---|---|
| WT | 16.7±23 |
| P55G | 31.3±2.4 |
| R58A | 15.7±25 |
| P59G | 48.5±4.7 |
| P64A | 18.4± 1.5 |
Discussion
TGF-β play vital roles in coordinating wound repair and in regulating the adaptive immune system—functions essential for the long-term survival of humans and other higher vertebrates. TGF-β regulate these indispensable functions, without apparent interference from other members of the superfamily, by signaling through a highly restricted subset of receptors, known as TβR-I and TβR-II. TGF-β's high specificity for TβR-II arises from two hydrogen-bonded ion pairs formed by Arg/Lys and Asp/Glu residues conserved among TGF-β and TβR-II, but not other ligands or type II receptors of the superfamily.34,35 TGF-β specificity for TβR-I likely arises from its pre-helix extension, an exposed loop that binds in the cleft between TGF-β and TβR-II, but this has not been investigated.
The present results show that the unbound form of TβR-I is structurally similar to the bound form not only in terms of the β-strand framework and the five disulfide bonds that stabilize it but also in terms of the pre-helix extension and the two 310 helices that flank it. The results further show that the pre-helix extension and the two flanking 310 helices are rigid on the nanosecond-to-picosecond timescale, with the most flexible residue being Arg58 at the tip of the extension with a Lipari–Szabo order parameter of 0.68 (Fig. 5). The accompanying purified component binding studies showed that substitution of Pro55, Arg58, and Pro59 within the extension perturbs binding and recruitment of TβR-I, whereas substitution of Pro64, a residue outside the extension and binding interface, does not. The Arg58 variant, R58A, diminished the Kd for TβR-I recruitment by about 65-fold, whereas the Pro55 and Pro59 variants, P55G and P59G, diminished the Kd even more than this (Fig. 7, Table 2).
The accompanying one-dimensional 1H NMR spectra clearly demonstrate that each of these variants is folded, although, as noted, they differ in how closely their patterns match WT, with P64A and R58A (the variants least perturbed in their binding) matching more closely than P55G and P59G (the variants most perturbed in their binding) (Supplementary Material, Fig. 3). The differences in the one-dimensional 1H spectra of P55G and P59G are probably due to structural changes arising from the substitutions that are propagated through the structure, rather than from a mispaired disulfide or other folding defects, since parallel results were obtained when the substitutions were studied in the context of cultured epithelial cells (Fig. 8, Table 3). The finding that large decreases in the measured affinity for TβR-I recruitment by the TGF-β/TβR-II complex translate into a much smaller decrease or no detectable decrease in the cell-based assays has been previously observed11,36 and is likely due to a combination of factors, including membrane localization effects that compensate for the weaker binding between the extracellular domain of the receptor and the TGF-β/TβR-II complex and the demonstrated low inherent sensitivity of the lucif-erase reporter gene assay to reductions in signaling output.36,37 Together, these results show that the pre-helix extension is essential for the binding of TβR-I by the TGF-β/TβR-II complex, with Pro55 and Pro59 being absolutely essential and with Arg58 contributing, although to a lesser extent.
The importance of Pro55 likely stems from its cis peptide bond that is essential for accommodating the extension within the cleft between TGF-β and TβR-II. The interactions that stabilize Pro55 in the cis configuration in the unbound form of the protein are not known but, as mentioned, may arise from restrictions in conformational space imposed by the 310 helices that flank the extension and the Cys24-Cys47 and Cys76-Cys62 disulfides that serve as rigid anchors on the N-terminal side of 310-1 and the C-terminal side of 310-2, respectively. The large disruption in binding brought about by the substitution of Pro55 with glycine is probably due to the glycine binding in the trans configuration and compromising native-state interactions that are dependent on the close complementarity between the extension and the cleft into which it binds.
The fact that substitution of Pro59 is just as disruptive as the substitution of Pro55 suggests that this residue also plays an important role in binding. This may be due to the disruption of the hydrophobic pocket on the surface of TβR-I that accommodates Val22 and Phe24 from the TβR-II N-terminal tail, but it may also be due to indirect effects on Pro55. The latter is suggested by the packing between Pro55 and Pro59 in the unbound form, as shown by close interproton distances between Hδ1, H δ2 of Pro55, and Hα of Pro59 (Fig. 3c), and that substitution of Pro59 appears to disrupt TβR-I recruitment more than elimination of the TβR-II N-terminal tail.11
The finding that substitution of TβR-I Arg58 contributes to binding, but to a lesser degree, is consistent with the prior finding that the residue with which Arg58 pairs, TβR-II Asp118, also contributes to recruitment, but to a limited degree (3-fold reduction in Kd for TβR-I recruitment).11 There are two additional residues within the extension, Arg56 and Asp57: Arg56 might contribute to binding by ion pairing with TGF-β3 Lys97, while Asp57 has no obvious partner and extends into the solvent. These residues, however, were not examined owing to the significant effort required to refold and purify TβR-I variants, especially those that lack detectable activity in the native gel assay.
The pre-formed conformation of the extension, including cis-Pro55, presumably contributes to binding by diminishing the degree of ordering that the extension undergoes as it binds and by pre-positioning residues within the extension to engage TGF-β and TβR-II. This initial complex, stabilized by interactions between TβR-I Arg58 and TβR-II Asp118 and between hydrophobic portions of the extension and hydrophobic residues on the TGF-β fingers, is then presumably further stabilized by the binding-induced folding of the TβR-II N-terminal tail, with TβR-II Val22 and Phe24 binding into the hydrophobic pocket on the surface of TβR-I.
TGF-β's specificity for binding and recruiting TβR-I has been extensively investigated, and while ample data show that TβR-I is the primary receptor for TGF-β,33,38 other type I receptors bind and signal in place of TβR-I.18–20 The most extensively studied is Alk1, which is expressed predominantly in endothelial cells and forms a mixed receptor complex with TGF-β, TβR-II, and TβR-I.19 This leads to the activation of Smads 1, 5, and 8, in addition to Smads 2 and 3, and has been proposed to underlie TGF-β opposing effects on the migration of endothelial cells. This ‘lateral signaling' phenomenon has also been shown to occur in the context of several different normal and transformed cell lines with the type I receptors Alk2 and Alk3.18,20 The fact that these type I receptors are capable of substituting for TβR-I and transducing signals in response to TGF-β, albeit with significantly reduced efficiency, may reflect their ability to transiently bind into the space between TβR-II and TGF-β, become phos-phorylated by TβR-II, and signal. This presumes, of course, that these receptors retain sufficient affinity to bind even though they lack the critical pre-helix extension. Although further experimentation is required, this seems plausible given that elimination of the extension, on one hand, would be expected to greatly impair binding, while, on the other hand, the drastic reduction in affinity might be compensated for by membrane localization effects that promote receptor binding and signaling.
The activin type I receptor, ActR-Ib, also includes a pre-helix extension within its extracellular domain, yet functional studies with TβR-I-deficient mink lung epithelial cells show that ActR-Ib is not capable of substituting for TβR-I and transducing signals in response to TGF-β.38,39 This is unexpected given the importance of the pre-helix extension to the binding and recruitment of TβR-I and the high level of similarity of the extension in the two receptors, –PRDRP– in TβR-I and –PAGKP– in ActR–Ib (Fig. 1c). The most likely explanation for this apparent contradiction is that ActR-Ib's extension either is more flexible (due to its internal glycine residue) or adopts a conformation distinct from that of TβR-I. This would impair or prevent ActR-Ib from binding into the cleft between TGF-β and TβR-II and thus greatly attenuate any additional interactions that stabilize the complex. The possibility that ActR-Ib's extension might have increased flexibility or might adopt an alternate conformation seems plausible, given that the environment into which the extension binds is expected to be entirely distinct. This follows, since the extension is expected to contact activin on the edges of the ligand fingers, as in the TGF-β receptor complex,40,41 yet the activin type II receptor binds on the ligand “knuckles,” rather than “fingertips,” as in the TGF-β complex,42 leaving ActR-Ib without direct contact with its type II receptor. This mixed mode of receptor binding, with a BMP-like manner of type II receptor binding and a TGF-β-like manner of type I receptor binding, may have been a crucial step in contributing to a membrane-independent highly cooperative recruitment mechanism peculiar to TGF-β ligand–receptor complexes.
These results have shown that the manner by which TGF-β binds and recruits its type I receptor, TβR-I, is very different from the manner by which BMPs bind their type I receptor, BMPR-Ia. TβR-I's principal interaction element, the pre-helix extension, is ‘pre-ordered' and does not undergo any significant conformational changes on binding, including the critical cis-Ile54-Pro55 peptide bond. This, together with its overall rigidity and pre-ordered conformation, is likely important for promoting the binding of TβR-I into the TGF-β receptor complex by minimizing the change in configurational entropy. The high complementarity between the extension and the cleft into which it binds is also likely important in minimizing the binding of other type I receptors, particularly BMPR-Ia, which lacks the extension, but also the activin type Ib receptor, which includes the extension but may adopt a different conformation. BMPR-Ia's principal interaction element, the 1.6-turn α-helix structurally conserved with respect to TβR-I's 310-2 helix, is, in contrast, largely structurally disordered in the unbound form and undergoes a disorder-to-order transition upon binding, with the two residues most essential for binding (Phe85 and Gln86) undergoing a large-scale reorientation to engage the ligand.17 This flexibility in the binding site for the ligand on the type I receptor has been proposed to be necessary for enabling promiscuity in binding, an essential feature for BMPs due to the large number of ligands in comparison to the limited number of receptors.43
Materials and Methods
Protein purification
Human TβR-I ED was expressed in Escherichia coli using a construct in which the coding sequence for residues 7–91, following the predicted signal peptide cleavage site,22 was inserted between the NdeI site and the BamHI site in plasmid pET15b (Novagen, Madison WI). This construct, termed TβR-I 7–91, was expressed and isolated using the procedure previously reported for the full-length extracellular domain, TβR-I 1–101.11,21 Briefly, this entailed expression at 37 °C, refolding in the presence of a glutathione redox couple at pH 8.0, cleavage with thrombin to remove the N-terminal histidine tag, and sequential fractionation on high-resolution cation-exchange (Source S; GE Healthcare) and C18 reverse-phase (Jupiter C18 2 μM; Phenomenex) columns. Human TβR-II ED was expressed in E. coli, refolded, and purified as previously described.44
NMR samples
Samples of TβR-I 7–91 for NMR spectroscopy were prepared in a buffer consisting of 25 mM sodium phosphate, 0.02% sodium azide, and 5% 2H2O (pH 7.2), and were placed in 5-mm susceptibility-matched thin-wall microcells (Shigemi). Samples uniformly labeled with either 15N or 15N and 13C were prepared by culturing the cells on M9 minimal medium with isotopically labeled growth substrates following the procedure outlined by Marley et al.45 Fractionally 13C-labeled TβR-I 7–91 was prepared using M9 medium enriched with 0.03 g/L [13C] glucose and 0.27 g/L unlabeled glucose.46
NMR spectrometers
All NMR experiments were performed at 27 °C on Bruker 600-MHz and 700-MHz spectrometers with cryogenically cooled 5-mm 1H probes equipped with 13C and 15N decoupler and pulsed-field gradient coils. All spectra were processed using NMRPipe 47 and analyzed using the program NMRView.48
Resonance assignments
Backbone resonance assignments of TβR-I 7–91 were obtained by collecting and analyzing sensitivity-enhanced triple-resonance data sets, including HNCA,49 HNCACB,50 CBCA(CO)NH,51 and HNCO.52 Aliphatic 1H and 13C assignments were obtained by collecting and analyzing HBHA(CO)NH,51 (H)CC(CO)NH,53 H(CC)H correlated spectroscopy,54 and H(CC)H total correlated spectroscopy54 data sets. Aromatic ring assignments were obtained from a CB(CGCD)HD data set.55
Structural restraints
Interproton distance restraints were obtained by recording 3D 15N-edited and 13C-edited NOESY spectra at 700 MHz using a mixing time of 120 ms. Backbone φ and ψ restraints were obtained by an analysis of the assigned chemical shifts using the program TALOS.56 φ was additionally restrained by measuring 3JHNHa couplings using an HNHA experiment.57 Orientational restraints for the backbone 1H–15N bond vectors were obtained from the difference in the measured 1H–15N splittings in the absence and in the presence of 10 mg/mL Pf1 phage (Hyglos GmbH).58 The couplings themselves were measured using a two-dimensional in-phase anti-phase (IPAP) HSQC experiment modified to suppress signals arising from –NH2.59
Structural calculations
NOE distance restraints were initially derived by manually assigning the 13C-edited and 15N-edited 3D NOESY data sets. Initial structures were calculated using CNS 1.160 with the manually assigned NOEs, HN–HαJ-couplings, the TALOS-derived dihedral angles, and 1H–15N RDCs as restraints. Final refined structures were calculated using ARIA 1.2 with the protein_allhdg force field.30 Restraints used in the ARIA calculations were those noted above, but with approximately 30% more NOEs identified by automated assignment within ARIA. Fifty starting structures were generated based on a linear template molecule with randomly associated velocities for all atoms. For iterations 0–7, for which 50 structures were calculated, the NOE distance restraints were recalibrated by ARIA based on the 10 lowest-energy structures. The violation tolerance was progressively reduced to 0.1 Å in iteration 8, in which 200 structures were calculated. For the structure calculations, a four-stage SA protocol that employed torsion-angle dynamics was used. The high-temperature stage consisted of 10,000 steps at 10,000 K, followed by three cooling stages: 8000 steps to 2000 K, 20,000 steps to 1000 K, and 15,000 steps to 50 K. During the SA protocol, the force constant for the NOE restraints was set to 0, 10, 10, and 50 kcal/mol/Å2. The final 20 lowest-energy structures were further refined with explicit water.61
Measurement of backbone 15N relaxation data
Backbone amide 15N T1, 15N T2, and{1H}–15N NOE relaxation parameters were measured in an interleaved manner at 300 K at a 15N frequency of 60.8 MHz using 1H-detected pulse schemes previously described.62 The T1 and T2 data sets were each collected using 12 delay times, varying between 8 and 1320 ms and between 8 and 192 ms, respectively. The T1 and T2 relaxation times were obtained by fitting the relative peak intensities as a function of T1 or T2 delay time to a two-parameter decaying exponential.{1H}–15N NOE values were obtained by taking the ratio of peak intensities from experiments performed with 1H presaturation to peak intensities from experiments performed without 1H presaturation and by applying a correction factor to account for the incomplete recovery of both 15N and 1H magnetization.63
Analysis of backbone relaxation data
The overall correlation time and the degree of diffusional anisotropy were determined by maximizing the agreement between the experimentally measured 15N T1/T2 ratio and the calculated 15N T1/T2 ratio for an axially symmetric ellipsoid using the fitting procedure described by Tjandra et al.31 Amide bond vector orientations were obtained from the five lowest-energy structures, and the criterion given by Barbato et al. was used to identify and eliminate from the calculations any residue undergoing large-amplitude motion on the nanosecond-to-picosecond timescale or exchange.64 Internal dynamics were assessed by analyzing the experimental 15N relaxation parameters using the extended model-free formalism,65–67 with the overall correlation time and parameters relevant to diffusional anisotropy derived from the analysis described above. Internal motional parameters were derived using the program ModelFree4, which employs F-statistics for model selection.32 Five different models for internal motion were considered: S2 (model 1); S2 and τe (model 2); S2 and Rex (model 3); S2, τe, and Rex (model 4); and S2, , and τe (model 5).
TβR-I variants and characterization of their binding properties
Plasmids encoding TβR-I 7–91 P55G, R58A, P59G, and P64A variants were generated by QuikChange (Stratagene) site-directed mutagenesis and verified by sequencing over the length of the cloned gene. The variants were expressed, refolded, and purified as performed for the WT protein; however, because no activity could be detected with native gels for three of the four variants, it was necessary to divide the initial eluate from the cation-exchange profile into several sections and to fractionate each of these using C18 reverse-phase chromatography. The fractions corresponding to each of the major peaks in the reverse-phase column eluates were subjected to one-dimensional 1H NMR analysis to identify natively folded species.
The binding affinities of the TβR-I 7–91 variants for the TβR-II/TGF-β3 binary complex were measured using a Biacore 3000 SPR instrument, as previously described.11 Briefly, this was achieved by immobilizing TGF-β3 on the surface of a carboxymethylated dextran sensor chip (CM5; GE Healthcare) and by injecting increasing concentrations of the WT and variant receptors over the sensor chip in the presence of a near-saturating concentration (2 μM) of the purified TβR-II ED. Saturation with TβR-II ED was accomplished by adding it to the running buffer and to the injected samples. Brief injections (16 s) of 4 M guanidine hydrochloride were used between cycles to regenerate the surface. Instrument noise was removed by referencing the data against three or more buffer blank injections, while background signal was eliminated by referencing the data against a blank flow cell. Kd values were determined by fitting the equilibrium binding response, Req, as a function of the injected receptor concentration, [R], to Req=(Rmax [R])/(Kd + [R]) using the program Profit (Quantum Soft).
Cell-based reporter gene assay and Western blot analysis
The gene encoding full-length human TβR-I was inserted between the HindIII site and the NotI site in plasmid pRC/CMV (Invitrogen). Plasmids encoding TβR-IP55G, R58A, P59G, and P64A variants were generated by QuikChange (Stratagene) site-directed mutagenesis and verified by sequencing over the length of the cloned gene. L17-R1b mink lung epithelial cells, which do not express TβR-I,68 were plated on 24-well plates at 5 × 104 cells/well in minimal essential medium supplemented with nonessential amino acids and 10% fetal calf serum. After 24 h, cells were transfected with 80 ng/well WT and variant TβR-I constructs, along with CAGA12 luciferase (0.25 mg/well)69 and β galactosidase reporters (0.175 mg/well), using LT-1 transfection reagent (Mirus). Four hours after transfection, the medium was replaced with TGF-β3 containing minimal essential medium with 0.2% fetal calf serum. Luciferase production was quantified 48 h later using the Luciferase Assay System (Promega) and normalized with β-galactosidase activity using the β-Galactosidase Enzyme Assay System (Promega). Western blot analyses were performed by running a constant amount of total protein (10 μg), normalized by the β-gal transfection efficiency, from the protein lysates prepared from the transiently transfected L17-R1b cells on a reducing 12% SDS gel. Protein was transferred to a nitrocellulose membrane, blocked with 5% non-fat dried milk, and then probed with a rabbit TβR-I polyclonal antibody (catalog number SC-398; Santa Cruz Biotechnology). Blots were developed by incubation with a horseradish-peroxidase-conjugated secondary antibody and enhanced chemiluminescent detection (ECL+; GE Healthcare).
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health grants GM58670 and RR13879 awarded to A.P.H., National Institutes of Health grant CA54174 awarded to the University of Texas Health Science Center at San Antonio Cancer Therapy and Research Center, and Robert A. Welch Foundation grant AQ-1431 awarded to A.P.H. The authors would also like to acknowledge Dr. Joan Massagué for kindly providing the L17-R1b cells.
Abbreviations used
- TGF-β
transforming growth factor β isoforms
- TβR-I
TGF-β type I receptor
- TβR-II
TGF-β type II receptor
- BMP
bone morphogenetic protein
- GDF
growth and differentiation factor
- R-Smad
receptor-mediated Smad protein
- BMPR-I
BMP type I receptor
- BMPR-Ia ED
BMPR-Ia extracellular domain
- TβR-I ED
TβR-I extracellular domain
- HSQC
heteronuclear single-quantum coherence
- SPR
surface plasmon resonance
- TβR-II ED
TβR-II extracellular domain
- 3D
three-dimensional
- NOESY
nuclear Overhauser enhancement spectroscopy
- NOE
nuclear Overhauser enhancement
- IPAP
in-phase anti-phase
- SA
simulated annealing
- RDC
residual dipolar coupling
- WT
wild type
- PDB
Protein Data Bank
Footnotes
Accession numbers: Chemical shifts assignments for TβR-I 7–91 have been deposited in BioMagResBank under accession number 17276. The 10 lowest-energy structures satisfying all the experimental distance, dihedral angle, and RDC restraints have been deposited under Protein Data Bank (PDB) code 2L5S.
Supplementary materials related to this article can be found online at doi:10.1016/j.jmb.2011.07.046
References
- 1.Massagué J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 2.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 3.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 4.Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 5.Wrana JL, Attisano L, Wieser R, Ventura F, Massagué J. Mechanism of activation of the TGF-beta receptor. Nature. 1994;370:341–347. doi: 10.1038/370341a0. [DOI] [PubMed] [Google Scholar]
- 6.Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–3584. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y, Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 8.Massagué J. How cells read TGF-beta signals. Nat Rev Mol Cell Biol. 2000;1:169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 9.Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci USA. 2006;103:7643–7648. doi: 10.1073/pnas.0602558103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenwald J, Groppe J, Gray P, Wiater E, Kwiatkowski W, Vale W, Choe S. The BMP7/ActRII extracellular domain complex provides new insights into the cooperative nature of receptor assembly. Mol Cell. 2003;11:605–617. doi: 10.1016/s1097-2765(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 11.Groppe J, Hinck CS, Samavarchi-Tehrani P, Zubieta C, Schuermann JP, Taylor AB, et al. Cooperative assembly of TGF-beta superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 12.Kirsch T, Sebald W, Dreyer MK. Crystal structure of the BMP-2–BRIA ectodomain complex. Nat Struct Biol. 2000;7:492–496. doi: 10.1038/75903. [DOI] [PubMed] [Google Scholar]
- 13.Radaev S, Zou Z, Huang T, Lafer EM, Hinck AP, Sun PD. Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily. J Biol Chem. 2010;285:14806–14814. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber D, Kotzsch A, Nickel J, Harth S, Seher A, Mueller U, et al. A silent H-bond can be mutationally activated for high-affinity interaction of BMP-2 and activin type IIB receptor. BMC Struct Biol. 2007;7:6. doi: 10.1186/1472-6807-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massagué J. A very private TGF-beta receptor embrace. Mol Cell. 2008;29:149–150. doi: 10.1016/j.molcel.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Keller S, Nickel J, Zhang JL, Sebald W, Mueller TD. Molecular recognition of BMP-2 and BMP receptor IA. Nat Struct Mol Biol. 2004;11:481–488. doi: 10.1038/nsmb756. [DOI] [PubMed] [Google Scholar]
- 17.Klages J, Kotzsch A, Coles M, Sebald W, Nickel J, Muller T, Kessler H. The solution structure of BMPR-Ia reveals a local disorder-to-order transition upon BMP-2 binding. Biochemistry. 2008;47:11930–11939. doi: 10.1021/bi801059j. [DOI] [PubMed] [Google Scholar]
- 18.Daly AC, Randall RA, Hill CS. Transforming growth factor beta-induced Smad1/5 phosphorylation in epithelial cells is mediated by novel receptor complexes and is essential for anchorage-independent growth. Mol Cell Biol. 2008;28:6889–6902. doi: 10.1128/MCB.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. Balancing the activation state of the endothelium via two distinct TGF-beta type I receptors. EMBO J. 2002;21:1743–1753. doi: 10.1093/emboj/21.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai D, Kim SW, McKeller MR, Dahia PL, Aguiar RC. Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis. Proc Natl Acad Sci USA. 2010;107:3111–3116. doi: 10.1073/pnas.0910667107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuniga JE, Groppe JC, Cui Y, Hinck CS, Contreras-Shannon V, Pakhomova ON, et al. Assembly of TbetaRI:TbetaRII:TGFbeta ternary complex in vitro with receptor extracellular domains is cooperative and isoform-dependent. J Mol Biol. 2005;354:1052–1068. doi: 10.1016/j.jmb.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Franzen P, ten Dijke P, Ichijo H, Yamashita H, Schulz P, Heldin CH, Miyazono K. Cloning of a TGF beta type I receptor that forms a heteromeric complex with the TGF beta type II receptor. Cell. 1993;75:681–692. doi: 10.1016/0092-8674(93)90489-d. [DOI] [PubMed] [Google Scholar]
- 23.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 24.Eghbalnia HR, Wang L, Bahrami A, Assadi A, Markley JL. Protein energetic conformational analysis from NMR chemical shifts (PECAN) and its use in determining secondary structural elements. J Biomol NMR. 2005;32:71–81. doi: 10.1007/s10858-005-5705-1. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 26.Hinck AP, Eberhardt ES, Markley JL. NMR strategy for determining Xaa-Pro peptide bond configurations in proteins: mutants of staphylococcal nuclease with altered configuration at proline-117. Biochemistry. 1993;32:11810–11818. doi: 10.1021/bi00095a009. [DOI] [PubMed] [Google Scholar]
- 27.Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J Biomol NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- 28.Torchia DA, Sparks SW, Young PE, Bax A. Proline assignments and identification of the cis K116/P117 peptide-bond in liganded staphylococcal nuclease using isotope edited 2-D NMR-spectros-copy. J Am Chem Soc. 1989;111:8315–8317. [Google Scholar]
- 29.Shen Y, Bax A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR. 2010;46:199–204. doi: 10.1007/s10858-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–316. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 31.Tjandra N, Feller SE, Pastor RW, Bax A. Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J Am Chem Soc. 1995;117:12562–12566. [Google Scholar]
- 32.Mandel AM, Akke M, Palmer AG., III Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 33.Laiho M, Weis FM, Boyd FT, Ignotz RA, Massagué J. Responsiveness to transforming growth factor-beta (TGF-beta) restored by genetic complementation between cells defective in TGF-beta receptors I and II. J Biol Chem. 1991;266:9108–9112. [PubMed] [Google Scholar]
- 34.Baardsnes J, Hinck CS, Hinck AP, O'Connor-McCourt MD. TbetaR-II discriminates the high- and low-affinity TGF-beta isoforms via two hydrogen-bonded ion pairs. Biochemistry. 2009;48:2146–2155. doi: 10.1021/bi8019004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Crescenzo G, Hinck CS, Shu Z, Zuniga J, Yang J, Tang Y, et al. Three key residues underlie the differential affinity of the TGFbeta isoforms for the TGFbeta type II receptor. J Mol Biol. 2006;355:47–62. doi: 10.1016/j.jmb.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 36.Huang T, David L, Mendoza V, Yang Y, Villarreal M, De K, et al. TGF-β signaling is mediated by two autonomously functioning TβRI: TβRII pairs. EMBO J. 2011;30:1263–1276. doi: 10.1038/emboj.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amatayakul-Chantler S, Qian SW, Gakenheimer K, Bottinger EP, Roberts AB, Sporn MB. [Ser77]transforming growth factor-beta 1. Selective biological activity and receptor binding in mink lung epithelial cells. J Biol Chem. 1994;269:27687–27691. [PubMed] [Google Scholar]
- 38.Carcamo J, Weis FM, Ventura F, Wieser R, Wrana JL, Attisano L, Massagué J. Type I receptors specify growth-inhibitory and transcriptional responses to transforming growth factor beta and activin. Mol Cell Biol. 1994;14:3810–3821. doi: 10.1128/mcb.14.6.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cash JN, Rejon CA, McPherron AC, Bernard DJ, Thompson TB. The structure of myostatin:follistatin 288: insights into receptor utilization and heparin binding. EMBO J. 2009;28:2662–2676. doi: 10.1038/emboj.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrison CA, Gray PC, Fischer WH, Donaldson C, Choe S, Vale W. An activin mutant with disrupted ALK4 binding blocks signaling via type II receptors. J Biol Chem. 2004;279:28036–28044. doi: 10.1074/jbc.M402782200. [DOI] [PubMed] [Google Scholar]
- 41.Harrison CA, Gray PC, Koerber SC, Fischer W, Vale W. Identification of a functional binding site for activin on the type I receptor ALK4. J Biol Chem. 2003;278:21129–21135. doi: 10.1074/jbc.M302015200. [DOI] [PubMed] [Google Scholar]
- 42.Thompson TB, Woodruff TK, Jardetzky TS. Structures of an ActRIIB:activin A complex reveal a novel binding mode for TGF-beta ligand: receptor interactions. EMBO J. 2003;22:1555–1566. doi: 10.1093/emboj/cdg156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nickel J, Sebald W, Groppe JC, Mueller TD. Intricacies of BMP receptor assembly. Cytokine Growth Factor Rev. 2009;20:367–377. doi: 10.1016/j.cytogfr.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 44.Hinck AP, Walker KP, III, Martin NR, Deep S, Hinck CS, Freedberg DI. Sequential resonance assignments of the extracellular ligand binding domain of the human TGF-beta type II receptor. J Biomol NMR. 2000;18:369–370. doi: 10.1023/a:1026775321886. [DOI] [PubMed] [Google Scholar]
- 45.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J Biomol NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 46.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 49.Yamazaki T, Lee W, Revington M, Mattiello DL, Dahlquist FW, Arrowsmith CH, Kay LE. An HNCA pulse scheme for the backbone assignment of N-15, C-13, H-2 labeled proteins— applications to a 37 kDa Trp repressor DNA complex. J Am Chem Soc. 1994;116:6464–6465. [Google Scholar]
- 50.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3-D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J Magn Reson Ser B. 1993;101:201–205. [Google Scholar]
- 51.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 52.Kay LE, Ikura M, Tschudin R, Bax A. 3-Dimensional triple-resonance NMR spectroscopy of isotopically labeled proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Grzesiek S, Anglister J, Bax A. Correlation of backbone amide and aliphatic side-chain resonances in C-13/N-15-enriched proteins by isotropic mixing of C-13 magnetization. J Magn Reson Ser B. 1993;101:114–119. [Google Scholar]
- 54.Kay LE, Xu GY, Singer AU, Muhandiram DR, Forman-Kay JD. A graidient-enhanced HCCH-TOCSY experiment for recording side-chain H-1 and C-13 correlations in H2O samples of proteins. J Magn Reson Ser B. 1993;101:333–337. [Google Scholar]
- 55.Yamazaki T, Forman-Kay JD, Kay LE. Two-dimensional NMR experiments for correlating carbon-13 beta and proton delta/epsilon chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J Am Chem Soc. 1993;115:11054–11055. [Google Scholar]
- 56.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 57.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNH.alpha) coupling constants in 15N-enriched proteins. J Am Chem Soc. 1993;115:7772–7777. [Google Scholar]
- 58.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 59.Ishii Y, Markus MA, Tycko R. Controlling residual dipolar couplings in high-resolution NMR of proteins by strain induced alignment in a gel. J Biomol NMR. 2001;21:141–151. doi: 10.1023/a:1012417721455. [DOI] [PubMed] [Google Scholar]
- 60.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Crystallogr Sect D: Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 61.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 62.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 63.Freedberg DI, Ishima R, Jacob J, Wang YX, Kustanovich I, Louis JM, Torchia DA. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002;11:221–232. doi: 10.1110/ps.33202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 65.Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple 2-parameter model-free approach to the interpretation of N-15 nulcear magnetic-relaxation of protein. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 66.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance in macromolecules: 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 67.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules: 2. Analysis and experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 68.Boyd FT, Massagué J. Transforming growth factor-beta inhibition of epithelial cell proliferation linked to the expression of a 53-kDa membrane receptor. J Biol Chem. 1989;264:2272–2278. [PubMed] [Google Scholar]
- 69.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.