Summary
The nuclear factor-κB (NF-κB) family of transcription factors plays important roles in various biological processes including apoptosis, stress response, immunity, and inflammation. NF-κB signaling is involved in both immune cell development and function, and it is critical in modulation of the immune response through the transcriptional regulation of cytokine and chemokine expression. An area of great interest in T-cell-mediated adaptive immunity is the ability of naive CD4+ T cells generated in the thymus to differentiate into various subsets including T-helper 1 (Th1), Th2, Th17, Th9, follicular helper T (Tfh), Th22 and regulatory T (Treg) cells, upon encountering different pathogens and microenvironments. In this review, we discuss the role of NF-κB pathway in the development and functional divergence of the different helper T-cell subsets as well as in regulatory T cells.
Keywords: NF-κB, T-cell subsets, T-cell development, T-cell signaling
Introduction
T cells are critical elements of the vertebrate adaptive immune responses, where they function as both regulators and effectors of the immune response. Naive conventional T (Tconv) cells develop in the thymus and migrate to the periphery, where they encounter antigen or particular environmental conditions leading to the differentiation into effector subsets (Fig. 1). Notably, the NF-κB family of transcription factors has been shown to regulate various aspects of T-cell development, activation, differentiation, and survival.
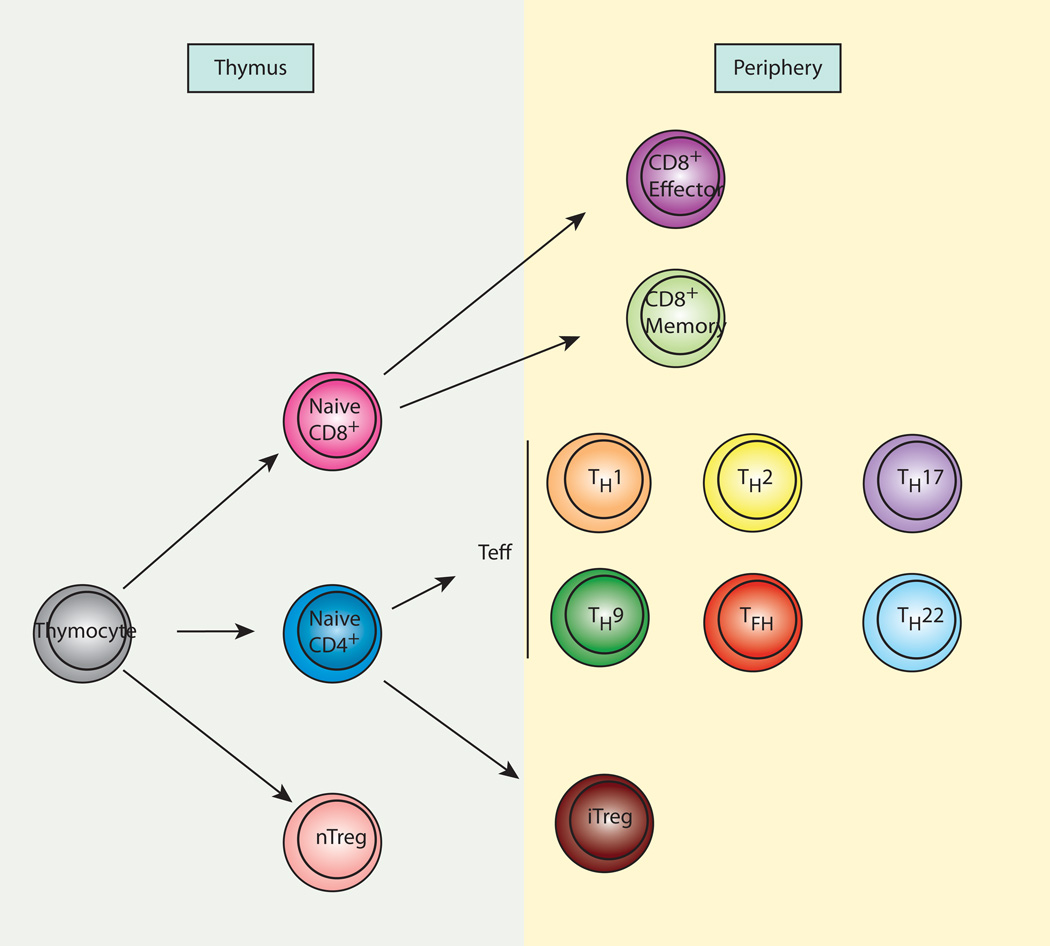
Fig. 1. CD4+ T-cell subsets.
In the thymus, naive CD4+ and CD8+ T cells develop from thymocytes. Naive CD4+ T cells from the thymus migrate to the periphery, where they can differentiate to various subsets of effector cells upon encountering specific antigens. The subsets include Th1, Th2, Th17, Th9, Tfh, and Th22. Naive CD8+ T cells also migrate to the periphery and differentiate to effector cells or memory cells upon encountering antigens. A small population of thymocytes differentiates into regulatory T cells (nTreg) in the thymus. In addition, some naive CD4+ T cells differentiate into regulatory T cells (iTreg) in specific microenvironments.
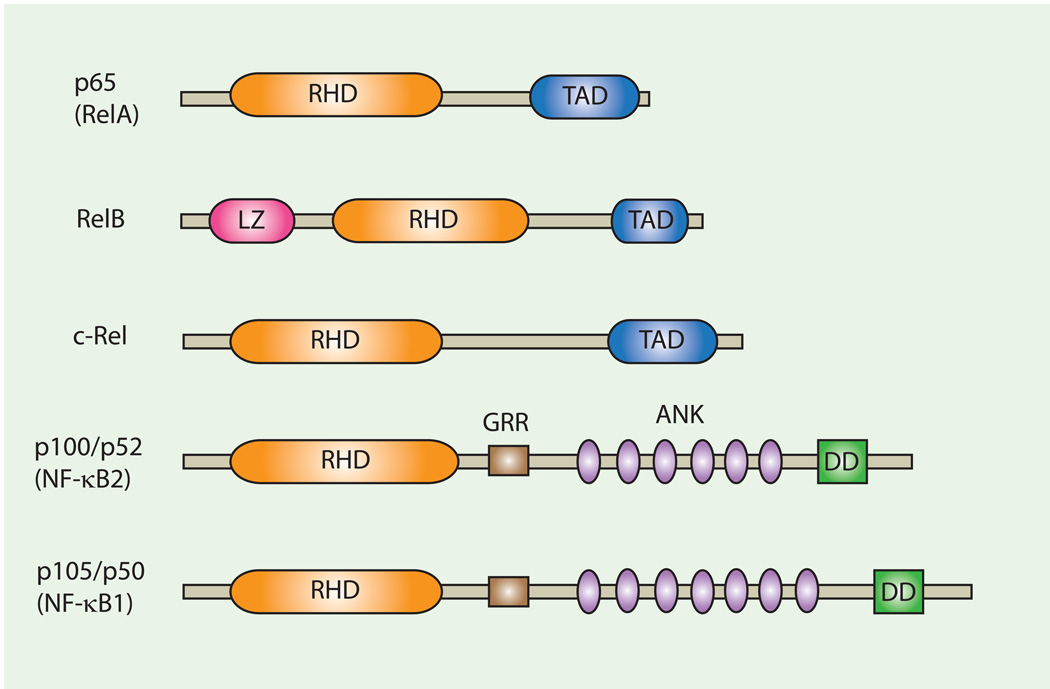
The NF-κB family is comprised of five subunits: RelA (p65), RelB, c-Rel, NF-κB1 (p105), and NF-κB2 (p100) (1) (Fig. 2). The different subunits share an N-terminal Rel-homology domain (RHD), which is responsible for DNA-binding and homo- or hetero-dimerization. The p65, c-Rel, and RelB subunits contain a transcription activation domain and are hence capable of driving transcription. Under resting conditions, NF-κB dimers are sequestered in the cytoplasm by the inhibitory IκB proteins. Upon stimulation, the IκB kinase (IKK), which is comprised of IKKα, IKKβ, and the regulatory subunit NEMO, phosphorylates the IκB proteins, leading to their subsequent ubiquitination and degradation. As a result, NF-κB dimers are released from IκB inhibition and translocate to the nucleus where they bind to κB-binding sites in the promoter or enhancer regions of target genes. In general, NF-κB activation occurs through two major pathways: the canonical and noncanonical pathways (Fig. 3). In the canonical pathway, IKKβ phosphorylation of IκBα leads to its degradation and the subsequent liberation of the NF-κB dimers. p65:p50 heterodimers are the major targets of the canonical pathway, but other combinations of dimers, including c-Rel containing dimers, can be involved in the pathway. In the noncanonical pathway, the NF-κB-inducing kinase (2) directly phosphorylates and activates IKKα. Activated IKKα phosphorylates p100, which results in proteosomal processing of p100 to p52. p52 forms a heterodimer with RelB and translocates to the nucleus to bind to κB-binding sites. Activation of the NF-κB pathway regulates expression of a plethora of immunomodulatory factors, including cytokines, chemokines, adhesion molecules, antimicrobial factors, cell cycle regulators, and cell survival factors.
Fig. 2. Members of the NF-κB/Rel family.
Schematic depiction of members of the NF-kB transcription factor family. RHD, Rel homolgy domain; TAD, transactivation domain; LZ, leucin zipper domain; GRR, glycine-rich region; DD, death domain; ANK, ankyrin repeats.
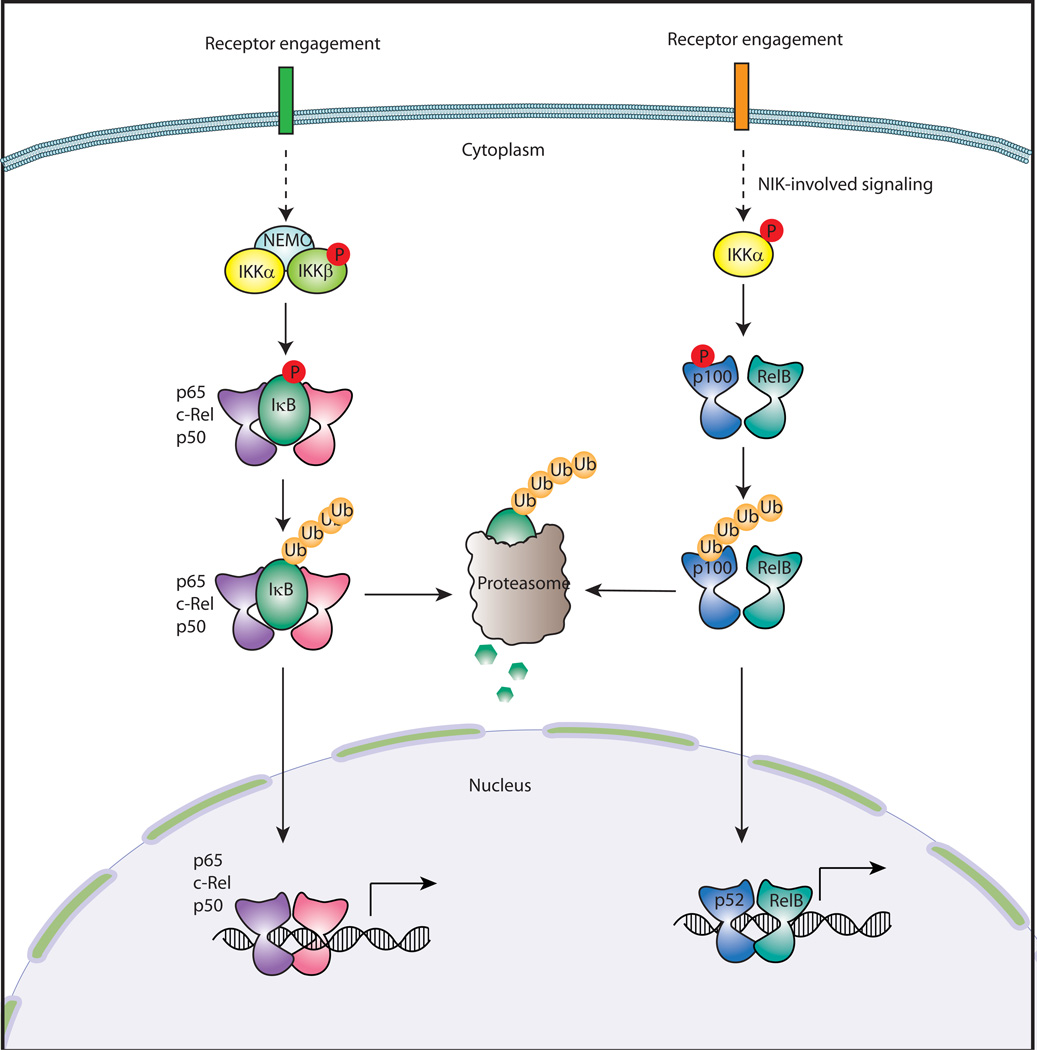
Fig. 3. Canonical and noncanonical pathways of NF-κB activation.
NF-κB activation occurs through two different pathways: canonical and noncanonical. In the canonical pathway, the IKK complex, composed of IKKα, IKKβ, and the regulatory subunit NEMO, phosphorylates IκB, leading to its ubiquitination and proteosomal degradation. A NF-κB dimer, which is comprised of p50, p65, or c-Rel, is released from IκB and translocates into the nucleus, where it binds to a κB-binding site for regulation of gene expression. In the noncanonical pathway, IKKα phosphorylates p100 resulting in its ubiquitination and partial processing into p52. A heterodimer of p52 and RelB translocates into the nucleus and binds to it binding sites. P, phosphorylation; Ub, ubiquitination.
Stimulation of T cells through engagement of the T-cell receptor (TCR) and the co-receptor CD28 induces expression of activation-associated cell surface molecules, such as the adhesion molecule leukocyte function-associated antigen-1 (LFA-1) (3), as well as cytokines important for T-cell proliferation and survival, such as interleukin-2 (IL-2) (4). TCR, and CD28 activation of the NF-κB pathway is thought to contribute to this transcriptional program. Initial activation is mediated by p65-containing NF-κB complexes and is followed by delayed and sustained activation of the pathway through c-Rel-containing complexes (4). Various genetic studies, employing inhibition or deficiency of specific NF-κB subunits in T cells, have implicated NF-κB activity in the protection from apoptosis and the production of IL-2 (1). Futhermore, genetic models in mice have demonstrated an essential role of NF-κB in T-cell activation, expansion, and effector function during infection (1).
Activation of naive T cells also results in the rapid proliferation and simultaneous differentiation into effector subsets. T lymphocytes are classified into subsets based on TCR (αβ or γδ) and coreceptor expression (CD4+ or CD8+), cytokine profile, and divergent functions during an immune response. For the purposes of this review, we focus our discussion on the development and function of the various CD4+ T-cell subsets.
CD4+ T cells recognize peptide-major histocompatibility complex (MHC) class II complexes presented by antigen-presenting cells (APCs) and differentiate into different effector T-cell subtypes or regulatory T cells upon activation. Among CD4+ T-helper cells, Th1 cells are responsible for immunity against intracellular pathogens primarily through secretion of interferon-γ (IFN-γ), whereas Th2 cells promote the humoral immune response against extracellular pathogens through secretion of interleukin-4 (IL-4) and other cytokines. Th17, a recently discovered CD4+ T-cell subtype, produces proinflammatory cytokines such as IL-17A, IL-17F, and IL-22, and mediates the immune response against extracellular bacteria. In some cases, particular T-cell subsets have also been shown to exacerbate disease conditions, e.g. Th17 cells contribute to autoimmune diseases such as multiple sclerosis. Another unique T-cell subset that has attracted tremendous interest in recent years is the regulatory T (Treg) cells, which are responsible for suppressing the immune response induced by effector T cells. Treg cells either develop directly in the thymus or differentiate from CD4+ naive T cells in the periphery in response to specific environmental cues (inducible Treg cells). Finally, Th9, follicular helper T (Tfh) cells, and Th22 represent the most recently discovered CD4+ subsets, which are actively being characterized at present.
Development of the different T-cell subsets is dependent on several known transcription factors: T-bet for Th1, GATA3 for Th2, forkhead box protein 3 (FoxP3) for Treg, and RORγt and RORα for Th17 cells. However, it is also becoming apparent that combinatorial activation of distinct signaling pathways and the concomitant mobilization of multiple transcription factors contributes to lineage determination. As previously mentioned, transcription factors of the NF-κB family are also involved in various aspects of T-cell development and function, and recent work has helped further define the unique functions of this pathway in the differentiation and functional divergence of the various T-cell subsets (5). In this review, we provide a comprehensive discussion of the current literature on the role and regulation of NF-κB in different T-cell subsets and speculate on the impact of NF-κB regulation in the overall immune response.
Differentiation of Th1 and T2 cells
CD4+ T cells produced in the thymus migrate to the periphery where they encounter antigens. Upon stimulation, naive CD4+ T cells differentiate into distinct subsets of effector cells, which allow for tailored immune responses against specific antigens. Th1 and Th2 cells are two major CD4+ effector T-cell subsets. As discussed earlier, the major role of Th1 cells is in immune response against intracellular viral or bacterial pathogens, predominantly through the production of IFN-γ. Th2 cells are involved in the response to extracellular pathogens, but they have also been implicated in allergic reactions. Th2 cells are characterized by their production of IL-4, as well as IL-5, IL-6, IL-9, IL-13, and IL-25.
Th1 cell differentiation is initiated by TCR engagement with a peptide-MHC class II complex presented by DCs or macrophages in response to infection with an intracellular pathogen. This TCR stimulation induces low-level expression of the transcription factors T-bet and GATA3. T-bet increases the surface expression of the IL-12 receptor, thus enhancing the sensitivity of cells to IL-12 and promoting further T-cell differentiation through IL-12 and IFN-γ produced by APCs. IL-12 stimulation of T cells activates the transcription factor signal transducer and activator of transcription 4 (STAT4). In addition, receptor engagement of IFN-γ induces activation of STAT1, which subsequently further increases the level of T-bet. Finally, cooperation of T-bet, STAT4 and STAT1 induces high level of IFN-γ production by the T cell (Th1). In addition to the previously described network of transcription factors, several studies have described a role of NF-κB in Th1 differentiation. In transgenic mice expressing a non-degradable form of IκB specifically in T cells, Th1 responses were significantly impaired due to diminished NF-κB activation (6). Also, c-Rel-deficient mice showed defective Th1-mediated immune responses and abrogated IFN-γ production by T cells (7). However, T-bet expression in those T cells was intact, implying that c-Rel might be downstream of T-bet and consequently be important for T-bet-induced Th1 differentiation. On the other hand, RelB-deficient T cells exhibited a defect in Th1 differentiation, but this was due to decreased expression of T-bet (8). RelB is thought to mediate STAT4 upregulation, which in turn, is responsible for IFN-γ induction of T-bet.
RelA (p65) is recruited to highly conserved non-coding sequences (CNSs) in the enhancer region in the IFN-γ locus and in the absence of RelA, IFN-γ production in Th1 cells is greatly impaired (9). Recruitment of RelA to the enhancer region is dependent on T-bet expression in Th1 cells. STAT4 is co-recruited to the CNS sites suggesting that RelA and STAT4 may cooperate to remodel the IFN-γ locus and induce the expression of IFN-γ. While these findings establish a critical role for RelA in Th1 differentiation and function, it still remains unknown whether RelA binds to the CNS regions as a heterodimer with p50 or c-Rel, or as a homodimer. According to chromatin immunoprecipitation (ChIP) analysis, p50 binding seems to be limited to only one CNS site suggesting that the other site might bind to a p65:c-Rel complex. Interestingly, p50-deficient CD4+ T cells are able to express T-bet and produce IFN-γ in Th1 differentiation, implying that RelA might work either as a homodimer or a heterodimer with c-Rel in the IFN-γ locus (10). Taken together, these studies establish an essential role for NF-κB members in the development and function of Th1 cells, especially in the production of IFNγ.
Th2 development is initiated by IL-4 stimulation, which leads to activation of the transcription factor STAT6. Furthermore, IL-4 signaling enhances the expression of the IL-4 gene via activation of NFAT, AP-1 and NF-κB, and induces expression of GATA3, a master regulator of Th2 differentiation. GATA3 governs the expression of another transcription factor, c-Maf, which also helps the induction of IL-4. However, overexpression of c-Maf alone in naive CD4+ T cells did not induce Th2 differentiation in Th1-skewing conditions (11) unlike the situation seen upon overexpression of GATA3 (12). Therefore, STAT6, GATA3, and c-Maf work cooperatively to induce a Th2 program and stabilize the features of Th2 cells. Importantly, NF-κB has been reported to be essential for the expression of the GATA3 gene (10). CD4+ T cells from p50-deficient mice were unable to express GATA3 under Th2-differentiating conditions in vivo and in vitro. Consequently, impaired Th2 development in p50-deficient mice led to a defective allergic airway inflammatory response. These findings demonstrate a crucial role for p50 in Th2 differentiation through the transcriptional regulation of GATA3 expression.
Expression of the IL-4 gene, which is crucial for Th2 differentiation, is known to be regulated by two mechanisms. First, GATA3-induced chromatin remodeling opens up the gene locus of IL-4 in the process of Th2 differentiation. Second, IL-4 is induced by TCR engagement in mature Th2 cells. NF-κB has been shown to bind two major enhancer sites in the IL-4 locus and it participates in induction of IL-4 in cooperation with nuclear factor of activated T cells (NFAT) (13). Since NF-κB is also involved in induction of GATA-3, it remains unclear whether the role of NF-κB in Th2 differentiation is a direct result of TCR-induced NF-κB activating IL-4 gene expression or if it acts primarily through its effect on GATA3 expression.
Development of natural regulatory T (nTreg) cells in the thymus
The immune system is a carefully balanced system with tight regulation of immune cell function. When the balance is disrupted, it can lead to disease. Since the original discovery of CD4+CD25+ Treg cells by Sakaguchi and colleagues (14), there has been much progress in understanding the role of Treg cells in immune homeostasis and function. The discovery of the Treg-specific transcription factor FoxP3 has accelerated this progress. FoxP3+ Treg cells are essential for suppressing excessive immune response against self or non-self-antigens. Human immunodysregulation, polyendocrinopathy, and enteropathy X-linked (IPEX) syndrome patients have mutations in the FoxP3 locus, resulting in a lack of functional FoxP3+ Treg cells. The symptoms of IPEX syndrome resemble those of Scurfy mice, which have a loss-of-function mutation in the FoxP3 gene. Scurfy mice have systemic lymphoproliferative disease in multiple organs and die within 2–3 weeks (15). IPEX patients suffer from various symptoms such as autoimmune endocrinopathies, eczema, diabetes, and severe enlargement of the secondary lymphoid organs (16).
In addition to playing a protective role against autoimmunity, Treg cells help maintain immune homeostasis in various settings including in response to commensal microbiota or food allergens at mucosal surfaces. Treg cells also suppress experimental graft-versus-host disease (GvHD) in mice (17), leading to the exploration of Treg modulation as a potential therapeutic targeting strategy. On the other hand, altered Treg function in some cancers facilitates immune evasion, thereby promoting the exploration of alternative approaches towards cancer immunotherapies (18). A recent publication from Rudensky and colleagues (19) reported a role of Treg cells in maternal-fetal immune tolerance and shed light on the contribution of Treg cells in situations such as pre-eclampsia during pregnancy. Since Treg cells are important in many immune disorders, it has been a major interest to study how Treg cells differentiate and function.
Treg cells are classified into two major classes based on their developmental origin. Treg cells can be generated in the thymus through the natural selection process and these cells are referred to as natural Treg (nTreg) or thymic Treg (tTreg) cells. Alternatively, conventional naive T cells can differentiate into FoxP3+ Treg cells in the periphery upon exposure to non-self-antigens such as commensal microbiota, food, or allergens. These Treg cells generated in the periphery are generally referred to as inducible Treg (iTreg) cells. However in this review, we primarily focus on the thymic development of nTreg cells.
T-cell progenitors are educated by exposure to various self-antigens presented by APCs in the thymus during positive selection. Only T cells that successfully detect MHCs bound to moderate-affinity peptides will receive a survival signal. However, T cells carrying high-affinity T-cell receptors (TCRs) against self-antigens are eliminated by apoptosis through negative selection. T cells escaping negative selection are released from the thymus and migrate to the periphery. However, a small number of self-reactive T cells with intermediate affinity for self-antigens can escape the elimination process during negative selection and differentiate into FoxP3+ Treg cells.
The mechanism of how Treg cells are selected in the thymus has been extensively studied, but several questions still remain to be answered. A two-step model has been suggested for thymic Treg development with the first step being TCR-dependent, while the second step is TCR-independent (20). TCR signaling in thymocytes triggered by the TCR-MHC class II interaction induces downstream signaling pathways that elicit NF-κB, NF-AT, and mammalian target of rapamycin (mTOR)/AKT activity. It has been suggested that TCR signaling in Treg development is different from the TCR signaling that occurs during the development of Tconv cells. Deficiency of several mediators of TCR signaling such as TAK1, BCL10, CARMA1, PKCθ, and IKKβ significantly reduced or abrogated the number of Treg cells generated in the thymus without affecting the generation of Tconv cells (21–25). These molecules are known to be important for signal transduction upstream of NF-κB activation, which suggests a possible role for NF-κB in thymic Treg development. Interestingly, thymic Treg generation in TAK1- or CARMA1-deficient mice can be rescued by crossing these strains to transgenic mice expressing a constitutively active mutant form of IKKβ under the control of a proximal Lck promoter (IKKEE-Tg), demonstrating that the defective Treg development is due to impaired NF-κB activation in these knockout mice (26). Also, overexpression of a constitutively active form of IKKβ generated thymic FoxP3+ Treg cells in RAG-deficient TCR-transgenic mice (OT-II and P14-TCR), which normally do not produce nTreg cells in the thymus. This demonstrates that forced activation of NF-κB can bypass the requirement of self-antigen recognition via TCR engagement. Together, these findings strongly suggest that that NF-κB activation is critical for thymic Treg development.
The mechanism of how the FoxP3 locus becomes more accessible to the transcription machinery during thymic selection has been a question of major interest. Enhancer regions in FoxP3 locus have been suggested to have an important role in inducing FoxP3 transcription. Three highly conserved non-coding sequences have been identified in the FoxP3 locus (32, 33): CNS1, CNS2, and CNS3. CNS1 contains a TGF-β-responsive element and binding sites for AP1, NFAT, and SMAD3, and it is known to be important for iTreg differentiation but not for thymic development of nTreg cells (34). CNS2 has binding sites for several transcription factors including CREB, CBFβ, and STAT5 and is necessary for maintenance of FoxP3 expression (34). The NF-κB subunit c-Rel binds to CNS3 and plays a major role in thymic and peripheral FoxP3 expression and Treg differentiation (26, 34). The CNS2 and CNS3 enhancer sequences have been reported to contain highly conserved CpG-rich regions which are fully demethylated in nTreg cells but highly methylated in Tconv cells as well as iTreg cells (32, 34). Demethylation in these CNS sequences is believed to be necessary for stable expression of FoxP3. It has been proposed that c-Rel binds to the CNS3 region as a homodimer, but it is not yet clear if c-Rel synergizes with other transcription factors at the FoxP3 locus. It will be exciting to further explore the interplay between transcription factor activation and the modification of chromosome structure that leads to FoxP3 expression and Treg differentiation.
NF-κB in Treg suppression of the immune response
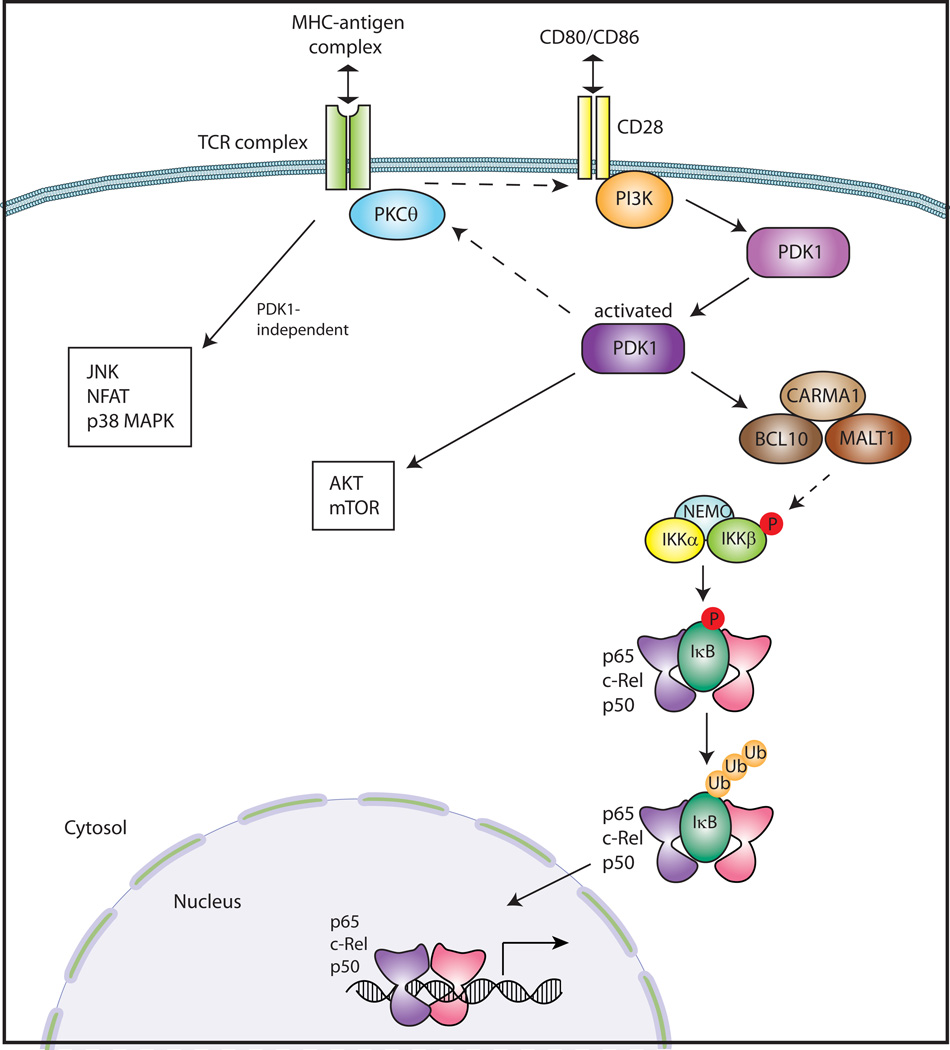
TCR stimulation is required for thymic Treg development and is also necessary for Treg function, but the signaling pathways governing the function of Treg cells are quite complex. PDK1 is an important protein kinase that integrates TCR stimulation and CD28 costimulation and induces activation of PKCθ, followed by the formation of CARMA1/MALT1/BCL10 complex, and subsequently NF-κB activation (47) (Fig. 4). PDK1 was conditionally ablated in CD4+ T cells using the Cre-Lox system in mice to study the role of PDK1 in CD4+ T cells. Interestingly, these mice spontaneously developed colitis, even though activation of effector CD4+ T cells was defective due to the absence of PDK1. It was shown that colitis was due to dramatically increased numbers of γδT cells in the colon, and crossing the PDK1-deficient mice with a TCRδ-knock-out mouse completely abolished the colitis (48). In line with these findings, PDK1-deficient Treg cells did not have suppressive activity both in vitro and in vivo in a transfer colitis model. As wildtype Treg cells are able to inhibit proliferation of γδT cells, the increased number of γδT cells in the PDK1-deficient mice was due to the lack of functional Treg cells. These studies suggested that PDK1 likely has an important role in Treg function. As the lack of PDK1 prevents NF- κB activation, it is possible that NF-κB might be important in the suppressive function of Treg cells. However, because PDK1 activation can induce other downstream signaling pathways such as mTOR/Akt (Fig. 4), it is possible that these other pathways are important in Treg function.
Fig. 4. TCR/CD28 signaling pathway.
A TCR complex on the surface of T cells recognizes a specific antigen-MHC complex on an antigen-presenting cell (APC). At the same time, a costimulatory receptor CD28 interacts with CD80 or CD86 on the APC. Upon TCR/CD28 stimulation, PI3K activates PDK1 leading to subsequent activation of PKCθ. This results in the formation of the complex consisting of CARMA1, BCL10, and MALT1, that activates the IKK complex. Phosphorylation of IκB by the IKK complex leads to its degradation, resulting in the liberation of NF-κB dimers. NF-κB dimers translocate into the nucleus and regulate the expression of target genes. PDK1 activates the Akt/mTOR pathway as well as NF-κB. TCR stimulation also leads to activation of JNK, NFAT, and p38 MAPK in a PDK-independent manner.
A recent paper from Sun and colleagues (49) showed that the Ubc13-IKK axis has a positive role in Treg suppressive function. Ubc13 is an E2 ubiquitin-conjugating enzyme that specifically conjugates Lys63-linked polyubiquitin chains, which are known to have a role in activation of IKK and NF-κB (50). Conditional ablation of Ubc13 in FoxP3+ Treg cells led to multi-organ inflammation due to uncontrolled activation of Tconv cells (49). Ubc13-deficient Treg cells could not suppress inflammatory colitis induced by adoptive transfer of naïve Tconv cells into lymphopenic recipients, demonstrating that Ubc13 depletion abrogates the suppressive function of Treg cells in vivo. When Ubc13 Treg-knockout mice were crossed to transgenic mice expressing a constitutively active form of IKKβ under the control of a loxP-flanked stop cassette, the suppressive function of Ubc13-deficient Treg cells was rescued, as indicated by a decrease in the frequency of memory and effector-like T cells in lymphoid organs (49). These results, coupled with the findings on PDK1-deficient mice (48), suggest that NF-κB might be a key regulator determining Treg function.
In contrast to the previous findings, Dustin and colleagues (51) reported that PKCθ, a downstream target of PDK1, has a negative role in Treg function. This is a surprising finding as PKCθ is activated upon TCR engagement and is believed to be critical for activation of NF-κB, NFAT, and mTOR/Akt pathways. Consistent with the fact that TCR signaling and NF-κB activation are required for nTreg differentiation, PKCθ-deficient mice showed reduced numbers of Treg cells. However, surprisingly, inhibition of PKCθ using a chemical inhibitor increased the suppressive capacity of Treg cells suggesting that PKCθ exerts a negative role in the suppressive function of Treg cells (22, 24). A possible explanation might be the different experimental approaches used in the two studies. Whereas PKCθ was blocked transiently using a chemical inhibitor, PDK1 was removed permanently using genetic ablation. However, further studies will be required to resolve the role of PKCθ in Treg cells.
NF-κB in the development of Th17 Cells
As mentioned previously, Th17 cells are a CD4+ T-cell subset that produces proinflammatory cytokines such as IL-17A, IL-17F, and IL-22, and mediates the immune response against extracellular bacteria. Recently, a possible role of NF-κB in the regulation of RORγt gene expression in the differentiation and function of Th17 cells was demonstrated (52). Earlier studies showed that c-Rel-deficient mice were resistant to experimental autoimmune encephalomyelitis (EAE), a mouse model for multiple sclerosis, which is mediated by T-helper cell subsets including Th17 cells (7). This finding suggested a possible role of c-Rel in the inflammatory Th17 response, but as c-Rel is involved in the development and function of various immune cells, the possible role of other cell types cannot be discounted. More recently, c-Rel- or p65-deficient T cells showed defective IL-17 gene expression and Th17 cell differentiation (52). In addition, gene expression of RORγt and RORγ, an isoform of RORγt, was significantly reduced in c-Rel-deficient T cells. RORγt and RORγ are encoded by a single gene, Rorg and share the last 9 exons. However, their first 100 nucleotides are different, which leads to different N-terminal sequences after translation. RORγt and RORγ share the same DNA-binding domain and ligand-binding domain. Like RORγt, RORγ was shown to bind to the Il17a promoter region, suggesting that RORγ might also be involved in Th17 differentiation. Gene expression of RORγt and RORγ are controlled by two different promoters, both of which were found to contain NF-κB-binding sites. A series of deletion and mutation experiments showed that c-Rel binds to these sites and that this binding was required for the expression of RORγt and RORγ. This was further confirmed by chromatin immunoprecipitation analysis in Th17 cells. p65 also co-localized with c-Rel at the same binding sites. These results indicate that c-Rel, possibly together with p65, activates the gene expression of RORγt and RORγ. However, evidence for c-Rel directly controlling IL-17a gene expression by binding to its promoter has not yet been provided. It has been hypothesized that the c-Rel:p65 dimers move into the nucleus and bind to the promoters of RORγt and RORγ as a complex with other transcription factors such as NFAT and STAT. A similar hypothesis has been put forward regarding the control of the FoxP3 promoter in Treg cells, where each transcription factor participates in various signaling pathways but together can have a unique role in a distinct pathway as a complex with multiple transcription factors, called an ‘enhanceosome’ (53).
In contrast to the previously mentioned findings, Visekruna et al. (54) reported that c-Rel is not required for Th17 cell differentiation, a discrepancy that has been ascribed to the absence of anti-IL-2 blocking antibody in their Th17 in vitro differentiation condition. NF-κB is known to be required for IL-2 production, so c-Rel-deficient T cells may produce less IL-2 than wildtype T cells. Since IL-2 is inhibitory for Th17 cell induction, the presence of IL-2 in the wildtype T cell cultures may have masked the difference between wildtype and c-Rel-deficient T cells in the in vitro Th17 differentiation assay in the study by Visekruna et al. (54).
Powolny-Budnicka et al. (55) reported a role of RelA (p65) and RelB in lymphotoxin (LT)-dependent IL-17 production in γδT cells. While they focused on γδT cells in this study, they also examined Th17 cells. They showed that differentiation of IL-17-producing γδT cells in the thymus is dependent on the LTβ receptor (LTβR), RelA, and RelB. RelA controls expression of LT ligands in accessory thymocytes to regulate IL-17 production in γδT cells. On the other hand, RelB may act downstream of LTβR and increase the mRNA level of RORγt, resulting in the differentiation of IL-17-producing γδT cells. However, they claimed that differentiation of Th17 cells is not dependent on either LTβR or activation of RelA or RelB. CD4+ T cells deficient in RelA or RelB did not show increased mRNA expression of RORγt in their in vitro Th17 differentiation assay. Chen and colleagues (52), on the other hand, showed that while the noncanonical NF-κB pathway including RelB is not involved in RORγt expression in Th17 differentiation, c-Rel and p65 (RelA) directly bind to the RORγt promoter region and upregulate its mRNA expression. Discrepancies between these two group’s studies could be explained by the experimental conditions of the in vitro Th17 differentiation assay. The in vitro differentiation cultures used in Powolny-Budnicka et al.’s study (55) were performed in the absence of anti-IL-2. As p65 (RelA) is known to be important in IL-2 production (56), the p65-deficient T cells may produce less IL-2 than wildtype T cells. Differences in Th17 differentiation might have been masked by the effect of inhibitory IL-2 in the culture of the wildtype cells. Ultimately, the role of the different NF-κB subunits in differentiation of Th17 and γδT cells is still not clear and needs to be further studied. Furthermore, the interplay between NF-κB and other transcription factors in controlling the differential expression of RORγt in T-cell subsets remains an area of great interest.
NF-κB in Th9 development
Th9 is a subset of T-helper cells producing IL-9, but their development and function remain poorly understood. A recent report suggested a mechanism of Th9 induction through OX40 signaling (57). OX-40 is a T-cell costimulatory molecule that belongs to the TNF-receptor family. Ligation of OX40 strongly induced Th9 differentiation in the presence of IL-4 and TGF-β and activated both canonical and noncanonical NF-κB pathways. However, Th9 induction was not affected when the canonical pathway was repressed by using transgenic mice overexpressing a nondegradable form of IκB or a chemical inhibitor of IKK activity (BAY 11–7082). On the other hand, p52-deficient T cells could not differentiate into Th9 cells. Also, when p52-RelB was overexpressed in wildtype CD4+ T cells by retrovirus-mediated gene transfer, cells were able to convert into Th9 cells even without OX40 ligation in the presence of IL-4 and TGF-β. OX40 signaling activates the ubiquitin ligase TRAF6, which activates NF-κB-inducing kinase (2). This results in the processing of p100 to generate p52, which then forms a heterodimer with RelB. These results suggest that the noncanonical NF-κB pathway via the RelB:p52 heterodimer might be important in Th9 differentiation. Finally, it has been suggested that IL-9 expression in T cells is regulated by NF-κB, thereby implicating the pathway in the function of these cells (58). While many questions concerning the Th9 subset of CD4+ T cells remain unanswered, these findings represent the first steps towards a better understanding of the development and function of Th9 cells.
Follicular helper T cells (Tfh)
In the germinal center (GC), B cells exposed to antigens differentiate and mature to produce antibodies against specific antigens. Another T-cell subset called follicular helper T cells (Tfh) is critical for the maturation of germinal center B cells. Tfh cells express the chemokine receptor CXCR5, which enables Tfh cells to migrate to B-cell follicles. Multiple signals from Tfh cells are needed to enable full differentiation of B cells. IL-21 produced by Tfh cells and CD40L, and programmed death-1 (PD1) expressed on the surface of Tfh cells, are important for B-cell differentiation. Also, Tfh cells express high level of ICOS, and the resulting costimulatory signal through the interaction between T-cell ICOS and B-cell ICOSL is critical for B-cell maturation (59).
For Tfh development, T cells first need to be initially activated by DCs in the T-cell zone of the follicle. Then, these Tfh cell precursors interact with B cells at the border of T-B cell zones. In addition to TCR/CD28 signals, ICOS costimulation through B cells has been shown to be required for Tfh development (60–63). Furthermore, defective Tfh development was reported to occur in NIK-deficient mice (64), but it was not due to a T-cell intrinsic deficiency. NIK was required for maintaining the high-level expression of ICOSL in B cells, which is essential for Tfh development. Noncanonical NF-κB members RelB and p52 have been shown to bind to a κB-binding site in the ICOSL promoter region and shRNA-mediated knock-down of RelB in B cells impaired ICOSL induction in response to BAFF stimulation (64). These results suggest that the noncanonical NF-κB pathway might play an important role in Tfh development by regulating the expression of ICOSL in B cells.
In another report using NF-κB1-deficient OT-II mice, Tfh development was seen to be defective upon immunization with alum-precipitated OVA peptides (65). NF-κB1 deficiency particularly impaired CXCR5 expression. In addition, there were lower numbers of Tfh cells in these mice, which resulted in development of fewer germinal center B cells in response to OVA immunization. These findings suggest a role of NF-κB in Tfh development.
In addition to its role in stimulating B cells to differentiate, IL-21 has been shown to be important in the development of Tfh cells, and differentiated Tfh cells are dependent on IL-21 for their growth and survival (66, 67). IL-21 production was impaired in c-Rel-deficient mice, and the development of Tfh and germinal-center B cells was consequently inhibited in those mice (68). c-Rel was shown to bind to the IL-21 promoter, implying that c-Rel directly regulates the transcription of IL-21. Taken together, these findings highlight the role of NF-κB family members in regulating various steps in the development and function of Tfh cells.
Perspectives
This review discusses the role of NF-κB in different CD4+ T cell subsets, including Th1, Th2, Treg, Th17, Th9, and Tfh cells. NF-κB family members are critical transcription factors that regulate a plethora of genes in different biological processes. While the molecular details of NF-κB activation downstream of TCR/CD28 signaling and the role of the NF-κB pathway following TCR activation have been extensively studied, the differential functions of NF-κB during development and activation of the specific T-cell subset is a newer field of study. The utilization of the different NF-κB subunits and the synergy of the NF-κB pathway with other signaling pathways contribute to the complex transcriptional programs that allow for the differentiation and functional divergence of precursor cells into the various T-cell subsets. A major question that stems from the studies discussed in this review is how a common stimulus can direct different gene expression profiles downstream of NF-κB in response to different environmental conditions, thereby creating the unique effector profiles of the different T-cell subsets. Recently, a number of studies have suggested that a coordinated network of multiple transcription factors could regulate the expression of specific sets of genes in response to specific stimulation and environmental conditions. By studying the interplay between signaling pathways and the resultant networks formed by NF-κB and other transcription factors in response to various stimuli, we hope to better understand the tight regulatory mechanisms involved in the development and function of immune cells. Consequently, we may be able to better anticipate conditions that could contribute to dysregulation of immune cell function and immune responses, and perhaps use our understanding of lineage decisions in the design of therapeutics that specifically target the dysregulated cell types or cell functions.
Acknowledgements
The work carried out in the author’s laboratory is supported by grants from NIH (R37-33443, RO1-68977) and institutional support from Columbia University. We also thank Dr. Sujatha Gurunathan for her help in editing the manuscript.
Footnotes
The authors have no conflicts of interest to declare.
Literature Cited
- 1.Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell Res. 2011;21:223–244. doi: 10.1038/cr.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Dustin ML. The cellular context of T cell signaling. Immunity. 2009;30:482–492. doi: 10.1016/j.immuni.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harbor Persp Biol. 2010;2 doi: 10.1101/cshperspect.a000182. a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 6.Aronica MA, et al. Preferential role for NF-kappa B/Rel signaling in the type 1 but not type 2 T cell-dependent immune response in vivo. J Immunol. 1999;163:5116–5124. [PubMed] [Google Scholar]
- 7.Hilliard BA, et al. Critical roles of c-Rel in autoimmune inflammation and helper T cell differentiation. J Clin Invest. 2002;110:843–850. doi: 10.1172/JCI15254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corn RA, Hunter C, Liou HC, Siebenlist U, Boothby MR. Opposing roles for RelB and Bcl-3 in regulation of T-box expressed in T cells, GATA-3, and Th effector differentiation. J Immunol. 2005;175:2102–2110. doi: 10.4049/jimmunol.175.4.2102. [DOI] [PubMed] [Google Scholar]
- 9.Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, Weaver CT. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity. 2010;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das J, Chen CH, Yang L, Cohn L, Ray P, Ray A. A critical role for NF-kappa B in GATA3 expression and Th2 differentiation in allergic airway inflammation. Nat Immunol. 2001;2:45–50. doi: 10.1038/83158. [DOI] [PubMed] [Google Scholar]
- 11.Ho IC, Lo D, Glimcher LH. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4-dependent and -independent mechanisms. The J Exp Med. 1998;188:1859–1866. doi: 10.1084/jem.188.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 13.Li-Weber M, Giaisi M, Baumann S, Palfi K, Krammer PH. NF-kappa B synergizes with NF-AT and NF-IL6 in activation of the IL-4 gene in T cells. Eur J Immunol. 2004;34:1111–1118. doi: 10.1002/eji.200324687. [DOI] [PubMed] [Google Scholar]
- 14.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25) Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 15.Appleby MW, Ramsdell F. Scurfy, the Foxp3 locus, and the molecular basis of peripheral tolerance. Curr Topics Microbiol Immunol. 2008;321:151–168. doi: 10.1007/978-3-540-75203-5_7. [DOI] [PubMed] [Google Scholar]
- 16.Wildin RS, Smyk-Pearson S, Filipovich AH. Clinical and molecular features of the immunodysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J Med Genet. 2002;39:537–545. doi: 10.1136/jmg.39.8.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Ianni M, et al. Tregs prevent GVHD and promote immune reconstitution in HLA-haploidentical transplantation. Blood. 2011;117:3921–3928. doi: 10.1182/blood-2010-10-311894. [DOI] [PubMed] [Google Scholar]
- 18.Curiel TJ. Tregs and rethinking cancer immunotherapy. J Clin Invest. 2007;117:1167–1174. doi: 10.1172/JCI31202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samstein RM, Josefowicz SZ, Arvey A, Treuting PM, Rudensky AY. Extrathymic generation of regulatory T cells in placental mammals mitigates maternal-fetal conflict. Cell. 2012;150:29–38. doi: 10.1016/j.cell.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh CS, Lee HM, Lio CW. Selection of regulatory T cells in the thymus. Nat Rev Immunol. 2012;12:157–167. doi: 10.1038/nri3155. [DOI] [PubMed] [Google Scholar]
- 21.Medoff BD, et al. Differential requirement for CARMA1 in agonist-selected T-cell development. Eur J Immunol. 2009;39:78–84. doi: 10.1002/eji.200838734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta S, Manicassamy S, Vasu C, Kumar A, Shang W, Sun Z. Differential requirement of PKC-theta in the development and function of natural regulatory T cells. Mol Immunol. 2008;46:213–224. doi: 10.1016/j.molimm.2008.08.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt-Supprian M, et al. Mature T cells depend on signaling through the IKK complex. Immunity. 2003;19:377–389. doi: 10.1016/s1074-7613(03)00237-1. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt-Supprian M, et al. Differential dependence of CD4+CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proc Natl Acad Sci USA. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan YY, Chi H, Xie M, Schneider MD, Flavell RA. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat Immunol. 2006;7:851–858. doi: 10.1038/ni1355. [DOI] [PubMed] [Google Scholar]
- 26.Long M, Park SG, Strickland I, Hayden MS, Ghosh S. Nuclear factor-kappaB modulates regulatory T cell development by directly regulating expression of Foxp3 transcription factor. Immunity. 2009;31:921–931. doi: 10.1016/j.immuni.2009.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Nishizuka Y, Sakakura T. Thymus and reproduction: sex-linked dysgenesia of the gonad after neonatal thymectomy in mice. Science. 1969;166:753–755. doi: 10.1126/science.166.3906.753. [DOI] [PubMed] [Google Scholar]
- 28.Asano M, Toda M, Sakaguchi N, Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184:387–396. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fontenot JD, Dooley JL, Farr AG, Rudensky AY. Developmental regulation of Foxp3 expression during ontogeny. J Exp Med. 2005;202:901–906. doi: 10.1084/jem.20050784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pacholczyk R, Kern J, Singh N, Iwashima M, Kraj P, Ignatowicz L. Nonself-antigens are the cognate specificities of Foxp3+ regulatory T cells. Immunity. 2007;27:493–504. doi: 10.1016/j.immuni.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat Immunol. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- 36.Wing K, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- 37.Qureshi OS, et al. Trans-endocytosis of CD80 and CD86: a molecular basis for the cell-extrinsic function of CTLA-4. Science. 2011;332:600–603. doi: 10.1126/science.1202947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fallarino F, et al. Modulation of tryptophan catabolism by regulatory T cells. Nat Immunol. 2003;4:1206–1212. doi: 10.1038/ni1003. [DOI] [PubMed] [Google Scholar]
- 39.Grohmann U, et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat Immunol. 2002;3:1097–1101. doi: 10.1038/ni846. [DOI] [PubMed] [Google Scholar]
- 40.Kobie JJ, Shah PR, Yang L, Rebhahn JA, Fowell DJ, Mosmann TR. T regulatory and primed uncommitted CD4 T cells express CD73, which suppresses effector CD4 T cells by converting 5'-adenosine monophosphate to adenosine. J Immunol. 2006;177:6780–6786. doi: 10.4049/jimmunol.177.10.6780. [DOI] [PubMed] [Google Scholar]
- 41.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borsellino G, et al. Expression of ectonucleotidase CD39 by Foxp3+ Treg cells: hydrolysis of extracellular ATP and immune suppression. Blood. 2007;110:1225–1232. doi: 10.1182/blood-2006-12-064527. [DOI] [PubMed] [Google Scholar]
- 43.Boissonnas A, et al. Foxp3+ T cells induce perforin-dependent dendritic cell death in tumor-draining lymph nodes. Immunity. 2010;32:266–278. doi: 10.1016/j.immuni.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Cao X, et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27:635–646. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Beeston T, Smith TR, Maricic I, Tang X, Kumar V. Involvement of IFN-gamma and perforin, but not Fas/FasL interactions in regulatory T cell-mediated suppression of experimental autoimmune encephalomyelitis. J Neuroimmunol. 2010;229:91–97. doi: 10.1016/j.jneuroim.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gondek DC, et al. Transplantation survival is maintained by granzyme B+ regulatory cells and adaptive regulatory T cells. J Immunol. 2008;181:4752–4760. doi: 10.4049/jimmunol.181.7.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park SG, et al. The kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-kappaB and activate T cells. Nat Immunol. 2009;10:158–166. doi: 10.1038/ni.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park SG, et al. T regulatory cells maintain intestinal homeostasis by suppressing gammadelta T cells. Immunity. 2010;33:791–803. doi: 10.1016/j.immuni.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang JH, et al. Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nat Immunol. 2012;13:481–490. doi: 10.1038/ni.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- 51.Zanin-Zhorov A, et al. Protein kinase C-theta mediates negative feedback on regulatory T cell function. Science. 2010;328:372–376. doi: 10.1126/science.1186068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruan Q, et al. The Th17 immune response is controlled by the Rel-RORgamma-RORgamma T transcriptional axis. J Exp Med. 2011;208:2321–2333. doi: 10.1084/jem.20110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ruan Q, et al. Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Visekruna A, et al. c-Rel is crucial for the induction of Foxp3(+) regulatory CD4(+) T cells but not T(H)17 cells. Eur J Immunol. 2010;40:671–676. doi: 10.1002/eji.200940260. [DOI] [PubMed] [Google Scholar]
- 55.Powolny-Budnicka I, Riemann M, Tanzer S, Schmid RM, Hehlgans T, Weih F. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in gammadelta T cells. Immunity. 2011;34:364–374. doi: 10.1016/j.immuni.2011.02.019. [DOI] [PubMed] [Google Scholar]
- 56.Lai JH, Horvath G, Subleski J, Bruder J, Ghosh P, Tan TH. RelA is a potent transcriptional activator of the CD28 response element within the interleukin 2 promoter. Mol Cell Biol. 1995;15:4260–4271. doi: 10.1128/mcb.15.8.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao X, et al. OX40 signaling favors the induction of T(H)9 cells and airway inflammation. Nat Immunol. 2012;13:981–990. doi: 10.1038/ni.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Early SB, Huyett P, Brown-Steinke K, Borish L, Steinke JW. Functional analysis of - 351 interleukin-9 promoter polymorphism reveals an activator controlled by NF-kappaB. Genes Immun. 2009;10:341–349. doi: 10.1038/gene.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crotty S. Follicular helper CD4 T cells (Tfh) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 60.Akiba H, et al. The role of ICOS in the CXCR5+ follicular B helper T cell maintenance in vivo. J Immunol. 2005;175:2340–2348. doi: 10.4049/jimmunol.175.4.2340. [DOI] [PubMed] [Google Scholar]
- 61.Bossaller L, et al. ICOS deficiency is associated with a severe reduction of CXCR5+CD4 germinal center Th cells. J Immunol. 2006;177:4927–4932. doi: 10.4049/jimmunol.177.7.4927. [DOI] [PubMed] [Google Scholar]
- 62.Nurieva RI, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu YL, Metz DP, Chung J, Siu G, Zhang M. B7RP-1 blockade ameliorates autoimmunity through regulation of follicular helper T cells. J Immunol. 2009;182:1421–1428. doi: 10.4049/jimmunol.182.3.1421. [DOI] [PubMed] [Google Scholar]
- 64.Hu H, Wu X, Jin W, Chang M, Cheng X, Sun SC. Noncanonical NF-kappaB regulates inducible costimulator (ICOS) ligand expression and T follicular helper cell development. Proc Natl Acad Sci USA. 2011;108:12827–12832. doi: 10.1073/pnas.1105774108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Serre K, et al. Selective effects of NF-kappaB1 deficiency in CD4(+) T cells on Th2 and TFh induction by alum-precipitated protein vaccines. Eur J Immunol. 2011;41:1573–1582. doi: 10.1002/eji.201041126. [DOI] [PubMed] [Google Scholar]
- 66.Ma CS, Deenick EK, Batten M, Tangye SG. The origins, function, and regulation of T follicular helper cells. J Exp Med. 2012;209:1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 68.Chen G, Hardy K, Bunting K, Daley S, Ma L, Shannon MF. Regulation of the IL-21 gene by the NF-kappaB transcription factor c-Rel. J Immunol. 2010;185:2350–2359. doi: 10.4049/jimmunol.1000317. [DOI] [PubMed] [Google Scholar]