Abstract
Aims
Atrial fibrillation (AF) is a frequent co-morbidity in heart failure (HF) associated with increased mortality, but little is known about the mechanisms underlying AF onset in HF patients. We evaluated the association of cardiovascular and genetic risk factors with AF in HF patients.
Methods and results
Individuals hospitalized for HF (n = 1040; 500 with AF) were identified from a large, population-based cohort study (n = 30 447; 2339 with AF). Genetic polymorphisms in the chromosomal regions 4q25 (rs2200733) and 16q22 (rs2106261) associated with AF in genome-wide association studies were genotyped. Association of cardiovascular risk factors and polymorphisms with AF was tested in HF patients and the entire cohort using both prospective and non-time-dependent models. Cardiovascular risk factors—hypertension, body mass index, sex, smoking, diabetes, and myocardial infarction—were associated with AF in the entire cohort but not in HF patients. In contrast, polymorphisms on chromosomes 16q22 and 4q25 were associated with AF both in the entire cohort and in HF patients, conferring 75% [95% confidence interval (CI) 35–126, P = 2 × 10−5] and 57% (95% CI 18–109, P = 0.002) increased risk of AF per copy in HF patients, respectively. In the entire cohort, AF risk in the presence of HF was multiplicatively magnified by genotype for 16q22 (P for interaction = 7 × 10−4) but not 4q25 (P = 0.83). In prospective analyses excluding patients with AF diagnosis prior to or simultaneously with HF diagnosis, 16q22 but not 4q25 remained robustly associated with AF (hazard ratio 1.96, 95% CI 1.40–2.73, P = 8 × 10−5). The proportion of AF diagnoses in HF patients attributable to polymorphisms was 19% and 12%, respectively.
Conclusions
A polymorphism in the ZFHX3 gene, encoding a cardiac transcription factor, was associated with increased AF risk in HF patients, and the genetic association with AF was more pronounced in HF patients than in the general population.
Keywords: Atrial fibrillation, Heart failure, Genetic association, Risk factors, Genetic predisposition
See page 244 for the editorial comment on this article (doi:10.1093/eurjhf/hft005)
Introduction
Heart failure (HF) is frequently complicated by atrial fibrillation (AF), the most common sustained cardiac arrhythmia.1 Onset of AF in HF patients is associated with increased morbidity and mortality, potentially mediated by a rapid ventricular rate, loss of atrial contraction, irregular ventricular filling time, or thrombo-embolism.2–5 The prevalence of AF rises steeply with the severity of HF. However, more than half of patients with severe HF do not develop recognized AF,2–6 and pathophysiological mechanisms linking the two diseases remain largely uncharacterized.7 We therefore aimed to examine the risk factors for AF in patients with HF. Importantly, genetic factors could theoretically explain at least part of the AF risk in HF, but studies examining the heritability of AF in HF patients are challenging because of the complexity of assembling multigenerational HF samples of sufficient size. As an initial test for a genetic basis of AF in the context of HF, we examined the association of genetic polymorphisms found to influence AF risk in genome-wide association studies8−10 with occurrence of AF in HF patients.
Methods
Study sample
The population-based Malmö Diet and Cancer study (MDCS) included 30 447 randomly selected men born between 1923 and 1945, and women born between 1923 and 1950 from Malmö, Sweden. Participants attended baseline examinations between 1991 and 1996 with sampling of peripheral venous blood and collection of cardiovascular characteristics as previously described.11–13 Briefly, participants underwent anthropometric measurements, measurement of blood pressure using a mercury column sphygmomanometer in the supine position after 10 min of rest, and filled out a questionnaire including information about current smoking, diabetes mellitus, and medication use. Diabetes mellitus at baseline was defined as self-reported physician's diagnosis, use of antidiabetic medications, or fasting blood glucose >6.0 mmol/L when available. Hypertension was defined as use of antihypertensive medications or blood pressure ≥140/90 mmHg. Informed consent was obtained from all participants, and the study was approved by the ethics committee of Lund University, Sweden.
Patient selection
Individuals diagnosed with HF, AF, or myocardial infarction were identified by register linkage of Swedish personal identification numbers14 to national Swedish registers [Swedish Cause of Death Register (CDR) and Swedish Hospital Discharge Register (HDR)] maintained by the Swedish National Board of Health and Welfare15 as described previously.11,12 Follow-up extended until 1 January 2009.
Heart failure was ascertained from the Swedish HDR using a primary diagnosis code of 427.00, 427.10, and 428.99 for International Classification of Diseases-8th Revision (ICD-8), 428 for the 9th Revision (ICD-9), and I50 and I11.0 for the 10th Revision (ICD-10). Atrial fibrillation was ascertained from the HDR or the CDR using diagnosis codes 427.92 (ICD-8), 427D (ICD-9), and I48 (ICD-10). Myocardial infarction was ascertained from the HDR or the CDR using diagnosis codes 410 (ICD-8 and -9) and I21 (ICD-10). High case validity in the HDR and CDR has previously been reported for these definitions of HF,16 AF,11 and myocardial infarction.17
Genotyping
Two single nucleotide polymorphisms (SNPs) on chromosomes 4q25 near PITX2 (rs2200733) and 16q22 in ZFHX3 (rs2106261) associated with AF risk in genome-wide association studies and subsequently replicated in independent samples8−10 were genotyped in individuals from the MDCS who contributed DNA.18 DNA was extracted from peripheral blood cells and assigned to batches without regard to AF or HF status. Batches were genotyped with the same set of reagents using real-time polymerase chain reaction reagents and protocols on an ABI 7900HT (Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Genotype calls were obtained using SDS 2.3 software (Life Technologies, Carlsbad, CA, USA) and were manually curated.
Statistical analysis
As the exact onset of AF and HF can be difficult to determine and the temporal sequence of diagnoses of AF and HF can vary,3–5 primary analyses were performed under non-time-dependent assumptions. Genetic polymorphisms and cardiovascular risk factors [hypertension, body mass index (BMI), history of myocardial infarction, diabetes mellitus, and current smoking] were tested for association with AF in HF patients using unconditional logistic regression analysis adjusted for baseline age and sex. P-values <0.05 were considered statistically significant. Genetic polymorphisms were primarily tested using additive genetic models as in previous studies,8–10 and population-attributable risk was estimated from these models,11 but genotype-specific risk models were also evaluated. In a first sensitivity analysis, the association of risk factors with AF was tested prospectively in patients diagnosed with AF only after HF, using Cox proportional hazards regression with censoring at death or emigration, and exclusion of individuals with a diagnosis of AF before or simultaneously with HF. Results were compared with logistic regression analyses, given the potential differences between onset date and diagnosis date. In a second sensitivity analysis, individuals diagnosed with AF or HF prior to baseline were excluded. In a third sensitivity analysis, exclusions in the other two sensitivity analyses were combined. Finally, in an analysis of all participants in the entire cohort with DNA, risk estimates for AF in HF patients were compared with estimates in the general population obtained from logistic regression analysis by including multiplicative interaction terms for genotype with HF. Interaction of genetic polymorphisms with age was also explored. All analyses were performed using SAS 9.2 (SAS Institute, Cary, NC, USA).
Results
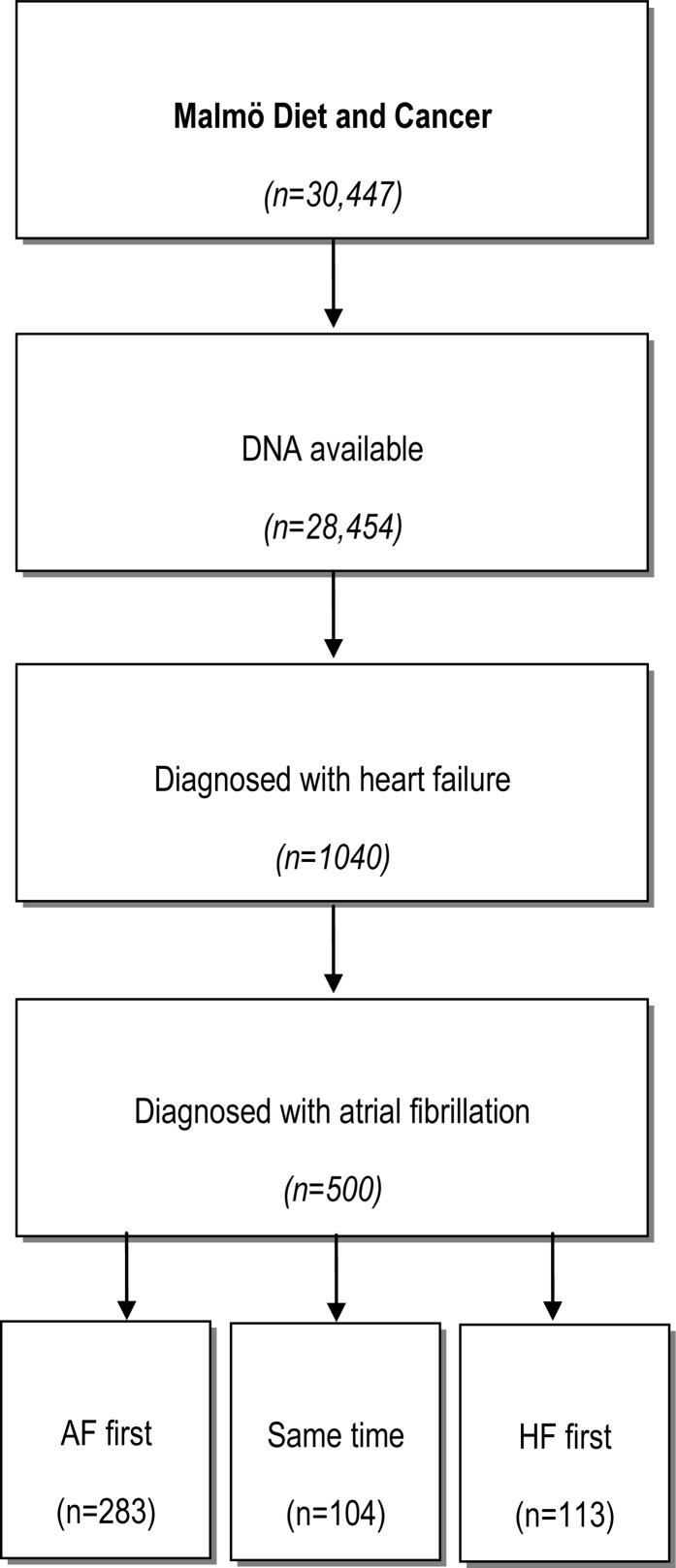
An outline of the cohort is shown in Figure 1. DNA was available for 28 454 individuals, of whom 2339 (8.2%) were diagnosed with AF prior to baseline or during follow-up. A total of 1040 individuals with DNA from the MDCS were diagnosed with HF prior to baseline or during follow-up, of whom 500 (48%) were also diagnosed with AF. Of participants with both AF and HF, 283 (57%) were diagnosed with AF first [median 992 days earlier, interquartile range (IQR) 199–2513], 113 (23%) were diagnosed with HF first (median 611 days earlier, IQR 132–1995), and 104 (21%) were diagnosed with both diseases simultaneously. Baseline characteristics for the HF cohort and the entire MDCS cohort are shown in Table 1. In contrast to the entire cohort, HF patients were predominantly male (58.0%), with an average age at diagnosis of 72.2 years. A large proportion (29.4%) of HF patients had a history of myocardial infarction at HF diagnosis.
Figure 1.
Description of the cohort. AF, atrial fibrillation; HF, heart failure.
Table 1.
Baseline characteristics
| Characteristic | MDCS (n =28 454) | Heart failure patients (n = 1040) |
|---|---|---|
| Baseline age, years | 58.0 (7.7) | 63.5 (6.7) |
| Age at HF diagnosis, years | – | 72.2 (8.1) |
| Men, % | 39.7% | 58.0% |
| Body mass index, kg/m2 | 25.8 (4.0) | 27.8 (4.7) |
| History of hypertension, % | 61.3% | 86.1% |
| History of myocardial infarction, % | 2.0% | 29.4% |
| History of diabetes, % | 3.1% | 11.6% |
| Current smoking, % | 28.8% | 32.7% |
| 4q25 (rs2200733, C/T) | 10.1 % | 11.8 % |
| 16q22 (rs2106261, G/A) | 17.5 % | 16.2 % |
Mean and standard deviation are given for quantitative variables. For history of myocardial infarction, numbers refer to the proportion of patients diagnosed prior to baseline or heart failure diagnosis, respectively. For genetic polymorphisms, major/minor alleles are shown within parentheses followed by minor allele frequencies.
HF, heart failure; MDCS, Malmö Diet and Cancer Study.
Cardiovascular risk factors for atrial fibrillation in heart failure patients
Of cardiovascular risk factors, age, sex, BMI, history of hypertension, myocardial infarction, and diabetes were associated with AF in the entire cohort (Table 2). In HF patients, increased age at diagnosis [odds ratio (OR) 1.22 per 10 years, 95% confidence interval (CI) 1.04–1.43, P = 0.02] but not history of hypertension, BMI, sex, smoking, or diabetes (P ≥0.05) was associated with increased risk of AF. A history of myocardial infarction was marginally associated with a decreased risk of AF (OR 0.75, 95% CI 0.57–0.99, P = 0.04) independently of age. In the first sensitivity analysis, prospective analyses excluding individuals diagnosed with AF prior to or simultaneously with HF, no risk factor was associated with AF in HF patients (all P > 0.05). In the second sensitivity analysis, excluding individuals diagnosed with HF or AF prior to baseline, only age remained associated with AF (OR 1.44 per 10 years, 95% CI 1.17–1.78, P = 6 × 10−4). In the third sensitivity analysis, with exclusions from both of the other sensitivity analyses, no risk factor was associated with AF in HF patients (all P > 0.05).
Table 2.
Association of cardiovascular risk factors with atrial fibrillation
| Risk factor | MDCS | Heart failure patients | ||
|---|---|---|---|---|
| OR (95% CI) | P-value | OR (95% CI) | P-value | |
| Age, 10 years | 2.33 (2.19–2.49) | <0.001 | 1.22 (1.04–1.43) | 0.02 |
| Male sex | 1.77 (1.62–1.94) | <0.001 | 1.30 (1.00–1.68) | 0.05 |
| Body mass index, kg/m2 | 1.06 (1.04–1.07) | <0.001 | 1.01 (0.99–1.04) | 0.33 |
| History of hypertension | 1.54 (1.37–1.72) | <0.001 | 1.15 (0.79–1.68) | 0.48 |
| History of myocardial infarction | 1.82 (1.48–2.25) | <0.001 | 0.75 (0.57–0.99) | 0.04 |
| History of diabetes | 1.31 (1.08–1.59) | 0.007 | 0.77 (0.51–1.15) | 0.20 |
| Current smoking | 1.09 (0.99–1.20) | 0.10 | 0.85 (0.65–1.11) | 0.24 |
Relative risk estimates from multiple regression analyses are presented with the corresponding confidence interval and P-value for the entire cohort and heart failure (HF) patients. Risk estimates are presented per 10 years of baseline age for the entire cohort and per 10 years of age at diagnosis in HF patients. Risk estimates for myocardial infarction are presented for prevalent myocardial infarction at the baseline visit for the entire cohort and for prevalent myocardial infarction at the time of HF diagnosis in HF patients. Body mass index and history of hypertension, diabetes, and smoking refer to the baseline visit.
CI, confidence interval; MDCS, Malmö Diet and Cancer Study; OR, odds ratio.
Genetic risk factors for atrial fibrillation in heart failure patients
Both SNPs on chromosomes 4q25 and 16q22 were genotyped in MDCS, with a call rate >0.95.18 Minor allele frequencies (Table 1) were similar in the entire cohort and in the HF cohort, and similar to previous AF studies (4q25, 0.17–0.23; 16q22, 0.17–0.19)8,10,19 and the European sample (CEU) in the third phase of the HapMap Project (4q25, 0.12; 16q22, 0.16).
Both SNPs were associated with increased risk of AF in HF patients in additive models (16q22 OR 1.75 per risk allele, 95% CI 1.35–2.26, P =2 × 10−5; 4q25 OR 1.57 per risk allele, 95% CI 1.18–2.09, P = 0.002). Risk of AF by genotype and in additive models is shown in Table 3. Adjustment for established risk factors shown in Table 1 did not attenuate the SNP associations (data not shown). We observed no significant interaction of polymorphisms with age at diagnosis (P for interaction > 0.05). The AF risks in HF patients attributable to polymorphisms were 19% and 12% for SNPs on 16q22 and 4q25, respectively.
Table 3.
Association of genetic polymorphisms with atrial fibrillation
| Genotype | MDCS | HF patients | Participants without HF | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n (%) | OR (95% CI) | P-value | n (%) | OR (95% CI) | P-value | n (%) | OR (95% CI) | P-value | |
| 4q25 (rs2200733) | |||||||||
| Major homozygote (CC) | 21 927 (80%) | 1.00 | – | 772 (78%) | 1.00 | – | 21 155 (81%) | 1.00 | – |
| Heterozygote (CT) | 4982 (19%) | 1.62 (1.46–1.81) | 2 × 10−19 | 213 (21%) | 1.59 (1.17–2.17) | 0.003 | 4769 (18%) | 1.61 (1.44–1.81) | 7 × 10−16 |
| Minor homozygote (TT) | 265 (1%) | 2.13 (1.48–3.08) | 6 × 10−5 | 11 (1%) | 2.00 (0.58–6.91) | 0.28 | 254 (1%) | 2.18 (1.46–3.27) | 1 × 10−4 |
| Additive | 27 174 | 1.58 (1.44–1.73) | 6 × 10−22 | 996 | 1.57 (1.18–2.09) | 0.002 | 26 178 | 1.58 (1.42–1.75) | 3 × 10−18 |
| HF interaction | 0.83 | ||||||||
| 16q22 (rs2106261) | |||||||||
| Major homozygote (GG) | 19 214 (68%) | 1.00 | – | 724 (70%) | 1.00 | – | 18 490 (68%) | 1.00 | – |
| Heterozygote (AG) | 8147 (29%) | 1.11 (1.01–1.22) | 0.03 | 280 (27%) | 1.60 (1.21–2.12) | 0.001 | 7867 (29%) | 1.08 (0.98–1.21) | 0.14 |
| Minor homozygote (AA) | 849 (3%) | 1.40 (1.12–1.77) | 0.004 | 27 (3%) | 2.86 (1.23–6.66) | 0.01 | 822 (3%) | 1.39 (1.08–1.79) | 0.01 |
| Additive | 28 120 | 1.14 (1.05–1.23) | 0.001 | 1031 | 1.75 (1.35–2.26) | 2 × 10−5 | 27 179 | 1.12 (1.03–1.22) | 0.01 |
| HF interaction | 7 × 10−4 |
Relative risk estimates are presented per genotype and per risk allele in an additive genetic model, with corresponding confidence interval and P-value. Risk estimates are presented in the entire cohort, in HF patients and in participants without HF. P-values are also presented for interaction of the linear genetic model with heart failure status in the entire cohort.
CI, confidence interval; HF, heart failure; MDCS, Malmö Diet and Cancer Study; OR, odds ratio.
In the first sensitivity analysis, individuals diagnosed with AF prior to or simultaneously with HF diagnosis were excluded, leaving 653 individuals with HF. During a median follow-up of 1.7 years, 113 individuals were diagnosed with AF. In this sample, the SNP on 16q22 remained strongly associated with AF risk [hazard ratio (HR) 1.96 per risk allele, 95% CI 1.40–2.73, P = 8 × 10−5] but not 4q25 (HR 1.14 per risk allele, 95% CI 0.75–1.75, P = 0.53). In prospective analyses, risk estimates were similar when obtained from Cox proportional hazards regression and logistic regression models. In a second sensitivity analysis, individuals diagnosed with HF (n = 87) or AF (n = 69) prior to baseline were excluded. Both SNPs remained associated with AF risk (4q25 OR 1.40 per risk allele, 95% CI 1.03–1.91, P = 0.03; 16q22 OR 1.76 per risk allele, 95% CI 1.35–2.30, P = 3 × 10−5). In a third combined sensitivity analysis, excluding individuals diagnosed with AF prior to or simultaneously with HF and individuals diagnosed with HF or AF prior to baseline, the SNP on 16q22 (HR 1.98 per risk allele, 95% CI 1.39–2.82, P = 1 × 10−4) but not that on 4q25 (HR 1.24, 95% CI 0.80–1.91, P = 0.34) remained associated with AF. Results from sensitivity analyses are shown in Supplementary material, Table S1.
Compared with HF patients with no risk alleles (n = 547), AF risk increased in a dose-dependent fashion in patients with one risk allele (n = 345, OR 1.46, 95% CI 1.13–1.89, P = 0.004) or two risk alleles (n = 89, OR 2.04, 95% CI 1.30–3.22, P = 0.002). The number of individuals with more than two risk alleles was small but, interestingly, all individuals with three (n = 5) or four (n = 1) risk alleles were diagnosed with AF (Fisher's exact P = 0.03 and P = 0.48, respectively). Larger samples will be necessary to study individuals with more than two alleles, and for formal tests of gene–gene interaction.
Interaction analyses in the general population
In the entire cohort, AF risk was higher in HF patients (OR 9.24, 95% CI 8.03–10.64, P = 1 × 10−210) and in risk allele carriers for SNPs on both 4q25 (OR 1.58, 95% CI 1.44–1.73, P = 6 × 10−22) and 16q22 (OR 1.14, 95% CI 1.05–1.23, P = 0.001) as shown in Table 3. AF risk estimates were similar to those reported in previous population-based studies for SNPs on 4q25 (OR 1.45, 95% CI 1.36–1.54) and 16q22 (OR 1.19, 95% CI 1.12–1.26).10 AF risk by genotype was significantly magnified in the presence of HF for 16q22 (P for interaction = 7 × 10−4) but not 4q25 (P for interaction = 0.83), as detailed in Table 3. Interaction terms of SNPs with age at HF diagnosis were not significant (P > 0.05)
Discussion
We examined risk factors for AF in a large population-based cohort of patients hospitalized for HF. Whereas the conventional risk factors for AF in the general population—hypertension, smoking, diabetes, and BMI11,12,18,20—were not associated with AF in HF patients, strong associations were observed with two genetic polymorphisms near genes encoding cardiac transcription factors. The SNP on chromosome 16q22 but not that on 4q25 remained robustly associated with AF in prospective analyses restricted to patients with AF diagnosed after HF. Moreover, our findings suggest context dependency on AF risk for the chromosome 16q22 polymorphism, with a substantially larger relative risk of AF in HF patients (75% per copy) than in the general population (14% per copy). Our findings in HF patients might provide new clues on mechanisms linking HF to AF, for which understanding is limited.
Cardiovascular risk factors for atrial fibrillation in heart failure patients
Principally, the increased occurrence of AF in HF patients could result from common aetiological factors acting independently on both ventricles and atria or from atrial effects of ventricular dysfunction, such as atrial stretch from increased filling pressure or increased neurohormonal activation. Although certain HF aetiologies such as infiltrative, infectious, or deposition diseases could potentially act on both atria and ventricles, the lack of association of the major AF risk factors with AF in HF patients in the present study argues against independent effects of these factors on atria and ventricles, and suggests that association between cardiovascular risk factors and AF could instead be mediated by ventricular dysfunction resulting in increased atrial stretch and neurohormonal activation. Further support for this view comes from studies of HF patients in which the prevalence of AF is strongly correlated with measures of HF severity and filling pressure, such as New York Heart Association (NYHA) class,2 and from population-based studies which have shown substantial overlap of conventional risk factors and echocardiographic measures of systolic ventricular dysfunction, increased left ventricular mass, and atrial dilation for AF risk.21 Consequently, in HF patients, other factors such as the severity of HF, timeliness of treatment, and potentially a genetic susceptibility could be expected to be more important determinants of AF. The lack of association of AF with conventional risk factors in HF patients could also reflect more limited power to detect associations in this smaller subcohort, potentially reduced further by regression dilution bias as discussed below.
Genetic polymorphisms and atrial fibrillation in heart failure patients
Our results demonstrate the relevance to AF in HF patients of genetic polymorphisms on chromosomes 4q25 and 16q22 found in previous studies to be associated with AF in the general population, providing initial evidence of a genetic component to AF in the context of HF. However, the mechanisms linking polymorphisms on chromosome 4q25 and 16q22 to risk of AF are currently unclear. The most proximal gene to the SNP on chromosome 4q25 is the PITX2 gene, encoding a transcription factor expressed in the developing heart during embryonic cardiogenesis. PITX2 has been found in animal models to play a role in the formation of the pulmonary vein myocardium,22 where ectopic impulse generation triggering AF onset has been reported in humans,23 and to inhibit left-sided atrial pacemaker specification, such that haploinsufficient mice showed increased expression of sinoatrial node-specific genes in the left atrium and increased propensity to atrial arrhythmia.24,25 Thus, risk allele carriers of the 4q25 SNP could have an increased propensity for ectopic impulse generation. The SNP on chromosome 16q22 is intronic to the gene Zinc Finger Homeobox 3 (ZFHX3), also encoding a transcription factor with cardiac expression. Little is known about the function of ZFHX3, but it has been shown to interact strongly with the Protein Inhibitor of Activated Stat3 (PIAS3), an inhibitor of STAT3,26 which in turn is a regulator of paracrine circuits in the heart essential for interstitial matrix deposition balance and capillary vasculature maintenance.27 Increased expression of STAT3 has been observed in animal models of AF and proposed to contribute to atrial matrix deposition.28,29 Thus, risk allele carriers of 16q22 could have an increased propensity to develop atrial fibrosis.
In the present study, SNPs on 16q22 but not on 4q25 were robustly associated with AF in prospective analyses of HF patients, and showed significant multiplicative interaction with HF for AF risk in the general population. Whereas SNPs on 4q25 have been shown to be strongly associated with lone AF and with AF following coronary artery bypass graft surgery,30 our observation of a stronger association for SNPs on 16q22 with AF in HF patients could be consistent with a more important role for atrial fibrosis than impulse generation in AF pathophysiology in the context of HF, which could have implications for selection of rhythm control strategies in these different settings.31 The lack of association for 4q25 with AF in prospective analyses and interaction tests could also reflect limited power to detect an association for this SNP with a lower allele frequency, although the lower risk estimate observed in the HF subsample supports a smaller effect. Experimental studies are needed to clarify the mechanisms linking polymorphisms with AF and the interaction of 16q22 with HF reported here.
Study strengths and limitations
The major strength of the present study was the very large size of the source population sample, which allowed detection of a large, population-based cohort of HF patients with AF (Figure 1). The use of a source population cohort also makes our study more representative of the general HF population than many previous studies of AF in HF patients which predominantly included younger, male participants of randomized controlled trials.2 Our cohort of hospitalized HF patients was similar to previous population-based studies of HF32,33 with a high age at diagnosis (mean 72 years), a slight majority of patients of male sex (58%), and a history of myocardial infarction in about a third of HF patients. However, our study has limitations which merit consideration. First, only patients hospitalized for a primary diagnosis of HF were detected. In contrast to diagnoses in the outpatient setting, such diagnoses have shown low inter-reader variability,16 but are likely to bias our sample towards more severe cases, as evidenced by the high rate of AF. Although generalizability to outpatients seems likely, additional studies need to address this question. Secondly, our study design did not allow information on left ventricular ejection fraction (LVEF), which is frequently used to subclassify patients into HF with reduced or preserved LVEF. It is possible that our results are specific to HF of one of these subclasses. We also did not have information on NYHA class or left ventricular filling pressures, which are likely to be strong and correlated determinants of AF risk, specific aetiologies of HF, or use of medications for HF. Additional studies in clinically well characterized cohorts are therefore warranted. Thirdly, undetected AF events may have resulted in underestimates of AF risk and reduced power to detect associations, but are not likely to have caused false-positive associations. Underestimation of AF events may be less pronounced in HF patients, who are frequently hospitalized and undergo electrocardiogram screening, resulting in relatively improved power to detect associations, but is unlikely to influence risk estimates. Differential underestimation by genotype would be expected to impact effect estimates, but appears unlikely. Fourthly, the possibility of undetected AF events makes the temporality of AF and HF diagnoses uncertain, as in previous studies on the temporality of AF in HF patients.3–5 Temporal misclassification might result in reduced precision in time-dependent sensitivity analyses, but is unlikely to result in false or inflated associations. Fifthly, as study participants did not attend follow-up examinations and cardiovascular risk factors were measured at baseline, it is possible that regression dilutions bias may have attenuated risk estimates for these risk factors. Finally, subsequent to the initiation of this study, SNPs in other genetic loci have been reported to be associated with AF, although with smaller effects.34,35 The association of these polymorphisms with AF in the context of HF remains to be determined.
Conclusions
Our results indicate a heritable component to the propensity of AF in HF patients, with increased risk of AF in HF patients carrying a common genetic variant in the ZFHX3 gene on chromosome 16q22 encoding a cardiac transcription factor. Furthermore, the association of this genetic polymorphism with AF was more pronounced in HF patients than in the general population. These findings may provide clues to the pathophysiology of AF onset in HF patients and have implications for rhythm control strategies in HF patients.
Supplementary material
"Supplementary material is available at European Journal of Heart Failure online.
Funding
The Malmö Diet and Cancer Study was supported by the Swedish Cancer Society, the Swedish Medical Research Council, the Swedish Dairy Association, the Albert Påhlsson and Gunnar Nilsson Foundations, and Malmö City Council. Genotyping was supported by governmental funding of clinical research within the Swedish National Health Service to P.G.P. The Swedish Heart-Lung Foundation (J.G.S.); the Swedish Medical Research Council and Lund University (O.M. and B.H.); the Medical Faculty of Lund University, Skåne University Hospital in Malmö, the Albert Påhlsson Research Foundation, the Crafoord Foundation, Swedish National Health Service, the Hulda and Conrad Mossfelt Foundation, the Ernhold Lundströms Research Foundation, the King Gustaf V and Queen Victoria Fund, the Lennart Hanssons Memorial Fund, the Marianne and Marcus Wallenberg Foundation, and the Knut and Alice Wallenberg Foundation (O.M.); National Institutes of Health (grants HL080025 and HL098283 to C.N.C.); a Doris Duke Charitable Foundation Clinical Scientist Development Award, and a Burroughs Wellcome Fund Career Award for Medical Scientists (C.N-C.); the Swedish National Health Service, Lund University Hospital and the Craaford Foundation (P.G.P.).
Acknowledgements
The authors wish to thank all participants in the Malmö Diet and Cancer Study for making this study possible.
Conflict of interest: none declared.
References
- 1.McMurray JJ, Adamopoulos S, Anker SD, Auricchio A, Bohm M, Dickstein K, Falk V, Filippatos G, Fonseca C, Sanchez MA, Jaarsma T, Kober L, Lip GY, Maggioni AP, Parkhomenko A, Pieske BM, Popescu BA, Ronnevik PK, Rutten FH, Schwitter J, Seferovic P, Stepinska J, Trindade PT, Voors AA, Zannad F, Zeiher A, Bax JJ, Baumgartner H, Ceconi C, Dean V, Deaton C, Fagard R, Funck-Brentano C, Hasdai D, Hoes A, Kirchhof P, Knuuti J, Kolh P, McDonagh T, Moulin C, Popescu BA, Reiner Z, Sechtem U, Sirnes PA, Tendera M, Torbicki A, Vahanian A, Windecker S, McDonagh T, Sechtem U, Bonet LA, Avraamides P, Ben Lamin HA, Brignole M, Coca A, Cowburn P, Dargie H, Elliott P, Flachskampf FA, Guida GF, Hardman S, Iung B, Merkely B, Mueller C, Nanas JN, Nielsen OW, Orn S, Parissis JT, Ponikowski P ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure 2012: The Task Force for the Diagnosis and Treatment of Acute and Chronic Heart Failure 2012 of the European Society of Cardiology. Developed in collaboration with the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail. 2012;14:803–869. doi: 10.1093/eurjhf/hfs105. [DOI] [PubMed] [Google Scholar]
- 2.Maisel WH, Stevenson LW. Atrial fibrillation in heart failure: epidemiology, pathophysiology, and rationale for therapy. Am J Cardiol. 2003;91:2D–8D. doi: 10.1016/s0002-9149(02)03373-8. [DOI] [PubMed] [Google Scholar]
- 3.Wang TJ, Larson MG, Levy D, Vasan RS, Leip EP, Wolf PA, D'Agostino RB, Murabito JM, Kannel WB, Benjamin EJ. Temporal relations of atrial fibrillation and congestive heart failure and their joint influence on mortality: the Framingham Heart Study. Circulation. 2003;107:2920–2925. doi: 10.1161/01.CIR.0000072767.89944.6E. [DOI] [PubMed] [Google Scholar]
- 4.Chamberlain AM, Redfield MM, Alonso A, Weston SA, Roger VL. Atrial fibrillation and mortality in heart failure: a community study. Circ Heart Fail. 2011;4:740–746. doi: 10.1161/CIRCHEARTFAILURE.111.962688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smit MD, Moes ML, Maass AH, Achekar ID, Van Geel PP, Hillege HL, van Veldhuisen DJ, Van Gelder IC. The importance of whether atrial fibrillation or heart failure develops first. Eur J Heart Fail. 2012;14:1030–1040. doi: 10.1093/eurjhf/hfs097. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell JC, Contractor H, Petkar S, Ali R, Clarke B, Neyses L, Mamas MA. Atrial fibrillation is under-recognized in chronic heart failure: insights from a heart failure cohort treated with cardiac resynchronization therapy. Europace. 2009;11:1295–1300. doi: 10.1093/europace/eup201. [DOI] [PubMed] [Google Scholar]
- 7.Platonov PG. Substrate for development of atrial fibrillation in patients with congestive heart failure: are we close to the answer? Heart Rhythm. 2009;6:452–453. doi: 10.1016/j.hrthm.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njølstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbäumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjörnsdóttir S, Valdimarsson EM, Løchen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D'Agostino RB, Sr, Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiríksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WH, Agarwal SK, Stricker BH, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Köttgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann HE, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kääb S, Ellinor PT, Witteman JC. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith JG, Platonov PG, Hedblad B, Engstrom G, Melander O. Atrial fibrillation in the Malmo Diet and Cancer study: a study of occurrence, risk factors and diagnostic validity. Eur J Epidemiol. 2010;25:95–102. doi: 10.1007/s10654-009-9404-1. [DOI] [PubMed] [Google Scholar]
- 12.Smith JG, Newton-Cheh C, Almgren P, Struck J, Morgenthaler NG, Bergmann A, Platonov PG, Hedblad B, Engström G, Wang TJ, Melander O. Assessment of conventional cardiovascular risk factors and multiple biomarkers for the prediction of incident heart failure and atrial fibrillation. J Am Coll Cardiol. 2010;56:1712–1719. doi: 10.1016/j.jacc.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berglund G, Elmstahl S, Janzon L, Larsson SA. The Malmo Diet and Cancer Study. Design and feasibility. J Intern Med. 1993;233:45–51. doi: 10.1111/j.1365-2796.1993.tb00647.x. [DOI] [PubMed] [Google Scholar]
- 14.Ludvigsson JF, Otterblad-Olausson P, Pettersson BU, Ekbom A. The Swedish personal identity number: possibilities and pitfalls in healthcare and medical research. Eur J Epidemiol. 2009;24:659–667. doi: 10.1007/s10654-009-9350-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludvigsson JF, Andersson E, Ekbom A, Feychting M, Kim JL, Reuterwall C, Heurgren M, Olausson PO. External review and validation of the Swedish national inpatient register. BMC Public Health. 2011;11:450. doi: 10.1186/1471-2458-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ingelsson E, Arnlöv J, Sundström J, Lind L. The validity of a diagnosis of heart failure in a hospital discharge register. Eur J Heart Fail. 2005;7:787–91. doi: 10.1016/j.ejheart.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Hammar N, Alfredsson L, Rosén M, Spetz CL, Kahan T, Ysberg AS. A national record linkage to study acute myocardial infarction incidence and case fatality in Sweden. Int J Epidemiol. 2001;30(Suppl 1):S30–S34. doi: 10.1093/ije/30.suppl_1.s30. [DOI] [PubMed] [Google Scholar]
- 18.Smith JG, Newton-Cheh C, Almgren P, Melander O, Platonov PG. Genetic polymorphisms for estimating risk of atrial fibrillation in the general population: a prospective study. Arch Intern Med. 2012;172:742–744. doi: 10.1001/archinternmed.2012.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith JG, Almgren P, Engström G, Hedblad B, Platonov PG, Newton-Cheh C, Melander O. Genetic polymorphisms for estimating risk of atrial fibrillation: a literature-based meta-analysis. J Intern Med. 2012 doi: 10.1111/j.1365-2796.2012.02563.x. doi:10.1111/j.1365-2796.2012.02563.x. Published online ahead of print 12 June 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamin EJ, Levy D, Vaziri SM, D'Agostino RB, Belanger AJ, Wolf PA. Independent risk factors for atrial fibrillation in a population-based cohort. The Framingham Heart Study. JAMA. 1994;271:840–844. [PubMed] [Google Scholar]
- 21.Vaziri SM, Larson MG, Benjamin EJ, Levy D. Echocardiographic predictors of nonrheumatic atrial fibrillation. The Framingham Heart Study. Circulation. 1994;89:724–730. doi: 10.1161/01.cir.89.2.724. [DOI] [PubMed] [Google Scholar]
- 22.Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, Christoffels VM. Pitx2c and Nkx2-5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 23.Haïssaguerre M, Jaïs P, Shah DC, Takahashi A, Hocini M, Quiniou G, Garrigue S, Le Mouroux A, Le Métayer P, Clémenty J. Spontaneous initiation of atrial fibrillation by ectopic beats originating in the pulmonary veins. N Engl J Med. 1998;339:659–666. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci USA. 2010;107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 26.Nojiri S, Joh T, Miura Y, Sakata N, Nomura T, Nakao H, Sobue S, Ohara H, Asai K, Ito M. ATBF1 enhances the suppression of STAT3 signaling by interaction with PIAS3. Biochem Biophys Res Commun. 2004;314:97–103. doi: 10.1016/j.bbrc.2003.12.054. [DOI] [PubMed] [Google Scholar]
- 27.Hilfiker-Kleiner D, Hilfiker A, Fuchs M, Kaminski K, Schaefer A, Schieffer B, Hillmer A, Schmiedl A, Ding Z, Podewski E, Podewski E, Poli V, Schneider MD, Schulz R, Park JK, Wollert KC, Drexler H. Signal transducer and activator of transcription 3 is required for myocardial capillary growth, control of interstitial matrix deposition, and heart protection from ischemic injury. Circ Res. 2004;95:187–195. doi: 10.1161/01.RES.0000134921.50377.61. [DOI] [PubMed] [Google Scholar]
- 28.Tsai CT, Lin JL, Lai LP, Lin CS, Huang SK. Membrane translocation of small GTPase Rac1 and activation of STAT1 and STAT3 in pacing-induced sustained atrial fibrillation. Heart Rhythm. 2008;5:1285–1293. doi: 10.1016/j.hrthm.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 29.Burashnikov A. Are there atrial selective/predominant targets for ‘upstream’ atrial fibrillation therapy? Heart Rhythm. 2008;5:1294–1295. doi: 10.1016/j.hrthm.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Body SC, Collard CD, Shernan SK, Fox AA, Liu KY, Ritchie MD, Perry TE, Muehlschlegel JD, Aranki S, Donahue BS, Pretorius M, Estrada JC, Ellinor PT, Newton-Cheh C, Seidman CE, Seidman JG, Herman DS, Lichtner P, Meitinger T, Pfeufer A, Kääb S, Brown NJ, Roden DM, Darbar D. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2:499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 2010;55:747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 32.Bhatia RS, Tu JV, Lee DS, Austin PC, Fang J, Haouzi A, Gong Y, Liu PP. Outcome of heart failure with preserved ejection fraction in a population-based study. N Engl J Med. 2006;355:260–269. doi: 10.1056/NEJMoa051530. [DOI] [PubMed] [Google Scholar]
- 33.Cowie MR, Wood DA, Coats AJ, Thompson SG, Poole-Wilson PA, Suresh V, Sutton GC. Incidence and aetiology of heart failure: a population-based study. Eur Heart J. 1999;20:421–428. doi: 10.1053/euhj.1998.1280. [DOI] [PubMed] [Google Scholar]
- 34.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel RB, Soliman EZ, Rice KM, Van Wagoner DR, Beckmann BM, van Noord C, Wang K, Ehret GB, Rotter JI, Hazen SL, Steinbeck G, Smith AV, Launer LJ, Harris TB, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Köttgen A, Moebus S, Newton-Cheh C, Li M, Möhlenkamp S, Wang TJ, Kao WH, Vasan RS, Nöthen MM, MacRae CA, Stricker BH, Hofman A, Uitterlinden AG, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Gudnason V, Psaty BM, Roden DM, Meitinger T, Wichmann HE, Witteman JC, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kääb S. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dörr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Völker U, Völzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjögren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kääb S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.