Abstract
Long noncoding RNAs (lncRNAs) are increasingly appreciated as regulators of cell-specific gene expression. Here, an enhancer-like lncRNA termed NeST (Nettoie Salmonella pas Theiler’s; cleanup Salmonella not Theiler’s) is shown to be causal for all phenotypes conferred by murine viral susceptibility locus Tmevp3. This locus was defined by crosses between SJL/J and B10.S mice and contains several candidate genes, including NeST. The SJL/J-derived locus confers higher lncRNA expression, increased interferon-γ abundance in activated CD8+ T cells, increased Theiler’s virus persistence and decreased Salmonella enterica pathogenesis. Transgenic expression of NeST lncRNA alone was sufficient to confer all phenotypes of the SJL/J locus. NeST RNA was found to bind WDR5, a component of the histone H3 lysine 4 methytransferase complex, and to alter histone 3 methylation at the interferon gamma locus. Thus, this lncRNA regulates epigenetic marking of IFNγ-encoding chromatin, expression of IFN-γ and susceptibility to a viral and a bacterial pathogen.
Introduction
Bioinformatic analysis of the chromatin marks in intergenic DNA regions and of expressed sequence tags (ESTs) predicts the existence of over 5,000 lncRNA genes in the human genome (Guttman et al., 2009; Khalil et al., 2009; Qureshi et al., 2010). However, it is currently unknown how many of these RNAs are functional. In a few well-studied cases, such as AIR, XIST and HOTAIR, lncRNAs have been shown to operate at the transcriptional level by binding to proteins in histone-modifying complexes and targeting them to particular genes (Chu et al., 2011; Jeon and Lee, 2011; Nagano et al., 2008). A role for lncRNAs in mammilian susceptibility to infection or the immune response to pathogens has not been previously described.
NeST, formally known as Tmevpg1, is a long noncoding RNA gene located adjacent to the IFN-γ-encoding gene in both mouse (Ifng) and human (IFNG). NeST was originally identified as a candidate gene in a susceptibility locus for Theiler’s virus: NeST abbreviates Nettoie Salmonella pas Theiler’s (cleanup Salmonella not Theiler’s). In both mouse and human genomes, NeST RNA is encoded on the DNA strand opposite to that coding for IFN-γ and the two genes are transcribed convergently (Fig 1A). In the mouse, NeST RNA contains six exons spread over a 45 Kb region (Vigneau et al., 2001; Vigneau et al., 2003). The most abundant splice variant is 914 nucleotides in length, is expressed in CD4+ T cells, CD8+ T cells and NK cells, and contains no AUG codons in translational contexts that appear functional. The orientation and location of human NEST is conserved, but the primary transcript encompasses the opposite strand of the entire IFNG gene (Fig 1A).
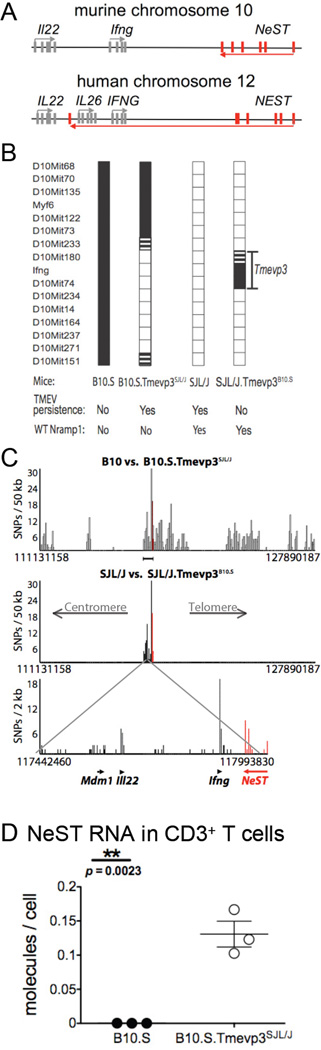
Figure 1. Genotypes of parental and congenic strains used to investigate NeST RNA and the Tmevp3 locus on murine chromosome 10.
(A) Schematic of the NeST-encoding genes in mouse chromosome 10 and human chromosome 12. The bars represent exons, arrows indicate the direction of transcription. NeST, previously termed Tmevpg1, is adjacent to both murine Ifng and human IFNG (Vigneau et al., 2003). The major transcript, shown in red, encodes six exons. In both mice and humans, the NeST and IFN-γ-encoding transcripts are convergently synthesized; in humans the transcribed regions overlap. (B) A diagram of the Tmevp3 locus on murine chromosome 10, as defined by the differential ability to clear persistent infection by Theiler’s virus observed between SJL/J and B10.S mice. The SJL/J.Tmevp3B10.S line, previously termed ‘C2’ (Vigneau et al., 2003), is congenic with SJL/J except for the region shown, from microsatellite marker D10Mit74 to the interval between D10Mit180 and D10Mit233. The B10.S.Tmevp3SJL/J strain is congenic with B10.S except for the region shown, between the D10Mit151/D10Mit271 interval and the D10Mit233/D10Mit73 interval. The Theiler’s virus (TMEV) persistence and clearance phenotypes and Nramp1 alleles for all four strains are listed. (C) Finer mapping of the polymorphic regions of the congenic lines. The x-axis indicates nucleotide number on mouse chromosome 10. The introgressed region of SJL/J in B10.S.Tmevp3SJL/J is up to 1.6×107 bp (top) whereas the introgression in SJL/J.Tmevp3B10.S is approximately 5.5×105 bp (middle and bottom). Each bar displays the number of single-nucleotide polymorphisms (SNPs) in the window size indicated. The most polymorphic region maps to the Tmevp3 locus and coincides with the region of introgression in SJL/J.Tmevp3B10.S; see Table S1 for lists of all genetic differences between the two Tmepv3 alleles. The physical locations and direction of transcription of the murine NeST, Ifng, Il22 and Mdm1 genes are indicated by arrows. (D) NeST RNA expression in CD3+ T cells. The abundance of NeST RNA in CD3+ splenocytes from B10.S mice and B10.S.Tmevp3SJL/J was determined by preparing total cellular RNA and determining the amount of RNA per cell using quantitative RT-PCR and standard curves of transcribed RNAs. The threshold of detection was 0.005 molecules of NeST RNA per cell. Mean values are shown with standard error.
Theiler’s virus, a picornavirus, is a natural pathogen of mice. The ability of inbred mice to clear Theiler’s infection varies greatly from strain to strain, and, because the phenotype can be conferred by bone marrow transfer (Aubagnac et al., 2002; Brahic et al., 2005; Vigneau et al., 2003), is likely to result from different immune responses to the pathogen. A major effect is conferred by the H2 locus. Two additional loci that affect Theiler’s virus clearance were mapped by crosses between H2s-bearing SJL/J and B10.S mice; while B10.S mice can clear the virus, SJL/J mice become persistently infected and develop demyelinating lesions similar to those observed in human multiple sclerosis (Aubagnac et al., 2002; Bureau et al., 1993).
One of these loci, Tmevp3 (Theiler’s murine encephalitis virus persistence 3; Fig. 1B), was mapped to a 550 Kb interval on murine chromosome 10 (Levillayer et al., 2007). Congenic mouse lines were developed by crossing SJL/J to B10.S and backcrossing to each parental line for 10–12 generations (Bihl et al., 1999; Bureau et al., 1993; Levillayer et al., 2007). The B10.S.Tmevp3SJL/J line is congenic with B10.S, but contains the Tmevp3 locus from SJL/J and is unable to clear persistent infections. Conversely, the SJL/J.Tmevp3B10.S line is congenic with SJL/J but contains the Tmevp3 locus from B10.S and successfully clears infections. Analysis of the single nucleotide polymorphisms in the smallest introgressed B10.S-derived region revealed a small number of polymorphic genes including those that encode Mdm1 (Chang et al., 2008), potent immune cytokines IL-22 and IFN-γ, and the long noncoding RNA (Fig. 1C).
Here, we show new phenotypes associated with the Tmevp3 locus. In addition to the failure to clear Theiler’s virus, the SJL/J-derived alleles also confer both resistance to lethal infection with Salmonella enterica Typhimurium and inducible synthesis of IFN-γ in CD8+ T cells. We show that NeST lncRNA is sufficient to confer these disparate phenotypes, demonstrating its crucial role in the host response to pathogens and illustrating a novel function for lncRNAs in immune regulation and susceptibility to infectious disease.
Results
Mapping the Tmevp3 locus of mouse chromosome 10
To refine the borders of the Tmevp3 locus, we utilized the JAX mouse diversity genotyping array, which employs 623,124 single nucleotide polymorphisms (SNPs) and 916,269 invariant genomic probes. We also sequenced cDNAs encoding IL-22, IFN-γ and NeST RNA from SJL/J and B10.S mice and added these findings to the Jackson microarray results (Fig. 1C) and the list of known polymorphisms in the locus (Table S1). Our results corroborated the presence of a unique introgressed region that contained the previously mapped Tmevp3 locus, and allowed us to refine its boundaries. The maximum sizes of the introgressed regions were 16×106 bp and 550×103 bp, respectively, for the B10.S.Tmevp3SJL/J and SJL/J.Tmevp3B10.S congenic lines (Fig. 1C).
These analyses identified Il22, Ifng, and NeST as the most likely candidates for the gene or genes responsible for the Tmevp3 locus phenotypes by virtue of their polymorphic character and their known expression patterns. In Figure 1C, the top and middle bar graphs represent the number of SNPs in a series of non-overlapping 50kb window regions. The regions of densest polymorphism between the congenic and parental lines can be seen in more detail in the bottom part of Figure 1C. The product of Mdm1 is expressed predominately in the retina (Chang et al., 2008), making it an unlikely candidate. The three most polymorphic genes are Ifng, NeST and Il22. The polymorphisms corresponding to NeST are shown in red and all polymorphisms in the locus are listed in Supplemental Table 1. We were especially interested in the lncRNA due to its potential novelty. As shown in Figure 1D, CD3+ T cells from B10.S. Tmevp3SJL/J mice displayed significantly higher amounts of NeST RNA than did those from B10.S mice. This result differs from that reported previously (Vigneau et al., 2003), a discrepancy that may result from differences in T cell preparation or in the previous use of saturating RT-PCR methods. Here, quantitative RT-PCR, the use of standard curves, and comparison between RNA abundances from identical numbers of cells show, repeatedly, that splenocytes from mice that contain an SJL/J-derived Tmevp3 allele accumulated substantially more NeST RNA than those from mice with a B10.S-derived Tmevp3 allele. Even so, the amount of NeST RNA that accumulated in total CD3+ T cells was, on average, only 0.15 molecules per cell (Fig. 1D). It is known that many lncRNAs are present at similarly low amounts but still are sufficient to cause epigenetic changes that are then self-propagating (reviewed in Guttman and Rinn, 2012). It was also possible that NeST RNA is more abundant in a subset of the CD3+ T cells. Indeed, higher abundance of NeST RNA was observed in CD8+ T cells (Fig. 3B) than in total CD3+ T cells (Fig. 1D).
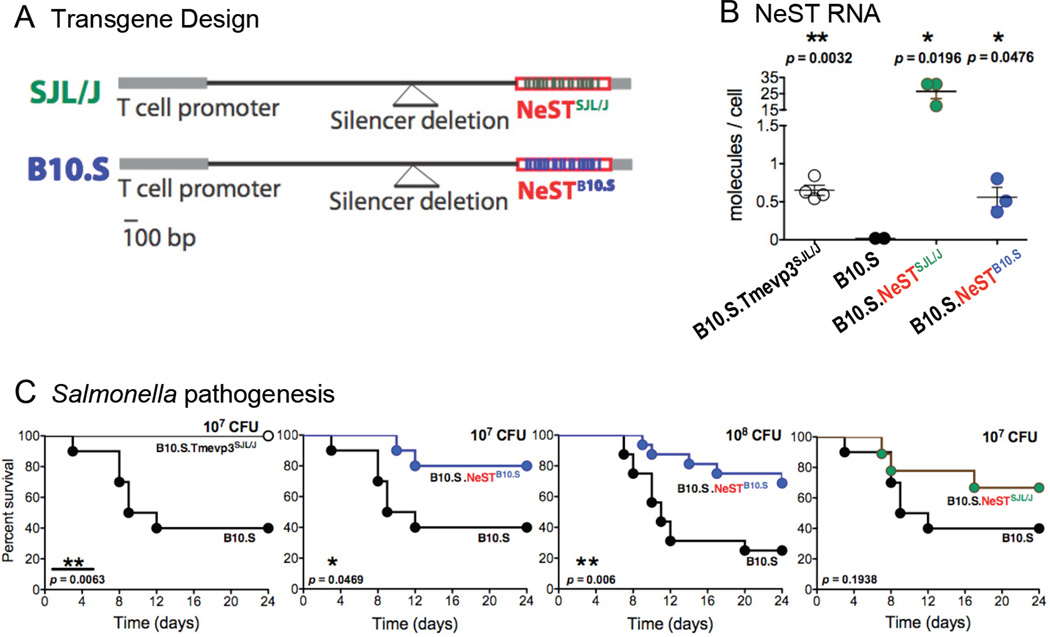
Figure 3. Effect of transgenically expressed NeST RNA on Salmonella pathogenesis.
(A) Schematic of transgenes introduced into B10.S mice. SJL/J NeST cDNA (Sea green) and B10.S NeST cDNA (Blue) were cloned downstream of a CD4+ and CD8+ T-cell specific promoter. The promoter-NeST transgene fragments were used for the construction of transgenic mouse lines in the B10.S background. (B) The abundance of NeST RNA was measured in CD8+ splenocytes from B10.S mice congenic for the SJL/J-derived Tmevp3 locus (B10.S.Tmevp3SJL/J), B10.S mice, B10.S mice containing the SJL/J NeST transgene (B10.S.NeSTSJL/J) and B10.S mice containing the B10.S NeST transgene (B10.S.NeSTB10.S). The amount of RNA per cell was determined using quantitative RT-PCR; in vitro transcribed NeST RNA was used to construct standard curves. (C) B10.S, B10.S.Tmevp3SJL/J, B10.S.NeSTSJL/J and B10.S.NeSTB10.S mice were orally inoculated with S. Typhimurium at dosages indicated and mortality was monitored. All panels with the 107 CFU/mouse were performed at the same time; the B10.S control is shown in these panels for clarity. Statistical significance was determined by the logrank test. See also Figure S1.
New pathogen phenotypes for the Tmevp3 locus
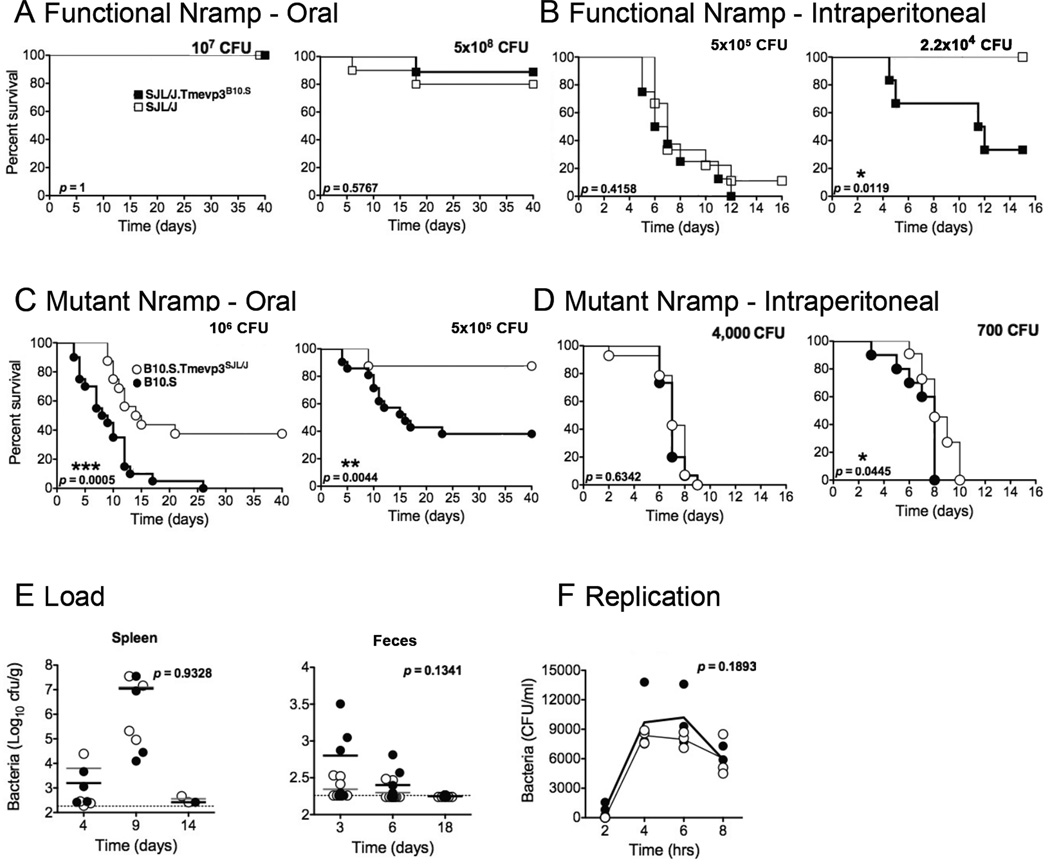
To determine whether Tmevp3 polymorphisms affected the outcome of another infection, we monitored their effects on the pathogenesis of Salmonella enterica Typhimurium, a pathogen which, like Theiler’s virus, grows in macrophages (Monack et al., 2004; Pena-Rossi et al., 1997) and is extremely sensitive to IFN-γ and CD8+ T cell control (Foster et al., 2005; Rodriguez et al., 2003). We began by comparing SJL/J and SJL/J.Tmevp3B10.S mice because the size of the introgressed region was smaller in this pair than in the B10.S and B10.S.Tmevp3SJL/J pair (Fig. 1B). Both the SJL/J mice and SJL/J.Tmevp3B10.S mice are homozygous for the functional allele of Nramp1, which encodes an ion channel that facilitates clearance of Salmonella (Frehel et al., 2002). As espected, both strains were resistant to oral inoculation (Fig. 2A). However, when subjected to the more potent intraperitoneal inoculation, both groups were susceptible but the SJL/J.Tmevp3B10.S mice showed significantly more mortality (Fig. 2B).
Figure 2. Effect of the Tmevp3 locus on Salmonella pathogenesis.
Strains SJL/J and SJL/J.Tmevp3B10.S were inoculated via oral (A) and intraperitoneal (B) routes with S. enterica Typhimurium. The Nramp1+/+ alleles expressed by SJL/J and SJL/J.Tmevp3B10.S mice render them relatively resistant to Salmonella infection. Strains B10.S and B10.S.Tmevp3SJL/J were also inoculated via oral (C) and intraperitoneal (D) routes with S. enterica Typhimurium at dosages indicated and mortality was monitored. The Nramp1169Asp/169Asp alleles render these mice highly sensitive to Salmonella pathogenesis. In both backgrounds, the SJL/J allele of the Tmevp3 locus reduced mortality after oral inoculation. Statistical significance was determined by the logrank test. (E) B10.S and B10.S.Tmevp3SJL/J were orally inoculated with S. Typhimurium at 106 CFU/mouse. Bacteria were monitored in spleen and feces at the indicated days. (F) Intracellular bacterial growth was monitored ex vivo in bone marrow derived macrophages from B10.S and B10.S.Tmevp3SJL/J mice. Lines represent mean of triplicate experiments and statistical significance was determined using the Student t-test. See also Figure S1.
B10.S and B10.S.Tmevp3SJL/J mice carry the Nramp1169Asp/169Asploss-of-function allele, which increases their susceptibility to Salmonella infection. When inoculated orally, B10.S mice displayed significantly more mortality than B10.S.Tmevp3SJL/J mice at several infectious dosages (Fig. 2C). Intraperitoneal inoculation was rapidly lethal for both mouse strains (Fig. 2D). The differences of phenotypes between SJL/J and SJL/J.Tmevp3B10.S and also between B10.S and B10.S.Tmevp3SJL/J mice strengthen the hypothesis that the Tmevp3 polymorphisms initially discovered by analysis of Theiler’s virus persistence have implications for general immune function. In subsequent experiments, we focused on B10.S and B10.S.Tmevp3SJL/J mice and for Salmonella pathogenesis, given that oral infection is the natural route.
To determine whether the differences in phenotype resulted from different bacterial loads, B10.S and B10.S.Tmevp3SJL/J mice were infected and the abundance of S. Typhimurium was monitored in spleen and feces. B10.S and B10.S.Tmevp3SJL/J mice were orally inoculated with 106 CFU and spleens were dissected 4, 9, and 14 days after inoculation. No significant differences in bacterial loads were observed in either spleen or feces at any time point (Fig. 2E). Interestingly, by day 14, both B10.S and B10.S.Tmevp3SJL/J mice had nearly resolved their infections even though mice from both groups continued to die. To test for differences in Salmonella growth in cultured macrophage, bone marrow-derived primary macrophage from B10.S and B10.S.Tmevp3SJL/J were infected and the amounts of intracellular Salmonella measured at various times after infection. No significant differences in bacterial growth within cells were observed (Fig. 2F). All these data are consistent with the hypothesis that lethality is not due to bacterial load per se, but due to the inflammatory response to bacterial infection (Miao and Rajan, 2011; Pereira et al., 2011; Strowig et al., 2012). In fact, the SJL/J-derived Tmevp3 locus also conferred increased resistance to the lethal inflammatory disease caused by LPS injection (Fig. S1).
Transgenic expression of NeST RNA reproduces the phenotype associated with the SJL/J Tmevp3 allele
We hypothesized that NeST RNA could play a causal role in the phenotypes conferred by the Tmevp3 locus. We developed B10.S transgenic mice that express either SJL/J- or B10.S-derived NeST RNA under the control of a promoter that directs constitutive expression in both CD4+ and CD8+ T cells (Fig. 3A; Sawada et al., 1994). Two transgenic mouse lines were obtained: B10.S.NeSTB10.S and B10.S.NeSTSJL/J. To test whether the transgenes had inserted near the endogenous Tmevp3 locus, genetic crosses were performed between the B10.S.NeSTB10.S and the B10.S.NeSTSJL/J transgenic mice and mice that bore neither marker. For both transgenic lines, the NeST transgenes and the endogenous locus showed no evidence of linkage (data not shown). Both transgenic NeST RNAs were expressed in CD8+ T cells (Fig. 3B), although at different amounts. The B10.S NeST transgene was expressed to an abundance similar to that of the endogenous NeST gene in the B10S.Tmevp3SJL/J line, whereas the SJL/J-derived transgene accumulated to much greater abundance (FIg. 3B).
To test whether the transgenic RNAs conferred protection against Salmonella pathogenesis, we inoculated B10.S mice, B10.S mice congenic at the Tmevp3 locus, and B10.S mice transgenic for each NeST allele orally with Salmonella. Mice that expressed the NeST B10.S transgene completely recapitulated the Tmevp3SJl/J survival phenotype (Fig. 3C). Mice that expressed the SJL/J NeST transgene also showed protection. These findings demonstrate that NeST RNA can function in trans to reduce Salmonella pathogenesis.
Transgenic NeST RNA expression prevents clearance of Theiler’s virus
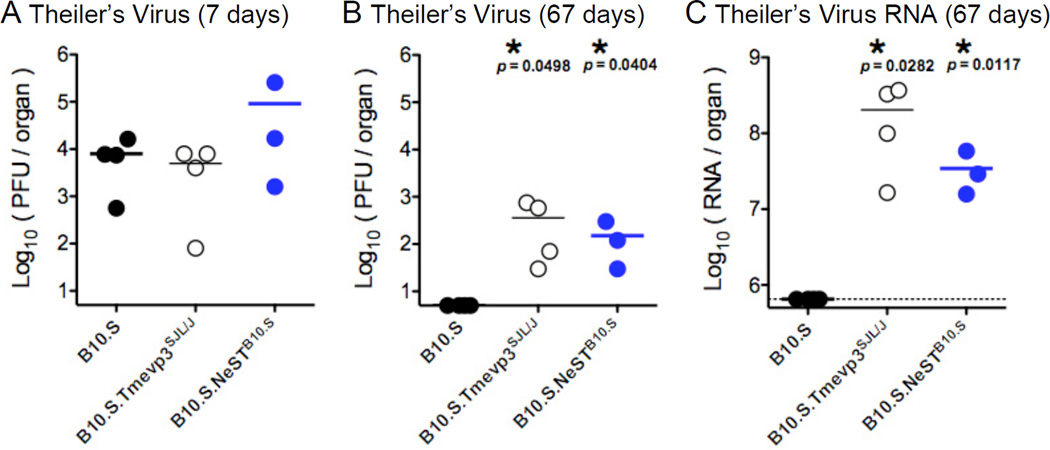
To test the role of NeST RNA in Theiler’s virus infection, the microbial susceptibility phenotype that led to its discovery, B10.S, B10.S.Tmevp3SJL/J, and B10.S.NeSTB10.S transgenic mice were inoculated by intracranial injection. Viral loads in the spinal cord were determined seven and 67 days after inoculation. At seven days, all strains displayed comparable viral titers (Fig. 4A), suggesting that NeST RNA plays little role during the acute phase of infection. However, 67 days after inoculation, infectious virus could only be recovered from mice that carried the NeST trasngene or the B10.S.Tmevp3 SJL/J locus (Fig. 4B). The amounts of viral RNA in the spinal cords of the transgenic mice and the B10.S.Tmevp3SJL/J mice were orders of magnitude higher than those found in the spinal cords of the non-transgenic B10.S parent (Fig. 4C). Thus, the susceptibility to Theiler’s virus persistence in spinal cord associated with the Tmevp3SJL/J allele was recapitulated by the expression of the NeST RNA transgene.
Figure 4. Effect of NeST RNA on Theiler’s virus persistence.
B10.S mice, B10.S mice congenic for the Tmevp3 locus from SJL/J mice (B10.S.Tmevp3SJL/J) and B10.S mice containing the B10.S NeST transgene (B10.S.NeSTB10.S) were inoculated by intracranial injections of 107 PFU of Theiler’s virus. Spinal cords were harvested at 7 days (A) and 57 days (B) post-inoculation and viral load was measured by plaque assay on BHK-21 cell monolayers. (C) The abundance of viral RNA in spinal cord from B10.S, B10.S.Tmevp3SJL/J and B10.S.NeSTB10.S mice was determined by preparing total cellular RNA from homogenized tissue and determining the amount per gram of tissue using quantitative RT-PCR. TMEV RNA was transcribed from cDNA-containing plasmid to construct standard curves. Means and standard error are shown.
Effect of the Tmevp3 locus on the expression of IFN- γ by CD8+ T cells
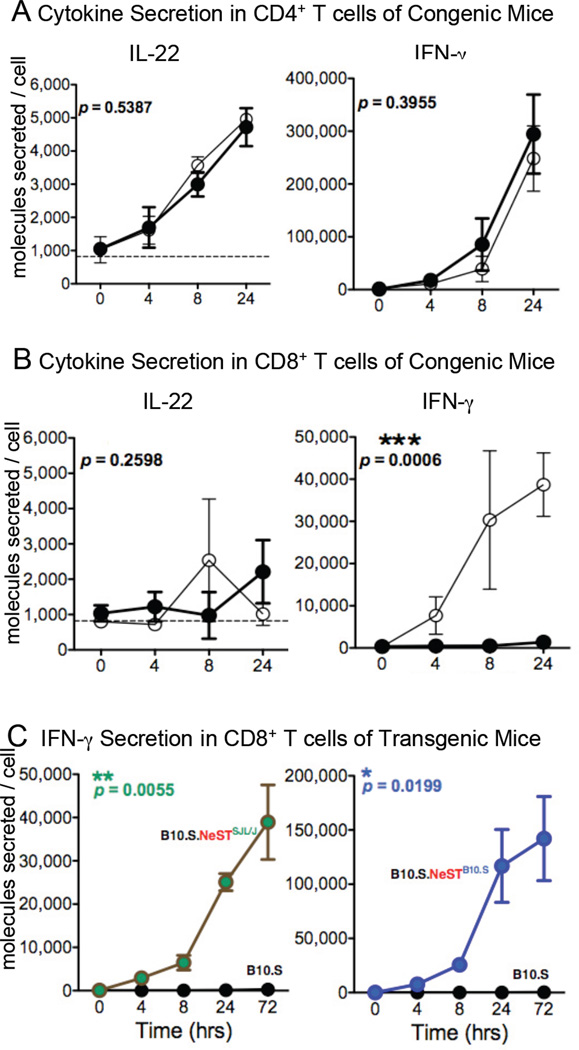
Several enhancer-like lncRNAs are known to activate neighboring genes, as exemplified by HOTTIP and Jpx (Orom et al., 2010; Tian et al., 2010; Wang et al., 2011). The physical proximity of Il22 and Ifng to NeST inspired us to test for differences in expression of these two genes in CD4+ and CD8+ T cells from B10.S and B10.S.Tmevp3SJL/J mice. Isolated CD4+ and CD8+ T cells were cultured for one day, stimulated with PMA and ionomycin, and monitored both for cytokine secretion (Fig. 5A, 5B) and intracellular RNA abundance (Fig. S2). In CD4+ T cells, ex vivo stimulation caused large but similar increases in the secretion of both cytokines in both B10.S and B10.S.Tmevp3SJL/J mice (Fig. 5A). Similarly, the Tmevp3 allele did not significantly affect the amounts of IL-22 secreted from CD8+T cells. However, while the amount of IFN- γ secreted from CD8+ T cells derived from B10.S mice was barely detectible, IFN- γ secretion from B10.S.Tmevp3SJL/J mice was robust after stimulation (Fig. 5B). The difference in IFN- γ production by CD8+ T cells coincided with the amounts of IFN- γ RNA and of NeST RNA (Fig S2B). These results show a strong correlation between the abundance of NeST RNA, IFN-γ RNA and IFN-γ protein in activated CD8+ T cells.
Figure 5. Effect of Tmevp3 locus and transgenically expressed NeST RNA on cytokine expression by T cell subsets.
Splenic (A) CD4+ and (B) CD8+ T cells were isolated from three B10.S (black circles) and three B10.S.Tmevp3SJL/J (white circles) mice and stimulated ex vivo with PMA and ionomycin. The abundance of IFN-γ and IL-22 protein secreted was determined by ELISA from supernatants collected at the indicated times. Means and standard error are indicated for each time point. Statistical significance was determined using a two-way ANOVA test; asterisks denote those values that differ significantly between T cells derived from B10.S and T cells derived from B10.S.Tmevp3SJL/J mice. (C) Splenic CD8+ T cells were isolated from B10.S (black), B10.S.NeSTSJL/J (sea green) and B10.S.NeSTB10.S (blue) mice and stimulated ex vivo with PMA and ionomycin. The abundance of secreted IFN-γ was determined by ELISA. Asterisks and p values refer to the comparisons between T cells derived from B10.S and T cells derived from each transgenic line. See also Figure S2.
Transgenic expression of NeST induces IFN-γ synthesis in activated CD8+ T cells
To determine whether the expression of NeST RNA alone could elicit the observed changes in IFN-γ expression in CD8+ T cells, we monitored the abundance of the cytokine in CD8+ T cells from B10.S, B10.S.NeSTSJL/J and B10.S.NeSTB10.S mice. As before, CD8+ T cells from the parental B10.S mice accumulated very little cytokine after ex vivo stimulation (Fig. 5C). However, the transgenic expression of either the B10.S or the SJL/J allele of NeST conferred the ability to induce IFN-γ secretion. Interestingly, the SJL/J-derived NeST RNA was less effective than the B10.S-derived RNA in mediating IFN-γ production, but both alleles caused statistically significant increases in IFN-γ expression upon CD8+ T cell activation. Subsequent experiments were designed to investigate the mechanism of these effects.
NeST is a nuclear lncRNA that can function in trans to affect its neighboring locus
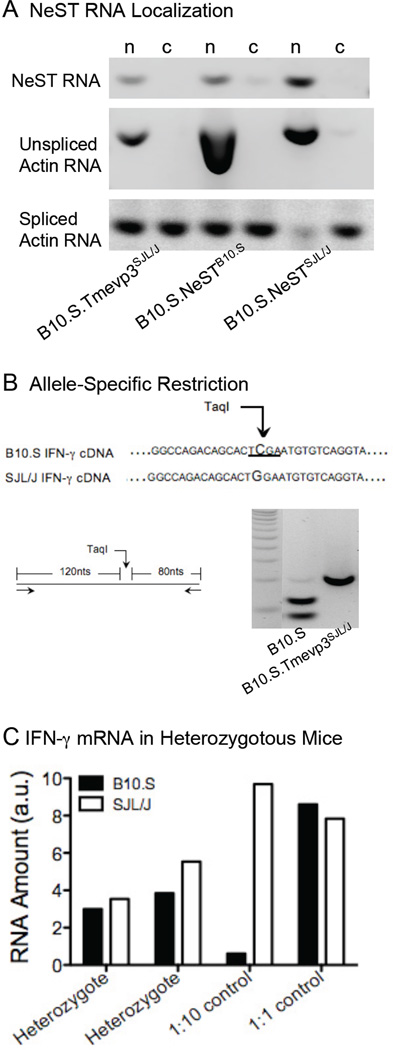
We hypothesized that, like several lncRNAs, NeST RNA affects IFN-γ accumulation at the transcriptional level by interacting with chromatin modification complexes. Consistent with this idea, most of the NeST RNA in either congenic or transgenic mice was found in the nuclear fraction of CD8+ T cells, co-fractionating with unspliced, but not spliced, actin mRNA (Fig. 6A).
Figure 6. NeST RNA localization and IFN-γ trans activation.
(A) Nuclear and cytoplasmic RNA from CD8+ T cells from B10.S.Tmevp3SJL/J, B10.S.NeSTB10.S and B10.S.NeSTSJL/J mice were fractionated by differential centrifugation (Huarte et al., 2010). NeST RNA, unspliced actin RNA (nuclear) and spliced actin RNA (cytoplasmic) from the nuclear and cytoplasmic fractions were assessed by RT-PCR and gel electrophoresis. (B) Quantitation of expression ratios of IFN-γ mRNA from the B10.S and the B10.S.Tmevp3SJL/J alleles. A natural SNP in the IFN-γ mRNA (coordinate 117882772, see Table S1) was amplified by RT-PCR (top and left panel). cDNAs from B10.S and B10.S.Tmevp3SJL/J were subjected to a B10.S allele-specific TaqI restriction digest (bottom, left) and fragments were analyzed by gel electrophoresis. (C) Splenic CD8+ T cells were isolated from two B10.S×B10.S.Tmevp3SJL/J heterozygous mice and stimulated with PMA and ionomycin. The proportion of B10.S and B10.S.Tmevp3SJL/J-derived IFN-γ mRNA was determined by densitometry of the allele-specific restriction fragments. Mixtures of in vitro transcribed RNAs at 1:10 and 1:1 ratios were used as controls.
The finding that two different transgenic NeST RNAs that were not genetically linked to the Ifng locus conferred the properties of the Tmevp3SJL/Jlocus to B10.S mice (Figs. 3–5) argues that this long noncoding RNA can function in trans. To determine if, indeed, NeST RNA can function in trans from its normal position of synthesis, we took advantage of the fact that NeST RNA is expressed in stimulated CD8+ T cells of B10.S.Tmevp3SJL/J mice but not in CD8+ T cells of B10.S mice (Fig. 3B and S2B). We developed a PCR assay to distinguish between the SJL/J- and B10.S-derived IFN-γ alleles (Fig. 6B). CD8+ T cells from two heterozygous B10.S/B10.S.Tmevp3SJL/J mice were stimulated, RNA was extracted, and the allele which the RNA was transcribed determined using the from the RT-PCR shown in Figure 6B. Approximately equal amounts of IFN-γ mRNA from the B10.S and the SJL/J alleles accumulated following stimulation, arguing that the single functional NeST gene in the heterozygous mice could stimulate transcription from the Ifng genes on both chromosomes.
NeST RNA binds to a subunit of the MLL/SET1 H3K4 methylase complex and increases chromatin modification at the Ifng locus
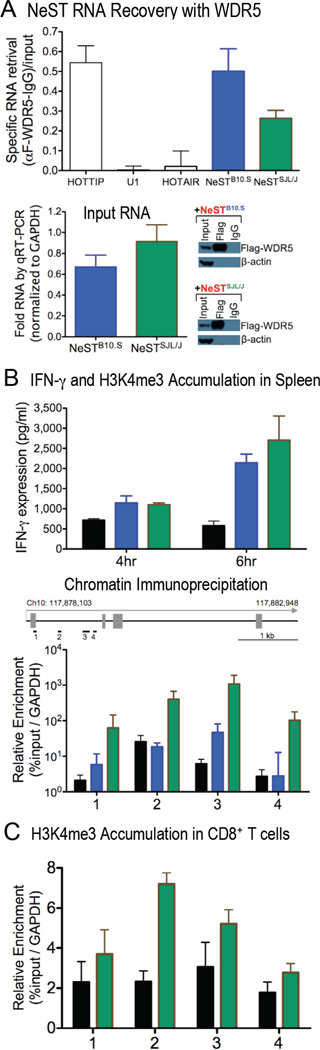
If NeST RNA were to influence the expression of IFN-γ directly, via chromatin modification, it should be an activating effect. Recently, a new class of enhancer-like lncRNAs was discovered (Orom et al., 2010; Wang et al., 2011). Among these, HOTTIP lncRNA has been found to bind WDR5 protein to recruit complexes that facilitate transcription (Wang et al., 2011). WDR5 is a core subunit of MLL1-4 and SET1A/1B complexes, which catalyze the methylation of histone H3 at lysine 4, a mark of active gene expression. To test whether NeST RNA physically interacts with WDR5, the epitope-tagged protein was co-expressed in combination with a variety of RNAs via transient transfection of 293T cells (Fig. 7A). Extracts were then prepared and WDR5 protein was immunoprecipitated and tested for associated RNAs by qRT-PCR. HOTTIP served as a positive control, and both HOTAIR lncRNA and U1 nuclear RNA served as negative controls. Immunoprecipitation of WDR5 specifically retrieved both NeST RNAs and HOTTIP, but not U1 or HOTAIR RNAs (Fig. 7A, B). The physical interaction between NeST and WDR5 raises the intriguing possibily that NeST may control H3K4 methylation at the Ifng locus.
Figure 7. NeST RNA physical association with WDR5 protein and effect on Histone 3 Lysine 4 trimethylation at the Ifng locus.
(A) RNA preparations from 293T cells that were co-transfected with FLAG-tagged WDR5 cDNA and either B10.S-derived NeST cDNA (blue) or SJL/J-derived NeST cDNA (sea green) were analyzed after immunoprecipitation with either anti-FLAG antibodies or anti-IgG control antibodies. NeST RNA retrieval was determined by measuring RNA input levels normalized to GAPDH (bottom left panel). Specific RNA retrieval was determined by subtracting NeST RNA retrieval with anti-IgG antibodies from the retrieval with anti-FLAG antibodies, followed by normalization to the amount of input RNA. Immunoblot analysis (bottom right panel) confirmed FLAG-WDR5 expression following transfection and the specificity of the anti-FLAG and anti-IgG antibodies. (B) IFN-γ production and H3K4me3 occupancy in spleen following immune challenge. B10.S, B10.S.NeSTB10.S and B10.S.NeSTSJL/J mice were injected intraperitoneally with 50 µg of lipopoylsaccharide (LPS); four and six hours later spleens were dissected. The abundance of IFN-γ protein was determined by ELISA in tissue homogenates (top panel) and the occupancy of Histone 3 Lysine 4 trimethylation at the Ifng gene was determined by ChIP-qPCR analysis (bottom panel). A schematic diagram of the positions of primers used for H3K4me3 is shown. Specific DNA retrieval was measured by normalization to the amount of input DNA and GAPDH. (C) ChIP-qPCR analysis of H3K4me3 at the Ifng locus in CD8+ T cells from B10.S and B10.S.NeSTSJL/J transgenic mice. CD8+ T cells were isolated from four B10.S and four B10.S.NeSTSJL/J mice and stimulated ex vivo with PMA and ionomycin. Occupancy of H3K4me3 at the Ifng gene was assayed 24 hours after stimulation by ChIP-qPCR at four different regions. For all pooled data, means and standard error are shown.
To examine the contribution of NeST RNA to IFN-γ production during immune challenge, we used a well-characterized mouse model of sepsis, the intraperitoneal injection of lipopolysaccharide (LPS). B10.S as well as B10.S.NeSTB10.S and B10.S.NeSTSJL/J transgenic mice were injected with LPS. By six hours post-injection, the presence of either transgenic NeST allele increased the amount of IFN-γ in splenic tissue compared to the B10.S control (Fig. 7B). An increase in H3K4me3 occupancy at the Ifng locus preceded this increased IFN-γ synthesis by two hours (FIg. 7B). Transgenic mice with the SJL/J-derived allele, which accumulate much more NeST RNA than those that express the B10.S allele (Fig. 3B), showed a larger amount of IFN-γ-encoding DNA with H3K4me3 modifications (Fig. 7B). Thus, increased NeST RNA abundance can result in more extensive H3K4me3 modification. Still, NeST RNA is extremely potent even at low abundance, either because the epigenic effects persist in its absence or because activation of only a subset of cells is necessary for the observed phenotypes.
The high occupancy of H3K4me3 at the Ifng locus in the B10.S.NeSTSJL/J transgenic mice allowed us to measure chromatin modification in isolated primary CD8+ cells in the presence and absence of NeST RNA. Following activation of B10.S- and B10.S.NeSTSJL/J-derived CD8+ T cells, we found that the presence of NeSTSJL/J RNA caused an increase in H3K4me3 at the Ifng locus (Fig. 7C). The NeST RNA-dependent increase in H3K4me3 activation in both total splenic cells and CD8+ T cells strongly suggests that, via binding to WDR5, NeST RNA is required to program an active chromatin state that confers inducibility to the Ifng gene.
Discussion
We have performed genetic analysis of a lncRNA texpressed in T cells. Mice that express NeST RNA, either in its natural chromosomal environment or by transgenic delivery, displayed increased resistance to Salmonella-induced pathogenesis but increased susceptibility to Theiler’s virus persistence. These disparate effects illustrate the role of balanced polymorphisms in susceptibility to infectious disease (Dean et al., 2002; Liu et al., 1996; Williams et al., 2005; Wang et al., 2010; Cagliani et al., 2011). Genes of the immune system are under purifying selection by challenges from a plethora of pathogens, and mutations that protect against one microbe may increase susceptibility to another. In the case of autoimmunity, the rs2076530-G allele of BTNL2, an MHC II-linked gene, confers increased susceptibility to rheumatoid arthritis and type 1 diabetes but decreased susceptibility to multiple sclerosis and autoimmune thyroiditis (Orozco et al., 2005; Sirota et al., 2009; Valentonyte et al., 2005).
A potential immunological mechanism for the disparate effects of NeST RNA on Theiler’s virus persistence and Salmonella pathogenesis might be that it alters the magnitude or the timing of inflammatory responses. CD8+ T cell populations are extremely heterogeneous (Davila et al., 2005; Joosten et al., 2007; Xystrakis et al., 2004); for example, the CD8+ Treg population is important in resolving inflammation and preventing autoimmunity (Frisullo et al., 2010; Sun et al., 2009; Trandem et al., 2011). Alternatively, NeST-dependent activaton of basal inflammation could serve to attenuate subsequent inflammatory events. Finally, NeST RNA may have targets in addition to the Ifng gene that contribute to its apparently anti-inflammatory effect (Fig. S2).
That the effects of NeST can be conferred by transgenic expression from ectopic loci, and to Ifng alleles on both chromosomes when NeST is expressed heterozygously, argue that NeST function, even on the adjacent IFN-γ-encoding locus, can be provided in trans. Although many lncRNAs, such as Xist and HOTTIP, exert their function on neighboring genes exclusively in cis, transacting lncRNA function has precedent in HOTAIR, linc-p21, and Jpx long noncoding RNAs (reviewed in Guttman and Rinn, 2012). Notably, Jpx is required to activate the expression of the adjacent Xist gene on the presumptive inactive X chromosome: this activation can occur whether Jpx RNA is supplied in cis or trans (Tian et al., Cell, 2010). Thus, there is increasing recognition in the field that lncRNA regulation of nearby genes can occur by trans-acting mechanisms; the increased demands made on these lncRNAs for target specificity are currently under investigation.
In the vicinity of the Ifng locus, many of the distal regulatory elements map to regions now known to encode NeST (Sekimata et al., 2009). For example, acetylation of histone 4 (H4Ac), a mark of active transcription, has been observed in discrete regions surrounding Ifng in activated CD4+ and CD8+ T cells. One peak in particular, which correlates well with the differentiation of both CD8+ and CD4+ T cells (Chang and Aune, 2005; Zhou et al., 2004), is located 59 Kb downstream of Ifng and coincides with the sixth exon of NeST (Fig. 1). Another noteworthy region critical for IFN-γ expression in CD4+ T cells maps 66 Kb downstream of murine Ifng and, in humans, 166 Kb downstream of IFNG. This regulatory element is also located in the NeST gene. It contains a lineage-specific DNase I hypersensitive site found in Th1 but not Th2 CD4+ T cells (Balasubramani et al., 2010). During lineage-specific induction of IFN-γ, proteins CTCF, T-bet and cohesin all localize to these sequences. Indeed, a recent study (Collier et al., 2012) has related the expression of NeST RNA (Tmevpg1) to the expression of IFN- γ in CD4+ T cells by a mechanism dependent on the simultaneous expression of T-bet. Simultaneous binding of cohesin, T-bet and CTCF results in a complex three-dimensional conformation predicted to bring the NeST and IFN- γ coding regions in direct proximity (Hadjur et al., 2009; Ong and Corces, 2011; Sekimata et al., 2009).
Humans express an RNA species homologous to NeST that also appears to be noncoding and is transcribed adjacent to the IFNG locus from the opposite DNA strand. Interestingly, polymorphisms that correlate with autoimmune and inflammatory diseases such as rheumatoid arthritis, Crohn’s disease and multiple sclerosis have been found in the DNA sequence that encodes the first intron of the IFN-γ-encoding gene (Goris et al., 2002; Latiano et al., 2011; Silverberg et al., 2009); this DNA region also encodes the fifth intron of the overlapping NeST RNA-encoding gene. As they do in mice, variations in human NeST RNA expression could contribute to differences in T cell response and disease susceptibility. Whether and how disease-associated SNPs alter human NeST expression or function will be addressed in future studies.
Natural polymorphisms, both in humans and in animal models, can yield subtle quantitative allelic effects that are more difficult to study, but more relevant to human medicine, than the effects of gene knock-out or other loss-of-function genetic techniques. The discovery of NeST RNA was the result of classical forward genetics. Our analysis of NeST RNA was based on the conceptual framework that regions thought to be “intergenic” encode functional RNA elements. Many genome-wide association studies have also pointed to intergenic regions as heritable causes of human disease (Libioulle et al., 2007; Sotelo et al., 2010). This study sets a precedent that some of these intergenic regions may encode functional lncRNAs that are critical for proper gene regulation. The promise of individualized medicine relies on our understanding of as many genetic polymorphisms as possible in the context of individual immunological and other environmental experiences.
Materials and Methods
Viral and bacterial strains and culture
The DA strain of Theiler’s virus was produced by transfecting the PTM762 plasmid into BHK-21 cells as previously described (McAllister et al., 1989). Salmonella enterica serovar Typhimurium SL1344 (Subbaiah and Stocker, 1964) strain was used. See supplemental info for Inoculation datails.
Mouse strains and transgenic line design
Congenic B10.S.Tmevp3SJL/J and SJL/J.Tmevp3B10.S mice were imported from the Pasteur Institute. B10.S and SJL/J mice were obtained from the Jackson Laboratory (Bar Harbor, ME). (Balb/c×129) F1 pseudopregnant female mice were provided by Drs. Hugh McDevitt and Grete Sonderstrup (Stanford, CA). All mice were bred at the Stanford Research Animal Facility (RAF) except for SJLJ/J mice, which were purchased at 4 weeks old and housed in the Stanford RAF for at least 6 weeks prior to all experiments.
B10.S mice that express NeST RNA transgenically were developed by pronuclear microinjections. NeST cDNA was cloned downstream of a CD4+ and CD8+ T cell-specific promoter and upstream of an SV40 polyadenylation signal (Sawada et al., 1994). Estrous cycle determination, recovery of single cell embryos, microinjection procedures and transfer to pseudopregnant females has been previously described (Singer et al., 1998).
Cell culture, infection and stimulation
Macrophage culture and infection has been previously described (Arpaia et al., 2011; Martinat et al., 2002). For T cell culture ex vivo, splenocytes were prepared and CD3+ T cell, CD4+ T cell or CD8+ T isolated using kits from Miltenyi Biotec (Auburn, CA). Nuclei were enriched as previously described (Huarte et al., 2010). For T-cell stimulation assays, cells were cultured for 10 hours prior to stimulation with 50 ng/ml phorbol 12-myristate 13-acetate and 1.5 µM ionomycin (Sigma-Aldrich).
WDR5 and chromatin immunoprecipitation
NeSTB10.S, NeSTSJL/J, HOTTIP, HOTAIR, and U1-encoding cDNAs were cloned into a eukaryotic gene expression plasmid and co-transfected with pcDNA3.1 that did or did not express FlagWDR5. Cells were lysed and immunoprecipitated as previously described, with modifications (Wang et al., 2011). Chromatin IP and quantitative PCR was carried out following the Farnham protocol (O’Geen et al., 2011).
RNA and cytokine quantitation
Protein quantitation was performed using commercially available ELISA kits (R&D Systems, Inc., Minneapolis MN) or Luminex (Affymetrix, Inc,. Santa Clara CA) according to manufacturer’s instructions. Quantitative RT-PCR was performed with total RNA from cells or tissues of interest and serial dilutions of known quantities of RNA. The three B10.S and 19 SJL/J polymorphisms (Supplemental Table 1) were introduced by site-directed mutagenesis.
Statistical Analysis
Mean values and significance were determined using student’s t-test. Survival curves were analyzed with the logrank test. The null hypothesis, that the strains compared were not different, was rejected when p values were less than or equal to 0.05. Instances when the observed differences could be reported with a confidence of 95% (*), 99% (**) or 99.9% (***) are denoted.
Supplementary Material
Highlights.
Dissection of viral susceptibility locus reveals role of long noncoding RNA, NeST
NeST RNA expression is required for inducible IFN-γ synthesis in CD8+ T cells
NeST RNA increases Theiler’s virus persistence and reduces Salmonella lethality
NeST binds to histone modification complex, changing epigenetic marks at Ifng locus
Acknowledgements
We thank Holden Maecker, Roberto Mateo and Peter Sarnow for comments on the manuscript. We are greatful to Nigel Killeen for provision of plasmids used for transgenic experiments, to Mary Vadeboncoeur and Grete Sonderstrup for their transgenics expertise, to Shyamali Roy for Salmonella titering, and to the Human Immune Monitoring Core for Luminex assays. Individuals involved in this work were supported by scholarships from the Gates Foundation and the Stanford University DARE program (J.A.G.), from the Stanford Medical Science Training Program (Y.W.Y) and from an NIH Training Grant (S.G). H.Y.C. is an Early Career Scientist of Howard Hughes Medical Institute. Research funding was provided by NIH grants (D.M.M., H.Y.C.), from the National Society for Multiple Sclerosis (K.K. and M.B.) and an NIH Director’s Pioneer Award (K.K.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Additional methods are included in Supplemental Information
References
- Arpaia N, Godec J, Lau L, Sivick KE, McLaughlin LM, Jones MB, Dracheva T, Peterson SN, Monack DM, Barton GM. TLR signaling is required for Salmonella typhimurium virulence. Cell. 2011;144:675–688. doi: 10.1016/j.cell.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubagnac S, Brahic M, Bureau JF. Bone marrow chimeras reveal non-H-2 hematopoietic control of susceptibility to Theiler's virus persistent infection. J Virol. 2002;76:5807–5812. doi: 10.1128/JVI.76.11.5807-5812.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramani A, Mukasa R, Hatton RD, Weaver CT. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol Rev. 2010;238:216–232. doi: 10.1111/j.1600-065X.2010.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihl F, Brahic M, Bureau JF. Two loci, Tmevp2 and Tmevp3, located on the telomeric region of chromosome 10, control the persistence of Theiler's virus in the central nervous system of mice. Genetics. 1999;152:385–392. doi: 10.1093/genetics/152.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahic M, Bureau JF, Michiels T. The genetics of the persistent infection and demyelinating disease caused by Theiler's virus. Annu Rev Microbiol. 2005;59:279–298. doi: 10.1146/annurev.micro.59.030804.121242. [DOI] [PubMed] [Google Scholar]
- Broz P, Newton K, Lamkanfi M, Mariathasan S, Dixit VM, Monack DM. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J Exp Med. 2010;207:1745–1755. doi: 10.1084/jem.20100257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau JF, Montagutelli X, Bihl F, Lefebvre S, Guenet JL, Brahic M. Mapping loci influencing the persistence of Theiler's virus in the murine central nervous system. Nat Genet. 1993;5:87–91. doi: 10.1038/ng0993-87. [DOI] [PubMed] [Google Scholar]
- Bureau JF, Montagutelli X, Lefebvre S, Guenet JL, Pla M, Brahic M. The interaction of two groups of murine genes determines the persistence of Theiler's virus in the central nervous system. J Virol. 1992;66:4698–4704. doi: 10.1128/jvi.66.8.4698-4704.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R, Riva S, Pozzoli U, Fumagalli M, Comi GP, Bresolin N, Clerici M, Sironi M. Balancing selection is common in the extended MHC region but most alleles with opposite risk profile for autoimmune diseases are neutrally evolving. BMC Evol Biol. 2011;11:171. doi: 10.1186/1471-2148-11-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Mandal MN, Chavali VR, Hawes NL, Khan NW, Hurd RE, Smith RS, Davisson ML, Kopplin L, Klein BE, et al. Age-related retinal degeneration (arrd2) in a novel mouse model due to a nonsense mutation in the Mdm1 gene. Hum Mol Genet. 2008;17:3929–3941. doi: 10.1093/hmg/ddn295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Aune TM. Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci U S A. 2005;102:17095–17100. doi: 10.1073/pnas.0502129102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier SP, Collins PL, Williams CL, Boothby MR, Aune TM. Cutting edge: influence of Tmevpg1, a long intergenic noncoding RNA, on the expression of Ifng by T cells. J. Immunol. 2012;189:2084–2088. doi: 10.4049/jimmunol.1200774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila E, Kang YM, Park YW, Sawai H, He X, Pryshchep S, Goronzy JJ, Weyand CM. Cell-based immunotherapy with suppressor CD8+ T cells in rheumatoid arthritis. J Immunol. 2005;174:7292–7301. doi: 10.4049/jimmunol.174.11.7292. [DOI] [PubMed] [Google Scholar]
- Dean M, Carrington M, O'Brien SJ. Balanced polymorphism selected by genetic versus infectious human disease. Annu Rev Genomics Hum Genet. 2002;3:263–292. doi: 10.1146/annurev.genom.3.022502.103149. [DOI] [PubMed] [Google Scholar]
- Foster N, Hulme SD, Barrow PA. Inhibition of IFN-gamma-stimulated proinflammatory cytokines by vasoactive intestinal peptide (VIP) correlates with increased survival of Salmonella enterica serovar typhimurium phoP in murine macrophages. J Interferon Cytokine Res. 2005;25:31–42. doi: 10.1089/jir.2005.25.31. [DOI] [PubMed] [Google Scholar]
- Frehel C, Canonne-Hergaux F, Gros P, De Chastellier C. Effect of Nramp1 on bacterial replication and on maturation of Mycobacterium avium-containing phagosomes in bone marrow-derived mouse macrophages. Cell Microbiol. 2002;4:541–556. doi: 10.1046/j.1462-5822.2002.00213.x. [DOI] [PubMed] [Google Scholar]
- Frisullo G, Nociti V, Iorio R, Plantone D, Patanella AK, Tonali PA, Batocchi AP. CD8(+)Foxp3(+) T cells in peripheral blood of relapsing-remitting multiple sclerosis patients. Hum Immunol. 2010;71:437–441. doi: 10.1016/j.humimm.2010.01.024. [DOI] [PubMed] [Google Scholar]
- Goris A, Heggarty S, Marrosu MG, Graham C, Billiau A, Vandenbroeck K. Linkage disequilibrium analysis of chromosome 12q14-15 in multiple sclerosis: delineation of a 118-kb interval around interferon-gamma (IFNG) that is involved in male versus female differential susceptibility. Genes Immun. 2002;3:470–476. doi: 10.1038/sj.gene.6363913. [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Yuan B, Flygare J, Lodish HF. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev. 25:2573–2578. doi: 10.1101/gad.178780.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten SA, van Meijgaarden KE, Savage ND, de Boer T, Triebel F, van der Wal A, de Heer E, Klein MR, Geluk A, Ottenhoff TH. Identification of a human CD8+ regulatory T cell subset that mediates suppression through the chemokine CC chemokine ligand 4. Proc Natl Acad Sci U S A. 2007;104:8029–8034. doi: 10.1073/pnas.0702257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latiano A, Palmieri O, Latiano T, Corritore G, Bossa F, Martino G, Biscaglia G, Scimeca D, Valvano MR, Pastore M, et al. Investigation of multiple susceptibility loci for inflammatory bowel disease in an Italian cohort of patients. PLoS One. 2011;6:e22688. doi: 10.1371/journal.pone.0022688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Levillayer F, Mas M, Levi-Acobas F, Brahic M, Bureau JF. Interleukin 22 is a candidate gene for Tmevp3, a locus controlling Theiler's virus-induced neurological diseases. Genetics. 2007;176:1835–1844. doi: 10.1534/genetics.107.073536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;2:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Martinat C, Mena I, Brahic M. Theiler's virus infection of primary cultures of bone marrow-derived monocytes/macrophages. J Virol. 2002;76:12823–12833. doi: 10.1128/JVI.76.24.12823-12833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister A, Tangy F, Aubert C, Brahic M. Molecular cloning of the complete genome of Theiler's virus, strain DA production of infectious transcripts. Microb Pathog. 1989;7:381–388. doi: 10.1016/0882-4010(89)90041-7. [DOI] [PubMed] [Google Scholar]
- Miao EA, Rajan JV. Salmonella and Caspase-1: A complex Interplay of Detection and Evasion. Front Microbiol. 2011;2:85. doi: 10.3389/fmicb.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med. 2004;199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- O'Geen H, Echipare L, Farnham PJ. Using ChIP-seq technology to generate high-resolution profiles of histone modifications. Methods Mol Biol. 2011;791:265–86. doi: 10.1007/978-1-61779-316-5_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco G, Eerligh P, Sanchez E, Zhernakova S, Roep BO, Gonzalez-Gay MA, Lopez-Nevot MA, Callejas JL, Hidalgo C, Pascual-Salcedo D, et al. Analysis of a functional BTNL2 polymorphism in type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus. Hum Immunol. 2005;66:1235–1241. doi: 10.1016/j.humimm.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Pena-Rossi C, Delcroix M, Huitinga I, McAllister A, van Rooijen N, Claassen E, Brahic M. Role of macrophages during Theiler's virus infection. J Virol. 1997;71:3336–3340. doi: 10.1128/jvi.71.4.3336-3340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira MS, Marques GG, Dellama JE, Zamboni DS. The Nlrc4 Inflammasome Contributes to Restriction of Pulmonary Infection by Flagellated Legionella spp. that Trigger Pyroptosis. Front Microbiol. 2011;2:33. doi: 10.3389/fmicb.2011.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M, Zoecklein LJ, Howe CL, Pavelko KD, Gamez JD, Nakane S, Papke LM. Gamma interferon is critical for neuronal viral clearance and protection in a susceptible mouse strain following early intracranial Theiler's murine encephalomyelitis virus infection. J Virol. 2003;77:12252–12265. doi: 10.1128/JVI.77.22.12252-12265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-gamma locus. Immunity. 2009;31:551–564. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, et al. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–220. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SM, Tisch R, Yang XD, Sytwu HK, Liblau R, McDevitt HO. Prevention of diabetes in NOD mice by a mutated I-Ab transgene. Diabetes. 1998;47:1570–1577. doi: 10.2337/diabetes.47.10.1570. [DOI] [PubMed] [Google Scholar]
- Sirota M, Schaub MA, Batzoglou S, Robinson WH, Butte AJ. Autoimmune disease classification by inverse association with SNP alleles. PLoS Genet. 2009;5:e1000792. doi: 10.1371/journal.pgen.1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, Stephens RM, Harris TJ, Munroe DJ, Wu X. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- Subbaiah TV, Stocker BA. Rough Mutants of Salmonella Typhimurium. I. Genetics. Nature. 1964;201:1298–1299. doi: 10.1038/2011298a0. [DOI] [PubMed] [Google Scholar]
- Sun J, Madan R, Karp CL, Braciale TJ. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med. 2009;15:277–284. doi: 10.1038/nm.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Sun S, Lee JT. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell. 2010;143:390–403. doi: 10.1016/j.cell.2010.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trandem K, Zhao J, Fleming E, Perlman S. Highly activated cytotoxic CD8 T cells express protective IL-10 at the peak of coronavirus-induced encephalitis. J Immunol. 2011;186:3642–3652. doi: 10.4049/jimmunol.1003292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentonyte R, Hampe J, Huse K, Rosenstiel P, Albrecht M, Stenzel A, Nagy M, Gaede KI, Franke A, Haesler R, et al. Sarcoidosis is associated with a truncating splice site mutation in BTNL2. Nat Genet. 2005;37:357–364. doi: 10.1038/ng1519. [DOI] [PubMed] [Google Scholar]
- Vigneau S, Levillayer F, Crespeau H, Cattolico L, Caudron B, Bihl F, Robert C, Brahic M, Weissenbach J, Bureau JF. Homology between a 173-kb region from mouse chromosome 10, telomeric to the Ifng locus, and human chromosome 12q15. Genomics. 2001;78:206–213. doi: 10.1006/geno.2001.6656. [DOI] [PubMed] [Google Scholar]
- Vigneau S, Rohrlich PS, Brahic M, Bureau JF. Tmevpg1, a candidate gene for the control of Theiler's virus persistence, could be implicated in the regulation of gamma interferon. J Virol. 2003;77:5632–5638. doi: 10.1128/JVI.77.10.5632-5638.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Baldassano R, Zhang H, Qu HQ, Imielinski M, Kugathasan S, Annese V, Dubinsky M, Rotter JI, Russell RK, et al. Comparative genetic analysis of inflammatory bowel disease and type 1 diabetes implicates multiple loci with opposite effects. Hum Mol Genet. 2010;19:2059–2067. doi: 10.1093/hmg/ddq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TN, Mwangi TW, Wambua S, Peto TE, Weatherall DJ, Gupta S, Recker M, Penman BS, Uyoga S, Macharia A, et al. Negative epistasis between the malaria-protective effects of alpha+-thalassemia and the sickle cell trait. Nat Genet. 2005;37:1253–1257. doi: 10.1038/ng1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xystrakis E, Dejean AS, Bernard I, Druet P, Liblau R, Gonzalez-Dunia D, Saoudi A. Identification of a novel natural regulatory CD8 T-cell subset and analysis of its mechanism of regulation. Blood. 2004;104:3294–3301. doi: 10.1182/blood-2004-03-1214. [DOI] [PubMed] [Google Scholar]
- Zhou W, Chang S, Aune TM. Long-range histone acetylation of the Ifng gene is an essential feature of T cell differentiation. Proc Natl Acad Sci U S A. 2004;101:2440–2445. doi: 10.1073/pnas.0306002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.