Abstract
The BIO2010 report challenged undergraduate institutions to prepare the next generation of researchers for the changing direction of biology that increasingly integrates advanced technologies, digital information, and large-scale analyses. In response, the Microbiology and Cell Science Department at the University of Florida developed a research-based course, “Bacterial Genome Sequencing.” The objectives were to teach undergraduates about genomics and original research by sequencing a bacterial genome, to develop scientific communication skills by writing and submitting the project results as a class effort, and to promote an interest in biological research, particularly genomics. The students worked together to sequence, assemble, and annotate the Enterobacter cloacae P101 genome. We assessed student learning, scientific communication skills, and student attitudes by a variety of methods including exams, writing assignments, oral presentations, pre- and postcourse surveys, and a final exit survey. Assessment results demonstrate student learning gains and positive attitudes regarding the course.
Since the sequencing of Haemophilus influenzae in 1995, whole genome analysis of microbes has expanded and is soon expected to reach 1,000 genomes (11, 25). To date, the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/genomes/lproks.cgi?view=1) features more than 500 complete microbial genomes. Genomics has enriched the understanding of microbial metabolism, diversity, structure, evolution, and potential of microbes as natural resources (34). Probing the microbial genome can lead to exciting applications like the rapid detection of pathogens in clinical settings, development of biofuels, environmental monitoring, protection from and detection of biological weapons, and the safe elimination of toxic waste, to name a few. The path to these discoveries begins with sequencing the genome. Whole genome sequencing is an example of the way biological research has shifted towards an integrative, data-rich strategy highlighted by acquisition and analysis of large datasets. Whole genome sequencing provides an excellent opportunity for undergraduates to participate in original research and receive training in creating and analyzing large datasets. Despite the prevalence of genomic sequencing in current research, however, few undergraduates learn about genomics let alone attain the necessary first-hand research experience so essential to their success as future researchers.
Recent reports and papers have made strong appeals and recommendations for a renovation of undergraduate biological education (7, 8, 16, 21, 28). In particular, the BIO2010 panel challenges institutions to revamp their undergraduate curricula by developing new courses that include discussion of cutting-edge fields and highlight the interdisciplinary nature of biological research—specifically recommending that students be familiar and adept at using computers in biological research (7). The panel also recommends that lab courses be rooted in real research problems that will capture the interest and motivation of students and engage them in a process of learning through scientific inquiry.
Active-learning exercises in the undergraduate science classroom enhance learning, increase student confidence and satisfaction, and generate enthusiasm as compared to more traditional, lecture-based approaches (22, 23, 29, 31, 35). In addition to engaging students in their own learning, educators need to activate the sense of inquiry that drives scientists by facilitating original undergraduate research in and out of the classroom (17). Several recent publications describe increased learning, critical thinking skills, interest, and enthusiasm when original research and/or inquiry-based activities were incorporated into the undergraduate biology classroom (1, 2, 10, 20).
Several institutions have established genomics education modules and courses to teach undergraduates about high-throughput data-rich fields that integrate informatics with biology (3, 9, 18, 19). There are impressive examples in the literature of modules and courses that integrate online data and web-based bioinformatic tools; original sequencing or functional genomics projects with the fly, HIV mutant bioinformatics, phylogenetic analysis with 16S rRNA sequences, and comparative genomic exercises (1, 4, 6, 12, 30, 32). Brad Goodner at Hiram College has made exceptional strides in integrating microbial genomics in undergraduate research and education by incorporating whole genome sequencing, comparative analysis, functional genomics, and annotation in undergraduate courses (13, 15).
With the intent of improving the undergraduate microbiology curriculum at the University of Florida by integrating original research and active learning in the emerging field of genomics, we developed a course centered on the sequencing of a microbial genome. Our long-term goal is to prepare undergraduates, as future biological researchers, to understand and engage in high-throughput research. To reach this goal, we began by developing a new course, “Bacterial Genome Sequencing,” in which 17 students worked collaboratively to analyze the sequence of Enterobacter cloacae P101, an endophytic bacterial strain. We had three major course objectives: (i) to learn about genome sequencing, assembly, annotation, comparison, and common bioinformatic tools and to appreciate the scope and power of genomics from transcriptomics and proteomics; (ii) to develop skills in science communication by writing an original research paper of publication quality; and (iii) to promote and strengthen an interest in biological research—particularly in genomics. Here, we describe the content and structure of the course and share the results of the pre- and postcourse surveys that queried students on their skills and attitudes towards research, genomics, and scientific writing.
METHODS
Class structure. The course, “Bacterial Genome Sequencing,” was open to undergraduates who had completed a prerequisite microbiology course and early career graduate students who were enrolled in the Microbiology and Cell Science graduate program. Fourteen undergraduate and three graduate students were enrolled in the initial offering, and the gender composition was thirteen female and four male students. Although enrollment included graduate students, the course content was targeted towards the undergraduate level. With the exception of DNA isolation, the genomics research was computer-based using in silico tools, so the class met in the Microbiology and Cell Science department’s computer laboratory (Laboratory for Genomics Education). The course met twice weekly for 3 hours each session. Typically, the first hour was devoted to a semiformal and interactive lecture. The second 2 hours were committed to research.
Sequencing project. The Enterobacter cloacae P101 genomic DNA was isolated using the Bio-Rad Genomic DNA isolation kit (Bio-Rad, Hercules, CA). Whole genome sequencing is feasible within the framework of an undergraduate course because of the University of Florida’s recent acquisition of the Genome Sequencer 20 (454 Life Sciences, Branford, CT). The Genome Sequencer 20 is a massively parallel pyrosequencer that can sequence 40 million bases of a genome in less than 5 hours (24). Because of this genome sequencing technology, the majority of the whole genome analysis is computer-based and a very small number of traditional Sanger sequencing reactions are necessary.
The sequence reads were aligned by the sequencer’s software and assembled into 228 contigs ranging in size from 515 bp to 270,020 bp. Four hundred and fifty-six specific primers were designed from the two ends of each contig and used in direct genome sequencing and PCR to amplify the inter-contig regions for sequencing and genome closure. Throughout this course, bioinformatics tools that are web based, user friendly, and open source or very inexpensive were chosen whenever possible. The goal is to give students access to and expertise with tools that are easily accessible and applicable in their future research. For example, BASys (http://wishart.biology.ualberta.ca/basys) delivered an automated annotation of open reading frames (33). Students used tRNA scan-SE (http://selab.janelia.org/tRNAscan-SE/) to annotate tRNAs, while rRNAs were annotated manually by searching for conserved rRNA sequences. Two databases were used frequently to assist with the genome analysis: Kyoto Encyclopedia of Genes and Genomes (www.genome.jp/kegg/), which links genomic information to biological pathways, and Profiling of E. coli Chromosome (http://www.shigen.nig.ac.jp/ecoli/pec/index.jsp), which provides a resource genome for comparison. Students also used the homology tool BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?) and the comparative tools at the Comprehensive Microbial Resource from The Institute of Genomic Research (http://cmr.tigr.org). Typically, the final 2 hours of the course were dedicated to hands-on student research that was prefaced by an introduction to and demonstration of the aforementioned tools. The sequence data was maintained and updated on a project website hosted by the University of Florida’s Interdisciplinary Center for Biotechnology Research. Since the sequence data is not yet published, the project website is limited to class accessibility, but all students were given access to the website for data entry and analysis. Further details and results of the genome sequencing project will be published in a separate manuscript with the students and instructors listed as authors.
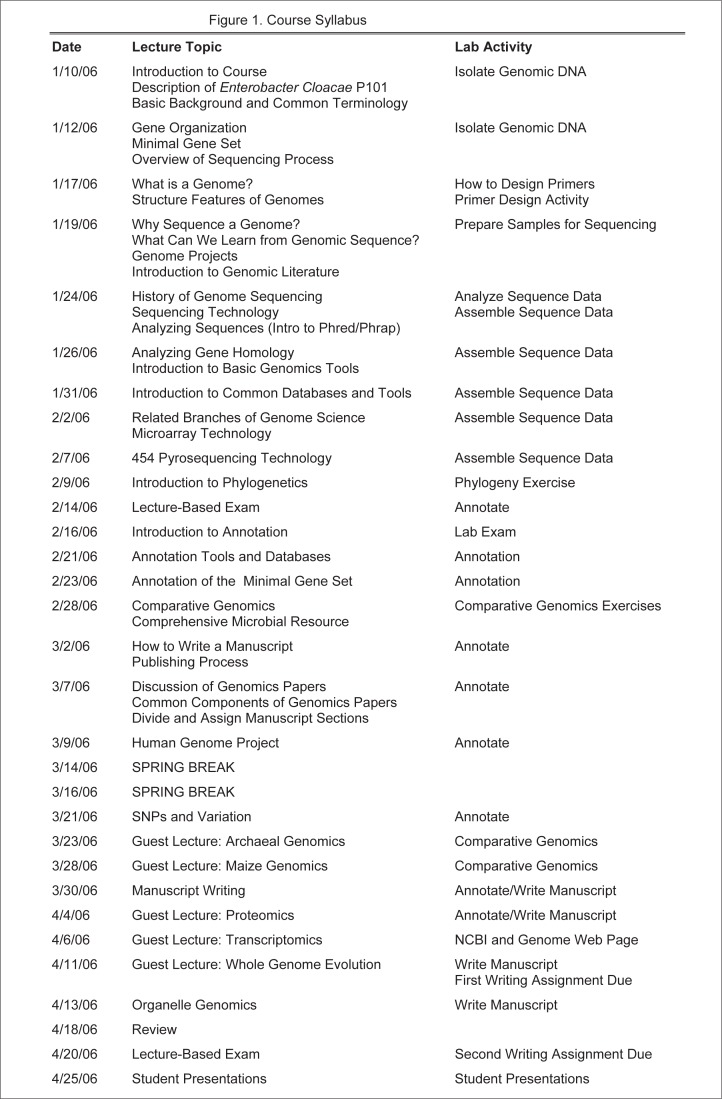
Course content and project organization. Biweekly lectures covered a variety of genomics related topics (Fig. 1). To emphasize the relevance of the class-wide sequencing project, the first day of lecture focused on the microorganism of study: Enterobacter cloacae P101. To date, no Enterobacter or endophyte genome has been sequenced, and unlocking the genetic information will lead to an increased understanding of how endophytes can provide benefits to their hosts. For example, plants inoculated with Enterobacter cloacae P101 demonstrate increased growth versus wild type plants (26, 27).
FIG. 1.
Course syllabus for “Bacterial Genome Sequencing.”
As there is no one textbook that fits the needs of this course, a variety of resources were used in preparing the course lectures and learning materials. Most of the information was drawn from the book A Primer of Genome Science(14) or gleaned from the primary literature and reviews to provide a strong connection to current and original research. Manuscript writing was a significant component of the pilot course. The class assembled a detailed outline of the manuscript and small groups each took responsibility for writing a section of the manuscript. Section drafts were exchanged for peer revision. The students completed the necessary literature searches and data analysis to make their section as complete as possible. Many of the manuscript sections were focused on different functional pathways in the Enterobacter genome such as virulence or chemotaxis genes. At the end of the semester, the students gave oral presentations summarizing the different sections and describing the results of different genetic pathways.
Course assessment. As this paper describes an entirely new course, the course evaluation strategy focused on measuring student learning gains in the subject matter, detecting increased skill and/or confidence in science writing and communication, and gauging overall student attitudes regarding this course and research in pre- and postcourse surveys. In addition to the surveys, students were given three exams, two lecture-based exams and one computer-based lab practical to assess learning, and two writing assignments and a final oral presentation to assess writing and communication skills.
The anonymous pre- and postcourse surveys asked students to use a Likert scale to indicate the degree to which they agreed with various statements (strongly agree = 5, agree somewhat = 4, neither agree nor disagree = 3, disagree somewhat = 2, strongly disagree = 1). The survey results were analyzed in two ways. First, the mean and standard deviation of the responses to each pre- and postcourse survey question were compared by the student’s t test. Although the averages presented an overview of student responses, analyzing the shifts along the Likert scale provided more information about the student’s responses. This was accomplished by comparing the proportion of students who selected a particular level of agreement in the precourse survey to the proportion of students who selected that same level of agreement in the postcourse survey and analyzing the results with Fisher’s exact test. The course assessment received approval from the University of Florida’s Institutional Review Board.
RESULTS
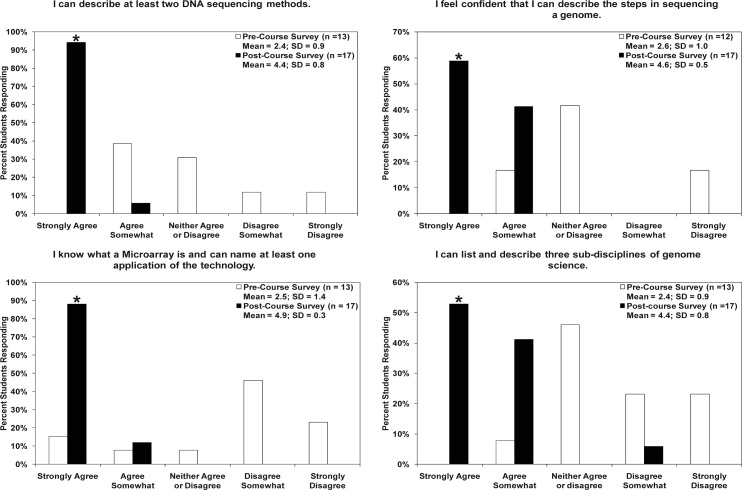
Student learning. As part of the survey, students were asked to describe two DNA sequencing methods, list the steps in sequencing a genome, give a definition and applications of a microarray, and indicate three subdisciplines of genome science (Fig. 2). For all four statements, students demonstrated a significant increase in genomics knowledge and skills on the postcourse survey versus the precourse survey. The means of the postcourse scores were all significantly higher than the precourse mean scores for all four questions (P value <0.05; student’s t test).
FIG. 2.
For all four statements, students demonstrated a significant increase in genomics knowledge and skills in the postcourse survey results versus the precourse survey. The precourse survey mean scores were significantly lower than the postcourse survey mean scores in all four categories (P <0.005) according to Student’s t test, two-sided analysis. The postcourse survey bar denoting the percentage of students who indicated that they “strongly agreed” with the above statements is significantly higher than the precourse results. In all four graphs, the P value is less than 0.003 according to Fisher’s exact test, two-sided analysis (denoted with an asterisk).
In addition to the pre- and postcourse surveys, student learning was assessed on two written exams and one lab-based practical. The exams contained a mix of multiple choice, true/false, short answer, and essay questions that were designed to cover concepts from the lectures and group discussions as well as determine the student’s proficiency in using in silico tools. Figure 3 displays an example of test questions and anonymous answers.
FIG. 3.
Sample questions and answers from course exams. For the first question shown, the average score earned was 5.8 out of 6 points total, with 76% of students earning full credit for their answer. For the second question, the average score was 4.8 out of 6 points with 60% of the students earning full credit for their answer.
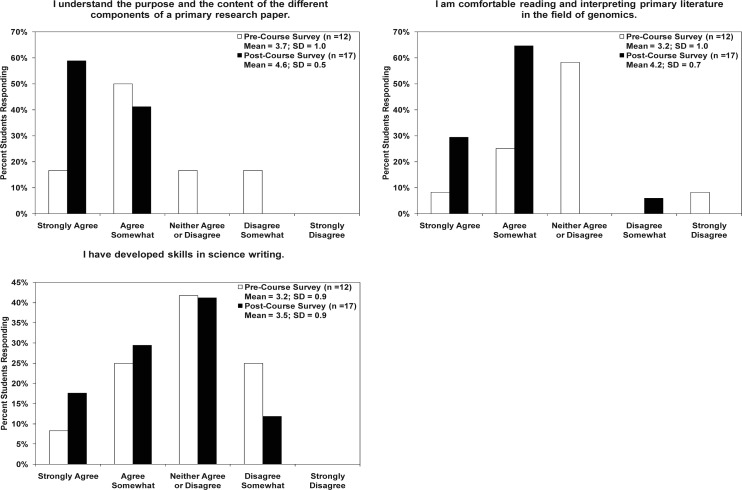
Scientific communication skills. A significant component of the course involved reading from the primary literature and collaboratively writing a publication-quality manuscript that described the genome sequencing project. We assessed the students’ abilities and comfort level with the primary literature and scientific writing (Fig. 4). The postcourse surveys indicated an increase in the understanding of the different sections of a primary paper (P = 0.003) with a significant shift in the “strongly agreed” category (59% versus 17%; P value = 0.05). Students also demonstrated a significant increase in their comfort with reading and interpreting genomic literature with a precourse mean of 3.2 versus a postcourse mean of 4.2 (P = 0.006). On the postcourse survey, 94% of the students “strongly agreed” or “agreed somewhat” that they felt comfortable reading and interpreting genomics literature as opposed to 33% in the precourse survey. Pre- and postcourse survey results demonstrate that students experienced only modest gains in science writing skills that were not statistically significant.
FIG. 4.
Assessment of student scientific reading and writing skills from course surveys. The postcourse survey results demonstrated a significant increase in the students’ ability to understand the sections of a primary research paper and to read and interpret genomic literature (P = 0.003 and 0.006, respectively). Based on the postcourse survey results, students experienced only modest gains in their science writing skills that were not statistically significant.
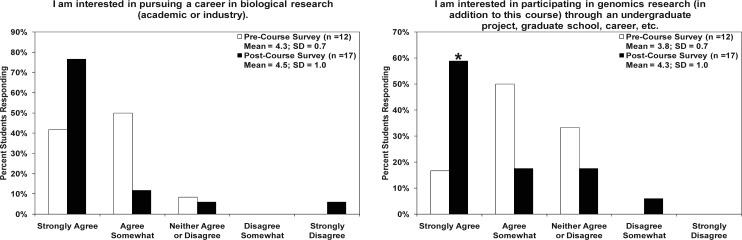
Student interest in research. Students were asked about their interest in pursuing a career in biological research and participating in future genomics research (Fig. 5). There were no statistical differences in the mean responses in the preand postcourse surveys when students were asked to agree with statements about their interest in pursuing future biological and genomic research. However, there was a marked trend towards an increase in the percentage of students who “strongly agreed” with an interest in a career in biological research and a statistically significant increase in those who “strongly agreed” with an interest in future genomics research (P values = 0.06 and 0.05, respectively).
FIG. 5.
Assessment of student interest in biological research and genomics research from course surveys. There were no significant differences in the mean responses in the pre and post-course surveys to both of the questions about student interest in research. However, when asked to agree with a statement about their interest in pursuing a career in biological research, there was a marked increase in the percentage of students who responded “Strongly agree” on the post-course survey versus the pre-course survey results (77% vs. 41%; P value = 0.06). In response to the statement regarding the student interest level in participating in future genomics research, there was also an increased level in the proportion of students who “strongly agreed” with the statement in the post-course survey as compared to the pre-course results (59% vs. 17%; P value = 0.05), denoted by an asterisk.
Final course survey—student attitudes towards the course. The students were asked in a final course evaluation to indicate the degree to which they agreed with various statements regarding their attitudes about the course (Table 1). Preparing and encouraging students for future research projects or careers was an important goal of the course, and 88% of students agreed (either strongly or somewhat) that they felt more prepared for future research projects. All of the students agreed that they learned how to use computer tools that will help them in their future research. Additionally, all of the students agreed that they gained an appreciation for the obstacles and challenges of research projects.
TABLE 1.
Final course survey resultsa
| Statement | Mean ± SD | Strongly agree | Agree somewhat | Neither agree nor disagree | Disagree somewhat | Strongly disagree |
|---|---|---|---|---|---|---|
| I learned how to use computer tools that will help me in my future research/career. | 4.5 ± 0.5 | 53 | 47 | 0 | 0 | 0 |
| I gained an appreciation for the obstacles or challenges of research projects. | 4.7 ± 0.5 | 71 | 29 | 0 | 0 | 0 |
| I feel more prepared for future research projects. | 4.5 ± 0.7 | 59 | 29 | 12 | 0 | 0 |
| I gained a sense of contributing to the genome field. | 3.9 ± 1.0 | 35 | 35 | 23 | 0 | 6 |
| I think that my previous courses provided adequate preparation for this course. | 3.5 ± 1.2 | 30 | 23 | 23 | 18 | 6 |
| I would recommend this course to someone else. | 4.2 ± 1.0 | 59 | 18 | 12 | 12 | 0 |
| I am satisfied with what I learned in this course. | 4.2 ± 1.1 | 53 | 35 | 0 | 12 | 0 |
| Overall, this course has been a positive experience for me. | 4.5 ± 1.0 | 77 | 12 | 6 | 6 | 0 |
Values are the percentage of seventeen students who indicated their level of agreement for each statement.
Over 70% of students agreed (35.3% strongly and 35.3% somewhat) that they gained a sense of contributing to the field of genomics, with another 23.5% harboring ambivalent feelings towards having contributed to the field. Student responses also varied considerably when asked if their previous courses prepared them for the genome sequencing course, with an average response score of 3.5 (SD = 1.2).
To ascertain their overall feelings towards the course, the postcourse evaluation asked students three questions: would they recommend the course to someone else, are they satisfied with what they learned, and has the course been a positive experience. Over 75% of the students agreed that they would recommend the course to someone else. Half of the students strongly agreed that they are satisfied with what they learned in the course; with another 35% agreeing somewhat that they are satisfied with the course. Finally, a majority of the students (76.5 %) “strongly agreed” that the course had been an overall positive experience.
DISCUSSION
One of the course goals was to teach students about the scope and power of genomics. Although the postcourse assessment demonstrates significant improvement and learning gains, the pre- and postcourse questions may not be the best predictor of learning since they are based upon student perception of ability. One way to correct for this is to use pre- and postcourse test questions that do not rely on student’s perceptions but on objective, scored responses to questions such as, “Please describe and compare two sequencing methods.”
In addition to written communication, small groups (two to three individuals) presented oral summaries of their contributions to the manuscript. The presentations were 15 to 20 minutes in length and varied widely in quality and content. Surprisingly, many students had never delivered a presentation in a science course or had no familiarity with presentation software such as PowerPoint. Despite their lack of experience, most of the students were enthusiastic about this assignment and carefully prepared their presentations. Typically, the students seemed more focused on the overall appearance and less focused on the content of the presentation. The decision to include an oral presentation was made midsemester and, although it was a useful component of the pilot course and will be included in future courses, there will be improvements. For example, the instructors will provide formative feedback to the students and provide clear evaluation criteria prior to the presentations.
As a group, the class developed a detailed manuscript outline. Each student chose a section of the manuscript to write. This activity is likely to have contributed significantly to their understanding of the components of the primary research paper. The lecture material was primarily drawn from the literature and students were expected to reference the primary literature within their assigned drafts. We plan to strengthen the writing aspect of the course by introducing the writing activities earlier in the semester and providing more frequent feedback on the students’ drafts. Several of the students did not understand the difference between primary and secondary literature and seemed unfamiliar with how and where to find research articles and how and when to cite papers within their drafts. We plan to devote more course time to teaching students how to search reference databases, where to find the papers, and how to cite appropriately. To date, the sequencing manuscript has not been submitted for publication and will require significant rewriting and editing from the instructors.
In offering a course that tightly integrates original research and learning, one of the objectives was to foster and encourage an interest in biological research, particularly in genomics. As the students self-selected by registering for this particular course, we anticipated a high degree of student interest in research at the start of the course, and indeed 50% of the students “agreed somewhat” with an interest in future biological research and an interest in genomics research specifically on the precourse survey. The postcourse survey indicated a considerable shift in the proportion of students who “strongly agreed” with those interests. These results are highly encouraging and underscore the importance of providing research opportunities to students at the undergraduate level to foster their interest in biological careers. Anecdotal evidence also supports these data: one student changed her career path from industry to graduate school after taking this course. In contrast, there was one student who “strongly disagreed” with an interest in pursuing biological research in the postcourse survey. Although this response was disappointing, one of seventeen students represents a small population. Since the surveys were anonymous, the identity of the student is not known; however, we suspect that this student may have marked the ambivalent category in the precourse survey and discovered that research did not match his/her personal interests.
As introducing students to common genomics analysis tools was an important goal for the course, it is very satisfying to report that all students agreed that the computer tools and skills will be helpful to them in their future research endeavors. Additionally, all of the students agreed that they gained an appreciation for the obstacles and challenges of research projects, which is not unexpected considering the difficulty with genome closure. Closing the gaps between the contigs in order to finish the genome was technically challenging but proved to be an excellent impetus for group discussions and strategizing. During the course, the students directed the closure efforts and divided into small groups of three to five individuals, with each group tackling a different closure strategy. To date, efforts continue to close the genome. However, to increase the genome sequencing efficiency, in the next course, we plan to sequence a smaller genome that is better suited to the sequencing technology, that will result in fewer contigs, and therefore, will require less time for closure. A smaller bacterial genome will also streamline the assembly and annotation process to allow course time for functional studies of selected genomes.
It is expected that not every student would agree that they felt more prepared for future research projects after taking this course. Students may not recognize how their skills in genomic sequencing may carry over to other research projects either directly through the use of specific tools or indirectly by improving their basic problem-solving skills. Interestingly, the same proportion of students (88%) who indicated an interest in pursuing future biological research also indicated on the postcourse survey that they felt more prepared for future projects.
A range of responses is understandable regarding the statement “I feel that I contributed towards a field of knowledge.” Since, as a class, the students did not submit a finished manuscript for publication by the end of the semester, many students may have felt that they did not contribute to the genomics field. However, we emphasized to the students that their work and time was valuable and would be incorporated and acknowledged through authorship when the manuscript was submitted.
The only course prerequisites were junior-level general microbiology and consent of the instructor. Although most of the students were juniors and seniors, they had a variety of academic backgrounds. The responses of students regarding their level of preparation from previous courses reflect the range of their backgrounds and experiences. In the written comments section of the course evaluation, some students specifically mentioned that they felt unprepared for this course because they had not taken genetics or organic chemistry. In the next course offering, we plan to spend more time at the beginning of the semester with a thorough review of molecular biology to ensure that the students are starting “on the same page” regardless of their varied academic backgrounds.
Overwhelmingly, the students reported positive attitudes toward the course. Written comments on evaluations revealed why a small proportion of students did not share the positive attitudes regarding the course. In many ways, this genomics sequencing course was a new experience for both the students and the instructors. Most students were excited to be a part of the research process (as indicated by written and personal comments). However, a few students were uncomfortable with the unscheduled and unanticipated directions that the research led us as a team. Overall, the course received high marks from students with regard to peer recommendation, course satisfaction, and a positive experience. These are indicators of a successful course, and, hopefully, foretell strong enrollment in a future genomics course as well as a sustained interest in genomics research.
The most significant concern regarding the expansion and future of the course is the cost of these genome projects. Is this course sustainable in the long term? Currently, the pyrosequencing technology generates 100 million bases per run with each run costing $7,000. However, the cost of sequencing continues to decline as the throughput (bases per run) increases (5). The era of the $1,000 human genome is fast approaching and with that will come affordable microbial genome research at the undergraduate level. It is our long-term goal to have a solidly constructed and well-assessed course in place to use this technology and these resources in the undergraduate classroom. By tightly integrating the research interests of the instructors into the course framework, original and timely research-based courses become more sustainable as they benefit the students and the instructors on multiple levels. In conclusion, the pilot course, “Bacterial Genome Sequencing,” provided a solid foundation to bring high-throughput research to the University of Florida undergraduate microbiology curriculum and produced publishable data (the manuscript is in preparation; the students and instructors will be listed as authors).
Acknowledgments
This research and educational effort was funded by grants from the National Science Foundation, MCB-0454030 and MCB-0412091. The authors acknowledge DNASTAR, Inc., (Madison, WI) for generously providing access to the Lasergene suite of sequence analysis tools for educational purposes.
REFERENCES
- 1.Boomer S, Dutton B. Bacterial diversity studies using the 16s rRNA gene provide a powerful research-based curriculum for molecular biology laboratory. Microbiol Educ. 2002;3:1–6. doi: 10.1128/me.3.1.18-25.2002. http://www.microbelibrary.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brodl MR. Tapping recent alumni for the development of cutting-edge, investigative teaching lab experiments. Bioscene: J. Coll. Biol. Teach. 2005;31:13–20. [Google Scholar]
- 3.Campbell AM. Meeting report: genomics in the undergraduate curriculum—rocket science or basic science. CBE Life Sci Educ. 2002;1:70–72. doi: 10.1187/cbe.02-06-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell AM. Public access for teaching genomics, proteomics, and bioinformatics. CBE Life Sci Educ. 2003;2:98–111. doi: 10.1187/cbe.03-02-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan EY. Advances in sequencing technology. Mutat Res. 2005;573:13–40. doi: 10.1016/j.mrfmmm.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Call GB, Beyer E, Bui C, Cespedes A, Chan A, Chan J, Chan S, Chhabra A, Dang P, Deravanesian A, Hermogeno B, Jen J, Kim E, Lee E, Lewis G, Marshall J, Regalia K, Shadpour F, Shemmassian A, Spivey K, Wells M, Wu J, Yamauchi Y, Yavari A, Abrams A, Abramson A, Amado L, Arson J, Bashour K, Bibikova E, Bookatz A, Brewer S, Buu N, Calvillo S, Cao J, Chang A, Chang D, Chang Y, Chen Y, Choi J, Chou J, Datta S, Davarifar A, Desai P, Fabrikant J, Farnad S, Fu K, Garcia E, Garrone N, Gasparyan S, Gayda P, Goffstein C, Gonzalez C, Guirguis M, Hassid R, Hong A, Hong J, Hovestreydt L, Hu C, Jamshidian F, Kahen K, Kao L, Kelley M, Kho T, Kim S, Kim Y, Kirkpatrick B, Kohan E, Kwak R, Langenbacher A, Laxamana S, Lee C, Lee J, Lee S, Lee T, Lee T, Lezcano S, Lin H, Lin P, Luu J, Luu T, Marrs W, Marsh E, Min S, Minasian T, Misra A, Morimoto M, Moshfegh Y, Murray J, Nguyen C, Nguyen K, Nodado E, O’Donahue A, Onugha N, Orjiakor N, Padhiar B, Pavel-Dinu M, Pavlenko A, Paz E, Phaklides S, Pham L, Poulose P, Powell R, Pusic A, Ramola D, Ribbens M, Rifai B, Rosselli D, Saakyan M, Saarikoski P, Segura M, Singh R, Singh V, Skinner E, Solomin D, Soneji K, Stageberg E, Stavchanskiy M, Tekchandani L, Thai L, Thiyanaratnam J, Tong M, Toor A, Tovar S, Trangsrud K, Tsang W, Uemura M, Unkovic M, Vollmer E, Weiss E, Wood D, Wu S, Wu W, Xu Q, Yackle K, Yarosh W, Yee L, Yen G, Alkin G, Go S, Huff D, Minye H, Paul E, Villarasa N, Milchanowski A, Banerjee U. Discovery-based science education: functional genomic dissection in Drosophila by undergraduate researchers. PLoS Biol. 2005;3(2):e59. doi: 10.1371/journal.pbio.0030059. http://biology.plosjournals.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Committee on Undergraduate Biology Education to Prepare Research Scientists for the 21st Century . BIO2010: transforming undergraduate education for future research biologists. National Research Council; Washington, DC: 2003. [Google Scholar]
- 8.Committee on Undergraduate Science Education . Science teaching reconsidered: a handbook. National Research Council; Washington, DC: 1997. [Google Scholar]
- 9.Dyer BD, LeBlanc MD. Meeting report: incorporating genomics research into undergraduate curricula. CBE Life Sci Educ. 2002;1:101–104. doi: 10.1187/cbe.02-07-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elwess NL, Latourelle SL. Inducing mutations in paramecium: an inquiry-based approach. Bioscene: J. Coll. Biol. Teach. 2004;30:25–35. [Google Scholar]
- 11.Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, McKenney K, Sutton GW, FitzHugh W, Fields C, Gocyne JD, Scott J, Shirley R, Liu L, Glodek A, Kelley JM, Weidman JF, Phillips CA, Spriggs T, Hedblom E, Cotton MD, Utterback TR, Hanna MC, Nguyen David T, Saudek DM, Brandon RC, Fine LD, Fritchman JL, Fuhrmann JL, Geoghagen NSM, Gnehm CL, McDonald LA, Small KV, Fraser CM, Smith HO, Venter JC. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Flowers SK, Easter C, Holmes A, Cohen B, Bednarski AE, Mardis ER, Wilson RK, Elgin SCR. Genome science: a video tour of the Washington University Genome Sequencing Center for high school and undergraduate students. CBE Life Sci Educ. 2005;4:291–296. doi: 10.1187/cbe.05-07-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forst S, Goodner B. Comparative bacterial genomics and its use in undergraduate education. Biol Control. 2006;38:47–53. doi: 10.1016/j.biocontrol.2005.11.006. [DOI] [Google Scholar]
- 14.Gibson G, Muse SV. A prime of genome science. Sinauer Associates Inc; Sunderland, MA: 2005. [Google Scholar]
- 15.Goodner B, Wheeler C. Functional genomics: using reverse genetics to test bioinformatics predictions. 2006. ASM MicrobeLibrary Curriculum Collection. http://www.microbelibrary.org.
- 16.Handelsman J, Ebert-May D, Beichner R, Bruns P, Chang A, DeHaan R, Gentile J, Lauffer S, Stewart J, Tilghman SM, Wood WB. Scientific teaching. Science. 2004;304:521–522. doi: 10.1126/science.1096022. [DOI] [PubMed] [Google Scholar]
- 17.Harwood WS. Course enhancement: a road map for devising active-learning and inquiry-based science courses. Int J Dev Biol. 2003;47:213–221. [PubMed] [Google Scholar]
- 18.Holtzclaw JD, Eisen A, Whitney EM, Penumetcha M, Hoey JJ, Kimbro KS. Incorporating a new bioinformatics component into genetics at a historically black college: outcomes and lessons. CBE Life Sci Educ. 2006;5:52–64. doi: 10.1187/cbe.05-04-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honts JE. Evolving strategies for the incorporation of bioinformatics within the undergraduate cell biology curriculum. CBE Life Sci Educ. 2003;2:233–245. doi: 10.1187/cbe.03-06-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard DR, Miskowski JA. Using a module-based laboratory to incorporate inquiry into a large cell biology course. CBE Life Sci Educ. 2005;4:249–260. doi: 10.1187/cbe.04-09-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenny RW, Boyer Commission on Educating Undergraduates in the Research University . Reinventing undergraduate education: a blueprint for America’s research universities. State University of New York—Stonybrook; Stonybrook, NY: 1998. [Google Scholar]
- 22.Klionsky DJ. Constructing knowledge in the lecture hall: a quiz-based group-learning approach to introductory biology. J Coll Sci Teach. 2002;31:246–251. [Google Scholar]
- 23.Malacinski GM. Student-oriented learning: an inquiry-based developmental biology lecture course. Int J Dev Biol. 2003;47:135–140. [PubMed] [Google Scholar]
- 24.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XY, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picoliter reactors. Nature. 2005;437:376–381. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Rückert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1,000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riggs PJ, Moritz RL, Chelius MK, Dong Y, Iniguez AL, Kaeppler SM, Casler MD, Triplett EW. Isolation and characterization of diazotrophic endophytes from grasses and their effects on plant growth. In: Finan TR, O’Brian MR, Layzell DB, Vessey JK, Newton WE, editors. Nitrogen fixation: global perspectives Proceedings of the 13th International Congress on Nitrogen Fixation. CABI; New York, NY: 2002. pp. 263–267. [Google Scholar]
- 27.Riggs PJ, Chelius MK, Iniguez AL, Kaeppler SM, Triplett EW. Enhanced maize productivity by inoculation with diazotrophic bacteria. Aust J Plant Physiol. 2001;28:829–836. [Google Scholar]
- 28.Rothman FG, Narum JL. Then, now, and in the next decade: a commentary on strengthening undergraduate science, mathematics, engineering and technology education. Project Kaleidoscope; Washington DC: 1999. [Google Scholar]
- 29.Smith AC, Stewart R, Shields P, Hayes-Klosteridis J, Robinson P, Yuan R. Introductory biology courses: a framework to support active learning in large enrollment introductory science courses. CBE Life Sci Educ. 2005;4:143–156. doi: 10.1187/cbe.04-08-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strong M, Cascio D, Eisenberg D. A web-based comparative genomics tutorial for investigating microbial genomes. Microbiol Educ. 2004;5:1–8. http://www.microbelibrary.org. [PMC free article] [PubMed] [Google Scholar]
- 31.Suchman E, Timpson W, Linch K, Ahermae S, Smith R. Student responses to active learning strategies in a large lecture introductory microbiology course. Bioscene: J. Coll. Bio. Teach. 2001;27:21–26. [Google Scholar]
- 32.Takayama K. Three-dimensional visualizations in teaching genomics and bioinformatics: mutations in HIV envelope proteins and their consequences for vaccine design. Microbiol Educ. 2004;5:1–11. doi: 10.1128/jmbe.v5.72. http://www.microbelibrary.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo AC, Dong XL, Lu P, Szafron D, Greiner R, Wishart DS. BASys a web server for automated bacterial genome annotation. Nucleic Acids Res. 2005;33(Suppl. 2):W455–W459. doi: 10.1093/nar/gki593. http://nar.oxfordjournals.org/archive/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward N, Fraser CM. How genomics has affected the concept of microbiology. Curr Opin Microbiol. 2005;8:564–571. doi: 10.1016/j.mib.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 35.Wyckoff S. Changing the culture of undergraduate science teaching. J Coll Sci Teach. 2001;29:409–414. [Google Scholar]