INTRODUCTION
Many student laboratory projects rely on the isolation, identification, and classification of bacteria from environmental or other sources. One major area of interest for classification of bacteria is the possession of virulence factors. Many bacteria utilize siderophores to help the process of ferric iron uptake in the environment. This process can be found across all three domains and is necessary for many microorganisms to obtain the environmental iron needed for essential processes. Siderophore production can also be a major factor in the ability of pathogens to cause disease because free iron is very limited, and often tightly bound to many proteins, such as hemoglobin, transferrin, and lactoferrin (6).
Siderophores are classified by the ligands used to chelate the ferric iron. These include the catecholates, hydroxamates, and carboxylates (6). Various assays have been developed to detect different phenotypes of siderophores (2–4). While these assays are useful for identifying various siderophores, numerous assays would have to be performed to detect all possible forms of siderophores. Schwyn and Neiland (8) developed a universal siderophore assay using chrome azurol S(CAS) and hexadecyltrimethylammonium bromide (HDTMA) as indicators. HDTMA has a moderate health and contact rating. Appropriate personal protective equipment should be used. The CAS/HDTMA complexes tightly with ferric iron to produce a blue color. When a strong iron chelator such as a siderophore removes iron from the dye complex, the color changes from blue to orange.
In Schwyn and Neiland’s original paper, the procedure given for making CAS agar is written in general terms and can be difficult to follow, especially for an individual who has limited experience making more complex media. Here, we give a step-by-step protocol for making the CAS agar plates, and we discuss how this media can be incorporated in a comprehensive project in a microbiology lab course for biology majors.
PROCEDURE
The following is a detailed, step-by-step procedure taken from Schwyn and Neilands (8). Preparing the CAS agar can be a tedious and difficult process. By providing a detailed protocol for doing this, we hope to reduce the difficulty.
Clean all glassware with 6M HCl to remove any trace elements, then rinse with ddH2O.
- Blue Dye:
- Solution 1:
- Dissolve 0.06 g of CAS (Fluka Chemicals) in 50 ml of ddH2O.
- Solution 2:
- Dissolve 0.0027 g of FeCl3-6 H2O in 10 ml of 10 mM HCl.
- Solution 3:
- Dissolve 0.073 g of HDTMA in 40 ml of ddH2O.
- Mix Solution 1 with 9 ml of Solution 2. Then mix with Solution 3. Solution should now be a blue color. Autoclave and store in a plastic container/bottle.
- Mixture solution:
- Minimal Media 9 (MM9) Salt Solution Stock
- Dissolve 15 g KH2PO4, 25 g NaCl, and 50 g NH4Cl in 500 ml of ddH2O.
- 20% Glucose Stock
- Dissolve 20 g glucose in 100 ml of ddH2O.
- NaOH Stock
- Dissolve 25 g of NaOH in 150 ml ddH2O; pH should be ∼12.
- Casamino Acid Solution
- Dissolve 3 g of Casamino acid in 27 ml of ddH2O.
- Extract with 3% 8-hydroxyquinoline in chloroform to remove any trace iron.
- Filter sterilize.
- CAS agar Preparation:
- Add 100 ml of MM9 salt solution to 750 ml of ddH2O.
- Dissolve 32.24 g piperazine-N,N’-bis(2-ethanesulfonic acid) PIPES.
- PIPES will not dissolve below pH of 5. Bring pH up to 6 and slowly add PIPES while stirring. The pH will drop as PIPES dissolves. While stirring, slowly bring the pH up to 6.8. Do not exceed 6.8 as this will turn the solution green.
- Add 15 g Bacto agar.
- Autoclave and cool to 50oC.
- Add 30 ml of sterile Casamino acid solution and 10 ml of sterile 20% glucose solution to MM9/PIPES mixture.
- Slowly add 100 ml of Blue Dye solution along the glass wall with enough agitation to mix thoroughly.
- Aseptically pour plates.
CONCLUSION
Since this media is very sensitive to variations in pH or FeCl3 concentrations, it may be useful to perform a quality control test prior to using the plates in the lab. A chelation change test can be performed on one plate of a batch. Prior to this test, it is important to visually identify that the media has a blue color to it. pH can change the color of the CAS/Fe complex preventing the CAS from changing color with the removal of the iron. If the plate appears too blue, then a plug of the media is removed. This can be achieved using a sterile 1 ml micropipette tip handled aseptically. The reverse (large opening) end is pressed into the agar then tilted up to remove a circular plug of the media. The well in the media created by the removal of this plug is filled with a saturated solution of 8-hydroxyquinoline. Within a few minutes, an orange halo should appear as the 8-hydroxyquinoline solution diffuses in the agar.
CAS agar plates are useful in the identification of siderophores in Gram-negative bacteria, but HDTMA, in high concentrations, is toxic to Gram-positive bacteria and fungi. A number of modified CAS assays have been developed to allow for the growth of these organism (1, 5, 7). One of the easiest is the O-CAS method, where different microorganisms can be grown on their optimal media and then overlaid with CAS agar (7).
We utilize CAS plates in part of a larger exploratory project in our teaching laboratory. Students are given a semester-long project where they are to isolate, identify, and characterize bacteria from the family Enterobacteraceae from retail ground meat (beef, turkey, or pork). Students spend a large amount of the semester isolating and identifying their unknown bacteria using conventional methods taught in most labs. After identifying their unknown bacteria, the students then conduct a variety of experiments to detect a variety of virulence traits, including serum resistance, acid tolerance, hemolysis, and siderophore productions using CAS plates. To test for the ability of an isolate to resist the bactericidal effects of serum, students inoculate their isolate into a tube containing 50% PG broth (1% peptone and 1% glucose) and 50% guinea pig serum. Tubes are incubated at 37oC and absorbance readings are taken at hours 1, 2, 3 and 24 to determine the OD600. Students construct a growth curve for inclusion in their lab write-up. To test for acid tolerance, students inoculate their isolate into five different tubes of nutrient broth, each tube at a different pH (pH 3, 4, 5, 6 and 7). Absorbance readings are taken as noted above. Alternatively, students can inoculate their isolates on nutrient agar at different pH and visually inspect the plate for growth after incubation for 24 hours. Lastly, to detect for hemolysis, students inoculate their isolate on blood agar, incubate overnight, and visually inspect the plates for hemolysis.
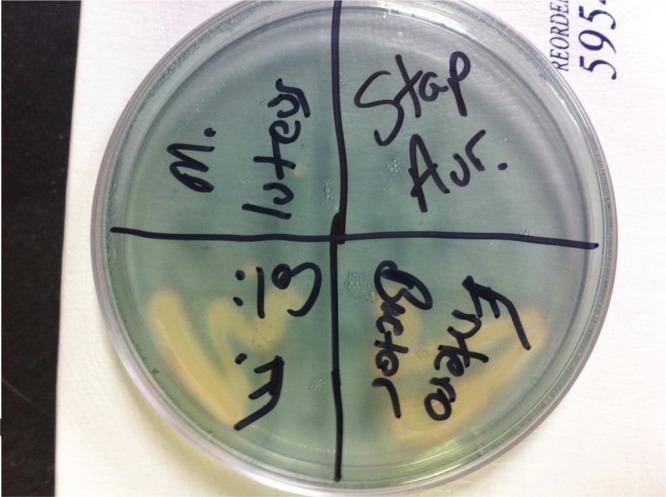
As with all experiments in our laboratory module, proper positive and negative controls are included with each experiment. While testing for any single trait would not indicate whether the identified bacterium is highly virulent or even pathogenic, testing for numerous traits could indicate the relative potential of the identified bacterium to cause disease. We have used CAS plates as part of the project for a few semesters with excellent results (see Figure 1).
FIGURE 1.
Example CAS plate with four different bacteria plated. Moving clockwise from top left: Micrococcus luteus, Staphylococcus aureus, Enterobacter aerogenes, Escherichia coli.
Overall, CAS plates provide a relatively easy-to-use selective and differential media that can be included in most microbiology laboratory classrooms.
Acknowledgments
The authors declare no conflict of interest.
REFERENCES
- 1.Ames-Gottfred NP, Christie BR, Jordan DC. Use of the Chrome Azurol S agar plate technique to differentiate strains and field isolates of Rhizobium leguminosarum biovar trifolii. Appl Environ Microbiol. 1989;55:707–710. doi: 10.1128/aem.55.3.707-710.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnold LD, Viswanatha T. The use of bis(mercaptoacetato-S,O)hydroxoiron(III) complex for the determination of hydroxamates. J Biochem Biophys Methods. 1983;8:307–320. doi: 10.1016/0165-022X(83)90005-2. [DOI] [PubMed] [Google Scholar]
- 3.Arnow LE. Colorimetric determination of the components of 3,4-Dihydroxyphenylalaninetyrosine mixtures. J Biol Chem. 1937;118:531–537. [Google Scholar]
- 4.Atkin CL, Neilands JB, Phaff HJ. Rhodotorulic acid from species of Leucosporidium, Rhodosporidium, Rhodotorula, Sporidiobolus, and Sporobolomyces, and a new alanine-containing ferrichrome from Cryptococcus melibiosum. J Bacteriol. 1970;103:722–733. doi: 10.1128/jb.103.3.722-733.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machuca A, Milagres AM. Use of CAS-agar plate modified to study the effect of different variables on the siderophore production by Aspergillus. Letters Appl Microbiol. 2003;36:177–181. doi: 10.1046/j.1472-765X.2003.01290.x. [DOI] [PubMed] [Google Scholar]
- 6.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Molecul Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-Miranda S, Cabirol N, George-Téllez R, Zamudio-Rivera LS, Fernández FJ. O-CAS, a fast and universal method for siderophore detection. J Microbiol Methods. 2007;70:127–131. doi: 10.1016/j.mimet.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Analytical Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]