SUMMARY
Cavefish and their conspecific surface-dwelling ancestors (Astyanax mexicanus) are emerging as a model system to study the microevolution of development. Here we describe attributes that make this system highly promising for such studies. We review how the Astyanax system is being used to understand evolutionary forces underlying loss of eyes and pigmentation in cavefish. Pigment regression is probably explained by neutral mutations, whereas natural selection is a likely mechanism for loss of eyes. Finally, we discuss several research frontiers in which Astyanax is poised to make significant contributions in the future: evolution of constructive traits, the craniofacial skeleton, the central nervous system, and behavior.
INTRODUCTION
Microevolution is defined as evolutionary changes of relatively small scale that operate within a single species (Dobzhansky 1951). In recent years, evolutionary developmental biologists have made considerable progress in understanding the molecular basis of developmental changes during evolution. In most cases, however, little is known about evolutionary forces that drive these changes. The use of appropriate model systems in microevolution will be necessary to determine these mechanisms.
What are the preferred characteristics of a model system for studying microevolution of development? First, the system should be comprised of populations of the same species that have diverged recently and are evolving independently. In this way, we can approach as closely as possible the point of original evolutionary divergence. Second, the divergent forms should show significant developmental differences that are genetically inherited. Third, the environmental conditions that drive phenotypic changes between the divergent forms should be well known. Finally, the model system should be suitable for experimental analysis. The suite of organisms should be easy to maintain in the laboratory, reproduce frequently and copiously, and be open to embryological, molecular, cellular, and genetic approaches. The Mexican tetra Astyanax mexicanus (sometimes called A. fasciatus and referred to here as Astyanax) has these attributes (Jeffery 2001).
The purpose of this article is to briefly review Astyanax as a model system for studying the microevolution of development. We will discuss the favorable attributes of this system, the environment and natural history of Astyanax cavefish, the regressive and constructive changes available for study, update current knowledge of the mechanisms of pigment and eye regression, and describe frontiers for future research.
ASTYANAX AS A MODEL SYSTEM
Astyanax is a teleost genus that contains a surface-dwelling form (surface fish) and many cave-dwelling forms (cavefish) (Fig. 1A). The divergence time between cavefish and surface fish is probably no greater than a few million years. Cavefish have lost their eyesight and pigmentation and gained some less obvious constructive features (Table 1). Thus, cavefish can be compared with surface fish in essentially the same way mutants are compared with wild-type phenotypes. The environmental cue leading to these phenotypic changes is the absence of light. Accordingly, adaptation to cave life involves changes in food finding, mating, and physical orientation in perpetual darkness.
Fig. 1.
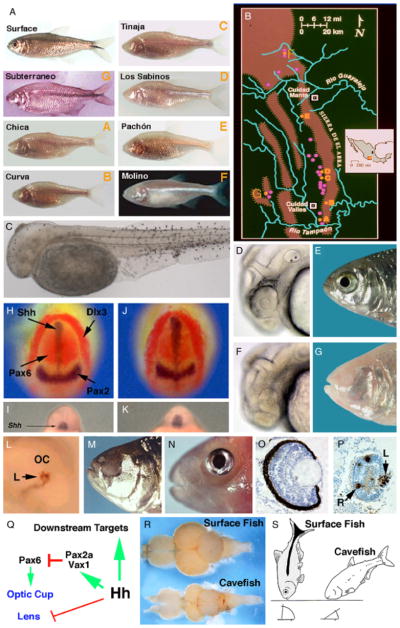
(A) Astyanax surface fish and some of the different cavefish populations (letters in orange indicate the location of caves harboring these populations; see (B)). (B) Sketch map of the Sierra de El Abra region showing approximate location of caves harboring cavefish populations (orange- or pink-filled circles). Orange-filled circles with letters indicate cavefish populations maintained in the Jeffery laboratory cavefish colony (see (A)). The inset in (B) is an outline of Mexico showing the region of the large map in black and Guerrero in the orange rectangle. (C) Albinitic Pachó n cavefish embryo with melanin positive melanoblasts after L-DOPA supplementation. (D, E) Surface fish embryo with eye primordium (D), and adult with eye (E). (F, G) Pachó n cavefish embryo with small eye primordium showing a reduced ventral sector (F), and eyeless adult (G). (H–K) Cavefish embryos show expanded shhA expression along the midline and contracted pax6 expression in the eye fields. (H, I) Surface fish. (J, K) Pachó n cavefish. (H, J). Neural plate stage viewed dorsally. (I, K) Ten somite stage viewed rostrally. Markers (dlx3) indicate the border of the neural plate and (pax2) boundary of the future midbrain and hindbrain region in the neural plate. (L, M) Overexpression of shh induces lens apoptosis (L) and a blind cavefish phenocopy (M) in surface fish. (N) Transplantation of a lens from an embryonic surface fish into the optic cup of a Los Sabinos cavefish rescues eye development. (O, P) Sections through embryonic surface fish and Pachó n cavefish eye primordia showing apoptosis in the cavefish (P) but not the surface fish (O) lens. Arrows in (P) indicate the lens (L) and retina (R). (Q) Diagram showing effects of expanded Hh signaling in cavefish. (R) Comparison of brain morphology in surface fish and cavefish. Dorsal views with anterior on the left. (S) Differences in bottom feeding posture in surface fish and cavefish. (A) is reproduced from Jeffery et al. (2003) and Protas et al. (2007), (C) from McCauley et al. (2004), (D, F) from Yamamoto and Jeffery (2000), (H–M) from Yamamoto et al. (2004), N from Jeffery et al. (2003), (O, P) from Strickler et al. (2007a), (R) from Soares et al. (2004), and (S) from Jeffery (2005).
Table 1.
Phenotypic changes in Astyanax cavefish
| Regressive changes |
| Eyes |
| Visual centers of the brain (e.g. optic tectum) |
| Visual behaviors (e.g. schooling, aggression, fright) |
| Pigment cells |
| Melanin biosynthesis |
| Number of thoracic vertebrae and ribs |
| Number of anal fin rays |
| Sclera ossification |
| Constructive changes |
| Jaw size |
| Tooth number (on maxillary bone) |
| Taste bud number and location |
| Solitary chemosensory cell number and location |
| Nostril size |
| Olfactory bulb size |
| Hypothalamus size |
| Cranial neuromast size |
| Fat content |
| Food sampling behavior |
| Other changes |
| Craniofacial skeleton |
| Brain shape and length |
| Body shape and length |
| Bottom feeding posture |
| Scale size and shape |
Astyanax is an excellent laboratory animal, which can be raised on a simple diet, spawns frequently and abundantly, and has a generation time of about 4–6 months. It has large, clear robust embryos that develop similarly to other teleosts and are suitable for experimental manipulations, such as the removal and transplantation of developing eye parts. The fairly close phylogenetic relationship between Astyanax and zebrafish permits cloning of genes identified in the latter, providing a host of potential molecular markers. Gene inhibition and overexpression methods are available in Astyanax, and cave-related phenotypes can be studied by forward genetic analysis. Surface fish and cavefish are interfertile (Sadoglu 1957a), and different cavefish populations can be crossed for genetic complementation analysis (Wilkens 1971; Borowsky 2008). Finally, the polarity of evolutionary change in Astyanax is certain: cavefish were derived from surface fish ancestors.
ENVIRONMENT AND NATURAL HISTORY
The cave environment is a natural laboratory for studying evolution (Poulson and White 1969). A diverse group of animals, including flatworms, arthropods, mollusks, and vertebrates, have adapted to life in dark caves by evolving a convergent suite of phenotypic changes (Culver 1982). These changes include loss of eyes and pigment, enhanced tactile sensitivity, lower metabolic rates, and increased longevity (Culver 1982). At least 86 known teleost species are obligate cave dwellers (Romero and Paulson 2001). Although often considered to be extreme habitats, caves are actually favorable for the propagation of these species. In most cases, cave animals have persisted long after the extinction of surface-dwelling ancestors exposed to more unpredictable environments. However, Astyanax is one of the rare cave-dwelling animals in which a prototype of the ancestral surface-dwelling form is still extant. Except for periodic flooding, cave environmental conditions are stable, and cavefish appear to be at the top of the predator–prey hierarchy.
Astyanax surface fish invaded Central America after the Isthmus of Panama was formed about 3Ma. Since then their range has gradually expanded north into Mexico and the southwestern United States. Surface fish probably entered limestone caves through entrapment, and subsequently lost their eyes and pigmentation. The hotspot of cavefish distribution is the Sierra de El Abra, a limestone escarpment of the Sierra Madre Orientale in the states of Tamaulipas and San Luis Potosí, Mexico (Mitchell et al. 1977). There is a single outlying cavefish population in the state of Guerrero, Mexico (Espinasa et al. 2001). Mexican blind cavefish were originally described as three distinct species, Anoptichthys jordani (Hubbs and Innes 1936), A. antrobius (Alvarez 1946), and A. hubbsi (Alvarez 1947). Based on interfertility and other features, they are now considered to be a single species (Avise and Selander 1972). About 30 different Astyanax cavefish populations with varying degrees of eye and pigment reduction have been discovered (Fig. 1, A and B) (Mitchell et al. 1977; Espinasa et al. 2001), and some of these populations may have been derived independently from surface fish ancestors (Espinasa and Borowsky 2001; Dowling et al. 2002; Strecker et al. 2003, 2004; Borowsky 2008).
Astyanax cavefish populations are named after their cave of origin. For example, Pachó n, Molino, Subterráneo, and Granadas cavefish reside in La Cueva de El Pachón (Tamaulipas), Sótano de El Molino (Tamaulipas), La Cueva de la Subterráneo (San Luis Potosí), and La Cueva de las Granadas (Guerrero), respectively (Fig. 1, A and B). Several cavefish populations are currently undergoing large-scale introgression with surface fish (Mitchell et al. 1977). The best example is Chica cavefish, which hybridizes with surface fish entering the cave from the nearby Rio Tampaón (Breder 1943). Cavefish originally collected from La Cueva Chica are often available for purchase in pet stores, but not recommended for research because of their extensive hybrid background. In this article, unless another population is named specifically, reference to cavefish will imply the Pachón population, which has been the subject of most extensive experimental analysis.
CHALLENGES
Cavefish and other cave animals present two challenges to evolutionary developmental biologists: (1) to understand the developmental mechanisms underlying phenotypic changes related to cave life and (2) to determine the evolutionary forces responsible for driving these developmental changes. Focusing primarily on regressive traits, several different mechanisms have been proposed for the evolution of cave phenotypes (Culver 1982; Wilkens 1988; Jeffery 2005; Protas et al. 2007). The first is direct selection, the process by which constructive changes are likely to have evolved. In contrast to constructive traits, the benefits of eye and pigment reduction are less obvious, although energy conservation is a reasonable possibility. The second mechanism is indirect selection based on antagonistic pleiotropy between regressive and beneficial traits. For example, the visual system could be lost by this mechanism if it is negatively linked to a positive trait, such as enhancement of other sensory systems that are adaptive in the cave environment. The third mechanism is accumulation of neutral mutations in the absence of natural selection. We frame our subsequent discussion around these mechanisms.
LOSS OF MELANOPHORES AND MELANIN
Surface fish have three types of pigment cells: light reflecting iridophores, yellow-orange xanthophores, and black melanophores. Pigmentation normally functions in protection from the damaging effects of sunlight, species and sex recognition, and camouflage. However, selective pressure for retaining these functions is relaxed in the absence of light. As a result, different cavefish populations show variously decreased numbers of all pigment cell types (Wilkens 1988; McCauley et al. 2004). The ability to interbreed cavefish and surface fish provides a means for identifying the genes involved in the loss of pigmentation and mutations underlying this process. Genetic analysis indicates that multiple genes control melanophore regression (Wilkens 1988). By quantitative trait locus (QTL) analysis, Protas et al. (2007) have recently identified 18 QTL responsible for the loss of differentiated melanophores. If natural selection were the evolutionary force responsible for this process all of these QTL variants should result in decreased melanophores. Instead, there are individual QTL that increase as well as decrease melanophores, suggesting that recurrent neutral mutations and genetic drift account for this trait. However, one can also conceive of positive benefits of losing pigmentation, such as reducing the risk of spontaneous melanomas. The role of neutral mutation as the only force responsible for melanophore reduction needs to be confirmed by identifying the mutated genes within the melanophore QTL. The recent construction of Astyanax BAC libraries (DiPalma et al. 2007) should help accelerate this identification.
In addition to reduced melanophore numbers, the ability to synthesize melanin is lost in some Astyanax cavefish. Melanin is synthesized within the melanosome via a series of well-characterized biochemical reactions. The essential amino acid L-tyrosine is transported from the cytoplasm into the melanosome, where it is converted to L-DOPA by the multifunctional enzyme tyrosinase. L-DOPA is then converted into melanin by a series of enzymatic reactions, the first of which is also catalyzed by tyrosinase. Albino cavefish have melanoblasts with active tyrosinase and other melanogenic enzymes but they are unable to produce L-DOPA in their melanosomes. Accordingly, melanin synthesis can be rescued by supplying exogenous L-DOPA to cavefish (Fig. 1C; McCauley et al. 2004). The existence of melanophore precursors with the potential to synthesize melanin suggests that elimination of the pigment cell lineage is not an option in cavefish. Body pigment cells are derived from the neural crest, which is responsible for the production of a myriad of cell types essential to life inside or outside of caves, and thus inflexible to gross evolutionary change.
A single recessive gene controls cavefish albinism (Sadoglu 1957b; Borowsky and Wilkens 2002). Furthermore, crosses between Pachón and Molino cavefish do not complement the phenotype, indicating that the same gene is mutated in both cavefish (Protas et al. 2006). QTL mapping followed by a candidate gene approach has identified pink-eyed dilution/ oculocutaneous albinism2 (p/oca2) as the gene responsible for cavefish albinism (Protas et al. 2006). P/OCA2 is an integral membrane protein, which appears to regulate the supply of L-tyrosine substrate within the melanosome, perhaps by serving as a transport channel or pH modulator. Different deletions in the p/oca2-coding region are responsible for albinism in Pachón and Molino cavefish, indicating a role for convergent evolution. The reason that cavefish oca2 is a frequent target of mutation is unknown. Extrapolating from the mammalian p/oca2 gene (Rosenblatt et al. 1994; Yi et al. 2003), possibilities are its unusually large size, its chromosomal localization in a region that may be a mutational hotspot, and its possible function exclusively in melanogenesis.
LOSS OF EYES
Although cavefish lack functional eyes as adults, eye primordia are formed during embryogenesis (Fig. 1, D–G). The lens vesicle, optic cup, and neural retina are initially formed in cavefish but the embryonic eye subsequently arrests in growth, degenerates, and disappears into the orbit (Fig. 1, O and P; Langecker et al. 1993; Jeffery et al. 2000; Yamamoto and Jeffery 2000). The cavefish eye primordium is smaller that its surface fish counterpart, and a ventral reduction of the optic cup is observed in several different cavefish populations (Jeffery et al. 2003). The cavefish lens vesicle undergoes extensive apoptosis and eventually disappears or is retained as a tiny vestige in the adult (Fig. 1, O and P; Jeffery and Martasian 1998; Soares et al. 2004). Cells within the retinal layers also die, including most of the photoreceptor cells (Langecker et al. 1993; Yamamoto and Jeffery, 2000; Alunni et al. 2007; Strickler et al. 2007a). Optic development begins and then regresses in this way even in the oldest cavefish populations, suggesting that early steps cannot be eliminated without disrupting critical embryonic processes. When a surface fish lens is transplanted into the cavefish optic cup eye degeneration is prevented and a structurally complete eye is present in adults (Fig. 1N; Yamamoto and Jeffery 2000; Jeffery et al. 2003). The ability to restore the eye suggests that all genes involved in late steps of optic development are potentially functional in cavefish.
Genetic analysis shows that multiple genes are responsible for cavefish eye degeneration (Wilkens 1988; Borowsky and Wilkens 2002), and 12 QTL affecting lens or eye size have been discovered (Protas et al. 2007). Some of the gene loci underlying eye loss are distinct among various types of cavefish. Crosses between several different cavefish populations can generate a low number of F1 and F2 progeny that recover vision, and this proportion is increased in compound hybrids created by crossing the progeny of different cavefish/surface fish F1 hybrids (Borowsky 2008). Although the genes within the 12 eye loss QTL have yet to be identified, the development of small eyes in the F1 progeny of a cavefish × surface fish cross suggests that some of them have dominant effects. Accordingly, in a candidate gene analysis, more genes were found to be upregulated than downregulated in cavefish relative to surface fish (Jeffery 2005). A prime example is hsp90α, a potential pro-apoptotic factor, which is activated in the cavefish lens vesicle (Hooven et al. 2004). Other upregulated genes are components of the Hedgehog (Hh) midline-signaling pathway.
The expression domains of two hh genes, shhA and shhB (formerly tiggy winkle hedgehog), are expanded along the anterior midline of early cavefish embryos (Fig. 1, H–K; Yamamoto et al. 2004). Negative regulation by Hh signaling leads to contraction of pax6 expression in the neural plate region fated to form optic vesicles (Strickler et al. 2001). Later, the expression domains of pax2a and vax1 in the cavefish optic vesicle, which are positively regulated by shhA, are expanded at the expense of pax6 (Yamamoto et al. 2004). Vertebrate optic vesicles are patterned by reciprocal transcriptional repression between the pax6 and pax2/vax1 genes (Schwarz et al. 2000; Take-uchi et al. 2003). Pax6 controls the portion of the optic vesicle devoted to optic cup, whereas Pax2 and Vax1 cooperate to regulate the part of the optic vesicle that will become the optic stalk. Hh hyperactivity reduces the size of the ventral sector of the cavefish optic primordium by altering the normal balance between these transcription factors (Fig. 1Q). Injection of excess shhA mRNA into surface fish embryos to increase Hh signaling induces lens apoptosis, producing a blind cavefish phenocopy (Fig. 1, M and N; Yamamoto et al. 2004). The effects of expanded midline signaling on lens and optic cup development are illustrated in Fig. 1Q.
Linkage has been observed between many cavefish eye QTL and traits that are enhanced in cavefish, such as jaw size and taste buds, which is suggestive of pleiotropy (Protas et al. 2007, 2008). Because shh is also required for development of constructive features, including taste buds, developmental tradeoffs between positively and negatively regulated targets of the Hh signaling pathway could underlie these pleiotropic effects (Jeffery 2005; Tian and Price 2005; Franz-Odendaal and Hall 2006). However, mutations in hh genes are unlikely to directly generate the cavefish eyeless phenotype; none of the likely candidates correspond to known eye QTL (Protas et al. 2007). Therefore, if Hh signaling has a role of the evolution of eye degeneration the mutated genes are likely to be upstream enhancers of the pathway.
According to the neutral mutation model, eye genes should be free to gradually accumulate mutations and eventually lose function if they do not have a critical role in other parts of the embryo or adult. However, downregulated genes involved in eye development have been difficult to identify in cavefish (Jeffery 2005). An exception is the αA-crystallin gene, which encodes an anti-apoptotic factor strongly downregulated in the lens of two different cavefish populations (Behrens et al. 1998; Strickler et al. 2007b). However, αA-crystallin protein coding and upstream regulatory sequences are virtually identical in cavefish and surface fish (Behrens et al. 1998), casting doubt on the possibility that changes in the structure of this gene lead directly to eye loss. In addition, some of the genes encoding major lens-specific proteins, other crystallins and membrane transport channels, which would be expected to be prime targets for neutral mutation, are expressed at relatively high levels in the cavefish lens vesicle (Jeffery et al. 2000; Strickler et al. 2007b). Similarly, genes that control retina development, rx1, vsx1, prox1, pax6, lhx2, lhx9, shh, and fgf8, are also expressed in the degenerating cavefish retina (Jeffery et al. 2000; Yamamoto and Jeffery 2000; Strickler et al. 2002; Alunni et al. 2007). These results do not support the neutral hypothesis for eye regression, although they also do not allow it to be rejected.
The negative polarity of all eye QTL supports natural selection as the most likely mechanism for cavefish eye degeneration (Protas et al. 2007). Although the adaptive advantages of eye loss remain to be determined, the possibilities are (1) creating morphological space for increased constructive features, (2) reducing the liability of exposing the eye, a potential entry point to the brain, to injury and disease, (3) conservation of energy required to maintain a functional eye, and (4) enhancement of non-visual sensory systems that have a negative genetic correlation with eyes. Considering the number of genes that are involved in eye loss, both positive selection for eye destruction and indirect selection against eye formation via antagonistic pleiotropy may be at work.
FRONTIERS IN CAVEFISH RESEARCH
Most studies have focused on trait regression in cavefish. This is because the absence of pigment and eyes are dramatic phenotypes whose ubiquity in cave animals marks them as one of the most pervasive convergences in evolutionary history. In contrast, constructive characters have received comparatively little attention, even though they are likely to be more straightforward examples of adaptive evolution. It will be important to learn more about the constructive characters, especially because some of them may be related to regressive characters through pleiotropy. Unfortunately, in contrast to pigment and eyes, there is considerably less general intuition about what genes might control the enhancement of constructive features, diminishing the value of candidate gene approaches. Therefore, for these traits it will be necessary to rely on QTL analysis for gene identification. Clearly, a sequenced Astyanax genome would be of great assistance in more quickly identifying the genes involved in the evolution of constructive (and other) traits, and this is a preferred component that is currently missing from the Astyanax system. As DNA sequencing becomes less expensive, it seems likely that this problem will be overcome.
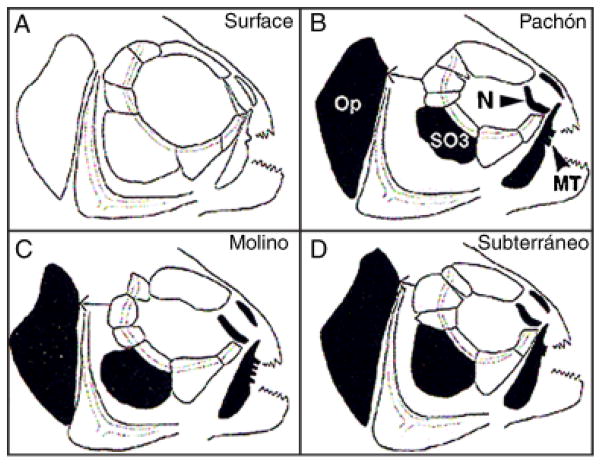
There are other areas of evo-devo in which Astyanax will be useful in breaking new ground. One of these is evolution of the craniofacial skeleton. Substantial craniofacial differences are observed in surface fish and cavefish, and different cavefish populations also show distinct craniofacial phenotypes (Fig. 2). Some, but not all, of these differences can be attributed to the presence or absence of eyes (Yamamoto et al. 2003). Lens transplantation and rescue of eye development changes the organization of cavefish suborbital bones and nasal bones to mimic surface fish. The presence of a lens and large external eye apparently has a large impact on craniofacial development. However, there are some craniofacial features that vary between the two Astyanax forms independently of eye development, namely the shape of the operculum and tooth bearing maxillary bones (Fig. 2). Cavefish are excellent experimental organisms to investigate the evolutionary forces that govern the morphology of these bones.
Fig. 2.
Astyanax surface fish and cavefish craniofacial skeletons. Features that are changed are shown in black. N, nasal bones; MT, maxillary teeth. SO3: Fractured third suborbital bone. Op, operculum. Modified from Yamamoto et al. (2003).
Another novel area is evolution of the central nervous system (CNS). Brain morphology varies considerably among vertebrate species (Butler and Hodos 1996), but how this diversity is established and the evolutionary forces that create it are largely unexplored. Surface fish and cavefish brains are remarkably different, much more than expected from phenotypic variation within the same species (Fig. 1R). As predictable from the loss of vision, the size of the optic tectum is decreased in cavefish. However, there also appears to be a functional change in the optic tectum. The cavefish optic tectum responds to somatosensory stimuli that spatially correspond to the visually receptive field in its surface fish counterpart (Langecker et al. 1995). Thus, different sensory modules appear to be re-wired during cavefish CNS evolution. In addition to changes in the optic tectum, the cavefish brain is more elongated, has enlarged olfactory bulbs, and contains a larger hypothalamus (not shown in Fig. 1R) than surface fish (Peters et al. 1993; Soares et al. 2004). As a response to Hh hyperactivity, the downstream patterns of gene expression are changed in the developing cavefish forebrain, and higher levels of cell proliferation are induced in the cavefish basal diencephalon and hypothalamus primordium (Menuet et al. 2007). The evolutionary forces that generate these changes are not understood. The cavefish system has the potential to help understand how structural and functional diversity is generated in the forebrain, hypothalamus, and other parts of the CNS during development.
How and why behavioral changes have evolved is almost completely uncharted territory. Surface fish and cavefish have many differences in behavior (Parzefall 1985; Espinasa et al. 2005). An intriguing example is feeding posture behavior (Fig. 1S). In darkness, surface fish feed inefficiently from the bottom of a tank by positioning the long axis of their bodies at a 90° angle to the substrate. In contrast, under the same conditions, cavefish position their bodies at a 45° angle and move steadily forward, which makes more effective use of their shovel-like lower jaws. The unique feeding behavior of cavefish is likely to be adaptive for cave life (Hüppop 1987). When a surface fish and cavefish are placed in a lighted tank and required to compete for limited food the surface fish obtains most of it and thrives, but in a dark tank the cavefish has much better food-finding ability and flourishes. Genetic crosses between cavefish and surface fish suggest that the regulation of feeding behavior is simple, perhaps under the control of only one or a few genes (Schemmel 1980). In general, behavioral changes are attributed to the additive effects of many genes. The relative simplicity of cavefish behaviors may reflect the short time since their original divergence, underscoring the importance of a microevolutionary perspective. Astyanax has the potential to make significant contributions to understanding the evolution of behavior at the molecular level.
Each of these frontiers is a new research territory ripe for modern analysis using the tools that we have discussed in Astyanax. Although progress has been rapid during the past ten years of cavefish evo-devo research, there is still a long way to go, and we anticipate many exciting finales.
Acknowledgments
The following individuals working in or associated with my laboratory have conducted critical experiments, fieldwork, and/or have influenced the direction of cavefish research: R. Aurigemma, C. Beard, B. Bemis, M. Byerly, H. Brzezowski, K. Dittmar de la Cruz, L. Espinasa, K. Famuditimi, S. Goricki, S. Guiney, D. Heyser, E. Hixon, T. Hooven, K. Hornaday, B. Huchins, D. Jeffery, D. Martasian, D. McCauley, A. Parkhurst, M. Porter, M. Protas, E. Rogers, A. Romero, D. Rossi, D. Soares, D. Stock, Y. Yamamoto, M. Yeager, and M. Yoshizawa. Cavefish research in my laboratory is supported by grants from NIH (R01-EY014619) and NSF (IBN-0542384).
References
- Alunni A, Menuet A, Candal E, Pénigault JB, Jeffery WR, Rétaux S. Developmental mechanisms for retinal degeneration in the blind cavefish Astyanax mexicanus. J Comp Neurol. 2007;505:221–233. doi: 10.1002/cne.21488. [DOI] [PubMed] [Google Scholar]
- Alvarez J. Revisión del genéro Anoptichthys con descriptión de una especie nueva (Pisc. Characidae) An Esc Cien Biol México. 1946;4:263–282. [Google Scholar]
- Alvarez J. Descripción de Anoptichthys hubbsi caracinindo ceigo de La Cueva de Los Sabinos. S L P Soc Mex Hist Nat. 1947;8:215–219. [Google Scholar]
- Avise JC, Selander RK. Evolutionary genetics of cavedwelling fishes of the genus Astyanax. Evolution. 1972;26:1–19. doi: 10.1111/j.1558-5646.1972.tb00170.x. [DOI] [PubMed] [Google Scholar]
- Behrens M, Wilkens H, Schmale H. Cloning of the αA-crystallin genes of the blind cave form and the epigean form of Astyanax fasciatus: a comparative analysis of structure, expression and evolutionary conservation. Gene. 1998;216:319–326. doi: 10.1016/s0378-1119(98)00346-1. [DOI] [PubMed] [Google Scholar]
- Borowsky R. Restoring sight in blind cavefish. Curr Biol. 2008;18:R23–R24. doi: 10.1016/j.cub.2007.11.023. [DOI] [PubMed] [Google Scholar]
- Borowsky R, Wilkens H. Mapping a cave fish genome: polygenic systems and regressive evolution. J Hered. 2002;93:19–21. doi: 10.1093/jhered/93.1.19. [DOI] [PubMed] [Google Scholar]
- Breder CM., Jr Descriptive ecology of La Cueva Chica, with special reference to the blind fish, Anoptichthys. Zoologica. 1943;27:7–16. [Google Scholar]
- Butler AB, Hodos W. Comparative Vertebrate Neuroanatomy, Evolution and Adaptation. Wiley-Liss; New York: 1996. [Google Scholar]
- Culver D. Cave Life Evolution and Ecology. Harvard; Cambridge: 1982. [Google Scholar]
- DiPalma F, Kidd C, Borowsky R, Kocher TD. Construction of bacterial artificial chromosome libraries for the Lake Malawi cichlid (Metriaclima zebra), and the blind cavefish (Astyanax mexicanus) Zebrafish. 2007;4:41–47. doi: 10.1089/zeb.2006.9996. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia; New York: 1951. [Google Scholar]
- Dowling TE, Martasian DP, Jeffery WR. Evidence for multiple genetic lineages with similar eyeless phenotypes in the blind cavefish, Astyanax mexicanus. Mol Biol Evol. 2002;19:446–455. doi: 10.1093/oxfordjournals.molbev.a004100. [DOI] [PubMed] [Google Scholar]
- Espinasa L, Borowsky RB. Origin and relationships of cave populations of the blind Mexican tetra, Astyanax fasciatus, in the Sierra de El Abra. Environ Biol Fishes. 2001;62:233–237. [Google Scholar]
- Espinasa L, Rivas-Manzano P, Espinosa Pérez H. A new blind cave fish population of the genus Astyanax: geography, morphology and behavior. Environ Biol Fishes. 2001;62:329–344. [Google Scholar]
- Espinasa L, Yamamoto Y, Jeffery WR. Non-optical releasers for aggressive behavior in blind and blinded Astyanax (Telesotei, Characidae) Behav Process. 2005;70:144–148. doi: 10.1016/j.beproc.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Franz-Odendaal TA, Hall BK. Modularity and sense organs in the blind cavefish, Astyanax mexicanus. Evol Dev. 2006;8:94–100. doi: 10.1111/j.1525-142X.2006.05078.x. [DOI] [PubMed] [Google Scholar]
- Hooven TA, Yamamoto Y, Jeffery WR. Blind cavefish and heat shock protein chaperones: a novel role for hsp90α in lens apoptosis. Int J Dev Biol. 2004;48:731–738. doi: 10.1387/ijdb.041874th. [DOI] [PubMed] [Google Scholar]
- Hubbs CL, Innes WT. The first known blind fish of the family Characidae: a new genus from Mexico. Occas Papers Mus Zool Univ Mich. 1936;342:1–7. [Google Scholar]
- Hüppop K. Food finding ability in cave fish (Astyanax fasciatus) Int J Speleol. 1987;18:59–66. [Google Scholar]
- Jeffery WR. Cavefish as a model system in evolutionary developmental biology. Dev Biol. 2001;231:1–12. doi: 10.1006/dbio.2000.0121. [DOI] [PubMed] [Google Scholar]
- Jeffery WR. Adaptive evolution of eye degeneration in the Mexican blind cavefish. J Hered. 2005;96:185–196. doi: 10.1093/jhered/esi028. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Martasian DP. Evolution of eye regression in the cavefish Astyanax: apoptosis and the Pax-6 gene. Am Zool. 1998;38:685–696. [Google Scholar]
- Jeffery WR, Strickler AG, Guiney S, Heyser D, Tomarev SI. Prox1 in eye degeneration and sensory organ compensation during development and evolution of the cavefish Astyanax. Dev Genes Evol. 2000;210:223–230. doi: 10.1007/s004270050308. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Strickler AG, Yamamoto Y. To see or not to see: evolution of eye degeneration in Mexican blind cavefish. Comp Integr Biol. 2003;43:531–541. doi: 10.1093/icb/43.4.531. [DOI] [PubMed] [Google Scholar]
- Langecker TG, Neumann B, Hausberg C, Parzefall J. Evolution of the optical releasers for aggressive behavior in cave-dwelling Astyanax fasciatus (Telesostei, Characidae) Behav Process. 1995;34:161–168. doi: 10.1016/0376-6357(94)00063-m. [DOI] [PubMed] [Google Scholar]
- Langecker TG, Schmale H, Wilkens H. Transcription of the opsin gene in degenerate eyes of cave dwelling Astyanax fasciatus (Teleostei, Characidae) and its conspecific ancestor during early ontogeny. Cell Tissue Res. 1993;273:183–192. [Google Scholar]
- McCauley DW, Hixon E, Jeffery WR. Evolution of pigment cell regression in the cavefish Astyanax: a late step in melanogenesis. Evol Dev. 2004;6:209–218. doi: 10.1111/j.1525-142X.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- Menuet A, Alunni A, Joly JS, Jeffery WR, Rétaux S. Shh overexpression in Astyanax cavefish: multiple consequences on forebrain development and evolution. Development. 2007;134:845–855. doi: 10.1242/dev.02780. [DOI] [PubMed] [Google Scholar]
- Mitchell RW, Russell WH, Elliot WR. Mexican eyeless characin fishes, genus Astyanax: Environment, distribution, and evolution. Spec Publ Mus Texas Tech Univ. 1977;12:1–89. [Google Scholar]
- Parzefall J. On the heredity of behavioral patterns in cave animals and their epigean relatives. Nat Spel Soc Bull. 1985;47:128–135. [Google Scholar]
- Peters V, Schnacht NV, Schmidt W, Wilkens H. Gehirn-proportionen und Aussprägungsgrad der Sinnesorgane von Astyanax mexicanus (Pisces, Characidae). Ein Vergleich zwischen dem Flussfisch und seinen Hölendiriveten “Anoptichthys”. Z Zool Syst Evolforschg. 1993;31:144–159. [Google Scholar]
- Poulson TL, White WB. The cave environment. Science. 1969;165:971–981. doi: 10.1126/science.165.3897.971. [DOI] [PubMed] [Google Scholar]
- Protas M, Conrad M, Gross JB, Tabin C, Borowsky R. Regressive evolution in the Mexican cave tetra, Astyanax mexicanus. Curr Biol. 2007;17:452–454. doi: 10.1016/j.cub.2007.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protas M, Tabansky I, Conrad M, Gross JB, Vidal O, Tabin CJ, Borowsky R. Multi-trait evolution in a cave fish, Astyanax mexicanus. Evol Dev. 2008;10:196–209. doi: 10.1111/j.1525-142X.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nature Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- Romero A, Paulson KM. It’s a wonderful hypogean life: a guide to the troglomorphic fishes of the world. Environ Biol Fishes. 2001;62:13–41. [Google Scholar]
- Rosenblatt S, Durham-Pierce D, Garner JM, Nakatsu Y, Brilliant MH, Orlow SJ. Identification of a melanosomal membrane protein encoded by the pink-eyed dilution (type II oculocutaneous albinism) gene. Proc Natl Acad Sci USA. 1994;91:12071–12075. doi: 10.1073/pnas.91.25.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoglu P. Mendelian inheritance in hybrids between the Mexican blind fish and their overground ancestors. Verh Dtsch Zool Ges. 1957a;1957:432–439. [Google Scholar]
- Sadoglu P. A Mendelian gene for albinism in natural cave fish. Experientia. 1957b;13:394. [Google Scholar]
- Schemmel C. Studies on the genetics of feeding behaviour in the cave fish Astyanax mexicanus f. Anoptichthys An example of apparent monofactorial inheritance by polygenes. Z Teirpsychol. 1980;53:9–22. doi: 10.1111/j.1439-0310.1980.tb00730.x. [DOI] [PubMed] [Google Scholar]
- Schwarz M, et al. Spatial specification of mammalian eye territories by reciprocal transcriptional repression of Pax2 and Pax6. Development. 2000;127:4325–4334. doi: 10.1242/dev.127.20.4325. [DOI] [PubMed] [Google Scholar]
- Soares D, Yamamoto Y, Strickler AG, Jeffery WR. The lens has a specific influence on optic nerve and tectum development in the blind cavefish Astyanax. Dev Neurosci. 2004;26:308–317. doi: 10.1159/000082272. [DOI] [PubMed] [Google Scholar]
- Strecker U, Bernachez L, Wilkens H. Genetic divergence between cave and surface populations of Astyanax in Mexico (Characidae, Teleostei) Mol Ecol. 2003;12:699–710. doi: 10.1046/j.1365-294x.2003.01753.x. [DOI] [PubMed] [Google Scholar]
- Strecker U, Faúndez VH, Wilkens H. Phylogeography of surface and cave Astyanax (Teleostei) from Central and North America based on cytochrome b sequence data. Mol Phylogenet Evol. 2004;33:469–481. doi: 10.1016/j.ympev.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Strickler AG, Byerly MS, Jeffery WR. Lens gene expression analysis reveals downregulation of the anti-apoptotic chaperone αA crystallin during cavefish eye degeneration. Dev Genes Evol. 2007b;217:771–782. doi: 10.1007/s00427-007-0190-z. [DOI] [PubMed] [Google Scholar]
- Strickler AG, Famuditimi K, Jeffery WR. Retinal homeobox genes and the role of cell proliferation in cavefish eye degeneration. Int J Dev Biol. 2002;46:285–294. [PubMed] [Google Scholar]
- Strickler AG, Yamamoto Y, Jeffery WR. Early and late changes in Pax6 expression accompany eye degeneration during cavefish development. Dev Genes Evol. 2001;211:138–144. doi: 10.1007/s004270000123. [DOI] [PubMed] [Google Scholar]
- Strickler AG, Yamamoto Y, Jeffery WR. The lens controls cell survival in the retina: evidence from the blind cavefish Astyanax. Dev Biol. 2007a;311:512–523. doi: 10.1016/j.ydbio.2007.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Take-uchi M, Clarke JD, Wilson SW. Hedgehog signalling maintains the optic stalk-retinal interface through the regulation of Vax gene activity. Development. 2003;130:955–968. doi: 10.1242/dev.00305. [DOI] [PubMed] [Google Scholar]
- Tian NMML, Price DJ. Why cavefish are blind. BioEssays. 2005;27:235–238. doi: 10.1002/bies.20202. [DOI] [PubMed] [Google Scholar]
- Wilkens H. Genetic interpretation of regressive evolutionary process. Studies of hybrid eyes of two Astyanax cave populations (Characidae, Pisces) Evolution. 1971;25:530–544. doi: 10.1111/j.1558-5646.1971.tb01913.x. [DOI] [PubMed] [Google Scholar]
- Wilkens H. Evolution and genetics of epigean and cave Astyanax fasciatus (Characidae, Pisces) Evol Biol. 1988;23:271–367. [Google Scholar]
- Yamamoto Y, Espinasa L, Stock DW, Jeffery WR. Development and evolution of craniofacial patterning is mediated by eye-dependent and—independent processes in the cavefish Astyanax. Evol Dev. 2003;5:435–446. doi: 10.1046/j.1525-142x.2003.03050.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Jeffery WR. Central role for the lens in cavefish eye degeneration. Science. 2000;289:631–633. doi: 10.1126/science.289.5479.631. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Stock DW, Jeffery WR. Hedgehog signalling controls eye degeneration in blind cavefish. Nature. 2004;431:844–847. doi: 10.1038/nature02864. [DOI] [PubMed] [Google Scholar]
- Yi Z, et al. A 122.r kilobase section of the P gene underlies the high prevalence of oculocutaneous albinism in the Navajo population. Am J Hum Genet. 2003;72:62–72. doi: 10.1086/345380. [DOI] [PMC free article] [PubMed] [Google Scholar]