Abstract
To compare cryo-EM images and 3D reconstructions with atomic structures in a quantitative way it is essential to model the electron scattering by solvent (water or ice) that surrounds protein assemblies. The most rigorous method for determing the density of solvating water atoms for this purpose has been to perform molecular-dynamics (MD) simulations of the protein-water system. In this paper we adapt the ideas of bulk-water modeling that are used in the refinement of X-ray crystal structures to the cryo-EM solvent-modeling problem. We present a continuum model for solvent density which matches MD-based results to within sampling errors. However, we also find that the simple binary-mask model of Jiang and Brünger (1994) performs nearly as well as the new model. We conclude that several methods are now available for rapid and accurate modeling of cryo-EM images and maps of solvated proteins.
Keywords: solvent, reconstruction, molecular dynamics simulation
Introduction
For the development of image-processing algorithms, for the interpretation of experimental cryo-EM density maps and for the docking of high-resolution structures into such maps, there is continuing interest in the quantitative simulation of cryo-EM images of macromolecules. A recent example is the use of a quantitative model of image formation to evaluate the potential of phase-contrast electron microscopy (Hall et al., 2011). Traditionally such simulations start with the placement of electron-scattering density according to the atomic coordinates of a protein molecule. Atoms are typically modeled as point-scatterers weighted by the neutral-atom, low-angle scattering amplitudes, and the summation of scattering effects is simplified by the use of the weak-phase approximation. One proposed enhancement to this modeling (Zhong et al., 2002) is to include the effects of molecular orbitals, which are likely to account for the discrepancies that have been observed between experimental scattering factors and the theoretical isolated-atom values (Grigorieff et al., 1996). Other enhancements are to include the effects of complex scattering amplitudes (Rullgård et al.) and to use multi-slice calculations to model the images from “thick” specimens (Kirkland, 2009).
For typical cryo-EM specimens the most important part of such modeling is to provide proper simulation of the water shell that surrounds a protein molecule. As water—or in this case, the vitreous ice--has on average about 75% of the scattering power of protein, there is a very strong contrast-matching effect of the water shell; this greatly reduces the signal in cryo-EM images at low resolution. Figure 1 demonstrates the large difference in a simulated projection image of protein in vacuum compared to a corresponding image including solvent. The contrast between protein density and vacuum seen in a projection image (Fig. 1A) is so large that features in the protein interior are washed out, while a very different appearance (Fig. 1B) is obtained with solvent density included. A slice through the three-dimensional density (Fig. 1D) demonstrates that although the mean protein density is larger than that of solvent, there are minima in the protein density that fall well below the solvent density.
Figure 1.
Illustration of the importance of solvent modeling. Projection images and sections are shown from simulated 3D density maps of a monomer of influenza neuraminidase (ribbon diagram at top left). A, the projection is shown of the scattering density of neuraminidase protein in a vacuum; B, the same with solvent included. C, a slice of the 3D density at the z-coordinate z2 of protein D. A similar slice with modeled solvent, which can be seen as the gray continuous density surrounding the protein. E, a 1-dimensional projection, equal to the density along the line z2 in A; it is also equal to the sum along the coordinate y of the slice in C. F, the corresponding projection from the density including the solvent model. Note that the solvent caused an overall increase in the integrated inner potential, but there is strong contrast-matching between protein and solvent. G, the two 1D projections are shown shifted and overlaid to match the highest peak density. H, the same two projections are superimposed after shifting and scaling to approximately match both minimum and maximum values. Maps were computed from the X-ray structure of influenza neuraminidase (Varghese et al., 1995), pdb entry 1NNC.
A comparison of one-dimensional projections of the structure (Figure 1 E and F) shows the large degree to which the solvent matches the protein density. A direct comparison between the projected densities (Fig. 1G) shows that the fine structure of peaks and valleys are recognizable in both. For the most part these peaks and valleys arise from secondary structure in the protein interior, rather than cavities in the protein surface that become filled with solvent.
Figure 1H demonstrates the kind of error that arises in directly comparing a density map of solvated protein (P+S) with a scaled map computed from protein alone (P). To match the overall magnitude of the P+S map, the scaling of the P map is smaller, and therefore its peaks and valleys due to secondary structure are attenuated—by a factor of two or more—compared to the P+S map. Thus the modeling of protein structures using a protein-alone map overemphasizes the molecular boundary, and underemphasizes the density variations in the protein interior.
One approach to modeling protein solvation is to compute directly the average density and distribution of water molecules surrounding a protein by molecular dynamics (MD) simulation (Wang et al., 2008; Hall et al., 2011). Such calculations are however very time consuming. We sought instead a way to provide a rapid and simple way to obtain a continuum description of the water shell.
Models for protein hydration have been in use for some time in X-ray crystallography, where simulation of the water shell is important for high-quality structure refinement. One approach is to model the water density as the complement of the protein density, that is, a fuzzy density that is high wherever the protein density is low. For crystallography this has been implemented in Fourier space (Fraser et al., 1978) and essentially the same principle, implemented in real space, has been applied to cryo-EM image simulation (Chang et al., 2010). A much better approach is to model explicitly the volume of bulk water surrounding the protein, with a border region of low density interposed between protein and water (Jiang and Brünger, 1994; Fenn et al., 2010). The present work is an adaptation of this approach. We use MD simulations of two small proteins to parameterize a function that describes the density of water molecules surrounding a protein surface, and incorporate this function into a simple algorithm to simulate the electron-scattering density of any water-solvated protein whose atomic coordinates are known. We compare the performance of this algorithm with a simplified version, and with the original binary-mask algorithm of Jiang and Brünger (1994).
Methods
Molecular dynamics simulations were performed of bovine trypsin inhibitor (BPTI; PDB code 6PTI, molecular weight 6.5 kDa) and influenza neuraminidase monomer (PDB code 1NNC, 46 kDa, with ligand atoms deleted). BPTI was chosen as an exemplary small but rigid protein; neuraminidase was chosen by Jiang and Brünger (1994) because its large substrate-binding cavity serves as a challenge for solvent modeling. Using the program VMD (Humphrey et al., 1996) each molecule was placed into a water box approximately 75 × 70 × 70 Å or 88 × 98 × 98 Å, respectively, so that the minimum thickness of water surrounding the protein was about 20 Å and NaCl was present at a concentration of 0.15 M. The non-hydrogen atoms of the protein were fixed during the simulation while water was mobile. Using NAMD (Phillips et al., 2005) the system was run at 277 K and a constant pressure of 1 atm. BPTI was equilibrated for 5 nanoseconds (timestep: 1 fs) and neuraminidase for 2 ns (timestep: 0.5 fs). After equilibration, we made use of 100 snapshots from the last 200 ps of each trajectory. A second 2 ns simulation of neuraminidase with no constraints on either protein or water was also carried out as a control for artifacts arising in the fixed-protein simulations.
Electron scattering was computed on the basis of the weak-phase approximation and neutral atoms. Under these assumptions the incompletely-shielded positive potential of the atomic nucleus—the so-called inner potential—is the only origin of electron scattering. Each atom was assigned a single potential value, obtained at the limit of low scattering angle from the parameterization of Kirkland (Kirkland, 2009). The values of the inner potentials Vi were computed to be 25, 108, 130, 97, 100 and 267 V·Å3, while the atomic radii ai were taken to be 0, 1.55, 1.7, 2.0, 1.8 and 1.8 Å for the elements H, C, N, O, S and P, respectively. The density maps were computed in units of volts so that the projected potential, in units of VÅ, can be converted to electron scattering amplitude (or equivalently the electron phase shift) through multiplication by the interaction parameter σ. The interaction parameter has the values σ = 0.92, 0.73 and 0.65 mrad /V·Å for electron energies of 100, 200 and 300 keV, respectively.
Results
A model for water density
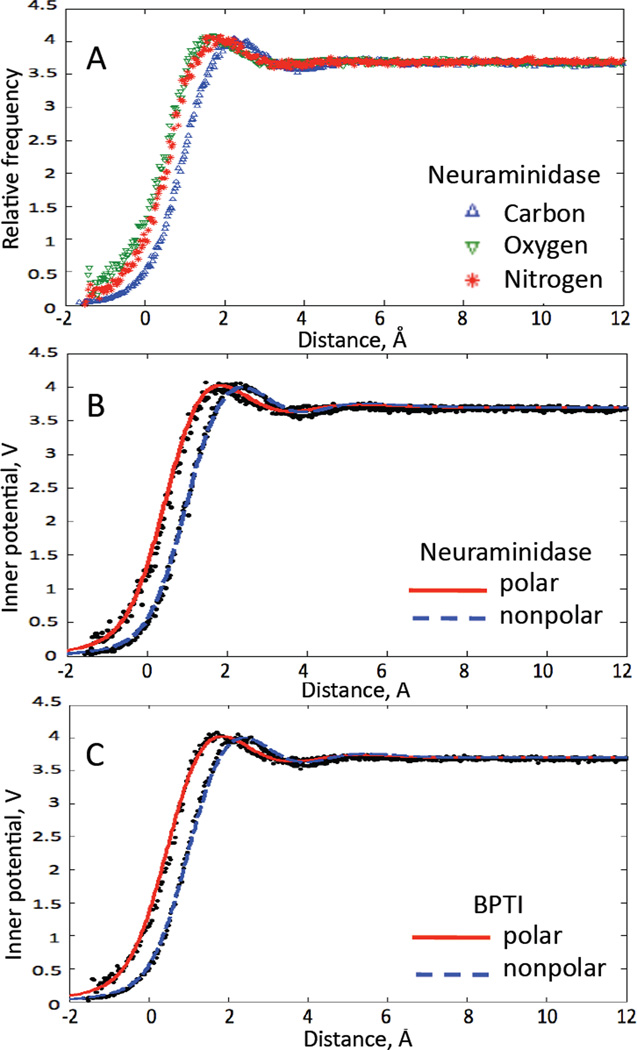
We first sought a parameterization of the density of water surrounding the protein. In these calculations we ignored hydrogen atoms, and considered only the positions of “heavy atoms” of the protein and the positions of the water oxygens. Using molecular-dynamics trajectories for two solvated proteins, BPTI and neuraminidase, we computed the density of water oxygens, normalized to the density in bulk water, as a function of distance from the van der Waals surface of the superficial protein atoms (Fig. 2). When plotted for individual atom types, we found that the function was essentially identical for all atoms except carbon (Fig. 2A). The approximately 0.5 Å longer distance from protein carbons to water reflects the predominantly nonpolar interactions (Lum et al., 1999), in contrast to the polar hydrophilic interactions involving the other atom types. The density functions are indistinguishable between the two proteins, and we expect them to be valid for protein complexes of any size, as the solvation density is very much a local property of the protein surface.
Figure 2.
Radial distribution functions. Plotted is the frequency of observing water oxygens at the given distance from the van der Wals radius of the nearest protein atom, scaled to yield the average inner potential. Distances are smoothed with a Gaussian function of 1 Å standard deviation. A, distances are shown for each atom type. Distances from carbon are larger by about 0.5 Å because these reflect nonpolar interactions. B, the fitted functions (eqn. 1 and Table 1) for polar (all atom types except carbon) and nonpolar interactions are superimposed on the frequency plot. The ordinate is the inner potential assumed for the continuum model for solvent. C, the functions are superimposed on data from the BPTI simulation.
We fitted the water density as convolved with a Gaussian function of 1Å standard deviation (Fig. 2B). The main term of the fitted function is a monotonic increase modeled by an error function. To this we added two Gaussian functions to simulate the overshoot and undershoot of the density resulting from the hydration shells. The distance r is taken as the distance from the nearest atom surface; thus the distance from the center of the closest atom i is the sum of r and the atom radius ai. The modeled normalized densities Ft, where t indicates the type of interaction (polar or nonpolar), are given by
| (1) |
and the parameters are given in Table 1. We call this the S3 model because it uses three terms to describe the solvent density function.
Table 1.
Parameter values for the water density distribution function (eqn. 1). The parameters r and σ are given in angstroms.
| Parameter | Polar | Nonpolar |
|---|---|---|
| a2 | 0.2 | 0.15 |
| a3 | −0.15 | −0.12 |
| r1 | 0.5 | 1.0 |
| r2 | 1.7 | 2.2 |
| r3 | 1.7 | 3.6 |
| σ1 | 1 | 1 |
| σ2 | 1.77 | 1.77 |
| σ3 | 1.06 | 0.85 |
The actual modeling of a solvated protein starts with the evaluation of the distance function d(r). This function gives at every point r in a volume the distance to the closest protein atom,
| (2) |
where q denotes the set either of polar (p) or nonpolar (n) atoms, and ri is the location of the center of atom i. The value of d is therefore obtained by computing the minimum of the distances to each atom; the identity of the closest atom is also stored. The solvent density at location r is then given by the density function (1) evaluated according to whether the nearest atom is nonpolar (that is, a carbon atom) or polar. In our implementation these functions are evaluated at discrete values r on a grid with 1 Å spacing.
A straightforward application of eqn. (1) to compute the solvent density provides good results for regions outside the protein boundary, but it predicts spurious solvent densities within the protein. (Jiang and Brünger, 1994) solved this problem by defining a boundary outside the protein using a large probe radius, and subsequently shrinking the boundary toward the protein to define the inner surface of the hydration shell. We created the binary mask m(r) in the same way to exclude protein-interior points. We found that a probe radius of 1.7 Å works best in excluding solvent from the protein interior while allowing water to fill cavities such as the deep one in neuraminidase. The implementation was as follows. Initially the value of m is set to 1 at every grid point. Then at any grid point closer than rprobe = 1.7 Å to the van der Waals surface of any protein atom, m is set to zero (Fig. 3A). Finally, the probe mask is shrunk around the protein as follows: for every grid point on the boundary that has the value 1, all grid points within a radius rshrink of that point are also set to 1 (Fig. 3B). The value rshrink is evaluated as the distance from the probe-mask point to the center of the nearest protein atom, a distance on average about 3Å. The resulting mask function m(r) is zero inside the protein but is equal to 1 at the centers of the protein surface atoms and beyond them into the bulk solution.
Figure 3.
Computing the binary mask. A, the first step: all points on a 1Å grid that are within the probe radius (1.7 Å) of the van der Waals surface of a protein atom are set to zero. B, in the second step, all grid points within rshrink of each boundary point is set back to one; rshrink is computed as the distance from each boundary point to the center of the closest protein atom, roughly 3 Å. The boundary points are those points falling within rprobe and rprobe + 1.8 Å of the van der Waals protein surface, where 1.8 Å is chosen to be greater than times the grid spacing.
With the inclusion of the mask function the complete S3 model for the normalized solvent density is
| (3) |
where s(r) indicates the identity of the protein atom closest to location r.
Hydrogen atoms are ignored in the distance calculations because their effective radius is nearly zero, and the experimental coordinates of hydrogen atoms are not always available. Hydrogen atoms however make a significant contribution to the electron-scattering density, so we added the inner potential of n hydrogen atoms to the potential for each heavy atom, where n=2 for water oxygens, and the values n= 1.13, 1.28, 0.08, 0.41, and 0 were chosen as representative numbers of hydrogens bound to the protein atoms N, C, O, S and P respectively. These values were obtained as the number of hydrogens bound to atoms in each type of amino-acid residue, averaged over all residue types with weights according to the prevalence of residues (Creighton, 1993). Each heavy protein atom i was then represented in the density map as a three-dimensional Gaussian function weighted by the corresponding potential value. To the summed protein contribution was added to the water (ice) density, taking the mean inner potential of bulk vitreous ice to be Vw = 3.6 volts.
The overall inner potential is computed as the sum of a water-density term and a protein density term. The latter is modeled by a Gaussian density centered at each protein atom position, so that the overall potential is given by
| (4) |
For simplicity we set the standard deviation σg of the Gaussian function equal to 1 Å. When evaluated on a 1 Å grid this Gaussian is sufficiently broad that there is negligible aliasing; that is, the discrete map preserves the centroid position and integrated inner potential of each atom to high precision. To obtain a map at lower resolution, this map can be low-pass filtered and resampled to the desired voxel size. Simulated images are then obtained by integrating V along the projection direction to yield a projected potential in units of V·Å.
Comparison with MD simulations
We first compared this S3 continuum model with the results of the MD simulations. Figure 4A shows cross-sections through the water density modeled as in eqn. (4) for the neuraminidase protein. These are compared with the mean density of water oxygens (Fig. 4B), averaged over the last 200 ps of the molecular-dynamics trajectory. In this trajectory protein atoms were held fixed at their crystallographic coordinates while waters were free to move. The occupancy by water of inner cavities is correctly described by the S3 model, and one-dimensional projections of the slices, corresponding to lines in a projection image, lie within the interquartile (25–75%) range of values observed in the snapshots of the MD simulation (Fig. 4C, D). In parts C and D of the figure can also be seen the relative contributions of solvent and internal protein structures to the peaks and valleys of projection profiles.
Figure 4.
Comparison of the continuum solvent model with MD simulation. Data are shown for slices at the three z-coordinates indicated by lines in Fig. 1A. A, slices through the 3D solvent model S3. B, slices through the corresponding MD water density, averaged over 200 ps. C, 1D projections along the Y direction of the sections in A and B, giving the total solvent density as a function of the X coordinate; D, total protein+solvent density. Bars indicate the 25–75% intervals of densities sampled from the 200ps MD simulation. Lines indicate the modeled densities in which the protein density is added to solvent density prediced by either the S3 model (red) or the Jiang and Brünger model (JB; black). Densities are given in units of integrated inner potential in volt-angstroms.
The MD simulations we used for analysis of solvent density were limited in that we fixed the location of the protein atoms. We therefore ran another simulation in which no restraints were applied to the protein-and-water system, and the results are shown in Fig. 5. Due to structural fluctuations the mean solvent density computed from the unconstrained simulation differed substantially from that in the constrained one (Fig. 5E). Nevertheless, to allow a comparison of the S3 model results and the solvent density computed directly from the MD simulation, we adopted the following strategy. For each of 100 snapshots of the MD trajectory, we computed the S3 solvent density and also the explicit solvent density from the snapshot coordinates. To each of these was added the protein density computed directly from the snapshot coordinates, and the two “total” 3D densities were averaged over the snapshots. The results are shown in Fig. 5F. We found that the deviations of the S3 model values were small and again lay within the interquartile range of the explicit density.
Figure 5.
Comparison of unconstrained and protein-fixed MD simulations. Protein molecules were aligned by rigid-body rotation and translation in each of 100 frames of the unconstrained MD simulation. A, three z-slices of protein density are shown from a frame of the unconstrained MD simulation. B, corresponding z-slices where the protein atoms were fixed at their X-ray coordinates. C, D snapshots of water density corresponding to A and B. E, the projected solvent density as obtained from slices of 100 frames by summing along the Y coordinate. Bars represent the 25 and 75 percentiles of density values computed from the frames. F, the average protein + solvent density is shown from the 100 frames. Bars indicate the 25 and 75 percentile values as computed directly from the atom density in the unconstrained simulation. For comparison, the red line shows the average values where, instead of using the water coordinates, the S3 model was used to estimate the solvent density for each frame.
Low and high-resolution models
The S3 solvent model that has been considered up to this point describes a peak and a valley in the water density profile (Fig. 2) corresponding to the first hydration shell. These occur on a distance scale, 3–4 Å, that is beyond the resolution of most cryo-EM maps. We therefore considered simplified models that should preserve the overall boundary of water outside the protein density without including the fine details of the water distribution. We first modeled the water density as a simple step function of the radius r. An appropriate distance for the water boundary was found from the first moment of the polar-interaction density function (1) to be r0 = .07 Å, and we ignored the distinction between polar and non-polar interactions. Then a simplified “step function” solvent density function, smoothed by a Gaussian of standard deviation σ0, is given by
| (4) |
We call this the S1 model as it contains a single component, and we combined it with the mask function as before. The use of the error function rather than a step function avoids aliasing artifacts and serves as a Gaussian low-pass filter whose half-power frequency is fc = 0.133 /σ0.
An even simpler model is the one proposed by Jiang and Brünger (1994) where the solvent density is represented as a binary function, simply one or zero, on a closely-spaced lattice. We call this the JB model, which must be evaluated on a finer lattice than our S3 model. An appropriate lattice spacing is 0.5 Å, and we compare this model, after filtering with a 1Å Gaussian filter, with the others in Figs. 4 and 6. In Fig. 4 the JB model is seen to follow the MD solvent less precisely, but most of its values fall within the interquartile intervals. In Fig. 6 A and B it is seen that the JB model shows higher solvent occupancy of internal cavities of the protein, but all three models give similar results. In line projections of slices of the 3D map the S3 model follows the MD densities the best, but the S1 and JB models show peak deviations of only a few percent (Fig. 6C). When the map is filtered to 8 Å resolution the differences are also small (Fig. 6D). We conclude that the simpler models are nearly as good as the S3 model for both high- and moderate-resolution work.
Figure 6.
Comparison of three solvation models. A, the binary solvent model of Jiang and Brünger was used to compute the solvent density for two of the slices shown in Fig. 4. The solvent density was computed on a 0.5 Å grid, and then filtered by convolution with a 1 Å Gaussian function. B, the corresponding S3 models (same as in Fig. 4A). C, from 3D maps filtered at 3Å, a comparison is made of the predictions of the JB model, the simplified S1 (step function) model, and the S3 model for the projection along the y coordinate of the slices shown. Values from the filtered, averaged MD map are shown as red dots. D, the same projections along slices after filtering the 3D maps with an 8Å filter.
At atomic resolution, that is resolutions better than 3 Å, the choices of functions and parameters we have made in the S3 model are expected to result in simulated density maps that are too smooth. We use Gaussians as the scattering profile of both protein atoms and water, with a somewhat arbitrarily chosen standard deviation of 1 Å. Not only does this fail to model the various radii of the atoms, but also the Gaussian function itself is a poor model for the sharply-peaked electron scattering of atoms at high resolution (Kirkland, 2009).
Application to an experimental map
Finally, we compared the model of protein and solvent with a cryo-EM map from a protein of known structure. From the refined X-ray crystal structure of GroEL (Braig et al., 1995) we computed density models from protein alone and with the S3 solvation model. In Fig. 7 we compare these models with the cryo-EM map obtained by single-particle methods by Stagg et al. (Stagg et al., 2008). Shown are models and projection images from one ring of the GroEL barrel (Fig. 7A) and from a 25 Å slice of the ring (Fig. 7B, C). Features of the experimental map are much better reproduced by the solvated model. When it is scaled to fit the experimental map the density peaks within the protein due to alpha helices are well reproduced. When a model without solvent is matched to the experimental map, the modeled excursions of density within the protein are too small. We believe that the discrepancy arises for the same reason as that demonstrated by the simulations in Fig. 1H.
Figure 7.
Comparison of simulations with an experimental cryo-EM structure. Protein-only and protein+solvent maps were computed from the E. coli GroEL X-ray structure (Braig et al., 1995, PDB entry 1OEL) and compared with a corresponding cryo-EM map (Stagg et al., 2008; EM Databank entry EMD-1458). Maps were Gaussian-filtered to a half-power frequency of 0.12 Å−1. A, projection images of one ring (7 subunits) of the GroEL barrel. B, projection of a 12 Å slice of the ring. C, plots of density along a line passing through alpha helices and solvent, as indicated by horizontal lines in B. The experimental cryo-EM density (rightmost plot in C) was scaled and shifted to approximately match the modeled densities. Note that the protein+solvent simulation matches the cryo-EM map better. Before the projection images were computed from the cryo-EM map, that map was filtered so that its radial power spectrum matched that of the protein+solvent model. Computation of the S3 model for the GroEL 3D map (27,078 atoms, 178 Å on a side) required 38 s of CPU time on a notebook computer using our Matlab code. Images are 220 Å on a side.
Conclusion
Using molecular-dynamics simulations as a reference, we obtained radial distribution functions of water molecules from protein surface atoms. Combining these functions with a binary mask, we obtain a continuum model for the interface between solvent and protein. We examine the discrepancies between this model and the mean solvent densities obtained by averaging over MD trajectories, and find that the discrepancies are on the order of the statistical errors in the MD averaging. Simpler models, including the binary solvent model of Jiang and Brünger, also provide a good description, especially at moderate resolution. A direct comparison was made of the modeled density based on the X-ray structure of GroEL and an experimental cryo-EM map. We expect that the algorithms described here will be useful in modeling cryo-EM images of proteins of known structure.
The algorithms were implemented in the Matlab environment (Mathworks, Natick, MA). Code and example scripts for the S1 and S3 models are available at mathworks.com/matlabcentral/fileexchange/33636.
Acknowledgments
This work made use of the public GroEL dataset provided by the National Resource for Automated Molecular Microscopy, and we are grateful to Lauren Fisher of NRAMM for providing us with database information. This work was supported by NIH grant NS21501 to F.J.S.
References
- Braig K, Adams PD, Brünger AT. Conformational variability in the refined structure of the chaperonin GroEL at 2.8 A resolution. Nat. Struct. Biol. 1995;2:1083–1094. doi: 10.1038/nsb1295-1083. [DOI] [PubMed] [Google Scholar]
- Chang W-H, Chiu MT-K, Chen C-Y, Yen C-F, Lin Y-C, Weng Y-P, et al. Zernike phase plate cryoelectron microscopy facilitates single particle analysis of unstained asymmetric protein complexes. Structure. 2010;18:17–27. doi: 10.1016/j.str.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Creighton TE. Proteins: Structures and Molecular Properties. W. H. Freeman; 1993. [Google Scholar]
- Fenn TD, Schnieders MJ, Brünger AT. A smooth and differentiable bulk-solvent model for macromolecular diffraction. Acta Crystallogr D Biol Crystallogr. 2010;66:1024–1031. doi: 10.1107/S0907444910031045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser RDB, MacRae TP, Suzuki E. An Improved Method for Calculating the Contribution of Solvent to the X-ray Diffraction Pattern of Biological Molecules. J. Appl. Crystallography. 1978;11:693–694. [Google Scholar]
- Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- Hall RJ, Nogales E, Glaeser RM. Accurate modeling of single-particle cryo-EM images quantitates the benefits expected from using Zernike phase contrast. J. Struct. Biol. 2011;174:468–475. doi: 10.1016/j.jsb.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jiang JS, Brünger AT. Protein hydration observed by X-ray diffraction. Solvation properties of penicillopepsin and neuraminidase crystal structures. J. Mol. Biol. 1994;243:100–115. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- Kirkland E. Advanced Computing in Electron Microscopy. Springer; 2009. [Google Scholar]
- Lum K, Chandler D, Weeks JD. Hydrophobicity at small and large length scales. J. Phys. Chem. B. 1999;103:4570–4577. [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rullgård H, Öfverstedt L-G, Masich S, Daneholt B, Öktem O. simulation of transmission electron microscope images of biological specimens. J Microsc. 243:234–256. doi: 10.1111/j.1365-2818.2011.03497.x. [DOI] [PubMed] [Google Scholar]
- Stagg SM, Lander GC, Quispe J, Voss NR, Cheng A, et al. A test-bed for optimizing high-resolution single particle reconstructions. J. Struct. Biol. 2008;163:29–39. doi: 10.1016/j.jsb.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese JN, Epa VC, Colman PM. Three-dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase. Protein Sci. 1995;4:1081–1087. doi: 10.1002/pro.5560040606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ounjai P, Sigworth FJ. Streptavidin crystals as nanostructured supports and image-calibration references for cryo-EM data collection. J. Struct. Biol. 2008;164:190–198. doi: 10.1016/j.jsb.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Dadarlat VM, Glaeser RM, Head-Gordon T, Downing KH. Modeling chemical bonding effects for protein electron crystallography: the transferable fragmental electrostatic potential (TFESP) method. Acta Crystallogr., A, Found. Crystallogr. 2002;58:162–170. doi: 10.1107/s0108767301020256. [DOI] [PubMed] [Google Scholar]