Abstract
Exploiting dielectrophoresis (DEP) to concentrate and separate biomolecules has recently shown large potential as a microscale bioanalytical tool. Such efforts however require tailored devices and knowledge of all interplaying transport mechanisms competing with dielectrophoresis (DEP). Specifically, a strong DEP contribution to the overall transport mechanism is necessary to exploit DEP of biomolecules for analytical applications such as separation and fractionation. Here, we present improved microfluidic devices combining optical lithography and focused ion beam milling (FIBM) for the manipulation of DNA and proteins using insulator-based dielectrophoresis (iDEP) and direct current (DC) electric fields. Experiments were performed on an elastomer platform forming the iDEP microfluidic device with integrated nanoposts and nanopost arrays. Microscale and nanoscale iDEP was studied for λ-DNA (48.5 kbp) and the protein bovine serum albumin (BSA). Numerical simulations were adapted to the various tested geometries revealing excellent qualitative agreement with experimental observations for streaming and trapping DEP. Both the experimental and simulation results indicate that DC iDEP trapping for λ-DNA occurs with tailored nanoposts fabricated via FIBM. Moreover, streaming iDEP concentration of BSA is improved with integrated nanopost arrays by a factor of 45 compared to microfabricated arrays.
Keywords: dielectrophoresis, DNA, protein, numerical simulation, trapping condition
1. Introduction
Manipulating biomolecules in homogeneous and inhomogeneous fields builds the basis of a variety of analytical separation techniques [1]. Inhomogeneous fields form the basis of gradient techniques, which can be based e.g. on magnetic, electric thermal or centrifugal forces [1,2,3]. Among the gradient techniques, dielectrophoresis (DEP) occurs in an electric field gradient while forces acting on a dipole result in migration upwards or downwards an electric field gradient. This migration phenomenon has recently attracted interest for the application to biomolecules, such as DNA [4,5] and proteins [6,7]. Acting as the dominating force, DEP can be applied for trapping [8,9,10], whereas overlaid with transport mechanisms such as hydrodynamic flow or electroosmostic flow (EOF), DEP can also be employed for separation, sorting and streaming concentration [11,12,13]. Knowledge of all transport mechanisms as well as experimental methods to develop suitable microfluidic devices are required to exploit DEP for biomolecules as a routine tool in analytical applications. In this contribution, we investigate micro- and nano-environments to improve our understanding of factors influencing streaming and trapping DEP for DNA and proteins.
The selectivity of DEP is based on the polarizability of biomolecules in a non-uniform electric field. The theoretical basis of DEP for large bio-particles such as cells and the scaling behavior for nanoparticles is well understood [14]. DEP manipulation of bio-particles in the nm scale is challenging since high electric fields are required to achieve sufficiently large forces for DEP trapping competing with diffusion [15] and dissipative electro-thermal forces [16]. Nonetheless, examples for biomolecule manipulation employing DEP have been demonstrated in the past. The first attempts reach back to the work of Washizu [17]. Further improvements demonstrated DEP manipulation with DNA and proteins [5,6,18]. Examples for DNA manipulation by DEP reach from Mbp down to ~40bp [12,16,19,20,21,22,23,24], whereas protein manipulation by DEP has only recently been explored [15,25,26,27,28,29,30,31].
Several experimental approaches have been proposed to create inhomogeneous electric field gradients evoking DEP while realized in microfluidic devices in the majority of cases [32]. Patterned microelectrodes achieve inhomogeneous field gradients, which can be integrated into microfluidic environments [32,33]. Integrating microelectrodes involves disadvantages, such as electrode reactions occurring in the DEP manipulation regions and field gradients localized around the microelectrodes [34,35]. 3-Dimensional glassy-carbon electrodes provide an alternative, as they allow electric fields gradients to be applied homogeneously throughout the entire microchannel depth [36]. Recent advances in the field of DEP have resulted in new approaches for characterizing the behavior of particles using direct current (DC) electric fields. In such approaches, spatial non-uniformities are created by embedding insulating obstacles in the channel to create non-uniform electric field gradients, which was demonstrated for the first time by Lapizco-Encinas et al. [37]. This emerging field of DEP is a 3D system commonly termed DC insulator-based dielectrophoresis (DC-iDEP) or simply insulator-based dielectrophoresis (iDEP). This technique provides several advantages over the traditional AC DEP approaches [30,37]. Insulators are less prone to fouling; that is, they generally retain their function despite surface changes. Further, insulators are robust and chemically inert compared to metallic electrodes avoiding undesired reactions at their surface. Depending on the material of the microfluidic device, electroosmotic flow (EOF) can be used for transporting analyte into the channel thus eliminating external pumps. Furthermore, the fabrication of iDEP devices is simpler as no metal deposition steps are required compared to the electrode based DEP devices. A variety of designs have been proposed and tested to create non uniform electric fields using either single insulating obstacles such as a rectangular posts [38,39,40,41], a triangular hurdle [40], a constriction in the depth of the channel [42], a single microchannel constriction [43], multiple insulating obstacles like insulating post [26,37,44], multiple rectangular blocks [45] or silica microbeads [46]. In addition, modified channel geometries were employed to achieve particle sorting or trapping such as aligned teeth [47], sawtooth channels [48], open-top microstructures [49] or serpentine microchannels [50].
iDEP has been used by several groups [30] including us [12,19,26] to manipulate biomolecules. Exploiting iDEP for separation, pre-concentration or fractionation is particularly favorable in these devices, as the necessary electric field gradients are homogeneous throughout the entire depth of a microchannel. DC-iDEP techniques however result in strong electrokinetic forces and interfere with DEP. Electrokinetic forces result from charged biomolecules, which are consequently transported by electrophoresis (EP). Moreover, EOF arises in the majority of microfluidic environments additionally acting as a bulk transport mechanism. As a result of electrokinesis, different transport regimes arise in iDEP [13,37]. First, if the DEP component is the dominating transport mechanism, the so-called trapping regime is reached. Molecules transported in the iDEP device are immobilized or trapped at specific locations in the microdevice. This regime is however difficult to achieve for small biomolecules, as large electric field gradients are necessary to evoke strong DEP forces over electrokinetic transport. The second regime refers to streaming DEP, where DEP still influences the transport mechanism. However, electrokinesis is dominating and trapping regions immobilizing particles are now replaced by regions of preferred streaming. This regime results in areas of streaming regions with increased concentration throughout a micropost array. For example, this streaming DEP behavior has recently been demonstrated for a 150 kDa and a 60 kDa protein in a DC iDEP device exhibiting a micropost array [26]. Hence, conditions in which DEP is preferably occurring for biomolecules, i.e. a trapping regime is apparent, are yet to be accomplished in DC iDEP and are important for exploiting DEP in analytical applications for biomolecules.
To achieve larger DEP contribution in DC iDEP, our approach involves the enhancement of electric field gradients to favor DC-iDEP over electrokinetic forces based on a combination of micro- and nanofabrication. It relies on replica molding of a nanostructured device, the master piece, transferring the nanostructure from the negative relief to the elastomeric material. This method is commonly termed soft lithography, and has been applied in similar approaches [51,52,53]. Here, we base the optimization of the master piece on the combination of standard photolithography techniques with focused ion beam milling (FIBM). Etching by FIBM is used for direct (mask free) processing of micro- and nanostructures [54]. However, FIBM has traditionally not been used for insulating substrates. A major problem in ion etching technology is related to the difficulty of calculating and selecting ion beam parameters for obtaining a preset surface geometry, especially in the case of dielectric targets with complex compositions [55,56]. Here, however, we present micro- and nanostructured patterns on SU-8 photoresist combining optical lithography and FIBM serving as the master piece to mold microfluidic devices for DEP manipulation of biomolecules. This approach thus builds on our previously developed iDEP devices obtained from optical lithography. The resulting microfluidic device has feature sizes of <1 µm well below the feature sizes reported by Shafiee et al. [57] realized in conjunction with deep reactive ion etching, and does not involve tedious multiple process steps such as those developed by Yamahata et. al. [58], Iliescu et. al. [59] and Liao et. al. [31].
With this novel approach, we investigate DC-iDEP for λ-DNA and the protein bovine serum albumin (BSA). We base this study on our previous work in which DC-iDEP streaming of proteins has been observed in an array of triangular microposts [26]. The integration of nanoposts and arrays thereof in the triangular iDEP device geometry was investigated to reveal the improvement in iDEP streaming concentration for DNA eventually leading to DC-iDEP trapping. For BSA, improved streaming concentration was observed. Theoretical considerations using a convection-diffusion model were in excellent agreement with our experimental study for DNA and confirm DC-iDEP trapping in distinct geometries.
2. Theory
Dielectrophoresis (DEP) is the migration of polarizable particles under the influence of an inhomogeneous electric field. Depending on the conductivity of the particle and the surrounding medium, the particle can move towards high electric field gradients, known as positive DEP, or away from regions of high electric field gradients as in negative DEP [14,60].
For DC voltages, the DEP force competes with electrokinetic forces in addition to diffusion. Our goal is to employ numerical simulations considering a convection-diffusion model to optimize the microfluidic channel geometries in DC-iDEP and thereby augmenting the dielectrophoretic component of biomolecule transport. For numerical simulations and as below investigated experimentally, we consider a large DNA molecule, namely λ-DNA (48.5 kbp), and the protein BSA.
The flux of particles, J⃗, in a microfluidic channel at steady state according to our convection-diffusion model is defined as:
| (1) |
where c is the concentration of the particles, u⃗EK is the electrokinetic velocity and u⃗DEP is the DEP velocity. D is the diffusion coefficient, which values 6.8×10−13 m2/s [19] and 6.1×10−11 m2/s [61] for λ-DNA and BSA, respectively.
The combined electrokinetic velocity results from EOF and electrophoresis as follows:
| (2) |
Here, µEOF is the electrosmotic mobility, µEP the electrophoretic mobility and E⃗ the electric field. The electrosmotic mobility in the employed microchannels in a dynamic or a static coating procedure of the tri-block-copolymer F108 has been determined to be 0.53×10−8 m2/Vs [62] and 1.5×10−8 m2/Vs [63], respectively. We note that the static coating procedure was employed for proteins, whereas the dynamic coating procedure was used for DNA. The electrophoretic mobility of λ-DNA (48.5 kbp) was previously determined as −3.5×10−8 m2/Vs [12]. In the case of BSA, we consider that µEP is considerably smaller than µEOF and of opposite sign. This is reasoned because our experimental observations reflected a strong cathodic EOF, confirming that electrophoresis counteracts electroosmosis only to a marginal extent. Therefore, we conducted simulations using an overall electrokinetic mobility of −3.0×10−8 m2/Vs for λ-DNA and 1.5×10−8 m2/Vs for BSA considering only µEOF contribution in equation (2).
The DEP velocity results from the equilibrium of DEP and drag forces and is described as [14,60]:
| (3) |
where µDEP is the dielectrophoretic mobility. This µDEP is the ratio of a particle’s polarizability, α, and it’s friction coefficient, f with [14]. Using the Einstein relation, we can determine the friction coefficient [64]:
| (4) |
where k is the Boltzmann constant and T the temperature. Thus, one can obtain µDEP for a biomolecule from known α and f. For a λ-DNA molecule, α amounts in 3.10×10−29 F/m2 as previously reported by Regtmeier et al. [19]. Therefore, we calculate a positive µDEP for λ-DNA of 2.61×10−21 m4/sV2 with the diffusion coefficient for λ-DNA as noted above. For the case of BSA, we assume a positive µDEP of 8.6×10−24 m4/sV2 as employed in our previous study [26].
Using COMSOL 4.2a software, we solve equation (1) at steady state. This model is useful to predict the concentration profile along a channel when the electrokinesis dominates over DEP, which is known as streaming behavior [13,30]. However, under trapping conditions where DEP forces dominate over electrokinesis, accumulation arises and [65]. Thus, under accumulation conditions, the steady-state condition via the convection-diffusion model cannot be found. Hence, we analyze the condition of trapping by calculating the electric field and ∇E2 in our microfluidic channel as previously described [66,67,68]. In the case of trapping, the particle migration is strongly governed by DEP and the direction of the flux perpendicular to E⃗ is null. It follows:
| (5) |
Substituting for J⃗ and omitting the diffusion term we write:
| (6) |
and
| (7) |
Equation (7) is useful to determine the trapping condition, i.e. the situation in which DEP dominates and accumulation is observed. When µDEP / µEK < 0, the condition of trapping is defined as:
| (8) |
Thus, from numerical simulations, we would expect the DEP trapping to occur when this condition is satisfied in a specific microfluidic design in the case of λ-DNA. If equation (8) reveals values smaller than unity, DEP streaming behavior is apparent. Consequently, when equation (8) is satisfied in a specific design, we plot these values for a nanostructured design. If however streaming DEP is predicted from numerical simulations for a specific nanostructured design, we further compute the concentration distribution obtained by solving equation (1) in the steady-state.
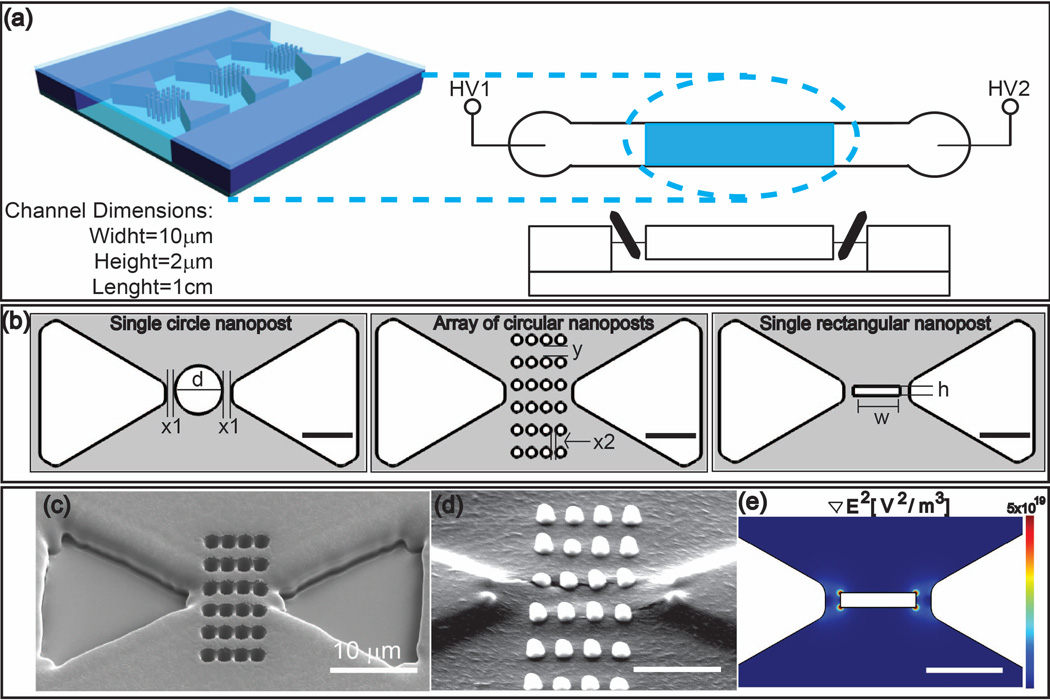
We explore different geometries varying in shape and dimensions as shown in Figure 1-b. Triangular microposts as shown in Figure 1a-c are fabricated by optical lithography. Using FIBM, we fabricate three different nanopost geometries in between the posts: a single circular post, an array of circular posts and a rectangular post, as shown in Figure 1-b.
Figure 1.
a) Schematic of the microfluidic device (not to scale): The upper right part shows a schematic top view with shaded areas indicating the position of the micropost array. The lower right part shows a cross section along the longitudinal axis with electrodes integrated in the reservoirs. The enlargement on the left shows a triangular micropost array with an integrated nanopost array. For simplicity, only one nanopost array per row is shown. b) Dimensions of the constrictions for a circular post in between triangles (left), circular array in between triangles (middle), and rectangular post in between triangles (right); detailed dimensions are found in Table 1. c) SEM image of the master piece after FIBM and optical lithography. d) Corresponding PDMS mold using the master piece in c) images with SEM. The height of the circular post array is ~2µm). e) ∇E2 is shown exemplarily for a rectangular nanopost design. The scale bar is 5 µm unless indicated in the figure.
3. Materials and Methods
3.1 Chemicals
Double-stranded 48.5kbp λ-phage DNA was obtained from Fermentas Inc. Alexa Fluor 488 labeled BSA and YOYO-1 iodide (excitation maximum 491 / emission maximum 509 nm) was acquired from Invitrogen (NY, USA). Na2HPO4, KH2PO4, poly(ethyleneglycol)-block-poly(propyleneglycol)-block-poly(ethylene glycol) (Pluronic F108) and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) were purchased from Sigma-Aldrich. Deionized water (Millipore, USA) was used for all buffers. Sylgard 184, composed of the silicone elastomer base and the curing agent for poly(dimethylsiloxane) (PDMS), was purchased from KR Anderson (AZ, USA). The negative photoresist SU-8 2007 and the developer were obtained from Microchem.
3.2 Device Fabrication
A schematic of the microfluidic device is shown in Figure 1-a. All channels were 2 µm high, 100 µm wide and 1 cm long. A combination of photolithography and FIBM was used to fabricate a master, from which a PDMS mold was formed. First, the master relief of SU-8 photoresist (2µm thickness) was patterned via photolithography on a silicon wafer using standard photolithography. This wafer was coated with a 20nm Cr layer using Cressington 308R Evaporator (Ted Pella, Inc., USA). After that, FIBM was used to mill the nanostructures as shown in Figure 1-b with a Nova 200 (FEI Company, USA). A current of 3 nA was used to mill the patterns. Characteristic dimensions of resulting posts are listed in Table 1 and defined in Figure 1-b. From this master wafer, a PDMS mold was formed resulting in channel cross sections of 2µm ×100 µm. Figure 1 c–d shows scanning electron microscopy images of the obtained master wafer and the resultant PDMS nano-posts respectively. Reservoir holes with a diameter of 2mm were punched at corresponding positions in the PDMS mold. Finally, the micro/nano structured PDMS mold was O2-plasma activated under vacuum (PDC-001 Harrick Plasma, Harrick, USA). Simultaneously, a cleaned glass slide was treated with O2-plasma under similar conditions. The glass slide and PDMS mold were assembled directly after plasma activation.
Table 1.
Dimensions of posts in the microfluidic channel as indicated in Figure 1-b.
| Geometry | d (µm) | x1(µm) | x2(µm) | y(µm) | w(µm) | h (µm) |
|---|---|---|---|---|---|---|
| Circular posts | 2.5 | 1 | NA | NA | NA | NA |
| Circular array of posts | 1 | 0.75 | 0.5 | 2.3 | NA | NA |
| Rectangular posts | NA | 1 | NA | NA | 5 | 1 |
3.3 Dielectrophoresis Experiments
Phosphate buffer (10 mM) was diluted with water to obtain a conductivity of 0.01 S/m resulting in pH=8.1. Before the experiment, PDMS channels and reservoirs were filled with 250µM F108 in phosphate buffer at 0.01 S/m for 10min under a low potential of 100 V. Next, 40pM λ-DNA in phosphate buffer containing YOYO-1 in a base pair ratio of 1:10 was added to one of the reservoirs. Potentials of 1500 V and 3000 V were employed to manipulate DNA by DEP.
Fluorescently labeled BSA was used in the same buffer as λ-DNA. However, the coating strategy was different. The channel was filled with 250µM F108 in buffer solution and incubated over night. Subsequently, the buffer was exchanged with a solution of 3mg/mL of CHAPS. Next, 7 nM BSA was added in one of the reservoirs and a potential of 1500 V was applied to manipulate BSA by DEP.
The DEP behavior of labeled DNA and BSA was observed by fluorescence microscopy on an inverted fluorescence microscope (IX 71, Olympus, USA) with filter sets for YOYO-1 and Alexa Fluor 488 fluorescence and a 60× objective (LUC Plan, Olympus, US). Fluorescence intensity was recorded with a CCD-Camera (Quantum 512SC, Photometrics, USA) at 100 ms exposure time. Image J software (version 1.43) was used for the data analysis. We analyzed the increase of intensities at either streamline or trapping areas at which biomolecules concentrated as obtained from the recorded images with the concentration factor R, which is given in equation 9:
| (9) |
where IM refers to the maximum intensity for either streamline or trapping areas at applied potential, IPDMS to the background intensity of PDMS outside the channel and IC to the intensity of the channel filled with analyte prior to the application of potential.
4. Results and Discussion
Three different geometries were used as schematically depicted in Figure 1-b to improve DC-iDEP concentration of DNA and proteins. We base this study on our previous work in which streaming DEP with proteins has been demonstrated. Hence, we use a well-characterized triangular micro-post array and study the improvement of iDEP with the integration of nanoposts by FIBM. We report the fluorescence distribution from the experimental work using the combined micro- and nanostructured device, reflecting the DNA and protein concentration, as well as the concentration profile obtained by numerical simulations using the above described convection-diffusion model.
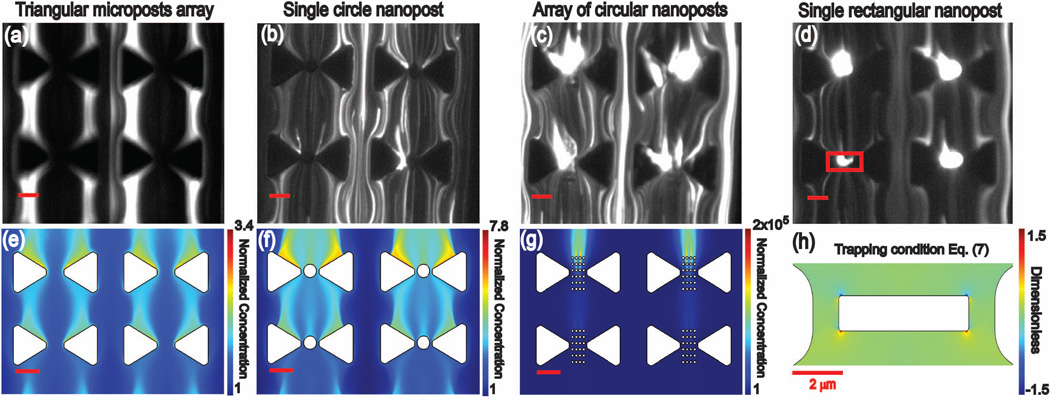
Figure 2 summarizes the experimental and numerical simulation studies for λ-DNA. In the case of triangular microstructures (no nanoposts), experimental results reveal streaming DEP along the triangular posts at 3000 V as shown in Figure 2-a. This streaming behavior for microposts leads to an increase in DNA concentration in regions close to the center of the microposts and can be assigned to biomolecules exhibiting positive DEP [26]. We thus refer to positive streaming behavior along the micro and nanopost arrays. This behavior is in accordance with previous simulations for streaming in the case of positive biomolecule DEP and observed positive DEP for similar buffer ionic strength and pH [12,19]. Note that negative streaming DEP would deplete regions that are concentrated in positive streaming DEP [26].
Figure 2.
Experimental results and numerical simulations for triangular microposts as well as the combination of micro- and nanostructures under DC-iDEP of λ-DNA. The flow direction in all cases is from bottom to top. Scale bar is 10µm unless indicated in the figure. The applied potential was 1500 V except for Figures a) and e) which were performed at 3000 V.
Figure a–d. Fluorescence microcopy snapshots obtained from video microscopy imaging. a) triangular microposts, b) single circular nanopost, c) array of circular nanoposts and d) single rectangular nanopost, the red rectangle marked in this figure is zoom for figure 2-h. Note that a) and b) indicate streaming iDEP while c) and d) figures indicate iDEP trapping.
Figure e–g. Simulation results under streaming regime. The concentration profile is plotted solving equation (1). e) triangular microposts f) a single circular nanopost, g) array of circular nanoposts post, and d) a single rectangular post.
Figure h. Equation (7) is plotted in the accumulation regime for the case of the rectangular nanopost, in the area indicated with the rectangle in figure 2-d. Note that the trapping condition is met for values >1 (red colors) according to eq. (8).
Similar results were observed when a single circular post is included in between the triangular posts, as shown in Figure 2-b, where streaming behavior is maintained. The DNA iDEP streaming behavior further changed significantly for integrated nano arrays of circular posts and an integrated rectangular post as demonstrated in Figures 2-c and d. In these two geometries, the characteristic profile of streaming DEP reduces significantly. We attribute this change to a transition from streaming to trapping DEP in the nanopost regions, as apparent in Figure 2-c, caused by larger DEP forces. Figure 2-c shows an even larger concentration around the nanopost, suggesting even stronger iDEP trapping. It is noteworthy that DEP trapping behavior was achieved for both the nanopost array and the rectangular nanopost geometry at half the electric field applied in comparison to Figure 2a (microposts only). Therefore, we note that trapping conditions were obtained at lower applied potentials through the integration of the nanoposts.
Table 2 summarizes the experimentally obtained concentration factor R as well as numerically calculated maximum ∇E2 values for the studied geometries. As shown in Table 2, a ~1.3-fold increase of fluorescence intensity is observed with the circular nano post compared to the triangular microposts, indicating an improvement of DNA streaming concentration. In the case of the circular nanopost array, the fluorescence intensity is further enhanced at the regions of the nanoposts by ~5-fold. This is augmented to ~30-fold for the rectangular case.
Table 2.
Maximum ∇E2 obtained by numerical simulations within different geometries and concentration factor R from experimental results for λ-DNA at 1500 V/cm unless indicated.
(n.d. = not determined, n.a.=not applicable)
| Geometry | ∇E2 max. [V2/m3] |
R for DNA | R for BSA | Rnm-posts/RTriangles for λ-DNA at 1500 V |
|---|---|---|---|---|
| Triangular array of microposts |
9.4×1016 | 35 ± 3.5 | 0.24** | n.a. |
| Triangular array of microposts* |
3.8×1017 | 87 ± 5.2 | n.d. | n.a. |
| Circular nanopost | 6.9×1018 | 46 ± 6.9 | n.d. | 1.3 |
| Circular array of nanoposts | 2.0×1018 | 173 ± 29.4 | 11 ± 0.9 | 5 |
| Rectangular nanopost | 5.2×1019 | 039 ± 145 | n.d. | 30 |
at 3000 V
from reference at 3000 V [26]
To further characterize this nanostructure-dependent DEP behavior, we carried out numerical simulations to reveal the acting ∇E2 (see Table 2) and concentration distribution in the iDEP channel solving equation (1), for cases in which the trapping condition (8) was not satisfied. Figure 1-e shows the ∇E2 exemplarity for rectangular nanopost. Note that the highest gradients are in between the triangular posts and the corners of the rectangular nanopost which is similar to the other nanopost geometries. Figures 2-(e–g) show the concentration profiles at steady-state for the case of λ-DNA and varying geometry. For triangular posts, we observe that our simulations match qualitatively well with our experimental results. They reflect the regions of streaming concentration obtained in experiments and show the concentration enhancement accordingly.
Next, we compare the experimental results for the single circular post compared to the numerical simulation. From Table 2, we calculate an enhancement of ∇E2 by 77% at 1500 V compared to the triangular microarrays, indicating a larger DEP force. Moreover, Figure 2-f shows a change in the position of the concentration profile, which is also observed experimentally in Figure 2-b. Although the streaming regime is maintained similarly to the case of triangular microposts, the concentration distribution is now located not only along the triangular posts but also around the single circular post. Numerical simulations reflect this effect demonstrating concentration along the microposts but also along the circular post (Figure 2-f).
In the case of the circular nanopost array, simulation results suggest that streaming behavior is still apparent but the shift of largest concentration to the nanopost regions is more pronounced (Figure 2-g). Table 2 also shows a higher ∇E2 value; hence, we attribute a larger DEP component to this nanopost array geometry even though streaming DEP is still dominating in simulations. However, experimental results (Figure 2-c) indicate DEP trapping at 1500 V. This discrepancy between simulation and experimental work may arise from irregularities in the FIBM process as well as from interactions of DNA molecules with posts or entanglement of DNA molecules at the nanopost regions that are not captured by simulations.
We further analyze the case of the rectangular nanopost. In this case, we plot the trapping regions according to equation (7) (see Figure 2-h) since equation (1) cannot be solved for the steady-state to obtain the concentration distribution. In Figure 2-h, the areas in which trapping occurs are represented by red colors (i.e. equation (8) is valid). Although this region is a very localized on a length scale of 100 nm, we suspect this area is sufficient to trap DNA molecules with persistence lengths of 50 nm [69,70]. Note that the experimentally observed trapping regions are much larger which we attribute to the molecular extension of the large λ-DNA molecules. Moreover, the rectangular nanopost increases the maximum ∇E2 by ~2.5 orders of magnitude (see Table 2), confirming largely augmented DEP forces. Our detailed study thus reveals conditions under which larger DNA molecules can be trapped by DC-iDEP.
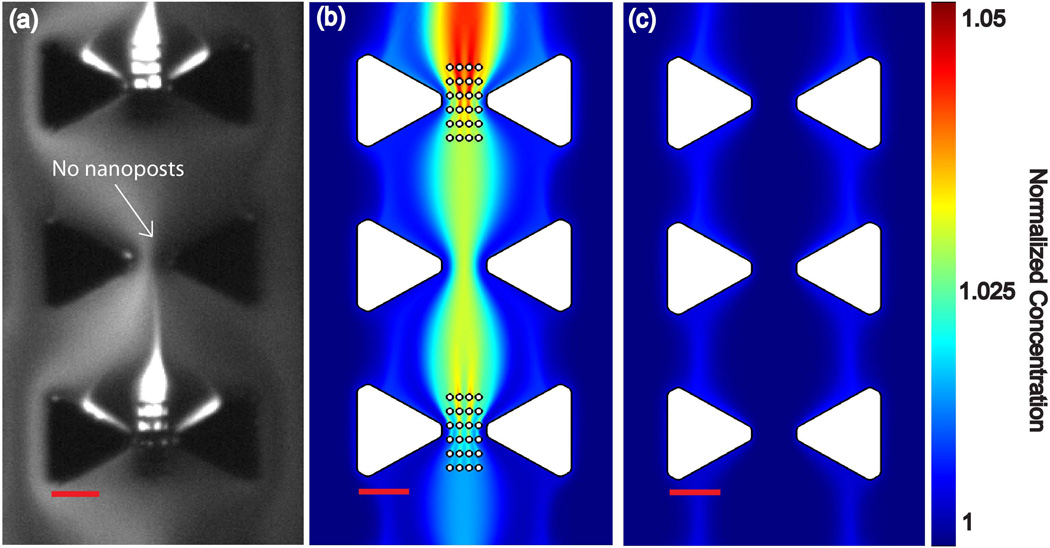
Finally, we investigated the improvement of iDEP-based concentration for proteins. Experimentally, we employed the protein BSA and compared to numerical simulations with adapted parameters (see theory section). Figure 3-a presents the iDEP streaming concentration of BSA, obtained with the nanopost array. We note that iDEP streaming is expected as µDEP for proteins is three orders of magnitude lower than for DNA (8.6×10−24 m4/V2s for protein vs 2.61×10−21 m4/V2s for DNA). Further, a concentration factor of 11 is obtained as shown in Table 2. This is a factor of ~45 more intense than observed in our previous work using only triangular microstructures [26]. Although numerical simulations in this geometry show an improved concentration by adding the nanopost array, the concentration factor according to the numerical simulation is only ~5% for the nanoposts array vs. ~2% for the microtriangular posts, see Figures 3 b–c. These differences in quantitative concentration between the numerical simulations and experimental may arise from irregularities in the FIBM process leading underestimated electric field gradients. Deviations in the estimation of µDEP, which was adapted to our previous study on DEP for an immunoglobulin G molecule [18,26] for BSA, are an additional uncertainty factor. We note that protein DEP is still little understood and factors contributing to µDEP are very likely to change for different proteins. Moreover, particle deformation and particle-particle interaction are additional factors that are not captured in our model and may contribute to the discrepancy in experimental observations and computations. Nonetheless, our theoretical study allows predicting changes in streaming iDEP of proteins due to variation in the device geometry.
Figure 3.
a) Experimental observation of streaming iDEP of BSA using the circular nanopost array in between the triangle microposts. Note that every other facing triangle was processed with a nanopost array. b) Concentration distribution obtained by numerical simulation solving eq. 1, qualitatively matching the experimental results. c) Concentration distribution obtained by numerical simulation without nanoposts solving eq. 1. The color scale for the concentration applies to both b and c. Scale bar is 10µm.
5. Conclusions
We conducted a thorough experimental and numerical study revealing parameters that improve iDEP-based concentration of λ-DNA and the protein BSA. While streaming DEP is observed in micropost arrays, the integration of nanoposts leads to an increase in concentration when a single circular nanopost is embedded due to the DEP enhancement. Moreover, when a nanopost array with closer spacing of posts or a rectangular nanopost is integrated, iDEP trapping is observed under DC conditions experimentally. These larger DEP components are shown through increased ∇E2 resulting from numerical simulations in the tested nanostructures. Moreover, numerical simulations according to a convection-diffusion model predict the concentration increase due to iDEP streaming in reasonable agreement with experimental observations. Discrepancies in the on-set of trapping between experiment and simulation remain for the nanopost array, which we attribute to the fact that the molecular structure of DNA is not adequately captured in our model as well as additional phenomena such as particle-particle interaction and particle deformation. Furthermore, we revealed that the integration of a rectangular array leads to iDEP trapping under DC conditions in excellent agreement with the trapping condition obtained theoretically.
While we predicted a DEP force for BSA is three orders of magnitude lower than for λ-DNA streaming behavior in the nanopost array is increased as observed experimentally and predicted by numerical simulation. Although there is a quantitative discrepancy with numerical simulations compared with experimental results, simulations predict an enhancement in concentration in the nanostructure array which was confirmed experimentally. In summary, our study provides details on the improvement of nanostructures for iDEP trapping under DC conditions. The results also reveal that iDEP trapping will require largely improved ∇E2 and further down-scaled geometries to achieve protein trapping under DC conditions. Compared to existing approaches for protein DEP, our approach involves a simple elastomer molding process to produce iDEP devices. We anticipate downscaling to dimensions lower than 100nm suitable for protein trapping by iDEP will be achieved in the future with improved FIBM.
Acknowledgements
The financial support by grants from the National Center for Research Resources (5R21RR025826-03) and the National Institute of General Medical Sciences (8R21GM103522-03) from the National Institutes of Health is gratefully acknowledged. The authors also thank the LeRoyEyring Center for Solid State Science at ASU for allowing us to use the focused ion beam facilities and Grant Baumgardner for his technical support on the FIBM.
Glossary
- EOF
Electroosmosis
- DEP
Dielectrophoresis
- iDEP
insulator-based Dielectrophoresis
- FIBM
Focused Ion Beam Milling
Biographies
Fernanda Camacho-Alanis earned her Ph.D. degree in Electrical Engineering in 2010 from the University of Virginia completing her thesis on the fabrication of molecular electronic devices on semiconductor substrates. She received her Mater’s degree as well as her BS from the National University of Mexico, Mexico City. Ms. F. Camacho-Alanis won an Award for Excellence in Scholarship as well as the Graduate Research Award at University of Virginia in 2009. She is currently a Postdoc in the Department of Chemistry and Biochemistry Arizona State University working in the area of microfluidics.
Lin Gan received her B.S. degree in Polymer Materials and Engineering in 2005 from College of Chemistry, Jilin University, Changchun, China. She is currently pursuing her Ph. D. Degree in Department of Chemistry and Biochemistry at Arizona State University. She is working with Dr. Alexandra Ros in her Bioanalytical Chemistry Lab, concentrating on the study of dielectrophoretic behavior of DNA structures in microfluidic systems.
Alexandra Ros received her Diploma in Chemistry from the Ruprecht-Karls University in Heidelberg, Germany, and her PhD from École Polytechnique Fédérale de Lausanne (EPFL) in Switzerland. She performed her post doc at Bielefeld University, Germany, where she subsequently received her habilitation and venia legendi in Biophysics in 2007. Since 2008, she is Assistant Professor in the Department of Chemistry and Biochemistry at Arizona State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meighan MM, Staton SJR, Hayes MA. Bioanalytical separations using electric field gradient techniques. Electrophoresis. 2009;30:852–865. doi: 10.1002/elps.200800614. [DOI] [PubMed] [Google Scholar]
- 2.Lin CC, Hsu JL, Lee GB. Sample preconcentration in microfluidic devices. Microfluidics and Nanofluidics. 2011;10:481–511. [Google Scholar]
- 3.Shackman JG, Ross D. Counter-flow gradient electrofocusing. Electrophoresis. 2007;28:556–571. doi: 10.1002/elps.200600592. [DOI] [PubMed] [Google Scholar]
- 4.Kuzyk A. Dielectrophoresis at the nanoscale. Electrophoresis. 2011;32:2307–2313. doi: 10.1002/elps.201100038. [DOI] [PubMed] [Google Scholar]
- 5.Hoelzel R. Dielectric and dielectrophoretic properties of DNA. IET Nanobiotechnology. 2009;3:28–45. doi: 10.1049/iet-nbt.2008.0014. [DOI] [PubMed] [Google Scholar]
- 6.Lapizco-Encinas BH, Rito-Palomares M. Dielectrophoresis for the manipulation of nanobioparticles. Electrophoresis. 2007;28:4521–4538. doi: 10.1002/elps.200700303. [DOI] [PubMed] [Google Scholar]
- 7.Garza-Garcia LD, Lapizco-Encinas BH. State of the art on protein manipulation employing dielectrophoresis. Rev. Mex. Ing. Quim. 2010;9:125–137. [Google Scholar]
- 8.Lewpiriyawong N, Yang C, Lam YC. Electrokinetically driven concentration of particles and cells by dielectrophoresis with DC-offset AC electric field. Microfluidics and Nanofluidics. 2012;12:723–733. [Google Scholar]
- 9.Henslee EA, Sano MB, Rojas AD, Schmelz EM, Davalos RV. Selective concentration of human cancer cells using contactless dielectrophoresis. Electrophoresis. 2011;32:2523–2529. doi: 10.1002/elps.201100081. [DOI] [PubMed] [Google Scholar]
- 10.Chen D, Du H. A microfluidic device for rapid concentration of particles in continuous flow by DC dielectrophoresis. Microfluidics and Nanofluidics. 2010;9:281–291. [Google Scholar]
- 11.Ajdari A, Prost J. Free-flow electrophoresis with trapping by a transverse inhomogeneous field. Proc. Natl. Acad. Sci. USA. 1991;88:4468–4471. doi: 10.1073/pnas.88.10.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regtmeier J, Duong TT, Eichhorn R, Anselmetti D, Ros A. Dielectrophoretic manipulation of DNA: Separation and polarizability. Anal. Chem. 2007;79:3925–3932. doi: 10.1021/ac062431r. [DOI] [PubMed] [Google Scholar]
- 13.Cummings EB. Streaming dielectrophoresis for continuous-flow microfluidic devices. IEEE Eng. Med. Biol. Magazine. 2003;22:75–84. doi: 10.1109/memb.2003.1266050. [DOI] [PubMed] [Google Scholar]
- 14.Jones TB. Electromechanics of Particles. Cambridge: Cambridge University; 1995. [Google Scholar]
- 15.Holzel R, Calander N, Chiragwandi Z, Willander M, Bier FF. Trapping single molecules by dielectrophoresis. Phys. Rev. Lett. 2005;95 doi: 10.1103/PhysRevLett.95.128102. [DOI] [PubMed] [Google Scholar]
- 16.Chaurey V, Polanco C, Chou CF, Swami NS. Floating-electrode enhanced constriction dielectrophoresis for biomolecular trapping in physiological media of high conductivity. Biomicrofluidics. 2012;6 doi: 10.1063/1.3676069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Washizu M, Suzuki S, Kurosawa O, Nishizaka T, Shinohara T. Molecular dielectrophoreisis of biopolymers. IEEE Trans. Ind. Appl. 1994;30:835–843. [Google Scholar]
- 18.Clarke RW, Piper JD, Ying LM, Klenerman D. Surface conductivity of biological macromolecules measured by nanopipette dielectrophoresis. Physical Review Letters. 2007;98 doi: 10.1103/PhysRevLett.98.198102. [DOI] [PubMed] [Google Scholar]
- 19.Regtmeier J, Eichhorn R, Bogunovic L, Ros A, Anselmetti D. Dielectrophoretic Trapping and Polarizability of DNA: The Role of Spatial Conformation. Anal. Chem. 2010;82:7141–7149. doi: 10.1021/ac1005475. [DOI] [PubMed] [Google Scholar]
- 20.Henning A, Bier FF, Hoelzel R. Dielectrophoresis of DNA: Quantification by impedance measurements. Biomicrofluidics. 2010;4 doi: 10.1063/1.3430550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tuukkanen S, Kuzyk A, Toppari JJ, Hakkinen H, Hytonen VP, Niskanen E, Rinkio M, Torma P. Trapping of 27 bp-8 kbp DNA and immobilization of thiol-modified DNA using dielectrophoresis. Nanotechnology. 2007;18 [Google Scholar]
- 22.Ying LM, White SS, Bruckbauer A, Meadows L, Korchev YE, Klenerman D. Frequency and voltage dependence of the dielectrophoretic trapping of short lengths of DNA and dCTP in a nanopipette. Biophys. J. 2004;86:1018–1027. doi: 10.1016/S0006-3495(04)74177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tegenfeldt JO, Prinz C, Cao H, Huang RL, Austin RH, Chou SY, Cox EC, Sturm JC. Micro- and nanofluidics for DNA analysis. Anal. and Bioanal. Chem. 2004;378:1678–1692. doi: 10.1007/s00216-004-2526-0. [DOI] [PubMed] [Google Scholar]
- 24.Chou CF, Tegenfeldt JO, Bakajin O, Chan SS, Cox EC, Darnton N, Duke T, Austin RH. Electrodeless dielectrophoresis of single- and double-stranded DNA. Biophys. J. 2002;83:2170–2179. doi: 10.1016/S0006-3495(02)73977-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lapizco-Encinas BH, Ozuna-Chacon S, Rito-Palomares M. Protein manipulation with insulator-based dielectrophoresis and direct current electric fields. J. Chromatogr., A. 2008;1206:45–51. doi: 10.1016/j.chroma.2008.05.077. [DOI] [PubMed] [Google Scholar]
- 26.Nakano A, Chao T-C, Camacho-Alanis F, Ros A. Immunoglobulin G and bovine serum albumin streaming dielectrophoresis in a microfluidic device. Electrophoresis. 2011;32:2314–2322. doi: 10.1002/elps.201100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunda NSK, Mitra SK. Modeling of dielectrophoretic transport of myoglobin molecules in microchannels. Biomicrofluidics. 2010;4 doi: 10.1063/1.3339773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clarke RW, White SS, Zhou DJ, Ying LM, Klenerman D. Trapping of proteins under physiological conditions in a nanopipette. Angew. Chem. 2005;44:3747–3750. doi: 10.1002/anie.200500196. [DOI] [PubMed] [Google Scholar]
- 29.Clarke RW, Piper JD, Ying L, Klenerman D. Surface conductivity of biological macromolecules measured by nanopipette dielectrophoresis. Phys. Rev. Lett. 2007;98 doi: 10.1103/PhysRevLett.98.198102. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava SK, Gencoglu A, Minerick AR. DC insulator dielectrophoretic applications in microdevice technology: a review. Anal. Bioanal. Chem. 2011;399:301–321. doi: 10.1007/s00216-010-4222-6. [DOI] [PubMed] [Google Scholar]
- 31.Liao K-T, Chou C-F. Nanoscale Molecular Traps and Dams for Ultrafast Protein Enrichment in High-Conductivity Buffers. J. American Chem. Soc. 2012;134:8742–8745. doi: 10.1021/ja3016523. [DOI] [PubMed] [Google Scholar]
- 32.Khoshmanesh K, Nahavandi S, Baratchi S, Mitchell A, Kalantar-zadeh K. Dielectrophoretic platforms for bio-microfluidic systems. Bios. Bioelectron. 2011;26:1800–1814. doi: 10.1016/j.bios.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 33.Zhang C, Khoshmanesh K, Mitchell A, Kalantar-zadeh K. Dielectrophoresis for manipulation of micro/nano particles in microfluidic systems. Anal. Bioanal. Chem. 2010;396:401–420. doi: 10.1007/s00216-009-2922-6. [DOI] [PubMed] [Google Scholar]
- 34.Wu J. Interactions of electrical fields with fluids: laboratory-on-a-chip applications. IET Nanobiotechnol. 2008;2:14–27. doi: 10.1049/iet-nbt:20070023. [DOI] [PubMed] [Google Scholar]
- 35.Regtmeier J, Eichhorn R, Viefhues M, Bogunovic L, Anselmetti D. Electrodeless dielectrophoresis for bioanalysis: Theory, devices and applications. Electrophoresis. 2011;32:2253–2273. doi: 10.1002/elps.201100055. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Duarte R, Renaud P, Madou MJ. A novel approach to dielectrophoresis using carbon electrodes. Electrophoresis. 2011;32:2385–2392. doi: 10.1002/elps.201100059. [DOI] [PubMed] [Google Scholar]
- 37.Lapizco-Encinas BH, Simmons BA, Cummings EB, Fintschenko Y. Dielectrophoretic concentration and separation of live and dead bacteria in an array of insulators. Anal. Chem. 2004;76:1571–1579. doi: 10.1021/ac034804j. [DOI] [PubMed] [Google Scholar]
- 38.Kang KH, Kang YJ, Xuan XC, Li DQ. Continuous separation of microparticies by size with direct current-dielectrophoresis. Electrophoresis. 2006;27:694–702. doi: 10.1002/elps.200500558. [DOI] [PubMed] [Google Scholar]
- 39.Kang KH, Xuan XC, Kang YJ, Li DQ. Effects of dc-dielectrophoretic force on particle trajectories in microchannels. J. Appl. Phys. 2006;99 [Google Scholar]
- 40.Kang Y, Li D, Kalams SA, Eid JE. DC-Dielectrophoretic separation of biological cells by size. Biomed. Microdevices. 2008;10:243–249. doi: 10.1007/s10544-007-9130-y. [DOI] [PubMed] [Google Scholar]
- 41.Kang Y, Cetin B, Wu Z, Li D. Continuous particle separation with localized ACdielectrophoresis using embedded electrodes and an insulating hurdle. Electrochim. Acta. 2009;54:1715–1720. [Google Scholar]
- 42.Hawkins BG, Smith AE, Syed YA, Kirby BJ. Continuous-flow particle separation by 3D insulative dielectrophoresis using coherently shaped, dc-biased, ac electric fields. Anal. Chem. 2007;79:7291–7300. doi: 10.1021/ac0707277. [DOI] [PubMed] [Google Scholar]
- 43.Zhu J, Tzeng T-RJ, Xuan X. Continuous dielectrophoretic separation of particles in a spiral microchannel. Electrophoresis. 2010;31:1382–1388. doi: 10.1002/elps.200900736. [DOI] [PubMed] [Google Scholar]
- 44.Cummings EB, Singh AK. Dielectrophoresis in microchips containing arrays of insulating posts: Theoretical and experimental results. Anal. Chem. 2003;75:4724–4731. doi: 10.1021/ac0340612. [DOI] [PubMed] [Google Scholar]
- 45.Lewpiriyawong N, Yang C, Lam YC. Dielectrophoretic manipulation of particles in a modified microfluidic H filter with multi-insulating blocks. Biomicrofluidics. 2008;2 doi: 10.1063/1.2973661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iliescu C, Xu G, Loe FC, Ong PL, Tay FEH. A 3-D dielectrophoretic filter chip. Electrophoresis. 2007;28:1107–1114. doi: 10.1002/elps.200600431. [DOI] [PubMed] [Google Scholar]
- 47.Pysher MD, Hayes MA. Electrophoretic and dielectrophoretic field gradient technique for separating bioparticles. Anal. Chem. 2007;79:4552–4557. doi: 10.1021/ac070534j. [DOI] [PubMed] [Google Scholar]
- 48.Chen KP, Pacheco JR, Hayes MA, Staton SJR. Insulator-based dielectrophoretic separation of small particles in a sawtooth channel. Electrophoresis. 2009;30:1441–1448. doi: 10.1002/elps.200800833. [DOI] [PubMed] [Google Scholar]
- 49.Jen CP, Chen TW. Trapping of cells by insulator-based dielectrophoresis using open-top microstructures. Microsystem Tech.-Micro-Nanosyst.-Information Storage and Processing Systems. 2009;15:1141–1148. [Google Scholar]
- 50.Zhu JJ, Tzeng TRJ, Hu GQ, Xuan XC. DC dielectrophoretic focusing of particles in a serpentine microchannel. Microfluidics and Nanofluidics. 2009;7:751–756. [Google Scholar]
- 51.Renault JP, Bernard A, Bietsch A, Michel B, Bosshard HR, Delamarche E, Kreiter M, Hecht B, Wild UP. Fabricating arrays of single protein molecules on glass using microcontact printing. J. Phys. Chem. B. 2003;107:703–711. [Google Scholar]
- 52.Park S-M, Liang X, Harteneck BD, Pick TE, Hiroshiba N, Wu Y, Helms BA, Olynick DL. Sub-10 nm nanofabrication via nanoimprint directed self-assembly of block copolymers. ACS Nano. 2011;5:8523–8531. doi: 10.1021/nn201391d. [DOI] [PubMed] [Google Scholar]
- 53.Pan T, Wang W. From Cleanroom to Desktop: Emerging Micro-Nanofabrication Technology for Biomedical Applications. Ann. Biomed. Eng. 2011;39:600–620. doi: 10.1007/s10439-010-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baglin JEE. Ion beam nanoscale fabrication and lithography-A review. Appl. Surf. Sci. 2012;258:4103–4111. [Google Scholar]
- 55.Ali MY, Hung W, Fu YQ. A review of focused ion beam sputtering. Int. J. Precision Eng. Manuf. 2010;11:157–170. [Google Scholar]
- 56.Evstrapov AA, Mukhin IS, Kukhtevich IV, Bukatin AS. Using focused ion beam for nanosized structure formation in microfluidic chips. Tech. Phys. Lett. 2011;37:956–959. [Google Scholar]
- 57.Shafiee H, Caldwell JL, Sano MB, Davalos RV. Contactless dielectrophoresis: a new technique for cell manipulation. Biomed. Microdevices. 2009;11:997–1006. doi: 10.1007/s10544-009-9317-5. [DOI] [PubMed] [Google Scholar]
- 58.Yamahata C, Collard D, Legrand B, Takekawa T, Kunternura M, Hashiguchi G, Fujita H. Silicon nanotweezers with subnanometer resolution for the micromanipulation of biomolecules. Journal of Microelectromechanical Systems. 2008;17:623–631. [Google Scholar]
- 59.Iliescu C, Yu L, Xu G, Tay FEH. A dielectrophoretic chip with a 3-D electric field gradient. J. Microelectromech. Syst. 2006;15:1506–1513. [Google Scholar]
- 60.Pohl H. Dieletrophoresis. Cambridge: Cambridge University; 1978. [Google Scholar]
- 61.Raj T, Flygare WH. Diffusion studies of bovine serum-albumin by quasi-elastic lightscattering. Biochemistry. 1974;13:3336–3340. doi: 10.1021/bi00713a024. [DOI] [PubMed] [Google Scholar]
- 62.Manchanda S. Master Thesis. Arizona State University; 2012. Surface modification of polydimethylsiloxane (PDMS) using polyethylene oxide polymers. [Google Scholar]
- 63.Hellmich W, Regtmeier J, Duong TT, Ros R, Anselmetti D, Ros A. Poly(oxyethylene) based surface coatings for poly(dimethylsiloxane) microchannels. Langmuir. 2005;21:7551–7557. doi: 10.1021/la0510432. [DOI] [PubMed] [Google Scholar]
- 64.Einstein A. The motion of elements suspended in static liquids as claimed in the molecular kinetic theory of heat. Ann. Phys. 1905;17:549–560. [Google Scholar]
- 65.Plawsky JL. Transport Phenomena Fundamentals. New York: Marcel Dekker, Inc.; 2001. [Google Scholar]
- 66.Davalos RV, McGraw GJ, Wallow TI, Morales AM, Krafcik KL, Fintschenko Y, Cummings EB, Simmons BA. Performance impact of dynamic surface coatings on polymeric insulator-based dielectrophoretic particle separators. Anal. Bioanal. Chem. 2008;390:847–855. doi: 10.1007/s00216-007-1426-5. [DOI] [PubMed] [Google Scholar]
- 67.Baylon-Cardiel JL, Lapizco-Encinas BH, Reyes-Betanzo C, Chavez-Santoscoy AV, Martinez-Chapa SO. Prediction of trapping zones in an insulator-based dielectrophoretic device. Lab on a Chip. 2009;9:2896–2901. doi: 10.1039/b906976c. [DOI] [PubMed] [Google Scholar]
- 68.Kwon J-S, Maeng J-S, Chun M-S, Song S. Improvement of microchannel geometry subject to electrokinesis and dielectrophoresis using numerical simulations. Microfluidics and Nanofluidics. 2008;5:23–31. [Google Scholar]
- 69.Ariel G, Andelman D. Persistence length of a strongly charged rodlike polyelectrolyte in the presence of salt. Phys. Rev. E. 2003;67 doi: 10.1103/PhysRevE.67.011805. [DOI] [PubMed] [Google Scholar]
- 70.Kassapidou K, Heenan RK, Jesse W, Kuil ME, Vandermaarel JRC. Effects of ionic-strength on the supramolecular structure in liquid-crystalline solutions of persistent length DNA fragments. Macromolecules. 1995;28:3230–3239. [Google Scholar]