Preface
The gut microbiota, the trillions of microbes inhabiting the human intestine, is a complex ecological community that through its collective metabolic activities and host interactions, influences both normal physiology and disease susceptibilities. Understanding factors underlying compositional and functional changes will aid in designing therapies that target the gut microbiota. This goal is formidable because of the immense diversity of the microbiota, interpersonal variation and temporal fluctuations in composition, especially during disease and early development. Here, we describe recent advances in understanding gut microbiota from an ecological perspective, and discuss how these insights might promote health by guiding therapeutic strategy development.
Most gut microbes are harmless or beneficial to the host. The gut microbiota protects against enteropathogens1,2, extracts nutrients and energy from our diets 3,4, and contributes to normal immune function5. Dysbiosis, disruption of the normal balance between the gut microbiota and host, has been associated with obesity6,7, malnutrition8, inflammatory bowel diseases (IBD) 9,10, neurological disorders11, and cancer12. Understanding how the gut microbiota affects health and disease therefore requires a shift in focus from individual pathogens, towards an ecological approach considering the community as a whole.
The first step towards understanding the symbiotic relationships of gut microbes with their hosts is to characterize the baseline healthy microbiota and differences that are associated with disease. Large-scale efforts including Meta-HIT 13 and the Human Microbiome Project (HMP) 14 have made substantial progress. Once we understand the desired compositional and functional states of the gut microbiota, we can determine which features, when disrupted, are associated with disease. However, the complexity of the microbiota, and intra- and inter-subject variability, complicate the definition of what a “desired” state may look like for a population or for a given individual.
Ecological principles have increasingly aided understanding of host-microbe interactions and specific gut microbial functions. Improved sequencing technologies and other “omics” technologies (such as proteomics and metabolomics), coupled with metabolic network modeling15,16, show how host and environmental factors affect gut microbial ecology over the human lifespan. The composition, diversity and function of gut microbial communities could potentially inform personalized nutritional and drug strategies (Fig. 1). In this review, we summarize recent progress towards characterizing the diversity and function of microbial communities in the healthy human gut, describe ways in which this ecosystem can go awry, and discuss prospects for ecosystem restoration.
Fig. 1. Maintaining our gut microbial lawn.
Maintaining a healthy microbiota is in some ways like lawn care: severe interventions like antibiotics can take the ecosystem back to bare earth, requiring it to be re-established from scratch. Although many people recover naturally, it is by no means guaranteed, and “weedy” species that are adapted to perturbed ecosystems often run wild. In this review, we discuss several current strategies for ecosystem restoration: probiotics (re-seeding with a few well-defined “good microbes”), prebiotics (adding compounds that are thought to specifically promote the growth of beneficial microbes), and fecal bacteriotherapy (transplanting the entire microbial ecosystem, e.g. from a stool sample). These strategies are analogous to using lawn seed, turf food, and sod respectively. An additional strategy, not shown, is to use specific drugs that target undesirable members of the microbial community such as narrow-spectrum antibiotics. Although we are beginning to learn what a healthy microbial community looks like and to recognize signs of weeds, our understanding of which strategies for altering the microbiota work best, and predicting which will work for a given individual, is still in its infancy.
Microbial diversity in the healthy gut
Even basic questions about the diversity of the gut microbiota remained unanswered until the recent advent of higher-throughput sequencing: How much diversity exists in the human microbiota and microbiome (the collection of genes represented in the microbiota) at the species level and at higher taxonomic levels? Which features of the microbiota, such as species or functions, are ubiquitous and which are unique to an individual? To what extent might we predict the functions in a community based on knowledge of the species present?
Taxonomic Diversity
Culture-based studies suggested that all healthy adults share most gut bacterial species, constituting a “core microbiota.” For example, Escherichia coli can be isolated from most people. However, culture-independent sequencing studies (Fig. 2) have repeatedly demonstrated a vast microbial diversity that is highly variable both over time and across human populations, defying the concept of a core. Each of us harbors an estimated >1000 species-level phylotypes (i.e. clusters of sequences with about as much diversity in their small subunit rRNA genes as in validly named species)17. Most of these phylotypes are bacteria belonging to just a few phyla. In adults, Bacteroidetes and Firmicutes usually dominate, whereas Actinobacteria, Proteobacteria, and Verrucomicrobia are frequent but generally minor constituents18 (Fig. 3). Our microbiota also contains methanogenic archaea (mainly Methanobrevibacter smithii), eukarya (mainly yeasts), and viruses (primarily phage)19. Despite the consistency of these major components, their relative proportions and the species present vary dramatically between individuals (Figs. 3, 4). Attempts to identify a core set of species-level phylotypes in the adult gut microbiota have yielded several “major players”, including Faecalibacterium prausnitzii, Roseburia intestinalis, and Bacteroides uniformis13, although even these species can be at <0.5% relative abundance in some individuals 20. As studies have expanded to include developing countries and a broad range of ages (from infancy to the ninth and tenth decade of life) 4,21, the notion that there is a core set of shared species in the human gut microbiota has been weakened further.
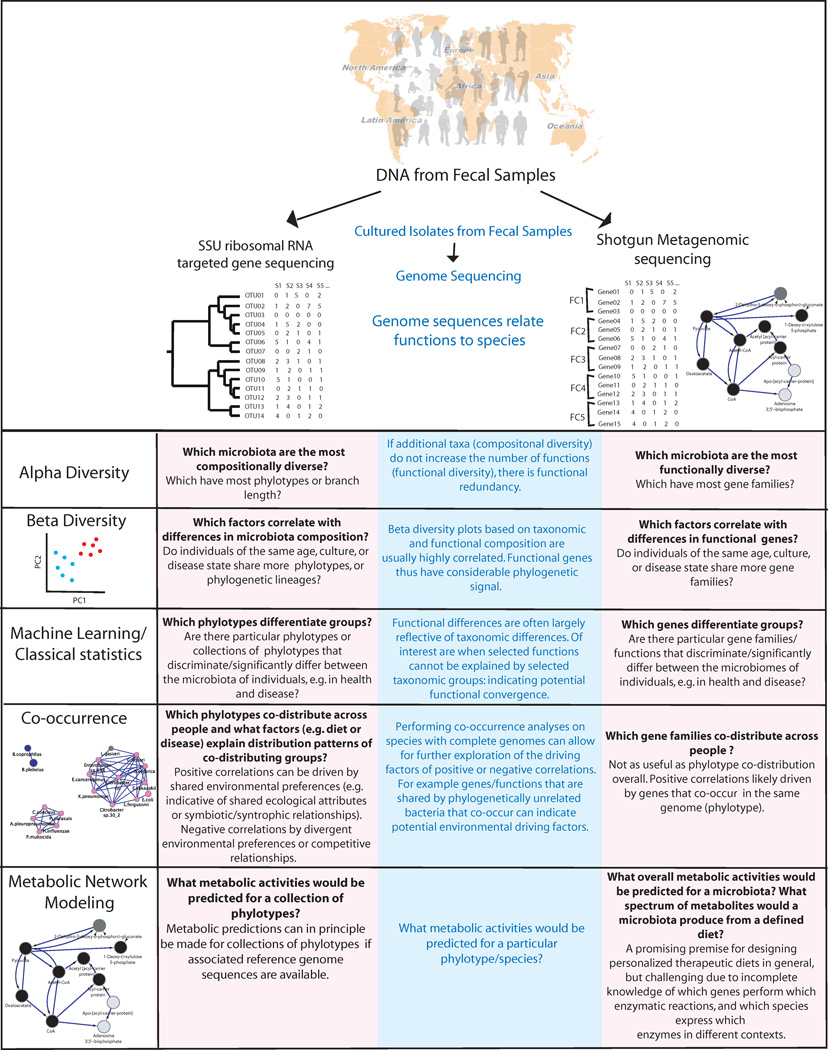
Fig. 2. Tools for understanding compositional and functional diversity of the microbiota, and for generating hypotheses about functionally important genes and how to modulate metabolic phenotypes.
Extracted DNA from fecal samples can be assessed using targeted sequencing of a phylogenetically informative gene (usually SSU rRNA) or random sequencing of all genes. Genome sequences from cultured isolates link these two datasets by indicating which species contain which genes, and therefore functions. Shotgun metagenomic data is thus substantially more useful as the number of reference genomes continues to increase with additional strain sequencing efforts. SSU rRNA gene sequences are usefully related to each other in phylogenetic trees, because related phylotypes (clusters of similar sequences defined by sequence similarity) generally have more similar functional attributes. Functional genes can be binned into functional categories (FC) that are a part of a functional ontology, but those encoding proteins that perform known enzymatic reactions are most usefully related to each other using metabolic networks, because genes that are adjacent in a particular metabolic pathway can produce a phenotype in concert with each other. Compositional and functional diversity patterns can inform each other. They are often highly correlated, but cases where these general correlations do not hold can be biologically or ecologically important. Predicting functions from the species assemblage present still remains an unsolved problem, although the fact that overall genome differences are highly correlated with differences in the SSU rRNA sequence suggests that such predictions may one day be possible. To date, the most powerful studies tend to combine SSU rRNA profiling to determine taxon abundance (the microbiota) with shotgun metagenomic profiling to understand the functions present (the microbiome). Supplementing these studies with mRNA, protein and metabolite level analyses of community samples (and of concurrently obtained host specimens, such as serum and urine) will be crucial so that we can move from in silico predictions of function to direct measurements of expressed community properties.
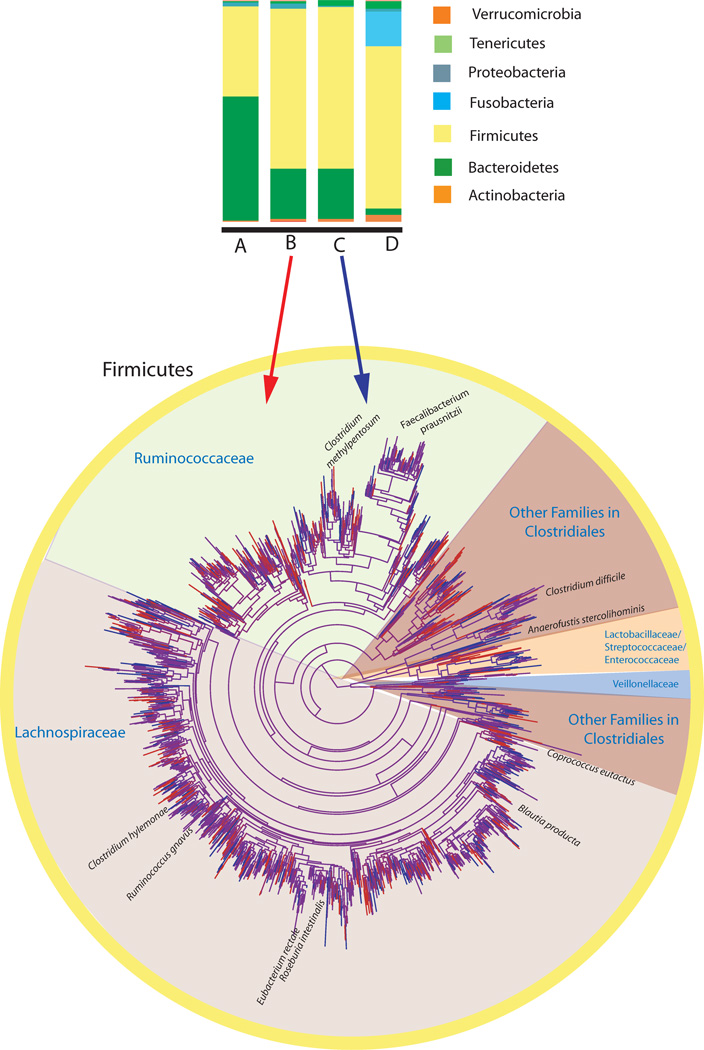
Fig. 3. Diversity of the human microbiota at different phylogenetic scales.
The human microbiota displays a remarkable degree of variation within and between individuals. Although this complexity can be simplified by evaluating communities at higher taxonomic levels, such as comparing relative abundances of phyla, the many species within each phylum have different biological properties, and significant changes detected at higher taxonomic levels are likely driven by only a subset of the species in those higher taxa. Here we illustrate the high diversity and variability among individuals, and the degree to which taxonomic grouping at high levels can mask this diversity, using 16S rRNA sequence data from four of the US adults previously described in ref. 4. We chose these four individuals to illustrate how phylum level diversity can vary dramatically even across healthy adults in the same population. Individual A has an unusually high proportion of Bacteroidetes, individual D unusually high Fusobacteria, and individuals B and C have more typical phylum level distributions for this cohort, dominated by Firmicutes and Bacteroidetes. However, even the apparently similar B and C differ at finer scales. The tree depicts the phylogenetic relationships between species-level phylotypes in just the Firmicutes phylum, by far the most diverse of the phyla, in individuals B and C. Branches specific to individual B are red, branches specific to individual C are blue, and shared branches are purple. Each individual has many unique phylotypes not found in the other. As described in many surveys of the human gut 14,18,40, the Ruminococcaceae and Lachnospiraceae families are particularly rich in phylotypes.
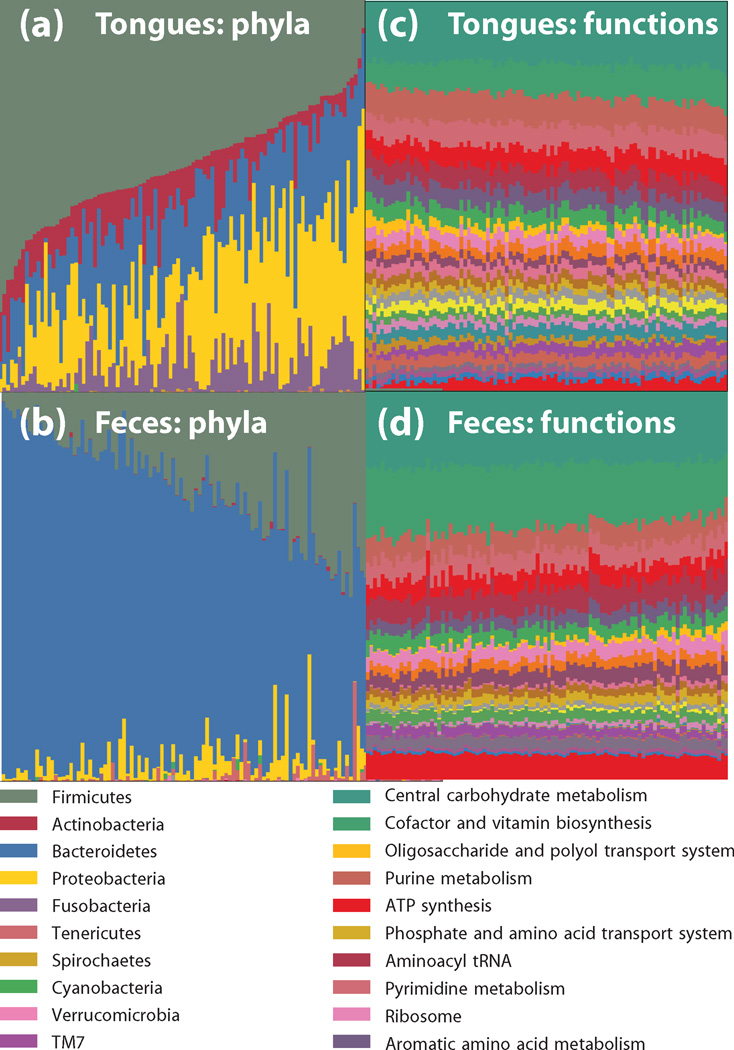
Fig. 4. Functional redundancy.
Microbial ecosystems exhibit a high degree of functional redundancy in microbial ecosystems may mirror that in macroecosystems. The HMP dataset illustrates this principle: oral communities (a) and fecal communities (b) show tremendous diversity in species abundance, yet remarkable similarities to one another in functional profiles obtained by shotgun metagenomics from the same samples (c) and (d) respectively.
Functional Diversity
Microbial community composition alone does not necessarily provide understanding of community function. Functional information comes in part from study of cultured isolates that are well-characterized in terms of in their genome content and ex vivo phenotypes and from sequencing community DNA. Functional screening by shotgun metagenomics relies on sequencing total microbial community DNA, including from uncultured members, and matching the sequences to those of known functional genes in databases (Fig. 2). Identification of genes involved in specific metabolic pathways can lead to predictions of functional capabilities but in lieu of mRNA, protein and metabolite profiling, these remain predictions. This reference-mapping process is improving as additional human gut microbial genomes are sequenced and annotated22 and as more complementary “omics” datasets become available 23,24.
Despite having highly divergent gut microbiota compositions, functional gene profiles are quite similar in different individuals (Fig. 4). This principle was first seen in 18 females who all shared >93% of ‘enzyme’ level functional groups, but few ‘genus-level’ phylotypes20; the HMP and MetaHit confirmed this result in much larger populations 13,14. Core functions of the gut microbiota include central metabolic pathways and pathways particularly important in the gut including carbohydrate and amino acid metabolism20. However, not all pathways are represented in the core, and grouping genes into broad functional categories can also conceal meaningful inter-individual differences in function that occur at finer scales. Variable functions restricted to species or strain, including pathogenicity islands, vitamin and drug catabolism, motility and nutrient transporters, are intriguing targets for personalized diets and therapeutic strategies.
Many genes are expressed only under specific conditions. Shotgun sequencing approaches that measure levels of mRNA (“metatranscriptomics”) or shotgun proteomics (“metaproteomics”) may uncover functional variation with disease, diet or other factors, that DNA studies overlook. For example, genes involved in carbohydrate metabolism and energy generation were expressed as proteins at higher levels than predicted from metagenome data, underscoring that these processes are important in the gut23.
The overall pattern from studies of the core is that we share a functional core microbiome, but not a core microbiota. This concept may be understood by analogy to macroecosystems. Rainforests in different parts of the world, for example, are highly similar visually and in many functional respects, yet are composed of different species that have independently evolved. For ecological studies of the gut, a key challenge is to understand functional redundancy, (i.e. which members of the community have similar functional niches and can substitute for one another). Although taxonomic gene composition is far more variable than functional gene composition, at least at a general process level, several studies have shown correlation between the two 4,14,25. Deviations from predictions about function from a microbiota may be critical for identifying and understanding functional components associated with altered physiological states (Fig. 2).
Factors driving normal variation
Having established that the normal gut microbiota is highly variable, we must next understand why it varies, so that we can use this information for tailored therapies or clinical trials. For example, the extent to which family members harbor similar microbes will determine whether family histories of microbiota-driven diseases is informative. The extent to which the microbiota varies with age or pregnancy should be taken into account when designing cohorts. The sensitivity of the microbiota to external factors such as diet, will inform the most promising strategies for treating microbiome-linked diseases. Recent studies have clarified how these factors affect the microbiota.
Role of age
Dramatic changes in the gut microbiota occur during early life, with an increase in diversity and stability over the first three years 4,26,27 (Fig. 5). The maturation of the human microbiota is thus an example of ecological succession 4,26,27, in which communities undergo consecutive compositional and functional changes following initial colonization until a relatively stable “climax community” is established.
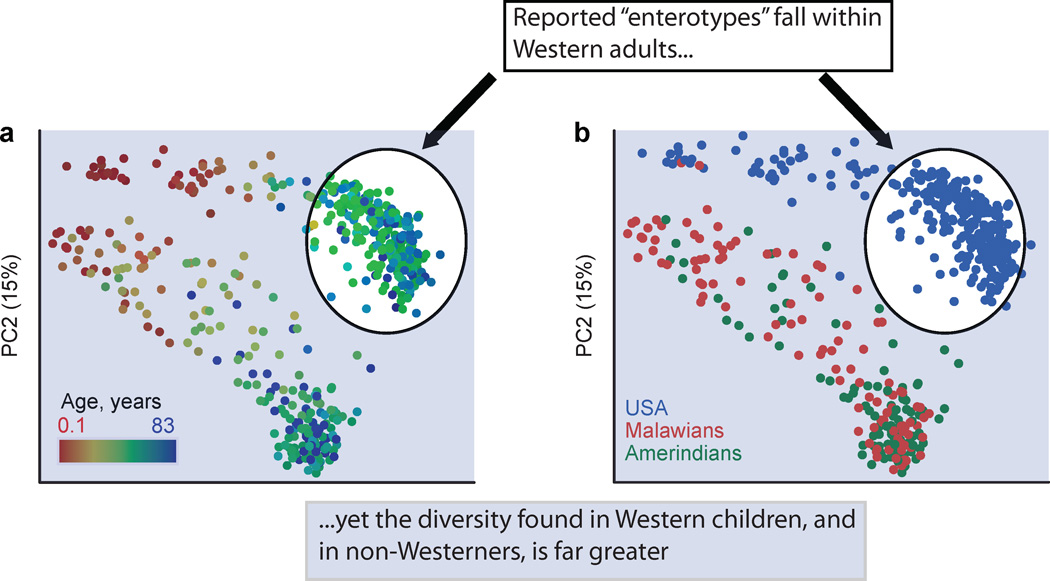
Fig. 5. Human microbial diversity and “enterotypes”.
The reported “enterotypes”31 were determined when evaluating only individuals from the US and Europe, yet including children from the US and children and adults from developing countries greatly expands the picture of human-associated microbiota diversity. We illustrate this here by showing the relationship between the microbiota of 531 healthy infants, children, and adults from Malawi, Venezuelan Amerindians, and the US that were evaluated using sequences from the 16S rRNA gene in fecal samples and a PCoA analysis of unweighted UniFrac distances (adapted from ref. 4 Fig. S2). Microbiota diversity is explained primarily by age (with infants differentiating strongly from adults) and next by culture (with adults from the US having distinct composition compared to adults from Malawi and Venezuelan Amerindians). The points from Western adults are circled in white, and the rest are shaded in blue.
The infant microbiota is relatively volatile. Interpersonal variation in both microbial communities and functional gene repertoires is greater in infants than adults. This observation is replicated in rural Malawian African populations and Venezuelan Amerindian populations, and in metropolitan populations of European and African ancestry in the USA 4. However, infant microbiomes share characteristic properties across individuals and populations. These characteristics are both compositional (many Bifidobacteria, and lower species richness, than adults) and functional (higher representation of genes encoding enzymes involved in folate biosynthesis 4).
Antibiotics, breastfeeding status, and delivery mode all have large effects on the infant microbiota, although whether microbiota differences in early life ultimately affect adult microbiota composition is not well understood28,29. Compositional differences driven by these factors in infancy, however, may affect susceptibility to immunologic diseases, including asthma and atopic diseases into adulthood29. A possible mechanism for this susceptibility has been demonstrated in mice. Germ-free GF) mice accumulate invariant natural killer T (iNKT) cells in the colonic lamina propria and lung, increasing morbidity in models of IBD and allergic asthma 5. Colonization of neonatal-but not adult-GF mice with a conventional microbiota protected the animals, indicating that infancy is a critical time for contact with the microbiota5. Further studies are needed to identify specific components of the human microbiota that shape our immune system in early life.
Role of genetics, environment and diet
The relative role of genetics and environment, including diet, in shaping the human microbiome is still unclear in part because these factors are often confounded. Related individuals, including twins and mother-daughter pairs, have more similar microbiota compositions, initially suggesting that human genetics influences the microbiota9,20. However, monozygotic and dizygotic adult twins share equally similar microbiota, so shared environment rather than genes may drive familial similarities20.
Characteristic differences separate the gut microbiota in different populations, including children in Italy and Burkina Faso30, and both children and adults in Malawi, Venezuela and the US 4. In the latter study, the US population was a clear outlier relative to the two populations in developing countries (Fig. 5) 4. Although genetically different, these populations also differ in many other factors potentially affecting the microbiota, including environmental exposures, hygiene, diet and antibiotic use. Cultural factors, especially diet, may thus be crucial in shaping the gut microbiota. The ratio of two major genera of gut bacteria, Prevotella and Bacteroides, correlates surprisingly well with overall diversity patterns across healthy adults (see discussion of enterotypes below) 31,32. Prevotella were enriched in children with a high-fiber diet in a rural African village of Burkina Faso30 and in children and adults in Malawi and Venezuelan populations with diets dominated by maize-, cassava-, and other plant-derived polysaccharides. In contrast, individuals from the US had more Bacteroides 4. Within healthy US adults, differences in long-term diet correlated with these same genera. Bacteroides were associated with a long-term diet rich in animal protein, several amino acids, and saturated fats (prevalent in the US and Europe), whereas Prevotella were associated with carbohydrates and simple sugars32 (prevalent in agrarian societies). The relative importance for health of microbiota changes due to diet versus other factors remains a subject of active investigation.
Understanding how varying human cultural traditions affect the microbiota should illuminate factors underlying dramatic differences in microbiota-associated disease incidence. For example, the incidence of IBD and allergy is greater in industrialized Western societies than in traditional agrarian cultures33. Deeper insight will require expanded studies that sample more populations and control for confounding factors. Perhaps we need a “Human Microbiome Diversity Project” to parallel the Human Genome Diversity Project34. Populations in which we can measure many variables that might correlate with microbiota diversity will be especially valuable. These variables include history of antibiotic usage, diet, and environmental exposures. The latter may require sampling not only from individuals but also from their environments. Because microbiota exposure in early life is particularly important for the development of diseases of an immunologic basis 5, prospective studies that enroll subjects in infancy, or even prenatally, will be especially valuable. Immigrant populations provide an opportunity to disentangle host genetics, geography, and culture when trying to determine disease etiology, because the incidence of some diseases matches that present in the place of destination rather than that in the place of origin 35.
Microbiota variability and human health and medicine
Studies of compositional and functional differences in the gut microbiota lay the foundation for relating these differences to human health. For example, differences in the microbiota and the microbiome may help explain interpersonal variations in gut metabolic processes, including metabolism of drugs and dietary substrates11,36. Because many of these metabolic pathways are outside the common functional core, they can underlie host-specific responses. For example, many health benefits of soy-rich diets, including positive outcomes for vasomotor symptoms, osteoporosis, prostate cancer, and cardiovascular disease, have been attributed to S-(-)equol produced from the soy isoflavone diadzein by bacterial rather than human enzymes37. Only 25–30% of the adult population of Western countries produce S-(-)equol when fed soy foods, compared to a 50–60% frequency in adults from Japan, Korea, or China38. Thus cancer-protective effects of soy described in Asian populations might not generalize to Westerners because of differences in key components of the microbiota. Similarly, gut microbiota indirectly determine whether acetaminophen will be metabolized to acetaminophen sulfate or acetaminophen glucuronide, potentially altering both efficacy and toxicity of this widely used analgesic36. Microbes mediate this metabolic phenotype by producing the compound p-cresol, which competes with acetaminophen for human enzymes catalyzing sulfonation36. Therefore, the results of drug trials conducted in one population (e.g. performed inexpensively in Africa or South Asia) may not generalize to a population that has a substantially different gut microbiota (e.g. Western populations). Understanding how the microbiota varies across the human population, and correlating this variability with specific microbial functions, is thus emerging as a component of personalized medicine.
Stable configurations of healthy microbiota
The landscape of stable states or “regimes” for the human gut is still unknown. Samples obtained over time from the same individual are more similar to one another compared to those obtained from other individuals, suggesting that each person has a relatively distinct, stable community 20,39–42. This temporal stability can be related to the concept that stable equilibrium states exist for the microbiota, in which disturbances, stochasticity, and temporal dynamics of individual microbes produce change yet the community is still drawn to a central attractor 43,44.
It would be especially convenient for clinical purposes if a few, clearly differentiable stable states could be used to stratify the gut microbiome across the human population. For example, if microbes in one state were known to metabolize a drug into harmful metabolites, a simple test could avoid problems associated with giving that drug to those patients, as in the case of acetaminophen and liver toxicity36. It would be especially convenient if the same states applied broadly so the same community features correlated with for example the generation of toxic metabolites from a drug also correlated with obesity.
The recently introduced concept of “enterotypes” proposes exactly this stratification. Based on 33 fecal shotgun metagenomes, the human microbiome was proposed to form three distinct host-microbial symbiotic states driven by groups of co-occurring species/genera, characterized by relatively high representation of the genera Bacteroides, Prevotella, or Ruminococcus, respectively31. Although the overall clustering structure in the original report was not statistically significant, these three enterotype clusters better described the real data than randomly generated datasets31. The authors reported similar patterns after re-analysis of existing 16S rRNA data from 154 Americans20 and metagenome data from 85 Danes 13. Subsequent studies of the microbiota of 98 healthy adults from the US32, 531 healthy infants, children, and adults from Malawi, Venezuelan Amerindians, and the US 4, and of 250 healthy adults from the US 14, failed to recover the same stratification. Variation among adults in these populations was, however, associated with a trade-off between Prevotella and Bacteroides, suggesting an important role for these taxa, or taxa that co-distribute with them, in structuring the microbiota. When infants were introduced into the analysis, variation was additionally associated with a trade-off between the infant-associated Bifidobacteria genus and lineages uniquely common to adults 4 (Fig. 5). The number of unique configurations forming functional, stable communities may thus be quite large and not easily classifiable into a manageable number of distinct “types”. This concept will be important to consider further in more extensive datasets from individuals representing different ages, cultural traditions/geographic locations, and physiologic or disease states.
Perturbation of stable states
Differences in the microbiota with disease
The composition of the microbiota at the community level differs with host physiological states. For example, obese persons harbor fewer types of microbes in their guts than lean persons, and lean and obese people differ significantly in abundances of specific taxa and functional genes6,7,45. Although community-level effects exists, and people can be classified as lean or obese with 90% accuracy based solely on their gut microbiota46,47, lean and obese individuals do not separate into distinct microbiota-based clusters on commonly used principal coordinates (PCoA) plots used to identify statistical differences between groups. Thus multiple statistical techniques are needed to fully reveal differences in microbiota correlated with different physiological states (Fig. 2).
Mouse studies show that some microbiota differences can contribute directly to disease states. “Gnotobiotic mice” raised germ-free then inoculated with the microbiota from an obese mouse gained adiposity more rapidly than those inoculated with a lean mouse’s microbiota 7,45. A given phenotype can emerge from different compositional backgrounds, perhaps indicating that specific components of the microbiota exert large effects or that many different changes lead to the same functional result.
Differences in fecal microbial community diversity, composition and function have also been correlated with IBD (Crohn’s disease and ulcerative colitis)9,10, irritable bowel syndrome (IBS)48, C. difficile associated disease (CDAD)49, and acute diarrhea50. One twin cohort study of IBD found marked and reproducible deviations in ileal Crohn’s patients relative to controls, and more subtle but characteristic changes with colonic Crohn’s patients 51; functional differences were observed based on metabolic profiling of the same samples24. Other diseases have less reproducible microbial correlations. For example, individuals with initial C. difficile infection and healthy controls had comparable phylum-level diversity. In contrast, individuals with recurrent CDAD had phylum-level diversity that diverged dramatically from healthy but that did not resemble each other 49. Many disease studies are confounded by extensive histories of antibiotic administration and other treatments that may obscure truly disease-associated changes. Prospective longitudinal studies that establish cause and effect are thus urgently needed.
Parallels between host physiological states
Most studies of the microbiota target one specific disease or state, but comparisons of the microbiota across diseases reveal common changes in the gut environment. For example, disturbed mucous layers lining the intestinal cell wall and concomitant inflammation are observed in individuals with IBD, celiac disease, HIV enteropathy, acute diarrhea, diverticulosis, carcinoma, and IBS52. Given these parallels, one might expect similar microbes to increase or decrease in abundance across these different disturbances 53, although elucidation of these differences may require detailed biogeographical studies along the length of the gut (once safe and reliable means for such comprehensive sampling are developed).
Perturbed adult gut microbial communities are intriguingly similar to infant gut microbial communities. Perhaps both systems represent successional communities where the same opportunistic or “weedy” species predominate 53. For example, C. difficile, a normal gut resident that can cause disease when antibiotics compromise stable adult gut communities, also colonizes 2–65% of infants, although most are asymptomatic54,55. Other Clostridium species, associated with the disturbed gut and systemic infections (Clostridium bolteae and Clostridium symbiosum) are found in the infant gut 53. Individuals with ileal Crohn’s disease also resemble infants in some respects; both have more Ruminococcus gnavus and Enterobacteraceae in their stools, and an underrepresentation of genera that are prevalent in healthy adults including Faecalibacterium and Roseburia 51. These examples underscore the importance of understanding whether generally opportunistic members of the gut microbiota have a selective advantage during early succession or during disruption caused by disease, and thus may be side-effects of disease rather than causal agents.
Resilience of stable states to perturbation
Resilience across scales
If the gut microbiota normally exists in a stable state, how resistant is this state to change in response to different perturbations? Resilience is the amount of stress or perturbation that can be tolerated before a system’s trajectory changes towards a different equilibrium state56. Several studies of macroecosystems illustrate how human interference can transform ecological systems into less productive or otherwise less desired states. Such interference includes resource exploitation, pollution, land-use change, and global warming56. For example, communities with extensive free-floating plant cover versus those in which submerged plants dominate represent alternate regimes in tropical lakes. The submerged-plant regime is preferred, because dense mats of floating plants create anoxic conditions that reduce animal biomass and diversity. Pollution can cause floating plants to predominate, however, because they better compete for light and can competitively exclude submerged plants when nutrient load is high. Understanding environmental drivers of conversion between states can enable interventions that induce regime change. For example a single harvest of the floating plants can induce a permanent shift to the submerged-plant-dominant state, but only if the nutrient loading is not too high57.
Environmental studies also provide examples of microbial responses to perturbations, and perhaps, insight into how gut microbiota might react. For example, during the recent Deepwater Horizon oil spill off the Gulf of Mexico, there was a shift in the microbial community structure and functional gene repertoire in the deep-sea oil plume, with a transient enrichment of microbes capable of hydrocarbon-degradation 58,59. This example may parallel the impact of an extreme dietary change on the gut microbiota of mice switched from a low-fat plant rich diet to a high-fat, high-sugar “Western” diet 7. In both cases, the microbial communities shift substantially and this shift is likely due to their exposure to new substrates that provide a selective advantage to specific members of the community, thus shifting to alternative community states. Whether a particular disturbance disrupts a formerly stable state depends on the resilience of the microbial community to a given type of perturbation.
Resilience to dietary changes
Understanding the resilience of the gut microbiota is critical for determining the efficacy of therapeutic diets. Studies in humans show that consuming carbohydrate- or fat-restricted low-calorie diets for 1 year6 or high-fat/low-fiber or low-fat/high-fiber diets for 10 days32 induce statistically significant changes in the gut microbiota. Nonetheless, these changes in species and/or gene content are small compared to baseline interpersonal variations. Long-term dietary surveys and cross-cultural comparisons suggest that dietary changes might lead to regime changes over longer time periods 4,32, perhaps “eroding” the landscape of alternative stable states to allow changes that short-term nudges cannot produce.
Resilience to antibiotic administration
Antibiotic administration can move the microbiota to alternate stable states. In healthy volunteers given two courses of ciprofloxaxin over a 10-month period, the fecal microbiota reached a stable state similar to yet distinct from the pre-treatment state41. The magnitude of disturbance following ciprofloxacin treatment, and the speed and extent of recovery to the pre-ciprofloxacin state, suggested that resilience of the microbiota varies across individuals and between ciprofloxacin treatments within an individual41.
Long-term studies of the microbiota following antibiotics indicate that post-antibiotic equilibrium states are themselves resilient. For example, clindamycin treatment affected gut Bacteroides up to two years following cessation of treatment60. Similarly, three individuals with dyspepsia given one week of metronidazole, clarithromycin, and omeprazole had a state shift that persisted up to four years without additional antibiotic treatment42. In both cases, significant increases in antibiotic-resistance genes persisted for years42,60, suggesting the post-disturbance state would likely be increasingly resilient against the same disturbance because a greater proportion of the microbiota would be resistant. This finding is consistent with the pattern found for one of the healthy subjects given the two courses of ciprofloxacin over a 10-month period41. In this case, initial recovery of the microbiota was slow and incomplete, stabilizing on a different interim state, but recovery was relatively fast after a second treatment. However, another individual in the same study had the opposite pattern, showing an essentially complete recovery following the first ciprofloxaxin treatment but stabilizing to a distinct state after the second, suggesting that the initial antibiotic treatment decreased resilience41. The impact of antibiotic disturbance on the resilience of microbiota to future antibiotic treatments can thus also vary considerably across individuals.
Resilience to invasion by new species
Resilience of the microbiota to challenge with exogenous microbes is also important. The gut microbiota generally exhibits “colonization resistance,” where the native microbiota prohibits establishment of harmful (pathogenic)1,2,61 and potentially beneficial (probiotic)62 microbes. Studies challenging the gut microbiota with a foreign microbiota rather than with specific pathogens or probiotics, however, suggest the native community may be less resilient to colonization by exogenous microbes than previously believed. When 14 conventionally raised Lewis rats were gavaged with the pooled microbiota of rats from the Sprague Dawley and Wistar strains, the recipients’ microbiota diversity increased substantially and changed to resemble that of the donors and phylotypes established from the donor persisted up to three months post-transplantation63. In contrast to ecological theory that disturbance eliminating native species facilitates establishment of exotic species64, transplantation after the resident microbiota were depleted with two broad-spectrum antibiotics reduced, rather than promoted, establishment of the donor microbiota63.
The success of microbiota transplantation in the treatment of recurrent CDAD further supports the plasticity of an established gut microbiota when challenged with a complex microbiota. In this technique, also called bacteriotherapy65, a fecal sample from a healthy donor is introduced as a homogenate by injection into the cecum using a colonoscope. In one case study, a patient with recurrent CDAD was cured of disease symptoms following fecal transplantation from a healthy donor. Her first well-formed bowel movement came only two days post-treatment, and one month later, no C. difficile was found in her feces. Prior to treatment, the recipient had a disturbed microbiota with Veillonella and Streptococcus predominating. In contrast, the donor microbiota was dominated by Bacteroides. The same donor species were found in the patient stool samples one month post-treatment, indicating that the donor microbiota persisted over this interval. Fecal transplantation for CDAD has been administered many times and appears to be highly effective. A recent survey of 317 patients treated across 27 case series reported disease resolution in 92% of cases, with a single treatment sufficing in 89%66. The success of bacteriotherapy suggests that prior antibiotic treatment reduced the resident microbiota to the extent that it enabled engraftment of a donor microbiota. However, because fecal transplantation is not a first-line therapy no data exist concerning its effectiveness without antibiotics (A. Khoruts, pers. comm.). Further studies are needed to understand the successional processes and microbial taxa that allow a healthy microbiota configuration to be established in fecal transplant recipients. These studies will allow development of a standardized way of formulating and analyzing the donor specimen, design and interpretation of dosing studies, and analysis of short- and long-term safety.
Mechanisms conferring resilience
Understanding mechanisms that confer resilience to stable states of the microbiota would allow us to devise strategies to increase resilience of healthy states, or decrease resilience of unhealthy states. For example, a healthy state with high resilience to exogenous microbes might enable one person at a dinner party to escape food poisoning while their companions fall ill. A degraded state with high resilience can contribute to chronic problems with diarrhea or inflammation, as is inherent with CDAD, IBD, and IBS.
Species and functional response diversity
In macroecosystems, several aspects of diversity are critical for conferring resilience, and the same features are likely important in microbial ecosystems including the human gut microbiota. One important parameter is species richness, i.e. the number of species present in a given system. (Note that in culture-independent studies, this number depends on sampling effort, i.e. the number of sequences collected per sample). Ecological theory predicts that species-rich communities are less susceptible to invasion because they use limiting resources more efficiently, with different species specializing to each potentially limiting resource64. Excess nutrient loading, or eutrophication, often causes decreased ecosystem diversity because a small number of species overgrow and outcompete everything else, with a concomitant decrease in resilience67. Consistent with this notion, decreased diversity has been linked with obesity and with a “Western” diet high in fat and sugar compared to those on low-fat plant-based diets 4,7, although whether this decreased microbiota diversity results in a decrease in resilience is not known. Low microbiota diversity also correlates with IBD51 and recurrent CDAD49 with unknown effects on microbiota resilience.
Another aspect of diversity that may be particularly important for promoting resilience is functional response diversity, defined as the degree to which species in a community that contribute to the same ecosystem function vary in their sensitivity to ecosystem changes68. High functional response diversity may, for instance, allow a relatively rare but functionally similar species to fill a niche when an abundant species is compromised by an environmental disturbance 68. In an example from macroecology, a regime change in coral reefs from a healthy state to an unhealthy, algal-dominated state is prevented by a diversity of algal grazers. One compromised reef only switched to an algal-dominated state after both algal-grazing fish were removed by overfishing and the sea urchin that had increased in numbers to fill this niche was compromised by a pathogen 68. The same principles likely apply to the gut microbiota. For instance, following antibiotic administration, a previously rare microbe may increase in abundance to fill an essential niche previously dominated by a microbe with higher antibiotic sensitivity, leading to persistence of the same stable state but with decreased resilience due to a decrease in functional redundancy.
High functional response diversity in human gut-adapted bacteria is likely because phylogenetically disparate microbes often perform similar metabolic functions. For example, in humans and mice, methanogenic Archaea, sulfate-reducing bacteria, and phylogenetically diverse acetogens all consume H2 generated by other microbes during fermentation69. The ecological diversity of butyrate-producers in the Clostridiales provides a second example of functional redundancy. Known butyrate producers in this family have different ecological strategies, including adaptation to different stages of community succession. Anaerostipes caccae abundance peaks in infancy, while Eubacterium hallii and R. intestinalis are more abundant in adults. These taxa also vary in oxygen tolerance, with A. caccae better able to survive 10–60 minute periods of exposure to air than E. hallii or R. intestinalis70. They also differ in their substrate preferences. For example, Eubacterium halli but not F. prausnitzii can utilize lactate as a substrate71.
Competition and feedback loops
Competition in the densely populated gut environment is also expected to be important. Microbes can compete to use the same resources, or inhibit each other directly using anti-microbial products. We might expect phylogenetically related bacteria to compete because of overlapping functional roles and/or habitats. Instead, phylogenetically similar species tend to appear in the same samples72. For example, a recent analysis of bacteria with complete/draft genome sequences across 124 Europeans found that related species within the genus Enterobacter, including E. coli, Salmonella enterica, Citrobacter koseri, and Enterobacter cancerogenus, were positively correlated in abundance in the same individuals 53. These related species may share environmental preferences that selects for all of them simultaneously. Mouse studies also show that abundances of closely related species can predict susceptibility to intestinal colonization by both pathogenic and commensal bacteria73.
Feedback loops, in which microbes affect their own abundance, may stabilize or destabilize the microbiota (Fig. 6). For example, stable physiological states are preserved by negative feedback, in which a change to the gut environment results in opposing changes that maintain homeostasis. These feedbacks are likely controlled by a tight interplay between microbial metabolic activities and host pathways. For example, microbial metabolites might induce changes in the expression of the host pathways that control gut retention time so that it rises above or below the optimum, causing diarrhea or constipation, respectively. Deviation from this optimum would likely induce host signaling pathways to correct it. This scenario is analogous to body temperature regulation. A rise above the optimum induces thermoregulatory mechanisms such as sweating to reduce it, and a descent below the optimum induces mechanisms such as shivering to increase it. Stability in physiological parameters controlled by negative feedback from the host could thus promote resilience of the microbiota.
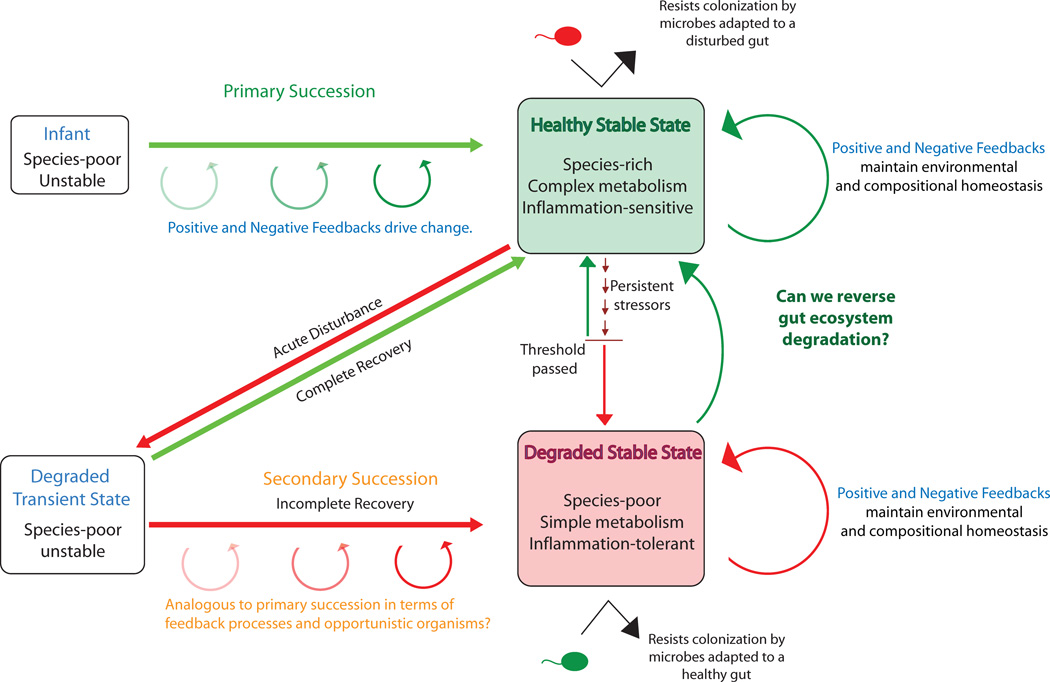
Fig. 6. Compositional transitions in the human gut microbiota.
During early development, the gut microbiota undergoes a systematic turnover of species (primary succession) until a stable adult state is reached. Positive and negative feedback loops likely play a role both in driving primary succession and in conferring resilience to healthy stable equilibrium states. Acute disturbances, such as antibiotic administration, generally are followed by an unstable state that progresses to a stable state through a process of secondary succession. In some cases, the stable state that returns highly resembles the pre-disturbance state, indicating a complete recovery, but sometimes the post-recovery stable state is distinct. Post-disturbance stable states may be both degraded and resilient, for instance as suggested the persistence of post-infectious irritable bowel syndrome (i.e. IBS that forms after an initial acute disturbance of the microbiota from an enteropathogen) in some individuals for years and even decades 78. Resilience of degraded states is likely driven by unique positive and negative feedbacks that occur both in concert with and independent of the host. Degradation to a stable state may also occur as a result of persistent stressors, such as poor diet, that slowly degrade resilience of a healthy state until a threshold is passed such that new feedbacks become important in maintaining community composition and stability. Developing therapies that encourage transition from degraded to healthy stable states, or complete recovery to a healthy stable state following disturbance, may involve identifying the species (or species combinations) and processes that are key drivers of these feedbacks. One critical unresolved question is whether interventions are more effective early in succession when communities are more unstable but may be stochastic, or later in succession when convergence to the end point is more certain but the trajectory may be more difficult to change.
Negative feedback loops that promote microbiota resilience could also operate independently of the host. For example, this would occur if when the abundance of a particular microbe exceeded a certain threshold, it would result in a change in the gut environment that would decrease that microbe’s growth relative to other species. Such negative feedback loops may involve the accumulation of phage specific to that microbe or the accumulation of a specific toxic metabolite. Modeling studies suggest that negative feedback promotes high ecosystem diversity which can promote resilience74.
Positive feedback is traditionally considered to induce ecosystem change, e.g. because a difference from a set point in one direction produces additional change in the same direction. Positive feedbacks, however, have the potential to support stability at the level of individual microbes or of microbial consortia that promote each other’s growth. For instance, microbes may, through their metabolic activities or interactions with host pathways, induce a physiologic state that favors their growth over those of potential competitors, thus promoting resilience. The microbiota is thus likely to contain both key functional drivers of physiological status as well as microbes that co-occur because they are able to thrive in such an environment. Invasion by microbes that do not thrive at that particular physiological state would be prevented. One example would be if a microbiota containing functional drivers of a low inflammatory state resists colonization from pathogens, and a microbiota with functional drivers of a relatively high inflammatory state resists colonization from beneficial microbes (Fig. 6).
Positive and negative feedbacks also likely play a strong role in destabilizing the microbiota during regime changes, such as during succession in early development and following a disturbance (Fig. 6). Negative feedbacks in which an organism’s activity alters the environment such that its fitness is decreased, can induce directional change when microbes induce a physiologic state that favors their competitors. For instance, a higher redox potential in the gut of infants is likely one of the factors explaining the relative success of facultative anaerobes such as Escherichia coli or certain Lactobacillus in early development75, but the reduction of oxygen that results from their metabolism favors their eventual replacement by a consortium dominated by strict anaerobes.
Feedbacks important for successional changes also likely involve a complex interplay with the microbiota and its host. As an example, the same changes in redox potential that directly affect microbial fitness, also affect the expression of host factors in the gut epithelium such as hypoxia-inducible factor (HIF) in both early development and in inflammatory diseases of the gut76. Understanding how to manipulate positive and negative feedback at the level of the host, the individual microbes and of the entire gut ecosystem will be critical for understanding how to maintain healthy stable states, and how to switch from an unhealthy to a healthy state.
Prospectus
Studies enabled by high-throughput sequencing over the past five years suggest that each individual’s microbiota has some resistance to perturbation, but that this resistance could potentially be overcome by therapies that alter the microbiota composition via diet, drugs, prebiotics or probiotics. For example, dietary changes may cause regime changes in the gut over long timescales. The surprising success of whole community transplants in healthy rats and in humans with CDAD reveals that exogenous microbes can colonize despite resistance from an entrenched microbiota. However, we do not yet understand which microbes will best colonize once introduced, or how particular microbial configurations and their functional attributes change in response to specific dietary components or exogenous microbes. Just like gardeners, we must learn what conditions promote the health of desired species and exclude undesirable weeds.
The landscape of stable states of the microbiota and its implications for resilience is an important research direction. Current evidence suggests that small perturbations, such as short-term dietary changes, may allow a return to the same state, but larger perturbations, such as antibiotic administration, may cause movement to a different state. The long-term implications for such changes for health are not yet well understood. Furthermore, perturbation of the landscape of stable equilibrium states of the gut microbiota through long-term changes, such as inflammation, diet, or repeated antibiotic administration, might make new states reachable even with smaller perturbations. Factors such as host genetics, the process of development, diet, and long-term drug administration might all contribute to differences in the landscape among individuals. Consequently, both the general landscape and the current community state may be important for determining individual responses to a given intervention.
The exhibited degree of resilience to diet, disturbances including antibiotics, and challenge with exogenous microbes has important implications for health care. The degree to which repeated applications of broad-spectrum antibiotics degrade the microbiota and its ability to provide ecosystem services needs to be studied, especially in children because early development is a crucial time for interactions between microbiota and host5. However, the identification of suitable controls is challenging given the large intra- and interpersonal variation in the gut microbiota during early development. As indicated by studies of resilience to dietary changes, regime change is not always instigated by acute disturbances and can occur gradually. Individuals with a ‘degraded’ microbiota from long-term consumption of a high-fat/high-sugar Western diet may need long-term dietary changes to restore their microbiota to a healthy state.
The decreased taxonomic diversity of individuals in Western cultures raises concern about the maintenance of important microbial symbionts in the broader population, and whether global trends in diet can result in the permanent loss/extinction of bacterial species that can provide important health benefits. Maintaining culture collections from individuals in the developing world or specifically in agrarian cultures may help to preserve potentially important components of the microbiota.
Given that gut microbes often produce unique states in the gut through their collective activities and cooperative metabolism, it will be important to understand associations between disease states and sets of species rather than single taxa 53. A central problem of culture-independent metagenomic analyses is that identified phylotypes (or collections of phylotypes) often represent species for which little is known biologically. Therefore, the field of gut microbial ecology has come full circle, with increasing attention being paid to developing methods for culturing the majority of diversity present in a community so that hypotheses about the contributions of community members can be further explored. Encouragingly, the human fecal microbiota is largely composed of readily cultured bacteria 77. In vitro experiments with these isolates will be extremely valuable for exploring specific hypotheses about the metabolic attributes of a particular microbe or set of microbes and the genes involved. However, because microbiota composition and function are also controlled by feedbacks from the host, in vivo studies in gnotobiotic mice will be particularly valuable. Indeed, the culturable component of the microbiota exhibits similar colonization dynamics, biogeographical distribution, and responses to dietary perturbations compared to the full microbiota when transplanted into gnotobiotic mice77. A translational science pipeline is thus developing where particular phylotypes and interactions between phylotypes that are important for health and/or that contribute to disease can first be identified based on distribution patterns with disease. Furthermore, biological attributes that may be driving these patterns can be predicted based on the expression or prevalence of functional genes in whole communities and in genomes. These specific hypotheses can then be tested and verified in culture and in animal models before application to humans. Tools for this pipeline are now in place. Although the path ahead will be difficult, the direction is becoming clearer.
Acknowledgements
We would like to thank Laura Parfrey, John Knight and Allison Knight for their comments on this manuscript.
References
- 1.Candela M, et al. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int J Food Microbiol. 2008;125:286–292. doi: 10.1016/j.ijfoodmicro.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Fukuda S, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 3.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 4.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olszak T, et al. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science. 2012 doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dicksved J, et al. Molecular analysis of the gut microbiota of identical twins with Crohn's disease. Isme Journal. 2008;2:716–727. doi: 10.1038/ismej.2008.37. [DOI] [PubMed] [Google Scholar]
- 10.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez A, et al. The mind-body-microbial continuum. Dialogues in clinical neuroscience. 2011;13:55–62. doi: 10.31887/DCNS.2011.13.1/agonzalez. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr. 2004;134:479–482. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- 13.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Consortium HMP. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borenstein E, Kupiec M, Feldman MW, Ruppin E. Large-scale reconstruction and phylogenetic analysis of metabolic environments. Proc Natl Acad Sci U S A. 2008;105:14482–14487. doi: 10.1073/pnas.0806162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freilich S, et al. Metabolic-network-driven analysis of bacterial ecological strategies. Genome Biol. 2009;10:R61. doi: 10.1186/gb-2009-10-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claesson MJ, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biagi E, et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5:e10667. doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson KE, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verberkmoes NC, et al. Shotgun metaproteomics of the human distal gut microbiota. Isme Journal. 2009;3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 24.Jansson J, et al. Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;4:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kozyrskyj AL, Bahreinian S, Azad MB. Early life exposures: impact on asthma and allergic disease. Curr Opin Allergy Clin Immunol. 2011;11:400–406. doi: 10.1097/ACI.0b013e328349b166. [DOI] [PubMed] [Google Scholar]
- 30.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. This paper introduces the concept of “enterotypes” for the human gut microbiota, i.e. that there are a limited number of well-balanced host–microbial symbiotic states that might respond differently to diet and drug intake. Although other studies have questioned the degree of stratification of the 3 particular enterotypes described here, that co-occurring microbial groups that can be associated with high Prevotella vs Bacteroides genus level abundance estimates is associated with major patterns of differentiation in the microbiota across people was an important observation.
- 32. Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. This study found a strong correlation between microbiota diversity and long term diets, as assessed using diet inventories, showing that the most important gradient found to structure diversity in adult populations (Prevotella/Bacteroides ratios defined by some as ‘enterotypes’) has a potential dietary basis. Changes associated with a 10 day controlled-feeding experiment were comparatively subtle.
- 33.Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology. 2004;126:1504–1517. doi: 10.1053/j.gastro.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 34.Cann HM, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 35.Bach JF, Chatenoud L. The hygiene hypothesis: an explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harb Perspect Med. 2012;2:a007799. doi: 10.1101/cshperspect.a007799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A. 2009;106:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson RL, Greiwe JS, Schwen RJ. Emerging evidence of the health benefits of S-equol, an estrogen receptor beta agonist. Nutr Rev. 2011;69:432–448. doi: 10.1111/j.1753-4887.2011.00400.x. [DOI] [PubMed] [Google Scholar]
- 38.Setchell KD, Clerici C. Equol: history, chemistry, and formation. J Nutr. 2010;140:1355S–1362S. doi: 10.3945/jn.109.119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caporaso JG, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. By applying detailed timeseries analysis of the microbiota of 3 healthy adults before, during, and after two 5 day long courses of the antibiotic ciprofloxacin 6 months apart, new insights into the resilience of the human microbiota in the face of repeated disturbance and the degree of baseline variation were obtained.
- 42.Jakobsson HE, et al. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beisner BE, Haydon DT, Cuddington K. Alternative stable states in ecology. Front Ecol Environ. 2003;1:376–382. [Google Scholar]
- 44.Walker B, Hollin CS, Carpenter SR, Kinzig A. Resilience, adaptability and transformability in social-ecological systems. Ecol Soc. 2004;9 [Google Scholar]
- 45.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 46.Sun Y, et al. Advanced computational algorithms for microbial community analysis using massive 16S rRNA sequence data. Nucleic Acids Res. 2010;38:e205. doi: 10.1093/nar/gkq872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knights D, Parfrey LW, Zaneveld J, Lozupone C, Knight R. Humanassociated microbial signatures: examining their predictive value. Cell Host Microbe. 2011;10:292–296. doi: 10.1016/j.chom.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carroll IM, et al. Molecular analysis of the luminal- and mucosal-associated intestinal microbiota in diarrhea-predominant irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2011;301:G799–G807. doi: 10.1152/ajpgi.00154.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang JY, et al. Decreased diversity of the fecal Microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 50.Young VB, Schmidt TM. Antibiotic-associated diarrhea accompanied by large-scale alterations in the composition of the fecal microbiota. J Clin Microbiol. 2004;42:1203–1206. doi: 10.1128/JCM.42.3.1203-1206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willing BP, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854. e1841. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 52.Swidsinski A, Loening-Baucke V, Herber A. Mucosal flora in Crohn's disease and ulcerative colitis - an overview. J Physiol Pharmacol. 2009;60(Suppl 6):61–71. [PubMed] [Google Scholar]
- 53.Lozupone C, et al. Identifying genomic and metabolic features that can underlie early successional and opportunistic lifestyles in human gut symbionts. Genome Res. 2012 doi: 10.1101/gr.138198.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Libby JM, Donta ST, Wilkins TD. Clostridium difficile toxin A in infants. J Infect Dis. 1983;148:606. doi: 10.1093/infdis/148.3.606. [DOI] [PubMed] [Google Scholar]
- 55.Yamamoto-Osaki T, Kamiya S, Sawamura S, Kai M, Ozawa A. Growth inhibition of Clostridium difficile by intestinal flora of infant faeces in continuous flow culture. J Med Microbiol. 1994;40:179–187. doi: 10.1099/00222615-40-3-179. [DOI] [PubMed] [Google Scholar]
- 56.Folke C, et al. Regime shifts, resilience, and biodiversity in ecosystem management. Annu. Rev. Ecol. Evol. Syst. 2004;35:557–581. [Google Scholar]
- 57.Scheffer M, et al. Floating plant dominance as a stable state. Proc Natl Acad Sci U S A. 2003;100:4040–4045. doi: 10.1073/pnas.0737918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hazen TC, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 59.Valentine DL, et al. Dynamic autoinoculation and the microbial ecology of a deep water hydrocarbon irruption. Proc Natl Acad Sci U S A. 2012 doi: 10.1073/pnas.1108820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. Isme Journal. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 61.van der Waaij D, Berghuis JM, Lekkerkerk JE. Colonization resistance of the digestive tract of mice during systemic antibiotic treatment. J Hyg (Lond) 1972;70:605–610. doi: 10.1017/s0022172400022464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McNulty NP, et al. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Science translational medicine. 2011;3:106ra106. doi: 10.1126/scitranslmed.3002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Manichanh C, et al. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010;20:1411–1419. doi: 10.1101/gr.107987.110. The high degree to which a complex ‘foreign’ microbiota (pooled from Spraque Dawley and Wistar rats) could colonize Lewis strain rats in this study indicates that indigenous microbiota may generally be more plastic than previously anticipated. The observation that antibiotic-pretreatment interfered with rather than promoted establishment of the donor community indicates that low species abundance/diversity alone cannot predict low colonization resistance.
- 64.Levine JM, D'antonio CM. Elton revisited: a review of evidence linking diversity and invasibility. Oikos. 1999;87:15–26. [Google Scholar]
- 65.Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360. doi: 10.1097/MCG.0b013e3181c87e02. [DOI] [PubMed] [Google Scholar]
- 66.Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 67.Hautier Y, Niklaus PA, Hector A. Competition for light causes plant biodiversity loss after eutrophication. Science. 2009;324:636–638. doi: 10.1126/science.1169640. [DOI] [PubMed] [Google Scholar]
- 68.Elmqvist T, et al. Response diversity, ecosystem change, and resilience. Front Ecol Environ. 2003;1:488–494. [Google Scholar]
- 69.Hansen EE, et al. Pan-genome of the dominant human gut-associated archaeon Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A 108 Suppl. 2011;1:4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- 71.Louis P, et al. Restricted distribution of the butyrate kinase pathway among butyrate-producing bacteria from the human colon. J Bacteriol. 2004;186:2099–2106. doi: 10.1128/JB.186.7.2099-2106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chaffron S, Rehrauer H, Pernthaler J, von Mering C. A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res. 2010;20:947–959. doi: 10.1101/gr.104521.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stecher B, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bever JD, Westover KM, Antonovics J. Incorporating the soil community into plant population dynamics: the utility of the feedback approach. Journal of Ecology. 1997;85:561–573. [Google Scholar]
- 75.Stark PL, Lee A. The Microbial Ecology of the Large Bowel of Breast-Fed and Formula-Fed Infants during the 1st Year of Life. Journal of Medical Microbiology. 1982;15:189–203. doi: 10.1099/00222615-15-2-189. [DOI] [PubMed] [Google Scholar]
- 76.Glover LE, Colgan SP. Hypoxia and metabolic factors that influence inflammatory bowel disease pathogenesis. Gastroenterology. 2011;140:1748–1755. doi: 10.1053/j.gastro.2011.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goodman AL, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dupont HL. Gastrointestinal infections and the development of irritable bowel syndrome. Curr Opin Infect Dis. 2011;24:503–508. doi: 10.1097/QCO.0b013e32834a962d. [DOI] [PubMed] [Google Scholar]