Abstract
Background
The plasminogen activator inhibitor-1 (PAI-1) is expressed in many cancer cell types and allows the modulation of cancer growth, invasion and angiogenesis. To date, studies investigated the association between a functional polymorphism in PAI-1 (4G/5G) and risk of cancer have shown inclusive results.
Methods
A meta-analysis based on 25 case-control studies was performed to address this issue. Odds ratios (OR) with corresponding 95% confidence intervals (CIs) were used to assess the association. The statistical heterogeneity across studies was examined with I2 test.
Results
Overall, a significant increased risk of cancer was associated with the PAI-1 4G/4G polymorphism for the allele contrast (4G vs. 5G: OR = 1.10, CI = 1.03–1.18, I2 = 49.5%), the additive genetic model (4G/4G vs. 5G/5G: OR = 1.21, CI = 1.06–1.39, I2 = 51.9%), the recessive genetic model (4G/4G vs. 4G/5G+5G/5G: OR = 1.11, CI = 1.04–1.18, I2 = 20.8%). In the subgroup analysis by ethnicity, the results indicated that individuals with 4G/4G genotype had a significantly higher cancer risk among Caucasians (4G/4G vs. 5G/5G: OR = 1.31, 95%CI = 1.09–1.59, I2 = 59.6%; 4G/4G vs. 4G/5G: OR = 1.12, 95%CI = 1.04–1.21, I2 = 3.6%; recessive model: OR = 1.12, 95%CI = 1.05–1.21, I2 = 25.3%).
Conclusions
The results of the present meta-analysis support an association between the PAI-1 4G/5G polymorphism and increasing cancer risk, especially among Caucasians, and those with 4G allele have a high risk to develop colorectal cancer and endometrial cancer.
Introduction
The urokinase plasminogen activator system is a serine protease family [1]. The included urokinase-type plasminogen activator (uPA)system provides the most substantial amount of activated plasminogen when tissues are being degraded and is involved in extracellular matrix (ECM) degradation [2], [3], hence it has been involved in numerous pathophysiological processes requiring the remodeling of basement membranes (BM) and ECM. Metastasis and invasion of malignant cancers require proteolytic degradation of the ECM, BM and infiltration of cancer cells into the surrounding tissues, the blood stream, or the lymphatic vessels. Studies revealing the uPA system, universal to all cancers, is associated with the process of cancer metastasis and progression by participating in the degradation and regeneration of the BM and ECM [4], [5], [6].
The plasminogen activator inhibitor-1 (PAI-1), a 52 kDa glyco-protein belong to the serine proteinase inhibitor super family, is a multifaceted proteolytic factor. It is the principal inhibitor of tissue and urinary plasminogen activators, and therefore constitutes an important regulatory protein in fibrinolysis [7], [8]. It is also involved in the regulation of cell adhesion, detachment and migration, playing an important role in cancer progression [9], [10], [11]. Indeed, PAI-1 is expressed in many types of cancer cell and allows the modulation of cancer growth, invasion and angiogenesis in a dose-dependent manner [12].
Genetic polymorphisms in the PAI-1 gene seem to contribute to the level of PAI-1 biosynthesis [13]. A single nucleotide insertion/deletion (4G/5G) polymorphism located at 675 base-pair (bp) upstream of the transcriptional start site in the PAI-1 promoter, is the most frequently studied variant because of its possible involvement in the regulation of PAI-1 transcription [14], [15], [16]. Based on the investigation by CDC (Centers for Disease Control and Prevention), the 4G/5G allele frequencies range in various populations from 26.7/73.3% to 52.5/47.5%,respectively. (http://www.cdc.gov/genomics/population/genvar/frequencies/SERPINE1.htm). The distribution of 4G/5G allele frequencies has been shown in Table S1.
Homozygosity of the 4G allele is considered to be a risk factor for developing deep vein thrombosis, myocardial infarction and high rate of miscarriage during pregnancy [17], [18], [19]. Many molecular epidemiological studies have been conducted to investigate the association between 4G/5G polymorphism and cancer risk in humans [20]–[43]. However, the results from these studies are to some extent divergent, but nevertheless intriguing, which may be owe to limitations in individual studies. To address this issue, we performed a meta-analysis with subgroup analysis from all eligible studies, to obtain a more precise estimation of the relationship between PAI-1 4G/5G polymorphism and cancer risk.
Table 1. Characteristics of studies included in the meta-analysis.
| First author | Ethnicity | Country | Cancer | Genotyping | Source ofControls | Sample size | HWE | |
| case | control | |||||||
| Turkmen 1997 | Caucasian | Germany | Ovarian cancer | PCR-RFLP | HB | 22 | 23 | Y |
| Smolarz 1999 | Caucasian | Poland | Breast cancer | Allele-specific PCR | HB | 37 | 53 | Y |
| Blasiak 2000 | Caucasian | Poland | Breast cancer | Allele-specific PCR | HB | 100 | 106 | Y |
| Loktionov 2003 | Caucasian | UK | Colorectal | PCR-RFLP | HB | 206 | 355 | Y |
| Castello 2006 | Caucasian | Spain | Breast cancer | Allele-specific PCR | HB | 104 | 104 | Y |
| Eroglu 2006 | Caucasian | Turkey | Breast cancer | PCR-RFLP | HB | 34 | 90 | Y |
| Sternlicht 2006 | Caucasian | UK | Breast cancer | PCR-RFLP | PB | 2539 | 1832 | Y |
| Eroglu 2007 | Caucasian | Turkey | Others | PCR-RFLP | HB | 125 | 180 | Y |
| Forsti 2007 | Caucasian | Sweden | Colorectal cancer | Taqman | PB | 304 | 581 | Y |
| Jorgenson 2007 | Mixed | USA | Prostate cancer | PCR-RFLP | PB | 638 | 478 | Y |
| Minisini 2007 | Caucasian | Italy | Breast cancer | Allele-specific PCR | HB | 193 | 142 | Y |
| Woo 2007 | Asian | Korea | Colorectal cancer | PCR-RFLP | HB | 185 | 304 | Y |
| Lei 2008 | Caucasian | Sweden | Breast cancer | Taqman | PB | 956 | 943 | Y |
| Bentov 2009 | Mixed | Canada | Ovarian cancer | MassARRAY | PB | 772 | 889 | Y |
| Palmirotta 2009 | Caucasian | Italy | Breast cancer | PCR-RFLP | HB | 99 | 50 | Y |
| Vairaktaris 2009 | Caucasian | Greece Germany | Oral cancer | PCR-RFLP | HB | 104 | 106 | Y |
| Ju 2010 | Asian | Korea | Gastric cancer | MassARRAY | PB | 252 | 406 | Y |
| Weng 2010 | Asian | Taiwan | Hepatocellular cancer | PCR-RFLP | HB | 102 | 344 | Y |
| Gilabert-Estelles 2011 | Caucasian | Spain | Endometrial cancer | Allele-specific PCR | HB | 212 | 211 | Y |
| Su 2011 | Asian | Taiwan | Endometrial | PCR-RFLP | HB | 134 | 302 | Y |
| Vossen 2011 | Caucasian | Germany | Colon cancer | Taqman | PB | 1059 | 1799 | Y |
| Vossen 2011 | Caucasian | Germany | Rectal cancer | Taqman | PB | 672 | 1799 | Y |
| Weng 2011 | Asian | Taiwan | Oral cancer | PCR-RFLP | HB | 253 | 344 | Y |
| Onur 2012 | Caucasian | Turkey | Others | Two parallel PCR | HB | 28 | 50 | Y |
| Tee 2012 | Asian | Taiwan | Cervical cancer | PCR-RFLP | HB | 75 | 336 | Y |
HB, hospital based; PB, population based; HWE,Hardy–Weinberg equilibrium.
Materials and Methods
Identification and Eligibility of Relevant Studies
All case-control studies on the association between PAI-1 polymorphisms and cancer risk published up to July 31, 2012 were identified through comprehensive searches using the PubMed and EMBASE database with the following terms and keywords: “plasminogen activator inhibitor-1″, “PAI-1” and “polymorphism”, “variation”, “mutation” and in combination with “cancer”, “tumor” and “carcinoma”. The search was limited to human studies and English language papers.
Inclusion Criteria
For inclusion in the meta-analysis, the identified articles have to meet the following criteria: (a) there is information on the evaluation of the PAI-1 4G/5G polymorphism and cancer risk, (b) using a case-control design, and (c) containing complete information about all genotype frequency. The exclusion criteria are as follows: (a) not for cancer research, (b) review articles, (c) reports without usable data and (d) duplicate publications.
Data Extraction
Information was carefully extracted from all the eligible publications independently by two researchers (SQ Wang and Q Cao) according to the inclusion criteria listed above. For conflicting evaluation, a consensus was reached by discussion. The following information was extracted from each included study using a standardized data collection protocol (File S1): the first author’s name, the year of publication, ethnicity, country of origin, cancer type, genotyping method and source of control groups (population- or hospital-based controls) and deviation from Hardy-Weinberg Equilibrium (HWE) of the control group. Different ethnic descents were categorized as African, Asian, European, or Mixed (composed of different ethnic groups). Meanwhile, different case-control groups in one study were considered as independent studies.
Statistical Methods
The strength of the association between the PAI-1 4G/5G polymorphism and cancer risk was measured by odds ratios (ORs) with corresponding 95% confidence intervals (CIs). The percentage weight determined by the precision of its estimate of effect and, in the statistical software in STATA and SAS, is equal to the inverse of the variance. The risks (ORs) of cancer associated with the PAI-1 4G/5G polymorphism were estimated for each study. In our study, the 5G allele was considered the reference genotype. The pooled ORs were performed for additive genetic model (4G/4G vs. 5G/5G and 4G/4G vs. 4G/5G), dominant model (4G/4G +4G/5G vs. 5G/5G) and recessive model (4G/4G vs. 4G/5G +5G/5G), respectively. Stratified analyses were also performed by cancer types (if one cancer type contained less than two individual studies, it was classified as other cancers group), ethnicity, source of controls and sample size (subjects ≥500 in both case and control groups or not). In consideration of the possibility of heterogeneity across the studies, a statistical test for heterogeneity was performed by a I2 test. I2value and its 95%/97.5CI were both calculated and shown in Table S2. A I2 smaller than 31% indicates lack of heterogeneity among the studies, and then the fixed-effects model (the Mantel-Haenszel method) was used to calculate the summary OR estimate of each study. Otherwise, the random effects model (DerSimonian and Laird method) was used. For each study, we examined whether the genotype distribution of controls was consistent with HWE using the χ2 test. One-way sensitivity analysis was performed to assess the stability of the results, namely, a single study in the meta-analysis was deleted each time to reflect the influence of the individual data set to the pooled OR. An estimate of potential publication bias was carried out by the funnel plot, in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was assessed by the method of Egger’s linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the OR. All statistical analyses were performed with the Stata software (version 12.1; StataCorp LP, College Station, TX, USA) and SAS software (Version 9.2; SAS Institute, Cary, NC, USA),using two-sided P-values.
Results
Characteristics of Studies
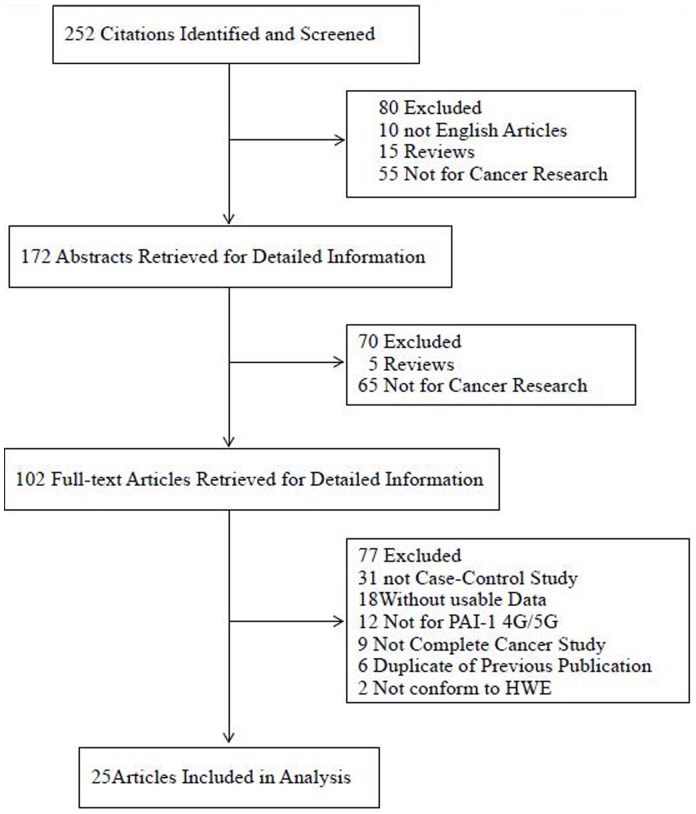
There were 25 studies retrieved on the basis of the search criteria (Fig. 1). Totally, 9,205 cases and 11,827 controls were included in the meta-analysis. Study characteristics were summarized in Table S1. Among the 25 case–control studies, there were 17 studies of Caucasians, 6 studies of Asians, and 2 studies of mixed descendents. There were 8 breast cancer studies, 5 colorectal cancer studies, 2 ovarian cancer studies, 2 endometrial cancer studies, 2 oral cancer studies, and the others were categorized into the “other cancer” group. Cancers were confirmed histologically or pathologically in most studies. Controls were mainly matched on sex and age, of which 17 were hospital based [21], [22], [23], [24], [29], [30], [31], [32], [33], [35], [36], [37], [38], [40], [41], [42], [43], 8 were population based [20], [25], [26], [27], [28], [34], [39]. Furthermore, 10 studies were conducted with subjects >500 in both case and control groups [20], [25], [26], [27], [28], [29], [34], [39], [40]. Diverse genotyping methods were used, including PCR–RFLP, TaqMan, allele-specific PCR, MassARRAY, and two parallel PCR. The distribution of genotypes in the controls of all studies was consistent with HWE.
Figure 1. Studies identified with criteria for inclusion and exclusion.
Quantitative Synthesis
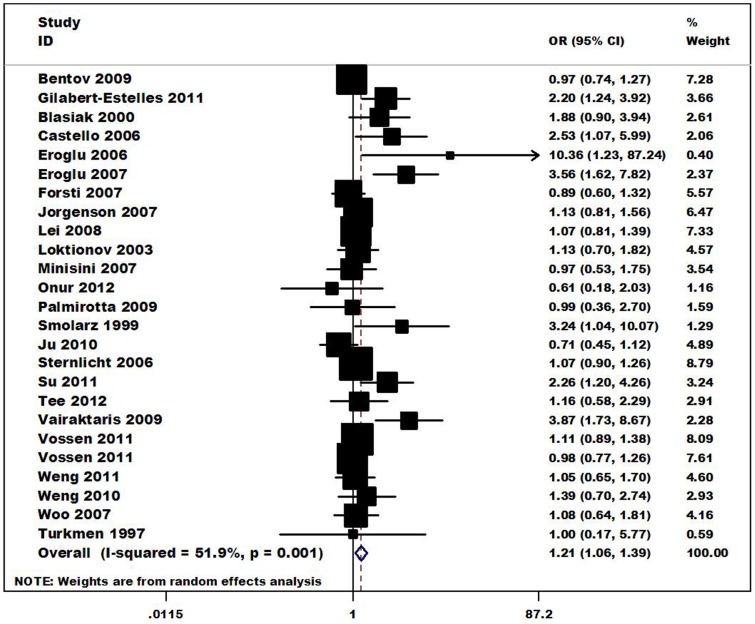
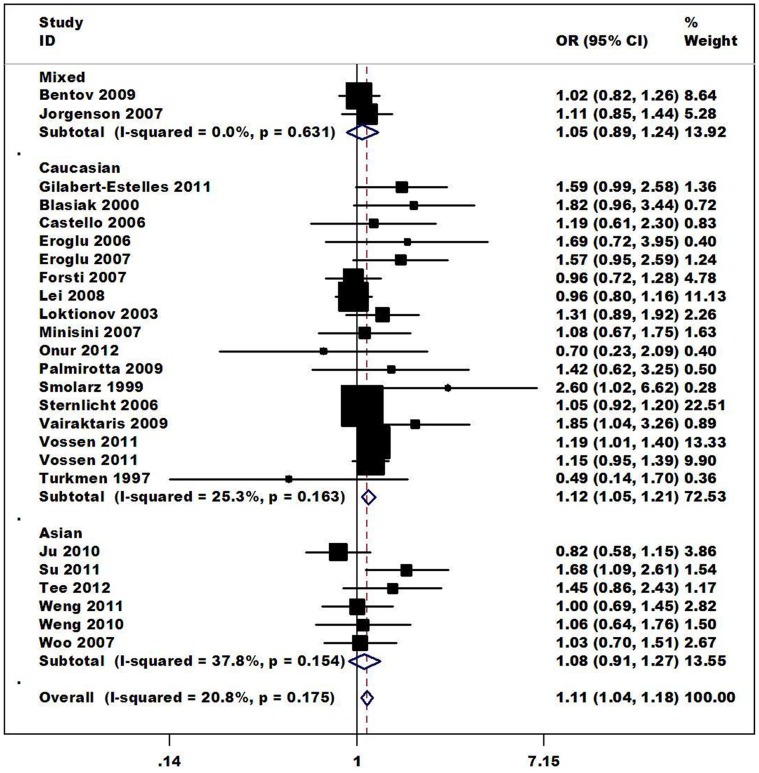
The relationship between the 4G/5G polymorphism in PAI-1 and the risk of different kinds of cancer are summarized in Table 2. Overall, a significantly increased risk of cancer was is associated with the PAI-1 4G polymorphism for the allele contrast (4G vs. 5G: OR = 1.10, CI = 1.03–1.18, I2 = 49.5%), the additive genetic model (4G/4G vs. 5G/5G: OR = 1.21 CI = 1.06–1.39, (Fig. 2); 4G/4G vs. 4G/5G: OR = 1.10 CI = 1.03–1.18), the recessive genetic model (4G/4G vs. 4G/5G+5G/5G OR = 1.11 CI = 1.04–1.18). In the subgroup analysis by ethnicity, the results indicated that individuals with 4G/4G genotype had a significantly higher cancer risks among Caucasians (4G/4G vs. 5G/5G: OR = 1.31, 95%CI = 1.09–1.59; 4G/4G vs. 4G/5G: OR = 1.12, 95%CI = 1.04–1.21; recessive model: OR = 1.12, 95%CI = 1.05–1.21), (Fig. 3). When restricting the analysis to the source of controls, significant associations were found in Hospital-based source (4G/4G vs. 5G/5G: OR = 1.59, 95%CI = 1.24–2.05; 4G/4G vs. 4G/5G: OR = 1.22, 95%CI = 1.07–1.40; dominant model: OR = 1.38, 95%CI = 1.10–1.74; recessive model: OR = 1.30, 95%CI = 1.14–1.48). In the stratified analysis by cancer types, significant associations were found for Endometrial cancer (4G/4G vs. 5G/5G: OR = 2.23, 95%CI = 1.45–3.42; 4G/4G vs. 4G/5G: OR = 1.45, 95%CI = 1.04–2.04; dominant model: OR = 1.74, 95%CI = 1.23–2.47; recessive model: OR = 1.64, 95%CI = 1.19–2.27), Colorectal cancer (4G/4G vs. 4G/5G OR = 1.19, 95%CI = 1.06–1.33; recessive model: OR = 1.14, 95%CI = 1.03–1.27). In the stratified analysis by sample size (both cases and controls), significant associations were found for <500 (4G/4G vs. 5G/5G: OR = 1.73, 95%CI = 1.31–2.31; 4G/4G vs. 4G/5G: OR = 1.24, 95%CI = 1.06–1.45; dominant model: OR = 1.49, 95%CI = 1.14–1.93; recessive model: OR = 1.36, 95%CI = 1.17–1.57).
Table 2. Stratification analyses of the PAI-1 4G/5G polymorphism on cancer.
| Variables | Sample size | 4Gvs5G | 4G/4Gvs5G/5G | 4G/4Gvs4G/5G | 4G/4Gvs4G/5G+5G/5G | ||||||
| Na | case | control | OR(95% CI) | I2(%) | OR(95% CI) | I2(%) | OR(95% CI) | I2(%) | OR(95% CI) | I2(%) | |
| Total | 25 | 9205 | 11827 | 1.10(1.03–1.18) | 49.5c | 1.21(1.06–1.39) | 51.9c | 1.10(1.03–1.18) | 0c | 1.11(1.04–1.18) | 20.8c |
| Tumor type | |||||||||||
| Breast cancer | 8 | 4062 | 3320 | 1.14(1.00–1.29) | 48.3 | 1.30(0.99–1.70) | 48.8 | 1.05(0.94–1.16) | 6 | 1.07(0.97–1.18) | 22.8 |
| Colorectal cancer | 5 | 2426 | 4838 | 1.03(0.96–1.11) | 0 | 1.04(0.90–1.19) | 0 | 1.19(1.06–1.33) | 0 | 1.14(1.03–1.27) | 0 |
| Ovarian cancer | 2 | 794 | 912 | 0.98(0.86–1.13) | 0 | 0.97(0.74–1.27) | 0 | 1.01(0.81–1.26) | 55.1 | 1.00(0.81–1.23) | 22.9 |
| Endometrial cancer | 2 | 346 | 513 | 1.45(1.19–1.77) | 0 | 2.23(1.45–3.42) | 0 | 1.45(1.04–2.04) | 0 | 1.64(1.19–2.27) | 0 |
| Oral cancer | 2 | 357 | 450 | 1.39(0.73–2.63) | 87.3 | 1.94(0.54–6.91) | 86.5 | 1.07(0.77–1.49) | 0 | 1.20(0.88–1.64) | 67.7 |
| Others | 6 | 1220 | 1794 | 1.08(0.90–1.30) | 57.8 | 1.18(0.79–1.78) | 63.2 | 1.07(0.89–1.28) | 0 | 1.08(0.91–1.28) | 24.6 |
| Ethnicity | |||||||||||
| Caucasian | 17 | 6794 | 8424 | 1.14(1.04–1.25) | 56.8 | 1.31(1.09–1.59) | 59.6 | 1.12(1.04–1.21) | 3.6 | 1.12(1.05–1.21) | 25.3 |
| Asian | 6 | 1001 | 2036 | 1.07(0.92–1.25) | 45.9 | 1.14(0.84–1.56) | 44.8 | 1.07(0.90–1.28) | 17.3 | 1.08(0.91–1.27) | 37.8 |
| Mixed | 2 | 1410 | 1367 | 1.02(0.92–1.13) | 0 | 1.03(0.84–1.27) | 0 | 1.06(0.89.1.27) | 0 | 1.05(0.89–1.24) | 0 |
| Control source | |||||||||||
| Hospital based | 17 | 2013 | 3100 | 1.25(1.11–1.40) | 43 | 1.59(1.24–2.05) | 48.1 | 1.22(1.07–1.40) | 0 | 1.30(1.14–1.48) | 0 |
| Population based | 8 | 7192 | 8727 | 1.02(0.97–1.07) | 0 | 1.03(0.94–1.13) | 0 | 1.07(0.99–1.15) | 8.4 | 1.06(0.99–1.13) | 0 |
| Sample size(both cases and controls) | |||||||||||
| <500 | 15 | 1554 | 2401 | 1.30(1.14–1.48) | 40 | 1.73(1.31–2.31) | 46.5 | 1.24(1.06–1.45) | 0 | 1.36(1.17–1.57) | 0 |
| ≥500d | 10 | 7651 | 9426 | 1.02(0.98–1.07) | 0 | 1.03(0.94–1.13) | 0 | 1.08(1.00–1.16) e | 5 | 1.06(0.99–1.14) | 0 |
Number of studies.
I 2The value of heterogeneity test.
Fix-effects model was used when I2 value for heterogeneity test <31%; otherwise, random-effects model was used.
Stratified according to subjects ≥500 in both case and control groups or not.
The exact value is 1.077(1.002–1.156).
Figure 2. Forest plot of cancer risk associated with the PAI-1 4G/5G polymorphism (4G/4G vs. 5G/5G).
The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Figure 3. Forest plot of cancer risk associated with the PAI-1 4G/5G polymorphism by Ethnicity (recessive model).
The squares and horizontal lines correspond to the study-specific OR and 95% CI. The area of the squares reflects the weight (inverse of the variance). The diamond represents the summary OR and 95% CI.
Test for Heterogeneity
There was significant heterogeneity for allele contrast (4G vs. 5G: I2 = 49.5%), homozygote comparison (4G/4G vs. 5G/5G: I2 = 51.9%), heterozygote comparison (4G/5G vs. 5G/5G : I2 = 48.7%), dominant model comparison (4G/4G+4G/5G vs. 5G/5G: I2 = 53.9%), recessive model comparison (4G/4G vs. 4G/5G+5G/5G: I2 = 20.8%). Then, we used a meta-regression analysis to explore the source of heterogeneity for homozygote comparison (4G/4G vs. 5G/5G) by Ethnicity, cancer types, source of controls and sample size. We found that the sample size (τ2 = 0, P = 0.001) contributed to substantial altered heterogeneity, which could account for 100% source of heterogeneity. Also, control source (τ2 = 0, P = 0.005) contributed to 100% source of heterogeneity. However, we did not find cancer types (τ2 = 0.074, P = 0.615), or ethnicity (τ2 = 0.075, P = 0.947) contributed to source of heterogeneity.
Sensitivity Analysis
The sensitivity analysis was conducted by leaving out certain studies, such as the study that did not conform to HWE. The omission of individual studies did not materially alter the results, although on some occasions, the I2 value for heterogeneity was reduced. The sensitivity analysis thus confirmed that the results of this meta-analysis were statistically robust. This procedure proved that our results were reliable and stable. Furthermore, when excluding the studies that were not in HWE, the estimated pool OR still did not change at all.
Publication Bias
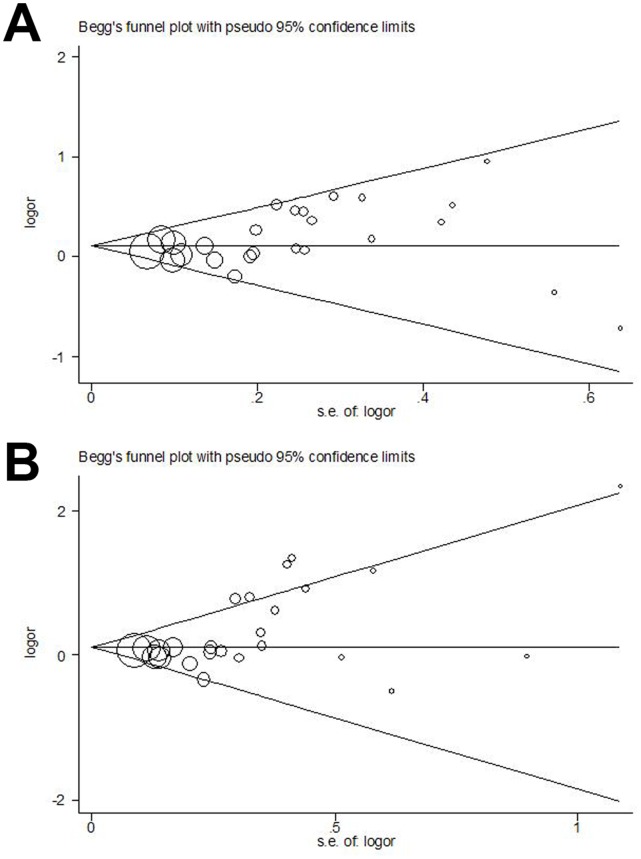
Begg’s funnel plot and Egger’s test were performed to assess the publication bias of literatures. As shown in the Fig. 4, the shapes of the funnel plots seems symmetrical in the recessive genetic model (4G/4G vs. 4G/5G+5G/5G), but not for homozygote model(4G/4G vs. 5G5G). Thus, the Egger’s test was used to provide statistical evidence of funnel plot symmetry. For recessive genetic model, the results did not show any evidence of publication bias (t = 1.96, P = 0.097 for 4G/4G vs. 4G/5G+5G/5G). However, the homozygote model showed significant publication bias (t = 2.99 and P = 0.014). To adjust for this bias, a trim-and-fill method developed by Duval and Tweedie [44] was used to both identify and correct for funnel plot asymmetry arising from publication bias.We trimmed off the asymmetric outlying part of the funnel after estimating how many studies were in the asymmetric part with the help of Stata software. The result showed only one study should be trimmed after three times of iterations. We then estimated the ture center of the funnel and then replaced that trimmed study and its missing counterpart around the center. The final estimate of the true mean, and also its 95%CI, were then based on the filled funnel plot. The OR estimates and 95%CI in fixed-effect model before and after trim-and-fill were 1.119, (1.032–1.213) and 1.115, (1.029–1.209). Also, for random-effect model, the results were 1.214, (1.057–1.394) and 1.204, (1.049–1.392). Meta-analysis with or without the trim-and-fill method did not draw different conclusions, indicating that our results were statistically robust.
Figure 4. Begg’s funnel plot of publication bias test.
(A) 4G/4G vs. 4G/5G+5G/5G. (B) 4G/4G vs. 5G/5G. Each point represents a separate study for the indicated association. Log(OR), natural logarithm of OR. Horizontal line, mean effect size.
Discussion
The present meta-analysis, including 9,205 cases and 11,827 controls from 25 case-control studies, explored the association between the PAI-1 4G/5G polymorphism and cancer risk. Our results indicated that the variant 4G/4G genotype was associated with an increased risk of cancers, especially of colorectal cancer and endometrial cancer.
In recent years, many studies have been conducted to investigate the associations between the PAI-1 4G/5G polymorphisms and disease risk across different countries. The results remain inconclusive. A new manuscript in Blood by Huang et al. [45] just published validated the role of the 4G/5G polymorphisms in circulating PAI-1 levels using GWAS data. However, they revealed no association between PAI-1 4G/5G and type 2 diabetes (T2D) and coronary artery disease (CAD), despite enormous sample sizes.
Many researchers investigated the relationship between PAI-1 blood concentrations and diseases risk. Palmirotta et al. [32] reported plasma PAI-1 levels in breast cancer patients were approximately two fold higher than those observed in control subjects and were strongly dependent on cancer size, suggesting that cancer-related factors might be responsible for PAI-1 up-regulation. However, the exact concentration of PAI-1 in these breast cancer patients blood stayed 27.2 ng/ml (16.5–35.0), and Huang et al. [45] revealed cumulative effect of all common alleles explained extremely low blood levels of PAI-1 in their GWAS data. How would such a seemingly small influence of genotype on PAI-1 levels be expected to modify cancer risk? Given the important roles of PAI-1 in multiple biological functions, such as regulation of cell adhesion, detachment and migration, it is biologically plausible that the PAI-1 4G/5G polymorphism may modulate the risk of cancers.Functional studies on this polymorphism have shown that the 4G allele binds only an activator, while the 5G allele binds a repressor as well as an activator, therefore results in reduced transcription of PAI-1 [46]. It suggests that the 4G allele is associated with reduced inhibition of the plasminogen activators and, consequently, increased plasminogen conversion to plasmin, increased activation of MMPs and decreased adhesive strength of cells for their substratum [17], [18], [46]. Consistent with these observations, our meta-analysis showed that individuals carrying 4G/4G genotype were associated with a higher cancer risk than subjects carrying at least one 5G allele.
In addition, our results showed that the 4G allele may be a risk factor for colorectal cancer and endometrial cancer but not for breast cancer, ovarian cancer, oral cancer, or hepatocellular cancer. One factor that would contribute to the discrepancy among different studies is that this polymorphism might play a different role in different cancer sites. However, even at the same cancer site, considering the possible small effect size of this genetic polymorphism to cancer risk and the relatively small sample size in some studies, the discrepancy will become apparent since some of these studies may be underpowered to detect a small but real association. For endometrial cancer, there were only two studies included in the analysis with limited sample sizes, therefore, the results should be interpreted with caution.
In the subgroup analysis by ethnicity, an increased risk in 4G carriers was found among Caucasians but not Asians or Mixed. One explanation for this result may be that the studies using Mixed ethnicity participants enrolled them from various countries with diverse cultural, environmental and genetic characteristics. It is expected that these factors affected the synthesis results. On the other hand, the sample size and numbers of studies in Asian group were not adequate to evaluate the association. Other factors such as selection bias and different matching criteria may also play a role.
The genetic models were summarised in Table 2 including allele contrast model, homozygote model, heterozygote model and recessive model. Because of the strong heteogeneity in allele contrast and homozygote model, though, the results of these two shows significantly different, we do not suggest any one of these two as the best-fit model to represent the whole genetic models. There is a relatively low heterogeity (I2 = 20.8%) in recessive model, the OR value and the confidence interval shows significantly different. As a result, the recessive model might be the best-fit model in this meta-analysis to reflect the whole results.
Furthermore, despite the overall robust statistical evidence generated through this analysis, some methodological limitations have been identified. Firstly, the relatively high heterogeneity and small sample size are the major defect in this meta-analysis. In the subgroup analyses by ethnicity and cancer type, the sample size of studies among Asians and among several cancer types is small and limited. As a result, the sample size accounted for most of the source of heterogeneity.Also, lacking the original data of the reviewed studies limites our further evaluation of potential interactions, because the interactions among gene-gene, gene-environment and even different polymorphic locis of the same gene may modulate cancer risk. Furthermore, the significant difference of results for hospital based control source should be interpreted in cautious. Accordingly, it is required that more studies be conducted to provide a more definitive conclusion that comprehensively explores the relationship between the PAI-1 4G/5G polymorphism and risk of cancer in the overall population.
Conclusions
In conclusion, the evidence of the results from the present meta-analysis support an association between the PAI-1 4G/5G polymorphism and increasing cancer risk, especially among Caucasians, and those with colorectal cancer and endometrial cancer or cancers identified in the other cancers group, though significant heterogeneity from included studies existed. To advance an understanding of this relationship, the following recommendations have been made: (1) Large studies using standardized unbiased methods, enrolling precisely defined cancer patients and well matched controls, with more detailed individual data is needed. (2) Studies conducted with ethnic groups other than Caucasians are required to gain a more comprehensive and generalizable conclusion. (3) More and larger studies, especially studies stratified for gene-environmental interaction, should be performed to clarify the possible roles of the PAI-1 4G/5G polymorphisms in the etiology of cancer.
Supporting Information
The genetype frequencies on each studies. A generalized distribution of genetype frequencies on each included studies are listed.
(DOC)
Stratification analyses of the I2 and 95%/97.5% confidence interval. If I2 = 0, the one-sided 97.5% CI is presented. Otherwise, a two-sided 95% CI is performed.
(DOC)
PRISMA 2009 Checklist.
(DOC)
Funding Statement
This work was supported by the Program for Development of Innovative Research Team in the First Affiliated Hospital of Nanjing Medical University, Provincial Initiative Program for Excellency Disciplines of Jiangsu Province, by the National Natural Science Foundation of China (grant numbers 81201571). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Duffy MJ (2004) The urokinase plasminogen activator system: Role in malignancy. Curr Pharm Des 10: 39–49. [DOI] [PubMed] [Google Scholar]
- 2. Sidenius N, Blasi F (2003) The urokinase plasminogen activator system in cancer: Recent advances and implication for prognosis and therapy. Cancer Metastasis Rev 22: 205–222. [DOI] [PubMed] [Google Scholar]
- 3. Blasi F, Carmeliet P (2002) uPAR: A versatile signalling orchestrator. Nat Rev 3: 932–943. [DOI] [PubMed] [Google Scholar]
- 4. Andreasen PA, Kjoller L, Christensen L, Duffy M (1997) The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer 72: 1–22. [DOI] [PubMed] [Google Scholar]
- 5. Morgan H, Hill P (2005) Human breast cancer cell-mediated bone collagen degradation requires plasminogen activation and matrix metalloproteinases activity. Cancer Cell Int 5: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choong PF, Nadesapillai AP (2003) Urokinase plasminogen activator system: A multifunctional role in tumor progression and metastasis. Clin Orthop Relat Res 415: S46–S58. [DOI] [PubMed] [Google Scholar]
- 7. Pepper MS (2001) Role of the matrix metalloproteinase and plasminogen activator-plasmin systems in angiogenesis. Arterioscler Thromb Vasc Biol 21: 1104–1117. [DOI] [PubMed] [Google Scholar]
- 8. Ulisse S, Baldini E, Sorrenti S, DArmiento M (2009) The urokinase plasminogen activator system: A target for anti-cancer therapy. Curr Cancer Drug Targets 9: 32–71. [DOI] [PubMed] [Google Scholar]
- 9. Dellas C, Loskutoff DJ (2005) Historical analysis of PAI-1 from its discovery to its potential role in cell motility and disease. Thromb Haemost 93: 631–640. [DOI] [PubMed] [Google Scholar]
- 10. Fersching DM, Nagel D, Siegele B, Salat C, Heinemann V, et al. (2012) Apoptosis-related biomarkers sFAS, MIF, ICAM-1 and PAI-1 in serum of breast cancer patients undergoing neoadjuvant chemotherapy. Anticancer Res 32: 2047–2058. [PubMed] [Google Scholar]
- 11. HanB,NakamuraM, Mori I, Nakamura Y, Kakudo K (2005) Urokinase-type plasminogen activator system and breast cancer. Oncol Rep 14: 105–112. [PubMed] [Google Scholar]
- 12. McMahon B, Kwaan HC (2008) The plasminogen activator system and cancer. Pathophysiol Haemost Thromb 36: 184–194. [DOI] [PubMed] [Google Scholar]
- 13. Steqnar M, Uhrin P, Peternel P, Mavri A, Salobir-Pajnic B, et al. (1998) The 4G/5G sequence polymorphism in the promoter of plasminogen activator inhibitor-1 (PAI-1) gene: relationship to plasma PAI-1 level in venous thromboembolism. Thromb Haemost 79: 975–979. [PubMed] [Google Scholar]
- 14. Zorio E, Gilabert-Estellés J, Espana F, Ramon LA, Cosin R, et al. (2008) Fibrinolysis: the key to new pathogenetic mechanisms. Curr Med Chem 15: 923–929. [DOI] [PubMed] [Google Scholar]
- 15. SartoriMT, Vettor R, DePergola G, De Mitrio V, Saqqiorato G, et al. (2001) Role of the 4G/5Gpolymorphismof PAI-1 gene promoter on PAI-1 levels in obese patients. Influence of fat distribution and insulin-resistance. Thromb Haemost 86: 1161–1169. [PubMed] [Google Scholar]
- 16. Burzotta F, Di Castelnuovo A, Amore C, D’Orazio A, Di Bitondo R, et al. (1998) 4G/5G promoter PAI-1 gene polymorphism is associated with plasmatic activity in Italians: a model of gene–environment interaction. Thromb Haemost 79: 354–358. [PubMed] [Google Scholar]
- 17. Grubic N, Stegnar M, Peternel P, Kaidre A, Binder BR (1996) A novel G/A and the 4G/5G polymorphism within the promoter of the plasminogen activator inhibitor-1 gene in patients with deep vein thrombosis. Thromb Res 84: 431–443. [DOI] [PubMed] [Google Scholar]
- 18. Sartori MT, Danesin C, Saggiorato G, Tormene D, Simioni P, et al. (2003) The PAI-1 gene 4G/5G polymorphism and deep vein thrombosis in patients with inherited thrombophilia. Clin Appl Thromb Hem 9: 299–307. [DOI] [PubMed] [Google Scholar]
- 19. Eriksson P, Kallin B, van’t Hooft FM, Bavenholm P, Hamsten A (1995) Allele-specific increase in basal transcription of the plasminogen activator inhibitor 1 gene is associated with myocardial infarction. Proc Natl Acad Sci USA 92: 1851–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bentov Y, Brown TJ, Akbari MR, Royer R, Risch H, et al. (2009) Polymorphic variation of genes in the fibrinolytic system and the risk of ovarian cancer. Plos One 4: e5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blasiak J, Smolarz B (2000) Plasminogen activator inhibitor-1 (PAI-1) gene 4G/5G promoter polymorphism is not associated with breast cancer. Acta Biochim Pol 47: 191–199. [PubMed] [Google Scholar]
- 22. Gilabert-Estelles J, Ramon LA, Braza-Boils A, Gilabert J, Chirivella M, et al. (2011) Plasminogen activator inhibitor-1 (PAI-1) 4G/5G polymorphism and endometrial cancer. Influence of PAI-1 polymorphism on tissue PAI-1 antigen and mRNA expression and tumor severity. Thromb Res 130: 242–7. [DOI] [PubMed] [Google Scholar]
- 23. Castello R, Espana F, Vazquez C, Fuster C, Almenar SM, et al. (2006) Plasminogen activator inhibitor-1 4G/5G polymorphism in breast cancer patients and its association with tissue PAI-1 levels and tumor severity. Thromb Res 117: 487–492. [DOI] [PubMed] [Google Scholar]
- 24. Eroglu A, Ulu A, Cam R, Akar N (2006) Plasminogen activator inhibitor -1 gene 4G/5G polymorphism in patients with breast cancer. J BUON 11: 481–484. [PubMed] [Google Scholar]
- 25. Forsti A, Lei H, Tavelin B, Enquist K, Palmqvist R, et al. (2007) Polymorphisms in the genes of the urokinase plasminogen activation system in relation to colorectal cancer. Ann Oncol 18: 1990–1994. [DOI] [PubMed] [Google Scholar]
- 26. Jorgenson E, Deitcher SR, Cicek M, Liu X, Plummer S, et al. (2007) Plasminogen activator inhibitor type-1 (PAI-1) polymorphism 4G/5G is associated with prostate cancer among men with a positive family history. Prostate 67: 172–177. [DOI] [PubMed] [Google Scholar]
- 27. Ju H, Lim B, Kim M, Noh SM, Kim WH, et al. (2010) SERPINE1 intron polymorphisms affecting gene expression are associated with diffuse-type gastric cancer susceptibility. Cancer 116: 4248–4255. [DOI] [PubMed] [Google Scholar]
- 28. Lei H, Hemminki K, Johansson R, Altieri A, Enquist K, et al. (2008) PAI-1–675 4G/5G polymorphism as a prognostic biomarker in breast cancer. Breast Cancer Res Treat 109: 165–175. [DOI] [PubMed] [Google Scholar]
- 29. Loktionov A, Watson MA, Stebbings WSL, Speakman CTM, Bingham SA (2003) Plasminogen activator inhibitor-1 gene polymorphism and colorectal cancer risk and prognosis. Cancer Lett 189: 189–196. [DOI] [PubMed] [Google Scholar]
- 30. Minisini AM, Fabbro D, Di Loreto C, Pestrin M, Russo S, et al. (2007) Markers of the uPA system and common prognostic factors in breast cancer. Am J Clin Pathol 128: 112–117. [DOI] [PubMed] [Google Scholar]
- 31. Onur E, Kurdal AT, Tugrul B, Iskesen I, Dundar P, et al. (2012) Is genetic screening necessary for determining the possibility of venous thromboembolism in cancer patients? Med Princ Pract 21: 160–163. [DOI] [PubMed] [Google Scholar]
- 32. Palmirotta R, Ferroni P, Savonarola A, Martini F, Ciatti F, et al. (2009) Prognostic value of pre-surgical plasma PAI-1 (plasminogen activator inhibitor-1) levels in breast cancer. Thromb Res 124: 403–408. [DOI] [PubMed] [Google Scholar]
- 33. Smolarz B, Blasiak J, Pytel J, Romanowicz-Makowska H, Zadrozny M (1999) 4G/5G polymorphism in the promoter of plasminogen activator inhibitor-1 (PAI-1) gene in subjects with node-negative and node-positive breast cancer. Med Sci Monitor 5: 833–837. [Google Scholar]
- 34. Sternlicht MD, Dunning AM, Moore DH, Pharoah PDP, Ginzinger DG, et al. (2006) Prognostic value of PAI1 in invasive breast cancer: Evidence that tumor-specific factors are more important than genetic variation in regulating PAI1 expression. Cancer Epidem Biomar 15: 2107–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Su CK, Yeh KT, Yeh CB, Wang PH, Ho ESC, et al. (2011) Genetic polymorphism of the plasminogen activator inhibitor-1 is associated with an increased risk of endometrial cancer. J Surg Oncol 104: 755–759. [DOI] [PubMed] [Google Scholar]
- 36. Tee YT, Wang PH, Tsai HT, Lin LY, Lin HT, et al. (2012) Genetic polymorphism of urokinase-type plasminogen activator is interacting with plasminogen activator inhibitor-1 to raise risk of cervical neoplasia. J Surg Oncol 106: 204–208. [DOI] [PubMed] [Google Scholar]
- 37. Turkmen B, Schmitt M, Schmalfeldt B, Trommler P, Hell W, et al. (1997) Mutational analysis of the genes encoding urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in advanced ovarian cancer. Electrophoresis 18: 686–689. [DOI] [PubMed] [Google Scholar]
- 38. Vairaktaris E, Serefoglou Z, Avgoustidis D, Yapijakis C, Critselis E, et al. (2009) Gene polymorphisms related to angiogenesis, inflammation and thrombosis that influence risk for oral cancer. Oral Oncol 45: 247–253. [DOI] [PubMed] [Google Scholar]
- 39. Vossen CY, Hoffmeister M, Chang-Claude JC, Rosendaal FR, Brenner H (2011) Clotting factor gene polymorphisms and colorectal cancer risk. J Clin Oncol 29: 1722–1727. [DOI] [PubMed] [Google Scholar]
- 40. Weng CJ, Lin CW, Chung TT, Tsai CM, Chen MK, et al. (2011) Impact of uPA system gene polymorphisms on the susceptibility of environmental factors to carcinogenesis and the development of clinicopathology of oral cancer. Ann Surg Oncol 18: 805–812. [DOI] [PubMed] [Google Scholar]
- 41. Weng CJ, Tsai CM, Chen YC, Hsieh YH, Lin CW, et al. (2010) Evaluation of the association of urokinase plasminogen activator system gene polymorphisms with susceptibility and pathological development of hepatocellular carcinoma. Ann Surg Oncol 17: 3394–3401. [DOI] [PubMed] [Google Scholar]
- 42. Woo M, Park K, Nam J, Kim JC (2007) Clinical implications of matrix metalloproteinase-1, -3, -7, -9, -12, and plasminogen activator inhibitor-1 gene polymorphisms in colorectal cancer. J Gastroenterol Hepatol 22: 1064–1070. [DOI] [PubMed] [Google Scholar]
- 43. Eroglu A, Ulu A, Cam R, Akar N (2007) Plasminogen activator inhibitor-1 gene 4G/5G polymorphism in cancer patients. J BUON 12: 135–136. [PubMed] [Google Scholar]
- 44. Duval S, Tweedie R (2000) Trim and fill: a simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 56: 455–463. [DOI] [PubMed] [Google Scholar]
- 45. Huang J, Sabater-Lleal M, Asselbergs FW, Tregouet D, Shin SY (2012) Genome-wide association study for circulating levels of plasminogen activator inhibitor-1 (PAI-1) provides novel insights into the regulation of PAI-1. Blood 120 (24): 4873–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dawson S, Hamsten A, Wiman B, Henney A, Humphries S (1991) Genetic variation at the plasminogen activator inhibitor-1 locus is associated with altered levels of plasma plasminogen activator inhibitor-1 activity. Arterioscler Thromb 11: 183–190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genetype frequencies on each studies. A generalized distribution of genetype frequencies on each included studies are listed.
(DOC)
Stratification analyses of the I2 and 95%/97.5% confidence interval. If I2 = 0, the one-sided 97.5% CI is presented. Otherwise, a two-sided 95% CI is performed.
(DOC)
PRISMA 2009 Checklist.
(DOC)