Abstract
There is a critical need for developing new malaria diagnostic tools that are sensitive, cost effective and capable of performing large scale diagnosis. The real-time PCR methods are particularly robust for large scale screening and they can be used in malaria control and elimination programs. We have designed novel self-quenching photo-induced electron transfer (PET) fluorogenic primers for the detection of P. falciparum and the Plasmodium genus by real-time PCR. A total of 119 samples consisting of different malaria species and mixed infections were used to test the utility of the novel PET-PCR primers in the diagnosis of clinical samples. The sensitivity and specificity were calculated using a nested PCR as the gold standard and the novel primer sets demonstrated 100% sensitivity and specificity. The limits of detection for P. falciparum was shown to be 3.2 parasites/µl using both Plasmodium genus and P. falciparum-specific primers and 5.8 parasites/µl for P. ovale, 3.5 parasites/µl for P. malariae and 5 parasites/µl for P. vivax using the genus specific primer set. Moreover, the reaction can be duplexed to detect both Plasmodium spp. and P. falciparum in a single reaction. The PET-PCR assay does not require internal probes or intercalating dyes which makes it convenient to use and less expensive than other real-time PCR diagnostic formats. Further validation of this technique in the field will help to assess its utility for large scale screening in malaria control and elimination programs.
Introduction
Four different Plasmodium species with different clinical implications infect humans in different combinations around the world, with recent studies demonstrating that P. knowlesi is also capable of causing human malaria due to zoonotic transmission [1]. Approximately 225 million cases of malaria and 781,000 deaths due to malaria occurred in 2009 [2]. Due to the apparent decline in the number of malaria cases and deaths in the last few years, there is a concerted global effort to control and eliminate malaria with the support of many public and private initiatives (reviewed in [3], [4]). Therefore, there is a clear need for diagnostic tools that are robust enough to accurately detect the species of infecting parasite(s), to identify the transmission foci of malaria reservoirs (often which may be submicroscopic and asymptomatic) and to monitor the success of malaria control and elimination programs.
The existing tools for malaria diagnosis include microscopy, parasite antigen/enzyme detection kits [commonly referred to as rapid diagnostic tests (RDTs)] and molecular tools (nucleic acid based tools). Microscopy remains the gold standard for the diagnosis of malaria in many malaria endemic countries. Although microscopy is relatively inexpensive and can be used to differentiate parasite species as well as provide quantitative data on the level of parasitemia, several limitations including poor sensitivity [5]–[9] begs for novel methods in the elimination era. RDTs have become an alternative tool for malaria diagnosis. While some RDTs are Plasmodium-specific, (pan) detecting the genus-specific aldolase and lactate dehydrogenase enzymes, the majority of the RDTs are specific for the P. falciparum histidine-rich protein –2 (Pf HRP-2). The recent discovery that some P. falciparum parasites in parts of South America and Africa have deleted the hrp-2 gene [10], [11] has brought into question the use of the HRP-2 based RDT tests due to potential false negatives. Another drawback of RDTs is the fact that they cannot be used to determine parasite densities and such quantification is increasingly essential with respect to both, antimalarial drug resistance surveillance and malaria control programs. With limits of detection at ∼100 parasites/µl (reviewed in [12]), RDTs can only be useful for routine diagnosis and treatment. With the limitations of both microscopy and RDTs, there is a clear realization that more sensitive diagnostic tools are needed for the detection of subclinical but transmissible infections to guide and monitor malaria elimination programs [13].
Molecular tools are far more sensitive in detecting low level infections and accurately detecting species of malaria parasite (reviewed in [5], [12]). There are many versions of molecular tools for malaria diagnosis ranging from conventional PCR-based assays, real-time PCR assays and more recently, isothermal amplification assays (reviewed in [7], [12]). When considering the development of tools for large scale field application, it is desirable to consider cost, robustness, and ease of use. The real-time PCR platform has some advantages for large scale screening as it requires no major post-PCR steps to acquire the results and a large number of samples can be analyzed at one time.
Real-time PCR assays depend on the use of fluorophores or DNA intercalating fluorescent dyes. As in the cases of TaqMan probes, molecular beacons and scorpion, sequence-specific oligonucleotide probes are dual labeled with a fluorescent dye and quencher. Although these molecular methods are robust and simpler than conventional PCR methods for large scale screening, the reagents are expensive and require special handling, which then creates practical challenges for field use in malaria endemic countries. A less complicated real-time PCR technique is the use of non-specific DNA intercalating dyes such as SYBR Green, SYTO-9, and calcein, which emit fluorescence signals when bound to double stranded DNA. Another drawback for these non-specific intercalating dyes is that they do not allow for multiplexing. Other alternatives for the detection of real-time PCR assays have been described including the direct labeling of one of the primers (forward or reverse) with a single fluorophore in a manner that facilitates self-quenching without the need of a quencher. These self-quenching primers facilitate the use of real-time PCR without the need for internal dual-labeled sequence specific probes. An example of this is the Light Upon Extension (LUX) primer that is labeled with an internal single fluorophore near the 3′ end of the primer in a hairpin structure which allows for the quenching before amplification [14]. These self-quenching, single labeled primers are less expensive compared to the dual labeled probe. However, the design of LUX primers requires the specific software, LUX Designer.
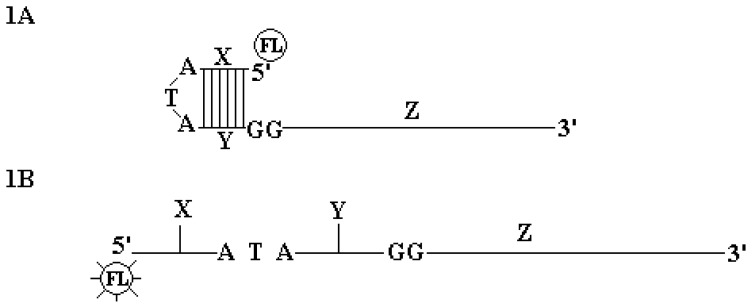
Here we describe a novel method for labeling real-time PCR primers based on the principle that was recently described in a patent filed by Jothikumar et al. (patent # PCT/US2008/084347). In this simplified technique, the 5′ end of one of the primers is modified by the addition of a 17-bases oligonucleotide tail that is labeled with a fluorophore (HEX or FAM) on its 5′ end, (Figure 1). In the absence of amplification, this tail forms a loop and remains in a closed conformation resulting in effective quenching of fluorescence due to close proximity of four G bases (i.e., the two overhang GG and two complementary GG residues in the hairpin formation) via photo-induced electron transfer (PET) mechanism; hence the name PET-PCR (not to be confused with the PET fluorescence dye from ABI). When participating in nucleic acid amplification, the stem loop structure of the PET primer opens up. The subsequent fluorescence increase is due to the de-quenching effect of the two guanosine residues located in the overhang positions and also due to the formation of the complementary DNA strand.
Figure 1. PET-PCR Primer design.
The photo-induced electron transfer (PET) primer consists of a quasi-universal tail sequence at 5′ and sequence specific region at 3′. This 5′ generic tail sequence consists of a 6 base pair stem, 3 nucleotide loop, a single fluorophore attached at 5′ end and two deoxyguanosine located in the opposite overhang region. The first two dangling end base on the PET primers are deoxyguanosine located at first and second overhang position respectively provided the greatest quenching in the absence of amplification. Circled FL is fluorescence attached to 5′. Z is sequence specific primer. X, ATA and Y together form a loop like structure. X and Y are complementary structure. Figure.1B. Fluorescence of PET primer increases during amplification when an extension of the complementary strand takes place. The result of fluorescence increase is due to dequenching effect of deoxyguanosine located in overhang positions and due to the formation of double strand.
To evaluate this concept, we designed PET-PCR primers for the specific amplification of the Plasmodium genus and P. falciparum species. We demonstrate the utility of this assay using 119 clinical samples of different malaria species infections, including mixed infections, and using well quantified Plasmodium species to estimate the limits of detection of this assay.
Materials and Methods
Ethics Statement
Some samples used in this study were obtained from a study conducted in Kenya. This study was approved by both the Kenya Medical Research Institute (KEMRI) and the CDC Institutional Review Board and informed written consent forms were obtained from each subject.
PET-PCR Primers
The design of the PET-PCR primers follows the same common rules for PCR primer design. In this study the genus specific primers were designed to amplify the 18S ribosomal RNA gene of Plasmodium. The primers were designed based on GenBank accession # GU815531; the P. malariae 18S small subunit ribosomal RNA gene. A BLAST search was performed to ascertain that the region selected for primer design was conserved in the rest of the Plasmodia species. The target for the P. falciparum specific primer set, Pfr364, was described previously [15]. In both cases, the 5′ end of the reverse primers was modified with the PET tag and labeled with FAM (for the genus) and HEX (for P. falciparum). These oligonucleotide primers for Plasmodium spp. and P. falciparum are shown in Table 1.
Table 1. Oligonucleotide primer sequences used in the real-time PET-PCR assay.
| Sequence (5′-3′) | Position | |
| Plasmodium genus-specific 18S ssrRNA * | ||
| Forward | GGCCTAACATGGCTATGACG | 305–324 |
| Reverse | FAM-aggcgcatagcgcctgg CTGCCTTCCTTAGATGTGGTAGCT | 395-372 |
| Plasmodium falciparum# | ||
| Forward | ACCCCTCGCCTGGTGTTTTT | |
| Reverse | HEX-aggcgcatagcgcctgg TCGGGCCCCAAAAATAGGAA | |
The target independent tail (underlined) is attached to the 5′end of the target specific sequences and do not show any homology to Plasmodium spp. sequences.
Position based on GenBank accession # GU815531 for 18S small subunit ribosomal RNA gene.
Designed based on a previously described P. falciparum specific target, Pfr364 [15].
Plasmodium Parasites and Clinical Samples
Different Plasmodium species available in our laboratories were used in this study: P. falciparum (3D7, FCR3, W2, D6, Dd2, V1-S, Nigeria, Santa Lucia), P. vivax (South Vietnam IV), P. malariae (Uganda I), P. knowlesi (Nuri) and P. ovale (Nigeria I). DNA from 50 clinical samples available from the CDC molecular diagnostic parasitology reference laboratory (Dr. A. DaSilva) (10 non-malaria samples, 13 P. falciparum, 9 P. vivax, 1 P. malariae, 11 P. ovale, 2 P. falciparum/P. malariae, 1 P. vivax/P. ovale, 2 P. falciparum/P. ovale mixed infections and 1 P. knowlesi; parasitemia data not available) were used. In addition, a set of 69 P. falciparum positive samples from a study in Kenya were tested anonymously (microscopic parasitemia range of 1230–231,040 parasites/µl).
DNA Extraction
DNA was isolated from all the samples using a QIAamp DNA Mini Kit (QIAGEN, Valencia, CA-(Qiagen method)). The DNA was aliquoted and stored at −20°C.
PET-PCR Method
Amplification of Plasmodium spp. or P. falciparum was performed in a 20 µl reaction containing 2X TaqMan Environmental Master Mix 2.0 (Applied BioSystems), 125 nM each forward and reverse primer, and 2 µl of DNA template. The reactions were performed under the following cycling parameters: initial hot-start at 95°C for 10 minutes, followed by 45 cycles of denaturation at 95°C for 10 seconds, annealing at 60°C. The correct fluorescence channel was selected for each fluorescently labeled primer set and the cycle threshold (CT) values recorded at the end of annealing step. Any sample with a CT value of 40.5 or below was considered positive. Both singleplex and multiplex assays were performed for all the samples.
Nested PCR and Real-time PCR Methods for Comparison
Due to its superior sensitivity over microscopy and other molecular tests, a nested PCR was chosen as a reference test to calculate the sensitivity and specificity of the PET-PCR. The nested PCR was performed with primers and cycling conditions as described by Singh et al. [16] with some modifications. Reactions were performed in a 20 µl total volume containing 1X buffer, 2.5 mM MgCl2, 200 µM dNTPs, 200 nM primers, and 1.25 units of Taq Polymerase (New England Biolabs, Ipswich, MA). The PCR amplified material was analyzed using gel electrophoresis (2% gel) to visualize the bands of appropriate size. Since the PET-PCR assay is a real-time PCR assay, a well-known real-time PCR diagnostic method developed by Rougemont et al. [17] (here after referred as Rougemont real-time PCR) was chosen as an additional method for making a quantitative comparison. The Rougemont real-time PCR is a duplex PCR, capable of detecting the four human infecting Plasmodium species in a set of two simultaneous separate duplex reactions (P. falciparum duplexed with P. vivax and P. malariae duplexed with P. ovale). For the comparison in our study, only one duplex reaction was performed (P. falciparum with P. vivax). This assay was performed as described by the authors. Briefly, a 25 µl reaction containing 12.5 µl of TaqMan Universal Master Mix (Applied Biosystems), 200 nM each of the Plasmodium specific forward and reverse primers and 80 nM of the species-specific probes was prepared. The assay was executed using the following cycling conditions: an initial step at 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 s and 60°C for 1 min. A cut-off CT value of 40 was used to indicate a positive result.
Test for Specificity, Sensitivity
The specificity of the primers was first tested by their ability to correctly amplify the gene of interest. P. falciparum, P. vivax, P. ovale, P. knowlesi and P. malariae DNA were used in the first round of testing. In addition, geographically-diverse strains for P. falciparum (3D7, FCR3, W2, D6, Dd2, and V1-S) and P.vivax were used to test for specificity of the primers.
Analytical Sensitivity
The analytical sensitivity (limits of detection) of both the genus- and falciparum-specific assays was determined using two well-quantified P. falciparum strains (Nigeria and Santa Lucia). The recommended WHO protocol for standards preparation used for quality control of RDT tests was used to prepare the P. falciparum standards (http://www.wpro.who.int/sites/rdt/using_rdts/qa/lot_testing.htm). The P. falciparum parasites were at the ring or early trophozoite stage of development when the sample was used (to exclude multinucleated parasites contributing to quantitative bias). The percent parasitemia was determined by three expert microscopists by counting the number of parasites in approximately 2000 erythrocytes examined. The total number of erythrocytes/µl was measured directly using a coulter counter and the number of parasites/µl was calculated from the total number of RBCs/µl and the percent parasitemia obtained. This standard sample was then diluted from the initial parasitemia to 2000 parasites/µl using uninfected whole blood and was then serial diluted fivefold to 0.64 parasite/µl using a 250 µl volume. DNA was extracted from each dilution point using 200 µl of sample. The analytical sensitivity of the PET-PCR assay was compared to that of the nested PCR [16] and a previously reported real-time PCR assay [17]. In addition, the limit of detection of the genus primer set to detect P. ovale, P. malariae and P. vivax was tested. These samples were kindly given to us by Dr. John Barnwell, Malaria Branch, Centers for Disease Control and Prevention (CDC).
Clinical Sensitivity and Specificity
The clinical sensitivity and specificity of the PET-PCR assay was calculated using 119 clinical samples: 69 microscopically positive P. falciparum samples from Kenya and 50 clinical samples available from the CDC molecular diagnostic parasitology reference laboratory which were previously tested for malaria using the Rougemont real-time PCR assay. Microscopy data was unavailable for these 50 clinical samples. A nested PCR assay [16] was used as the reference test. The percentage specificity and sensitivity were calculated as follows: Sensitivity = true positives/(true positives+false negatives) × 100. Specificity = true negatives/(true negatives+false positives) × 100. In addition, 95% Confidence Intervals (95% CI) for both sensitivity and specificity were calculated.
Results
Detection of Different Plasmodium Species Using the Genus-specific and P. falciparum-Specific PET- primers
In the initial round of testing we were able to amplify the five human-infecting Plasmodium species (P. falciparum, P. vivax, P. malariae, P. knowlesi and P. ovale) using the genus-specific primer set. The P. falciparum specific primer set amplified P. falciparum only. Both primer sets detected all the additional P. falciparum strains tested (3D7, FCR3, W2, D6, Dd2, and V1-S). In addition, the genus-specific primers detected 11 strains of P. vivax and 5 simian Plasmodium species, P. cynomolgi, P. inui, P. simiovale, P. hylobati and P. coatneyi.
Limits of Detection of PET-PCR Assay
The limits of detection of the genus and P. falciparum-specific primer sets was tested using serial dilution of calibrated DNA stock prepared from two P. falciparum strains (Nigeria and Santa Lucia). The parasitemia ranged from 2000, 400, 80, 16, 3.2 and 0.64 parasites/µl which was diluted using whole blood and DNA extracted for analysis. This was compared in parallel with the Rougemont real-time PCR and the nested PCR. Using a CT value of 40 as a cut off, both the PET-PCR and Rougemont real-time PCR assays detected as low as 3.2 parasites/µl. This was comparable to the nested PCR which consistently detected 3.2 parasites/µl and occasionally as few as 0.64 parasites/µl. The limits of detection of the genus-specific primer set for the other Plasmodium species was ascertained with DNA obtained from P. vivax, P. ovale and P. malariae infected blood and determined to be 5.0 parasites/µl, 5.8 parasites/µl, and 3.5 parasites/µl, respectively.
Quantitative Comparison of PET-PCR Assay and Rougemont Real-time PCR Assay
The quantitative nature of the PET-PCR method was compared in parallel with the Rougemont real-time PCR using the P. falciparum serial dilutions utilized in testing the limits of detection above. The obtained CT values were plotted against the log transformed parasitemia to determine any correlation between the two (Figure 2). Similar to the Rougemont real time PCR assay, the CT values obtained in the PET-PCR assay demonstrated reproducible linearity over the range of parasitemia tested in four replicate experiments. Both methods showed significant correlation with R2 values >0.990 and a significant correlation coefficient (p<0.0007) for the mean CT values and parasitemia implying that this method can be used to quantify parasitemia. No statistical difference in mean CT values was observed for the two different strains or for the two different methods (P>0.05).
Figure 2. Quantitative comparison of PET-PCR assay and Rougemont real-time PCR assay.
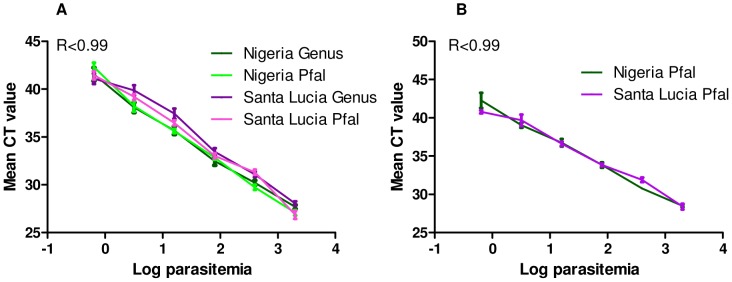
The PET-PCR and Rougemont real-time PCR assays were run using two well-quantified P. falciparum strains (Nigeria and Santa Lucia) used at six differing parasitemia levels (2000, 400, 80, 16, 3.2 and 0.64 parasites/µl). To determine the correlation between CT values and parasitemia, the mean CT values obtained in the PET-PCR assay (A) and in the Rougemont real time PCR assay (B) were plotted against the log transformed parasitemia (Log parasitemia). The CT values demonstrated reproducible linearity over the parasitemia range tested as both methods show significant correlation with R2 values >0.990. No statistical difference in mean CT values was observed for the two different strains or for the two different methods (P>0.05).
Multiplex Assay
To test the possibility of multiplexing the assay, the two primer sets were used in a duplex assay. A well-quantified P. falciparum strain (Nigeria strain) was used at six differing parasitemia levels (2000, 400, 80, 16, 3.2 and 0.64 parasites/µl). The reactions were performed both in the singleplex and multiplex formats. Both the singleplex and multiplex formats gave similar CT values for the different parasitemia levels tested, Table 2. In addition, the assay was performed using mock-mixed infections of P. falciparum/P. vivax, P, falciparum/P. ovale and P. falciparum/P. malariae with varying parasite densities of the P. falciparum. Both primer sets were able to detect all the varying concentrations in the mock mixed infections, Table 3.
Table 2. Reproducibility of the singleplex and multiplex PET-PCR assays for P. falciparum detection.
| Mean CT value ± SD | ||||||
| Target | 2000 (parasites/µl) | 400(parasites/µl) | 80(parasites/µl) | 16(parasites/µl) | 3.2(parasites/µl) | 0.64(parasites/µl) |
| Genus-S | 29.19±0.40 | 31.70±0.67 | 33.92±0.69 | 36.69±0.57 | 39.56±1.20 | 42.92±1.02 |
| Pf- S | 28.22±0.58 | 31.11±0.36 | 33.33±0.64 | 35.71±0.39 | 37.79±0.49 | 42.77±0.77 |
| Genus-M | 28.15±0.46 | 30.50±0.88 | 33.04±0.66 | 35.69±0.31 | 38.54±1.23 | 41.89±1.23 |
| Pf-M | 27.64±1.41 | 30.32±0.63 | 33.06±0.46 | 35.55±0.47 | 37.97±0.45 | 42.40±0.81 |
A well-quantified P. falciparum strain (Nigeria strain) was used at six differing parasitemia levels (2000, 400, 80, 16, 3.2 and 0.64 parasites/µl). The reactions using the genus-specific primers (Genus) and P. falciparum- specific primers (Pf) were performed both in the singleplex (Genus-S and Pf-S) and multiplex (Genus-M and Pf-M) formats. The mean CT values ± standard deviation (SD) obtained from three experiments each performed in duplicate are shown.
Table 3. Detection of mock mixed infections using the multiplex assay.
| P. falciparum/P. malariae (Parasites/µl) | P. falciparum (CT ±SD) | Genus (CT ±SD) |
| 2000/7 | 29.62±0.76 | 29.07±0.27 |
| 400/14 | 31.80±0.30 | 31.29±0.31 |
| 80/28 | 34.63±0.33 | 33.56±0.23 |
| 16/280 | 36.21±0.37 | 31.01±0.28 |
| 3.2/2800 | 38.61±0.53 | 27.80±0.21 |
| NTC | No CT | No CT |
| P. falciparum/P. vivax (Parasites/µl) | P. falciparum (CT ±SD) | Genus (CT ±SD) |
| 2000/5 | 29.98±0.18 | 29.46±0.44 |
| 400/10 | 31.88±0.43 | 31.63±0.15 |
| 80/100 | 34.23±0.33 | 33.35±0.85 |
| 16/1000 | 36.77±0.14 | 30.99±0.16 |
| 3.2/2000 | 39.70±0.84 | 27.57±0.53 |
| NTC | No CT | No CT |
| P. falciparum/P. ovale (Parasites/µl) | P. falciparum (CT ±SD) | Genus (CT ±SD) |
| 2000/5.8 | 29.56±0.29 | 29.31±0.31 |
| 400/11.6 | 31.01±0.13 | 30.41±0.47 |
| 80/23.1 | 34.08±0.42 | 32.92±0.41 |
| 16/231 | 35.51±0.39 | 30.92±0.27 |
| 3.2/2300 | 38.79±0.39 | 27.28±0.54 |
| NTC | No CT | No CT |
Mock mixed infections were prepared using a well-quantified P. falciparum strain (Nigeria strain) at five differing parasitemia levels (2000, 400, 80, 16 and 3.2 parasites/µl) combined with varying parasite densities of P. malariae, P. vivax and P. ovale (as shown in the table). The mean CT values for the P. falciparum and genus primers sets obtained from two experiments each performed in duplicate are shown. The multiplex assay was able to detect all the varying combinations of the mock-mixed infections. NTC = no template control; SD = standard deviation.
Clinical Sensitivity and Specificity of the Plasmodium Genus and P. falciparum Specific PET-primers
One hundred and nineteen clinical samples consisting of different malaria species and mixed infections were used to test the utility of the novel PET-PCR primers in diagnosis of clinical samples. As shown in Table 4 all the P. falciparum samples tested were shown to be positive by the P. falciparum–specific and the Plasmodium -specific primer sets. The genus-specific primers detected all other non-falciparum samples tested. No signal was detected with the 10 non-malaria human DNA. The sensitivity and specificity of these singleplex assays was calculated using the nested PCR as a reference test. Both primer sets showed 100% sensitivity and specificity. In the multiplex assays, the genus–specific primers were able to accurately detect all the Plasmodium spp. samples; however, the P. falciparum-specific primer set failed to detect two P. falciparum samples (Table 4). Microscopy data was only available for 69 P. falciparum samples. These samples (parasitemia range of 1230–231,040 parasites/µl) were all shown to be positive by both primers sets.
Table 4. Comparison of PET–PCR to nested PCR using clinical samples.
| Number of samples detected by each method (n) | ||||
| Nested PCR | Singleplex-PET | Multiplex-PET | ||
| P. falciparum | Genus | P. falciparum | Genus | |
| P. falciparum (82) | 80/80# | 81/81# | 81/82* | 82/82 |
| P. vivax (9) | 0/9 | 9/9 | 0/8# | 8/8# |
| P. ovale (11) | 0/9# | 9/9# | 0/11 | 11/11 |
| P. malariae (1) | 0/1 | 1/1 | 0/1 | 1/1 |
| P. knowlesi (1) | 0/1 | 1/1 | 0/1 | 1/1 |
| Pf/Po (2) | 2/2 | 2/2 | 1/2* | 2/2 |
| Pf/Pm (2) | 2/2 | 2/2 | 2/2 | 2/2 |
| Pv/Po (1) | 0/1 | 1/1 | ND | ND |
| Non-malaria (10) | 0/8# | 0/9# | 0/10 | 0/10 |
A total of 119 clinical samples consisting of different malaria species and mixed infections were used to test the utility of the PET-PCR primers in diagnosis of clinical samples.
Not all the samples tested by nested PCR were tested in all the PET-PCR assays due to insufficient sample volumes.
The P. falciparum primer (used in the multiplex assay) failed to detect two P. falciparum positive samples.
ND = not done.
Discussion
The use of self-quenching real-time PCR primers using a 17-base loop tail was recently described in a patent filed by Jothikumar et al. (patent # PCT/US2008/084347). In this proof-of-concept study, we designed PET-PCR primers for the detection of Plasmodium spp. and P. falciparum and successfully demonstrated that these primers can be duplexed to detect both Plasmodium genus and P. falciparum species in a single tube. The sensitivities of this assay are better than those reported for microscopy and RDTs (reviewed in [18]), and comparable to that of other real-time PCR assays (range from 0.7–10 parasites/µl) [17], [19], [20]. As reviewed by Okell [5], the existence of submicroscopic infections is well documented and are known to contribute to the infective reservoirs of malaria gametocytes [21]–[23]. Therefore, sensitive assays such as the PET-PCR described here, that are capable of detecting submicroscopic infections are valuable for any malaria control and elimination programs, as these will help monitor transmission foci and dynamics as well as control efforts. Moreover, this assay provides the option of analyzing a large number of samples in a short time.
We demonstrate that PET-PCR is comparable to Rougemont real-time PCR (a Taqman based assay which is widely used for malaria diagnosis) both in terms of high sensitivity and the ability to quantify parasitemia. To date, no self-quenching real-time PCR assay has been described for malaria diagnosis. The self-quenching in PET-PCR facilitates the use of real-time PCR without the need for probe designs involving internal quenching thereby reducing the cost of the assay by obviating the need for expensive probes from commercial vendors. In addition, high-performance liquid chromatography (HPLC) purification is not required for the single labeled primer, resulting in a high yield of product further reducing the costs as compared to the commonly used dual-labeled probes. Furthermore, the PET-PCR primers can be either stored in the refrigerator without loss of activity for up to a month or be lyophilized and reconstituted when the end user is ready to use them. These practical advantages together with the lower costs and the easy design of PET primers (no specific software is required) offers promising features that can enable the adoption of this method as an alternate real-time PCR for malaria detection for large scale screening of field samples in large malaria surveillance and control programs.
The genus-specific assay described here offers an efficient, high-throughput malaria detection tool, ideal for the screening of malaria, irrespective of the species. This is useful, for example, in surveillance/screening epidemiological studies. It can be used to both monitor and evaluate malaria control programs in which screening of malaria infection is necessary or as a confirmatory test for detecting malaria infection. If desired, the PET-PCR can be duplexed to detect genus and P. falciparum in one tube, albeit with some slight loss of sensitivity for the P. falciparum-specific primers. This is common with multiplex assays, which often require some compromise of the components and assay conditions to accommodate two or more primer sets which can lead to primer competition and loss of sensitivity. This caveat is not insurmountable as further improvements can be undertaken to address this issue. However, while duplexing these primer sets is a possibility, detection of mixed infections is not achievable because a positive result with the genus primer only indicates that Plasmodium spp. is present. Used together these two primer sets only indicates an infection with P. falciparum and does not necessarily indicate the lack of a mixed infection with other Plasmodium species. This is the same caveat encountered in many of the commonly used malaria RDTs that detect pan-Plasmodium antigens and the P. falciparum specific HRP-2 protein. Development of primers to detect non-falciparum species (P. vivax, P. malariae, P. ovale and P. knowlesi) will be required to make this method applicable in clinical laboratory settings.
Diagnostic tools for large scale field application need to be cheaper, robust, and easy to use. The PET-PCR assay described here meets these criteria and can be used for large scale screening in surveillance and epidemiological studies including as a confirmatory test in clinical laboratory settings.
Acknowledgments
We would like to thank the Malaria Branch, CDC, for supporting this project and Dr. John W. Barnwell for proving some of the samples used in this study. The use of trade names and names of commercial sources is for identification only and does not imply endorsement by the Centers for Disease Control and Prevention or the U.S. Department of Health and Human Services. The findings and conclusions in this presentation are those of the authors and do not necessarily represent those of the Centers for Disease Control and Prevention.
Funding Statement
This study was supported by the Atlanta Research and Education Foundation, Atlanta, Georgia. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, et al. (2004) A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363: 1017–1024. [DOI] [PubMed] [Google Scholar]
- 2.WHO (2010) World Malaria Report 2010. [Google Scholar]
- 3. Najera JA, Gonzalez-Silva M, Alonso PL (2011) Some lessons for the future from the Global Malaria Eradication Programme (1955–1969). PLoS Med 8: e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alonso PL, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, et al. (2011) A research agenda to underpin malaria eradication. PLoS Med 8: e1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Okell LC, Ghani AC, Lyons E, Drakeley CJ (2009) Submicroscopic infection in Plasmodium falciparum-endemic populations: a systematic review and meta-analysis. J Infect Dis 200: 1509–1517. [DOI] [PubMed] [Google Scholar]
- 6. McMorrow ML, Aidoo M, Kachur SP (2011) Malaria rapid diagnostic tests in elimination settings–can they find the last parasite? Clin Microbiol Infect 17: 1624–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McMorrow ML, Masanja MI, Abdulla SM, Kahigwa E, Kachur SP (2008) Challenges in routine implementation and quality control of rapid diagnostic tests for malaria–Rufiji District, Tanzania. Am J Trop Med Hyg 79: 385–390. [PMC free article] [PubMed] [Google Scholar]
- 8. Kahama-Maro J, D’Acremont V, Mtasiwa D, Genton B, Lengeler C (2011) Low quality of routine microscopy for malaria at different levels of the health system in Dar es Salaam. Malar J 10: 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ngasala B, Mubi M, Warsame M, Petzold MG, Massele AY, et al. (2008) Impact of training in clinical and microscopy diagnosis of childhood malaria on antimalarial drug prescription and health outcome at primary health care level in Tanzania: a randomized controlled trial. Malar J 7: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koita OA, Doumbo OK, Ouattara A, Tall LK, Konare A, et al. (2012) False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg 86: 194–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS ONE 5: e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Erdman LK, Kain KC (2008) Molecular diagnostic and surveillance tools for global malaria control. Travel Med Infect Dis 6: 82–99. [DOI] [PubMed] [Google Scholar]
- 13. mal ERACGoD (2011) Diagnostics (2011) A research agenda for malaria eradication: diagnoses and diagnostics. PLoS Med 8: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nazarenko I (2006) Homogeneous detection of nucleic acids using self-quenched polymerase chain reaction primers labeled with a single fluorophore (LUX primers). Methods Mol Biol 335: 95–114. [DOI] [PubMed] [Google Scholar]
- 15. Demas A, Oberstaller J, Debarry J, Lucchi NW, Srinivasamoorthy G, et al. (2011) Applied genomics: Data mining reveals species-specific malaria diagnostic targets more sensitive than 18S rRNA. J Clin Microbiol 49: 2411–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, et al. (1999) A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg 60: 687–692. [DOI] [PubMed] [Google Scholar]
- 17. Rougemont M, Van Saanen M, Sahli R, Hinrikson HP, Bille J, et al. (2004) Detection of four Plasmodium species in blood from humans by 18S rRNA gene subunit-based and species-specific real-time PCR assays. J Clin Microbiol 42: 5636–5643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH (2007) A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT). Am J Trop Med Hyg 77: 119–127. [PubMed] [Google Scholar]
- 19. Taylor SM, Juliano JJ, Trottman PA, Griffin JB, Landis SH, et al. (2010) High-throughput pooling and real-time PCR-based strategy for malaria detection. J Clin Microbiol 48: 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perandin F, Manca N, Calderaro A, Piccolo G, Galati L, et al. (2004) Development of a real-time PCR assay for detection of Plasmodium falciparum, Plasmodium vivax, and Plasmodium ovale for routine clinical diagnosis. J Clin Microbiol 42: 1214–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bousema T, Drakeley C (2011) Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin Microbiol Rev 24: 377–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Drakeley C, Sutherland C, Bousema JT, Sauerwein RW, Targett GA (2006) The epidemiology of Plasmodium falciparum gametocytes: weapons of mass dispersion. Trends Parasitol 22: 424–430. [DOI] [PubMed] [Google Scholar]
- 23. Ouedraogo AL, Bousema T, Schneider P, de Vlas SJ, Ilboudo-Sanogo E, et al. (2009) Substantial contribution of submicroscopical Plasmodium falciparum gametocyte carriage to the infectious reservoir in an area of seasonal transmission. PLoS ONE 4: e8410. [DOI] [PMC free article] [PubMed] [Google Scholar]