Abstract
Cell polarization occurs along a single axis that is generally determined by a spatial cue. Cells of the budding yeast exhibit a characteristic pattern of budding, which depends on cell-type-specific cortical markers, reflecting a genetic programming for the site of cell polarization. The Cdc42 GTPase plays a key role in cell polarization in various cell types. Although previous studies in budding yeast suggested positive feedback loops whereby Cdc42 becomes polarized, these mechanisms do not include spatial cues, neglecting the normal patterns of budding. Here we combine live-cell imaging and mathematical modeling to understand how diploid daughter cells establish polarity preferentially at the pole distal to the previous division site. Live-cell imaging shows that daughter cells of diploids exhibit dynamic polarization of Cdc42-GTP, which localizes to the bud tip until the M phase, to the division site at cytokinesis, and then to the distal pole in the next G1 phase. The strong bias toward distal budding of daughter cells requires the distal-pole tag Bud8 and Rga1, a GTPase activating protein for Cdc42, which inhibits budding at the cytokinesis site. Unexpectedly, we also find that over 50% of daughter cells lacking Rga1 exhibit persistent Cdc42-GTP polarization at the bud tip and the distal pole, revealing an additional role of Rga1 in spatiotemporal regulation of Cdc42 and thus in the pattern of polarized growth. Mathematical modeling indeed reveals robust Cdc42-GTP clustering at the distal pole in diploid daughter cells despite random perturbation of the landmark cues. Moreover, modeling predicts different dynamics of Cdc42-GTP polarization when the landmark level and the initial level of Cdc42-GTP at the division site are perturbed by noise added in the model.
Introduction
Cell polarization is essential for a variety of cellular processes and functions. Cdc42 is highly conserved from yeast to humans and plays a central role in polarity establishment [1], [2]. The budding yeast Saccharomyces cerevisiae provides a unique model to study the development of cell polarity owing to its pronounced cell polarization during growth and its experimental tractability. During vegetative growth, yeast cells choose a specific bud site depending on their cell type, which determines the axis of polarized cell growth. Haploid a and α cells bud in the axial pattern, in which both mother and daughter cells select a new bud site adjacent to their immediately preceding division site. In contrast, a/α cells (normal diploids) bud in the bipolar pattern, in which daughter cells usually bud at the pole distal to the previous division site (distal pole) and mother cells can choose a new bud site near the proximal pole (birth pole) or the distal pole (see Fig. 1A ) [3], [4], [5], [6]. These different budding patterns occur in response to cell-type-specific markers. The Rsr1 GTPase module, which is composed of Rsr1/Bud1, its GTPase activating protein (GAP) Bud2, and its GDP-GTP exchange factor (GEF) Bud5 [3], [7], [8], [9], [10], [11], links the spatial cues to the polarity establishment machinery including Cdc42. Cdc42 thus becomes polarized at the predetermined cortical site to trigger bud growth (see review [2] and references therein).
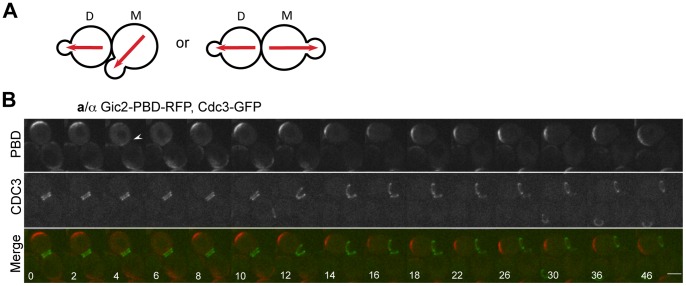
Figure 1. Time-lapse microscopy of Cdc42-GTP polarization in wild-type a/α diploids.
A. A schematics diagram of the bipolar budding pattern. M and D stand for mother and daughter cells, respectively. Red arrows depict the axis of cell polarity. B. Localization of Gic2-PBD-RFP and Cdc3-GFP in diploid wild-type cells (HPY2353). An arrowhead marks Gic2-PBD-RFP localized to the proximal pole in the daughter cell. Numbers indicate time (in min) from the first image. Size bars, 3 µm.
How do a/α cells select a bud site either at the distal or proximal pole? Previous studies uncovered a large number of genes affecting the bipolar budding pattern [12], [13], [14], [15]. These studies also indicate a close link between the cell cycle progression and the bipolar budding pattern [14], [16]. The bipolar pattern is dependent on transmembrane proteins including Bud8, Bud9, Rax1 and Rax2 [12], [17], [18], [19], [20]. Bud8 localizes to the distal pole of a newly born cell whereas Bud9 localizes to the bud side of the mother-bud neck (which becomes the proximal pole of a daughter cell) just before cytokinesis [20]. These localization patterns of Bud8 and Bud9 are consistent with their roles as putative distal and proximal pole markers, respectively. Rax1 and Rax2 localize to the tip of growing buds and to the mother-bud necks, and their presence at the division site is persistent throughout multiple generations [17], [18], [19]. Despite these interesting localization patterns of the putative bipolar landmarks, the mechanism by which the bipolar pattern is established remains largely unknown. One of the key questions is why daughter cells of a/α diploids choose predominantly the distal pole for their first budding despite the presence of Bud8 and Bud9 marking each pole. The complexity of the budding patterns led us to take a minimalist approach to address the question by combining mathematical modeling and live-cell imaging.
Recent studies in budding yeast have uncovered mechanisms by which Cdc42 becomes polarized in the absence of spatial cues via a process called 'symmetry breaking'. Two positive feedback mechanisms of symmetry breaking have been suggested – one involving the actin cytoskeleton and the other relying on a Cdc42 signaling network including the scaffold protein Bem1 and the Cdc42 GEF Cdc24 [21], [22], [23], [24]. Endocytosis- and GDI (Guanosine nucleotide dissociation inhibitor)-mediated recycling of Cdc42 and a negative feedback loop confer robust initiation of cell polarization [25], [26], [27], [28]. Using a stochastic mathematical model, an intrinsic stochastic mechanism involving linear positive feedback alone was shown to be sufficient to account for the spontaneous establishment of a single polarization site [29]. A Turing-type mechanism involving short-range excitation and long-range inhibition has also been proposed to explain the self-organized emergence of polarity [30], [31]. These models capture several features of cell polarization and provide a mechanistic insight into spontaneous polarization in the absence of spatial cues. However, some aspects of these mechanisms and their physiological relevance are still unclear and controversial [31], [32], [33]. More importantly, it had been unclear whether and how the spatial cues are recognized and amplified through these feedback mechanisms.
Here, we used computational modeling and live-cell imaging to explain cell polarization in diploid daughter cells. Because wild-type yeast cells undergo polarization in response to the cell-type-specific spatial cues, we considered these cues to understand distinct budding patterns. We report that both spatial landmarks and GTP hydrolysis of Cdc42 by Rga1 control the robust Cdc42-GTP polarization in diploid daughter cells.
Results and Discussion
A Mathematical Model of Cdc42 Polarization in Diploid Daughter Cells
Diploid a/α cells exhibit a strong bias toward the distal pole during their first and second bud-site selection [4], [12], [20] ( Fig. 1A ). To examine this preferential distal-pole budding event in daughter cells of diploids more closely, we monitored localization of Cdc42-GTP every 2 min in wild-type diploid cells expressing Gic2-PBD-RFP (tdTomato fused to the p21-binding domain of Gic2) as a reporter for Cdc42-GTP [34] and GFP fused to Cdc3, a component of septins, as a marker for the timing and site of cytokinesis. As expected, Gic2-PBD-RFP localized to the periphery of a growing bud until the end of the M phase, to the mother-bud neck (which becomes the proximal pole of daughter cells) during cytokinesis, and then to the distal pole in the daughter cells in the next G1 phase (100%, n = 15 movies) ( Fig. 1B ; Movie S1). While Cdc42 becomes enriched at the mother-bud neck at the division site [34], [35], the Gic2-PBD-RFP signal at the proximal pole was relatively weak presumably due to rapid hydrolysis of Cdc42-GTP by its GAP(s), consistent with a previous finding in haploids [34]. Nonetheless, our imaging was able to capture the daughter cells at an intermediate stage that exhibited Cdc42-GTP localization at both proximal and distal poles (see a cell marked with an arrowhead in Fig. 1B ). The dynamics of Cdc42-GTP polarization is thus consistent with the distal-pole budding of diploid daughter cells.
Why do daughter cells of diploids exhibit such dynamics of Cdc42-GTP despite the presence of spatial cues at both poles? Since our current knowledge of the bipolar landmark(s) does not provide a clear explanation for this time-evolved polarization of Cdc42-GTP in diploid daughter cells, we turn to mathematical modeling. We took into consideration several previous experimental observations and previous models for symmetry breaking. We assumed that the distal and proximal poles compete for Cdc42 or its effectors and regulators ( Fig. 2A ). Our model was built upon the positive feedback mechanism involving the Bem1 complex originally proposed by Goryachev and Pokhilko [30] and Lew and colleagues [24]. Importantly, our model included the Cdc42 GAPs to account for the weak Gic2-PBD-RFP localization at the division site and the spatial cues at both poles.
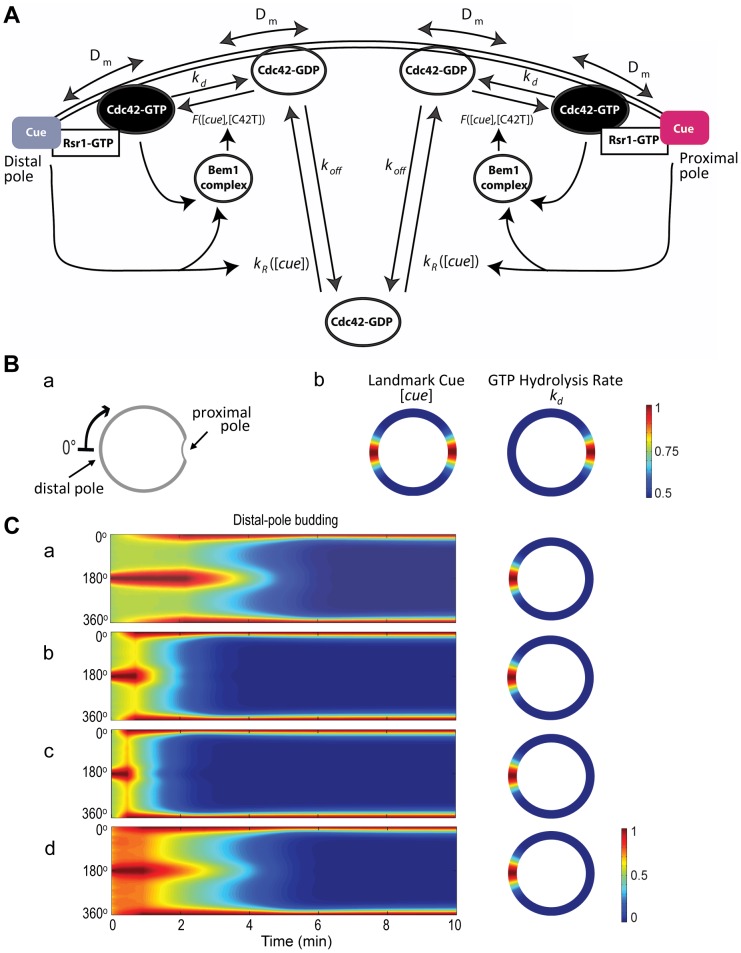
Figure 2. Mathematical modeling of Cdc42 polarization.
A. A schematic diagram of the reaction-diffusion model with the following parameters: Dm (the diffusion rate coefficient of Cdc42-GDP and Cdc42-GTP on the plasma membrane), kd (the inactivation rate coefficient of Cdc42 from the GTP- to the GDP-bound states), and koff (the rate at which the membrane-bound Cdc42-GDP is extracted into the cytoplasm). The function F([cue], [C42T]) represents the Bem1-mediated activation rate; and the function kR([cue]) is the landmark-signal-dependent recruitment rate of Cdc42 from the cytoplasm to the membrane. See details in Materials and Methods and the values of parameters in Tables 1 and 2 . Ba. Coordinate of the periphery of an a/α daughter cell. The cell periphery is parameterized by the radial angle (0°–360°) in a clockwise direction starting from the distal pole. Bb. Spatial distributions of the landmark cues and the GTP hydrolysis rate of Cdc42. Both quantities are normalized by their maximal values for better visualization, with the color map scale shown on the right. C. Spatiotemporal dynamics of Cdc42-GTP in a diploid daughter cell. The cell periphery is presented as a 1D vertical axis, with the proximal pole in the middle (180°) and distal pole at the top/bottom (0°/360°). The horizontal axis represents the time window from 0 to 10 min. The localization of Cdc42-GTP at steady state is shown on the 2D cell periphery (right). The level of Cdc42-GTP is normalized by its maximal value at steady state, and the color map is displayed. See parameters in Tables 1 and 2 for each simulation.
Specifically, several space-dependent rate parameters are included in our model as schematically shown in Fig. 2A . The establishment of Cdc42 polarization relied on the activation from its GDP- to GTP-bound state, which presumably depends on the pre-localized landmark signal and the Bem1-mediated feedback. This feedback was implemented in the activation rate of Cdc42 from the GDP- to the GTP-bound states (denoted by F), which depends on the levels of landmark cue (denoted by [cue]) and Cdc42-GTP, under the assumption that Bem1 is conserved (see Materials and Methods). The inactivation rate (kd) of Cdc42 from the GTP- to the GDP-bound states was space-dependent because it was assumed to vary with the level of the Cdc42 GAPs (which localize to the division site [34], [36], [37], [38]). The recruitment rate (kR) of Cdc42 from the cytoplasm to the membrane represents the association rate of cytoplasmic Cdc42-GDP with the membrane. The rate kR depends on the level of spatial cues because Rsr1 is likely to interact with Cdc42 to enhance its recruitment to the membrane in response to the landmark [39]. The landmark and the Rsr1 module were considered together as an upstream input to represent the spatial cue that triggers the initial localization of Cdc42. Thus kR was positively correlated with the level of the landmark signal in our simulations (see details in Materials and Methods). The parameters used in our simulations are listed in Tables 1 and 2 .
Table 1. Ranges of parameters in the simulations.
| Parameter | Value | Definition | Reference |
| R |
|
Radius of the cell | This study |
| Dm |
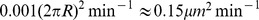
|
Lateral surface diffusion coefficient | [29], [30] |
| koff |
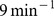
|
Rate coefficient from membrane to cytoplasm | [29] |
| k Rec |
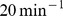
|
Rate parameter for recruitment from membrane to cytoplasm | [29] |
| KR | 0.1 | Parameter in recruitment rate | This study |
| kdL |
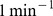
|
Inactivation rate coefficient of Cdc42 | This study |
| kdH | 1∼2 min −1 | ||
| kon | 0.1 min −1 | Activation rate coefficient of Cdc42 | [30] |
| C 0 | 0.1 | Basal level of [cue] | This study |
| C 1 | 0.15∼0.25 | Level of [cue] at proximal pole | This study |
| C 2 | 0.15∼0.25 | Level of [cue] at distal pole | This study |
| A 0 | 0.3 | Initial level of [C42D] | This study |
| A 1 | 0.005 | Basal level of initial [C42T] | This study |
| A2 | 0.005∼0.01 | Maximum level of initial [C42T] | This study |
| K | 0.015 | EC50 of the feedback | This study |
Table 2. Specific parameters used for simulations.
| Fig. 2. Ca | Fig. 2. Cb | Fig. 2. Cc | Fig. 2. Cd | Fig. 7. Ab | Fig. 7. Ac, top | Fig. 7. Ac bottom | Fig. 7B, top | Fig. 7B, bottom | |
| kdH | 2 min−1 | 2 min−1 | 2 min−1 | 2 min−1 | 1 min−1 | 1 min−1 | 1 min−1 | 2 min−1 | 2 min−1 |
| C 1 | 0.25 | 0.2 | 0.15 | 0.25 | 0.2 | 0.2 | 0.15 | 0.25 | 0.1 |
| C 2 | 0.15 | 0.2 | 0.25 | 0.15 | 0.2 | 0.25 | 0.25 | 0.1 | 0.25 |
| A 2 | 0.01 | 0.01 | 0.01 | 0.007 | 0.01 | 0.007 | 0.005 | 0.01 | 0.01 |
Our model involved two reaction-diffusion equations to describe the spatial dynamics of Cdc42-GTP and Cdc42-GDP (Eq. [1]–[2] in Materials and Methods) on a cross section of the cell membrane with a diameter of 4 µm. In this model, the spatially distributed landmark [cue] was assumed to be a function of the membrane periphery, which was parameterized by the angle x along the circle (0° ≤ x ≤360°) from the distal pole ( Fig. 2B , a). The function [cue] thus took maximal values locally at the proximal and distal poles ( Fig. 2B , b) to represent the localized landmark at these poles. Our model also involved the following reactions: lateral membrane diffusion of Cdc42-GTP and Cdc42-GDP, activation of Cdc42 to the GTP-bound state and its inactivation, recruitment of Cdc42 from the cytoplasm to the membrane and its reverse reaction, and GDI-mediated extraction of Cdc42-GDP into the cytoplasm (see Materials and Methods).
Our simulations started with a homogeneous level of Cdc42-GDP at initial time t = 0 and with Cdc42-GTP localized at the proximal pole of the cell, since Cdc42 is polarized to the division site ( Fig. 2C ). Fluctuations in the initial levels of these species due to naturally noisy background led to Cdc42-GTP clustering initially at both poles, which coexisted for a period of time. The Cdc42-GTP cluster at the proximal pole (180°) was gradually destabilized due to GTP hydrolysis by Cdc42 GAP(s) at the previous budding site, resulting in Cdc42 polarization at the distal pole. Indeed, the Cdc42-GTP cluster was consistently formed at the distal pole with any set of parameters within the ranges shown in Table 1 ( Fig. 2C , a–d), suggesting that the outcomes of competition are relatively insensitive to the concentration of spatial cues at each pole. Our modeling thus explains robust distal-pole budding of a/α daughter cells despite the competition between two poles for recruiting the Bem1 complexes and Cdc42-GTP.
Deletion of RGA1 Affects the Distal-pole Budding in Daughter Cells of a/α Diploids
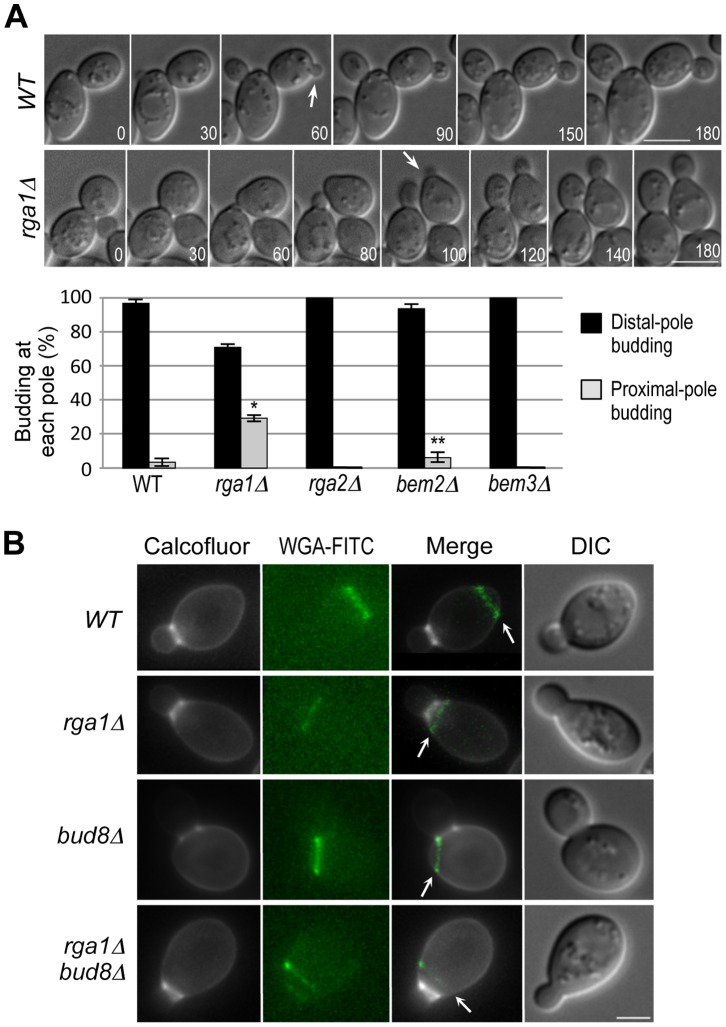
Because our modeling suggested that Cdc42 GTP hydrolysis rate at the division site contributes to robust distal-pole budding in a/α daughter cells, we wondered which Cdc42 GAP(s) play a role in this process. All predicted Cdc42 GAPs localize to the mother-bud neck at cytokinesis [34], [36], [37], [38]. We thus scored the position of the first bud of newly born daughter cells of diploid wild type and mutants deleted for a Cdc42 GAP such as Rga1, Rga2, Bem2, or Bem3. As expected, daughter cells rarely budded at the proximal pole in wild-type cells (3.4±2.3%, n = 106). In contrast, a significant number of daughter cells of an a/α rga1Δ homozygous diploid strain budded at the proximal pole (29.3±1.9%, n = 144; see daughter cells marked with arrows in Fig. 3A ), which is statistically significant (p<10−5). Deletions of RGA2 or BEM3 did not result in proximal-pole budding in daughter cells (0%, n = 56 and 53, respectively). While a bem2 deletion resulted in slightly increased proximal-pole budding (6.4±2.8%, n = 108), the difference between wild type and bem2Δ does not seem to be statistically significant (p = 0.22) ( Fig. 3A ). It is less clear whether Bem2, which is known as a GAP for Rho1, also functions as a GAP for Cdc42 in vivo [37], [40], [41], [42]. Taken together, these results suggest that among the Cdc42 GAPs, Rga1 is uniquely required for the preferential distal-pole budding of a/α daughter cells. We thus focused on Rga1 in subsequent studies.
Figure 3. Positions of the first bud in a/α daughter cells of wild type and mutants deleted for Cdc42 GAPs.
A. Time-lapse DIC images of diploid cells of wild type (YEF473) and rga1Δ (YEF1233). Arrows indicate budding events from daughter cells. Numbers indicate times (in min) from the first image. Size bars, 5 µm. (Histogram) The position of the first bud of daughter cells was scored in wild type (HPY1680), rga1Δ (HPY2205), rga2Δ (HPY2246), bem2Δ (HPY2384), and bem3Δ (HPY2426). The mean percentage ± SD of each budding pattern is shown from three or four independent countings of wild type (n = 106), rga1Δ (n = 144), rga2Δ (n = 56), bem2Δ (n = 108), and bem3Δ (n = 53). Statistical significance was determined by Student's t-test between proximal-pole buddings in wild type and rga1Δ or bem2Δ (marked with asterisks): *p<10−5 (rga1Δ) and **p = 0.02 (bem2Δ). B. The position of the first bud relative to the birth scar in diploid daughter cells. Cells were double stained with Calcoflour white and WGA-FITC as described in [44] from wild type (YEF473), rga1Δ (YEF1233), bud8Δ (YHH415), and rga1Δ bud8Δ (HPY2385). Arrows indicate birth scars. Size bar, 3 µm.
Because Rga1 is uniquely required for preventing budding at the division site [34], we wondered whether the diploid rga1Δ daughter cells that failed to bud at the distal pole also budded at the division site. Unlike mother cells, which have bud scars (chitinous scar tissue located at the division site), daughter cells have a much less conspicuous birth scar (which has little or no chitin) at the division site [43]. To examine more closely the first bud position in daughter cells relative to birth scar, we stained cells with Calcofluor, which stains bud scars as well as the base of a bud, and FITC-labeled wheat germ agglutinin (WGA-FITC), which stains both bud scars and birth scars [44]. As expected, almost all wild-type daughter cells formed a bud opposite to the birth scar (which is marked with an arrow in Fig. 3B ). In contrast, all of the rga1Δ daughter cells that failed to bud at the distal pole indeed budded within the birth scar (n = 65; note: this number includes some mother cells of rga1Δ because those mother cells that repeatedly budded within the birth scar could not be easily distinguished from daughter cells). As expected, almost all bud8Δ daughter cells budded at the proximal pole, but the position of a bud in bud8Δ was adjacent to, rather than within, the birth scar (97.4%, n = 39) ( Fig. 3B ). A small number of daughter cells of the diploid wild type (3.5%, n = 56) and bem2Δ mutant (6.3%, n = 63) also budded at the proximal pole, but these buds rarely appeared within the birth scar (data not shown). Interestingly, almost of all rga1Δ bud8Δ cells also budded within the birth scar (99.2%, n = 137; this counting is also likely to include some mother cells due to deletion of RGA1, see above). Taken together, these observations suggest that reduced distal-pole budding in the diploid rga1Δ daughter cells results from the increased Cdc42-GTP at the division site, consistent with a previous report [34].
Polarization of Cdc42-GTP in Diploid Daughter Cells Lacking RGA1
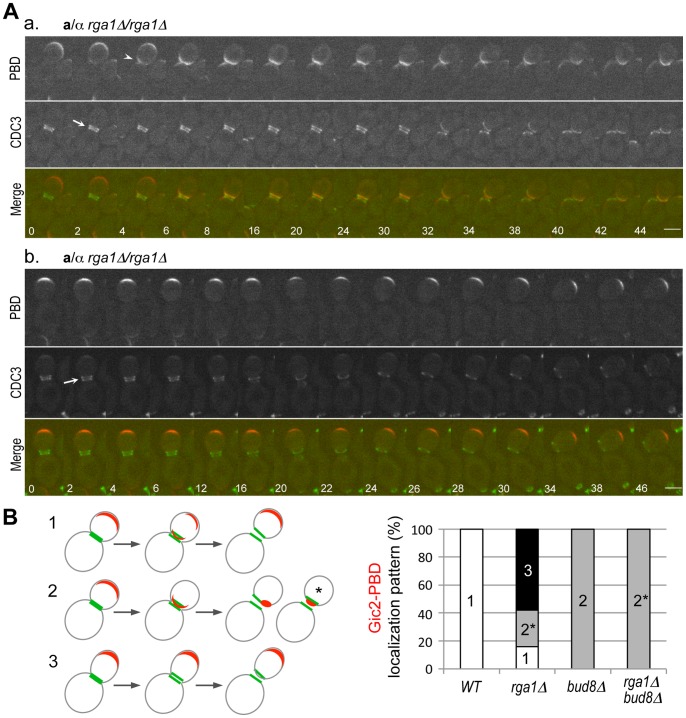
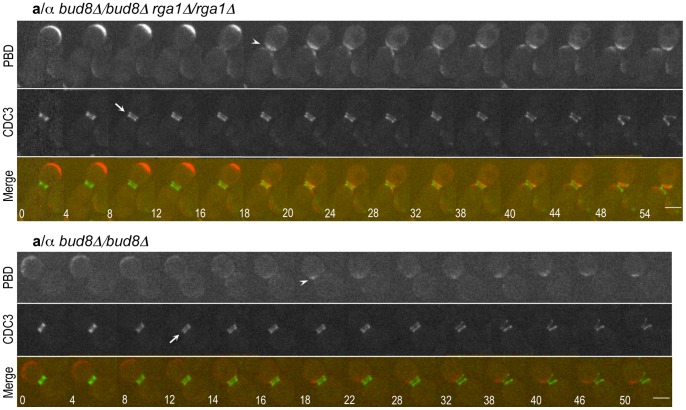
Although some diploid rga1Δ daughters budded within the birth scar, the majority of them (∼70%) still showed strong preference for distal-pole budding. To gain insight into this cellular behaviour, we monitored the localization of Cdc42-GTP (using Gic2-PBD-RFP) in diploid rga1Δ cells every 2 min. Gic2-PBD-RFP localized to the periphery of a growing bud in an rga1Δ mutant as in wild type until cytokinesis. During cytokinesis and in the next G1 phase, however, three different patterns of Gic2-PBD-RFP localization were observed in rga1Δ daughter cells (n = 19 movies): 1) Gic2-PBD-RFP localized to the proximal pole and then to the distal pole, as seen in wild type (15.8%; not shown); 2) Gic2-PBD-RFP remained at the proximal pole (26.3%; Fig. 4A , a; Movie S2); and 3) Gic2-PBD-RFP continuously localized to the distal pole in a large percentage of daughter cells (57.9%; Fig. 4A , b; Movie S3). Both the first and third patterns of Cdc42-GTP localization were expected to lead to the distal-pole budding in a/α rga1Δ daughter cells (see summary in Fig. 4B ). The localization patterns of Gic2-PBD-RFP are thus consistent with the observed budding patterns of the rga1Δ daughter cells (see Fig. 3 ).
Figure 4. Localization of Gic2-PBD-RFP and Cdc3-GFP in rga1Δ cells.
A. In rga1Δ cells (HPY2204), Gic2-PBD-RFP localized continuously to (a) the proximal pole or (b) the distal pole from cytokinesis to the next G1 phase. Arrows in (a) & (b) denote the Cdc3 ring splitting and an arrowhead in (a) denotes Gic2-PBD-RFP enriched at the division site (as well as the bud tip). Numbers indicate times (in min) from the first image. Size bars, 3 µm. B. Localization pattern of Gic2-PBD-RFP (red) prior to, during, and after cytokinesis (Cdc3-GFP in green) is summarized from time-lapse imagings of wild type (n = 15), rga1Δ (n = 19), bud8Δ (n = 7) and rga1Δ bud8Δ (n = 8). The proximal-pole localization pattern (marked with 2*) of rga1Δ or rga1Δ bud8Δ daughter cells is different from those seen in wild type and bud8Δ cells (see text for details).
While an increase of Cdc42-GTP at the proximal pole was expected given the lack of Cdc42 GAP activity at the division site in the rga1Δ mutant [34], it seemed counterintuitive that a significant percentage of rga1Δ daughter cells exhibited Cdc42-GTP polarization persistently at the distal pole. The one caveat is that our imaging was not fast enough to capture transient localization to the proximal pole in the third pattern ( Fig. 4B ). Nonetheless, these observations indicate that the dynamics of Cdc42-GTP in rga1Δ cells is different from that in wild type. Rga1 might thus have a unique role in Cdc42 polarization in diploid cells in addition to its role in clearing Cdc42-GTP at the division site (see below).
Bud8 is Necessary for Polarization of Cdc42-GTP to the Distal Pole in Diploid rga1Δ Daughter Cells
Since Bud8 functions as a distal pole marker important for normal bipolar budding pattern [20], we wondered whether the persistent distal-pole localization of Cdc42-GTP in the rga1Δ daughter cells is dependent on Bud8. Alternatively, Cdc42-GTP might be polarized to the distal pole independently of Bud8 as seen in the distal-pole budding of the rsr1 mutant during haploid invasive growth [45]. To distinguish these possibilities, we examined the Gic2-PBD-RFP localization in cells lacking both RGA1 and BUD8 by time-lapse microscopy. While Gic2-PBD-RFP still localized to the periphery of growing buds prior to cytokinesis in the rga1Δ bud8Δ cells, it always localized to the proximal pole during cytokinesis and remained at the proximal pole in the rga1Δ bud8Δ cells (100%, n = 8 movies) ( Fig. 5 , top panel; Movie S4). This observation indicates that Bud8 functions as a spatial cue for the enrichment of Cdc42-GTP at the distal pole of the rga1Δ daughter cells as in wild-type cells. Interestingly, Gic2-PBD-RFP localized to a site within the old Cdc3 ring (i.e., within the birth scar) in the rga1Δ bud8Δ daughter cells. In contrast, Gic2-PBD-RFP localized to the division site at cytokinesis but subsequently to a site adjacent to the old Cdc3 ring in bud8Δ cells (100%, n = 7 movies) ( Fig. 5 , bottom panel; Movie S5). These Cdc42-GTP polarization patterns are thus consistent with the first bud positions in daughter cells of these mutants (see Fig. 3B ).
Figure 5. Localization of Gic2-PBD-RFP and Cdc3-GFP in the diploids homozygous for bud8Δ rga1Δ and bud8Δ.
Imaging was performed as in Fig. 4 except in bud8Δ (HPY2370) and rga1Δ bud8Δ (HPY2371) cells. Arrows denote the Cdc3 ring splitting and arrowheads denote Gic2-PBD-RFP enriched at the proximal pole. Note: Gic2-PBD-RFP became enriched at a site adjacent to the Cdc3 ring in the bud8Δ daughter cell, whereas it appeared within the Cdc3 ring in rga1Δ bud8Δ daughter cell. Numbers indicate times (in min) from the first image. Size bars, 3 µm.
Why is it that the persistent enrichment of Cdc42-GTP at the distal pole was observed only in rga1Δ daughter cells (see Fig. 4B )? How might Rga1 control Cdc42-GTP polarization? At the early phase of the cell cycle, most growth is targeted to the tip of the bud in budding yeast. This ‘apical’ growth is switched to ‘isotropic’ growth in the G2 phase, during which growth is distributed diffusely within the bud, and then cells are repolarized at the site of cytokinesis [46]. It has been suggested that apical growth and repolarization during cytokinesis are critical for establishing spatial cues at the distal and proximal poles, respectively, and thus subsequent positioning of the division plane in diploid cells [14]. The rga1Δ cells have elongated bud morphology [47], [48], [49], [50], suggesting a delay in the transition from apical growth to isotropic growth. We thus speculated that the prolonged apical growth of the rga1Δ mutant might result in more efficient delivery of the distal-pole marker such as Bud8 to the distal pole. To test the idea, we examined Bud8 localization.
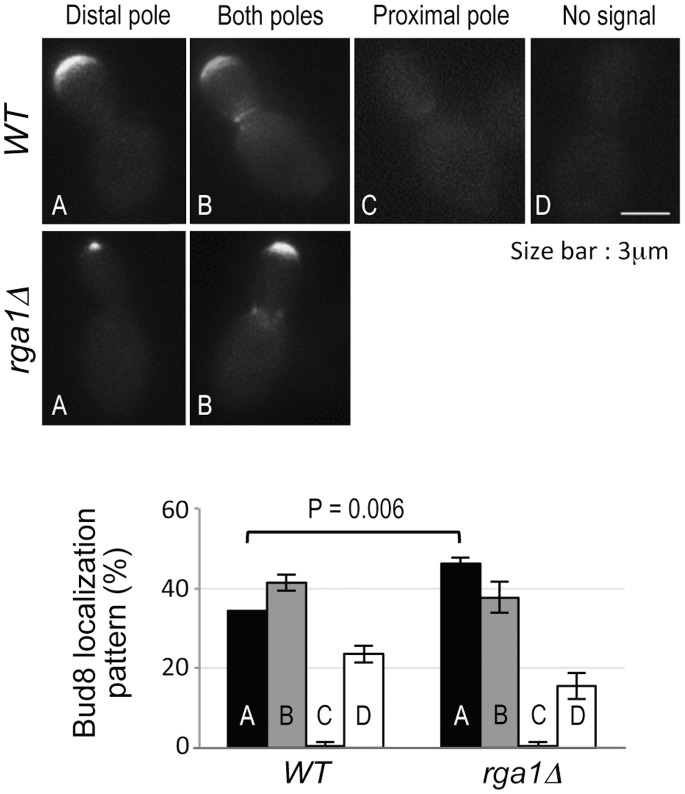
Bud8 localized to the bud tip of growing buds and the distal pole of wild-type daughter cells after division, as previously reported [20]. A significant percentage of large-budded cells also exhibited Bud8-GFP localization at both bud tips and the bud side of the mother-bud neck, although the latter was often weaker [19], [20], [51]. Interestingly, more large-budded rga1Δ cells exhibited Bud8-GFP localization to the bud tip (46.2±1.5%, n = 165) compared to wild type (34.5±0.1%, n = 174) ( Fig. 6 ), and this difference appeared to be statistically significant (p = 0.006). Bud8-GFP often appeared to be confined to the extreme bud tip in these rga1Δ cells ( Fig. 6 ). A minor difference of Bud8 localization was also observed in unbudded cells of rga1Δ compared to wild type (data not shown). These observations are thus consistent with the idea that Bud8 is more efficiently targeted to the bud tip (which becomes the distal pole of daughter cells) in rga1Δ cells, perhaps due to longer apical growth. However, it is unclear whether this different pattern of Bud8 localization solely accounts for persistent Cdc42-GTP polarization to the distal pole of rga1Δ cells. Indeed, we observed robust Cdc42-GTP polarization at the bud tip in large-budded cells of the bud8Δ rga1Δ mutant until cytokinesis (and even in bud8Δ cells, although Gic2-PBD-RFP appeared more broadly at the periphery of the buds in these cells) (see Fig. 5 ), suggesting that this Cdc42-GTP polarization prior to cytokinesis is independent on Bud8. Rga1 might also affect the targeting of Bud9 to the proximal pole or a component of the polarisome such as Spa2 or Ste20 at the bud tip [2], [14], which might affect Cdc42-GTP polarization prior to cytokinesis via a feedback mechanism (see below). Further investigation is necessary to understand the underlying mechanism involved in polarized growth and selection of a growth site in diploids.
Figure 6. Localization of Bud8 in large-budded cells of the wild type (HPY1680) and rga1Δ (HPY2205) carrying YEpGFP-BUD8F.
Representative images are shown for each pattern (A-D) and the percentage (mean ± SD) of each pattern is shown from three independent experiments (n = 160–230). Student's t-test was performed to compare the distal-pole localization in wild type and rga1Δ (P = 0.006).
Modeling Predicts Different Dynamics of Cdc42 Polarization Depending on the Levels of Spatial Cues as well as the GTP Hydrolysis of Cdc42
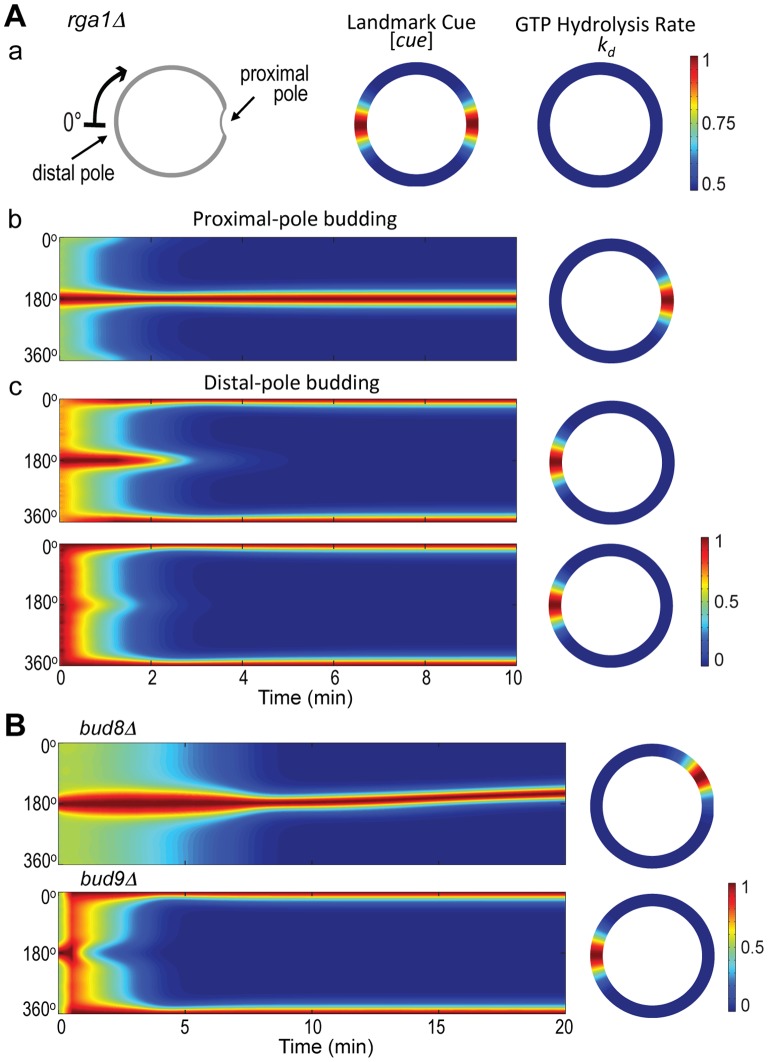
We then asked whether our mathematical modeling could account for these different types of Cdc42-GTP dynamics in the absence of a Cdc42 GAP or the spatial cues. In the absence of Rga1, the GTP hydrolysis rate kd would be spatially uniform; i.e., Cdc42 activity was no longer inhibited at the proximal pole ( Fig. 7A , a). We thus expected that Cdc42 was able to form a cluster at both proximal and distal poles marked by the landmarks and that its subsequent dynamics would be determined by the initial level of Cdc42-GTP at the division site and landmark cues that are subject to random perturbation. Our simulation showed that rga1Δ daughter cells could indeed bud at both poles: First, the high initial localization of Cdc42-GTP inhibited the formation of cluster at the distal pole, so that only one cluster was formed at the proximal-pole budding in the entire process ( Fig. 7A , b). Second, if the initial localization of Cdc42-GTP to the division site was reduced and the level of the distal-pole landmark was increased, the Cdc42-GTP cluster eventually formed at the distal pole ( Fig. 7A , c). Interestingly, the time window when Cdc42-GTP localized to both proximal and distal poles changes depending on the initial level of Cdc42-GTP at the division site and the strength of landmark cues. Within some parameter range that strength of landmark cue at the proximal pole is slightly less than that at the distal pole (see Table 2 ), Cdc42-GTP localization coexisted at both poles for a substantial time window ( Fig. 7A c, top). However, when the ratio of strength of landmark cue at the distal pole to that at the proximal pole increases beyond that range, Cdc42-GTP localization to the proximal pole could be barely monitored ( Fig. 7A c, bottom), and this scenario would account for the persistent distal-pole localization of Cdc42-GTP observed in over 50% of the rga1Δ daughter cells (see above). Our simulations thus predict that the relatively higher landmark at the distal pole or lower landmark at the proximal pole in the absence of Rga1 might result in persistent distal pole budding. These different patterns may thus arise from natural variations in the efficiency of delivery of these cues to the poles; in other words, the level of landmark cue or initial level of Cdc42-GTP in our model may be subject to substantial perturbation so that the parameters could fall in various ranges. While the exact mechanism remains unknown, a negative feedback loop involving Rga1 might be involved to buffer the level of Cdc42-GTP and thus to stop the polarity cluster from growing too large, as recently suggested by Howell et al. [28].
Figure 7. Mathematical modeling of Cdc42 polarization in diploid daughter cells deleted for RGA1, BUD8 or BUD9.
Aa. Coordinates are the same as in Fig. 2 (wild type) but the GTP hydrolysis rate of Cdc42 in the rga1Δ mutant is assumed to be about the same along the perimeter. See parameters in Table 2. Ab–Ac. Spatiotemporal dynamics of Cdc42-GTP leading to budding at (b) the proximal pole or (c) the distal pole in rga1Δ daughter cells. The horizontal axis represents the time window from 0 to 10 min. The 2D steady-state distribution of Cdc42-GTP is displayed on the right to each simulation. B. Spatiotemporal dynamics of Cdc42-GTP in diploid bud8Δ (top) and bud9Δ (bottom) mutants. The horizontal axis represents the time window from 0 to 20 min. The 2D steady-state distribution of Cdc42-GTP is displayed on the right to each simulation. Note: Cdc42-GTP became polarized at a site adjacent to the center of the proximal pole in bud8Δ (see 20 min time point), unlike in rga1Δ (see Fig. 7A, b).
Next, we asked whether our modeling could recapitulate the behavior of bud8Δ and bud9Δ mutants, which bud exclusively at the proximal and distal poles, respectively [12]. We used similar parameters except that the landmark cue [cue](x) is high only at either the proximal or distal pole in bud8Δ or bud9Δ, respectively. Our simulations indeed indicated that Cdc42-GTP polarized to the proximal pole in a bud8Δ mutant ( Fig. 7B , top) and to the distal pole in a bud9Δ mutant ( Fig. 7B , bottom). It is noteworthy that Cdc42-GTP polarization developed eventually at a site adjacent to the division site in bud8Δ, unlike that in rga1Δ, consistent with the bud position in the daughter cells of these mutants (see Figs. 3B & 5 ). Taken together, our computational modeling indicated different dynamics of Cdc42-GTP polarization when the levels of landmark and Cdc42-GTP were perturbed by noise in the model.
In summary, our mathematical modeling with limited parameters predicted the dynamics of Cdc42-GTP polarization, which accounts for robust distal-pole budding in diploid daughter cells. Live-cell imaging indicates that the distal-pole budding was dependent on Bud8 and GTP hydrolysis of Cdc42 by Rga1. While further investigation is necessary to fully understand the underlying mechanism, this study suggests that a Cdc42 GAP, not only the distal and proximal pole markers, affects the dynamics of Cdc42 polarization, contributing to selection of a growth site in diploid daughter cells.
Materials and Methods
A Mathematical Model of Cdc42 Polarization in Response to the Landmark Cues in Diploid Daughter Cells
The dynamics of the Cdc42-GTP and Cdc42-GDP on the cell membrane, with their particle densities denoted by [C42T] and [C42D], respectively, can be described by reaction-diffusion equations:
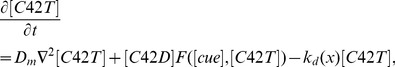 |
(1) |
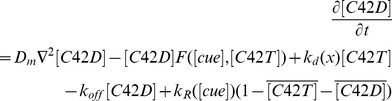 |
(2) |
The terms 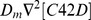 and
and 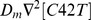 represent the surface lateral diffusion of Cdc42-GDP and Cdc42-GTP on the cell membrane, with
represent the surface lateral diffusion of Cdc42-GDP and Cdc42-GTP on the cell membrane, with 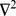 being the surface diffusion Laplacian operator and Dm the diffusion rate. The level of the landmark cues ([cue]) is a function of the angle x which parameterizes the membrane periphery (0° ≤ x ≤360°) from the distal pole:
being the surface diffusion Laplacian operator and Dm the diffusion rate. The level of the landmark cues ([cue]) is a function of the angle x which parameterizes the membrane periphery (0° ≤ x ≤360°) from the distal pole:
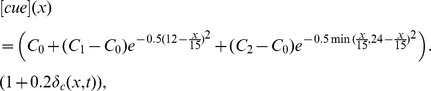 |
Where δc(x,t) is a random variable from standard normal distribution to model the fluctuations from natural background. We remark here that in the absence of random fluctuation, [cue](x) is a function with basal level C 0 and has two peaks, with maximal levels C 1 and C 2, at the proximal and distal poles, respectively. Other choices of the functional form with the same property will lead to similar results.
The Bem1-mediated feedback, implemented by the activation rate F, takes the form:
| (3) |
In equation [3], |M| denotes the total area of membrane surface, and the integral is taken over the cell membrane M, while the denominator represents the conservation of the total amount of Bem1 complex. We assume that the dynamic of Bem1 complex is much faster than that of Cdc42. We obtain the particle density of Bem1 complex at every time t by considering the quasi steady state solution of particle density of Bem1 complex, which is equal to the term
The detailed derivation can be found in the next section. In equations [1] and [2], the parameter kd represents the inactivation rate of Cdc42 from the GTP- to the GDP-bound states, which is space-dependent because it varies with the level of the Cdc42 GAPs. We define it to be in the following form, with higher level at the proximal pole (at 180°):
The parameters kdH and kdL are the maximal and minimal inactivation rates. Fig. 2B b shows the spatial distribution of [cue] and the GTP hydrolysis rate in wild type a/α daughter cells, in which kdH is assumed to be much larger than kdL. In Fig. 7A , kdH is taken to be equal to kdL, and therefore kd appears constant.
In equation [2], 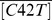 and
and 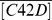 respectively represent the average amount of [C42T] and [C42D] over the membrane, that is, the integral of [C42T] and [C42D] over the cell membrane divided by the cell surface area. Thus, the recruitment of Cdc42 from the cytoplasm to the membrane is modeled by
respectively represent the average amount of [C42T] and [C42D] over the membrane, that is, the integral of [C42T] and [C42D] over the cell membrane divided by the cell surface area. Thus, the recruitment of Cdc42 from the cytoplasm to the membrane is modeled by 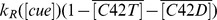 , where kR([cue]) is the landmark-signal-dependent coefficient and
, where kR([cue]) is the landmark-signal-dependent coefficient and 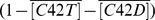 stands for the fraction of the cytoplasmic Cdc42. We remark here that to ensure
stands for the fraction of the cytoplasmic Cdc42. We remark here that to ensure 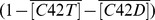 being between 0 and 1 to represent a fraction, the initial value for
being between 0 and 1 to represent a fraction, the initial value for  needs to be less than 1, which is true with the initial conditions and the associated parameters used in our simulations. Here we also assume that Cdc42 is uniformly distributed throughout the cytoplasm because cytoplasmic Cdc42 diffuses fast enough to reach a homogeneous state. In our simulations, we define the spatial-cue-dependent parameter kR([cue]) to be a function positively correlated with the function [cue], so that it has a similar spatial profile as the landmark cue. We choose to use the Michaelis-Menten form with power 1:
needs to be less than 1, which is true with the initial conditions and the associated parameters used in our simulations. Here we also assume that Cdc42 is uniformly distributed throughout the cytoplasm because cytoplasmic Cdc42 diffuses fast enough to reach a homogeneous state. In our simulations, we define the spatial-cue-dependent parameter kR([cue]) to be a function positively correlated with the function [cue], so that it has a similar spatial profile as the landmark cue. We choose to use the Michaelis-Menten form with power 1:
however, other function forms of kR, if properly scaled, can also produce the same results.
The parameter koff stands for the rate at which the membrane-bound Cdc42-GDP is extracted into the cytoplasm. This extraction of Cdc42-GDP away from the membrane is GDI-mediated, thus counteracting the recruitment of Cdc42.
For the initial values of our simulations, we assume that initially Cdc42-GDP is a constant and Cdc42-GTP is localized at the proximal pole of the cell, both of them with 20% perturbation from their basal levels. The initial values of [C42D] and [C42T] are defined as follows:
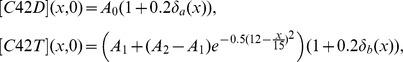 |
where δa(x) and δb(x) are the random variables from a uniform distribution between 0 and 1, and A 0 is the basal level for Cdc42-GDP, while A 1 and A 2 are the basal maximal and minimal levels for Cdc42-GTP. All the above parameters are listed in Tables 1 and 2 .
Derivation of Equation [3]
Let C(x,t) denote the particle density of Bem1 complex on the cell membrane, which is governed by
| (4) |
where α and β are constant parameters; 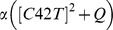 is the recruitment rate of Cdc24 from cytoplasm to membrane, depended on the particle density of Cdc42; Q is the spatial function representing the level of stimulation by the landmark cue and αQ represents the basal recruitment rate controlled by the landmark cue;
is the recruitment rate of Cdc24 from cytoplasm to membrane, depended on the particle density of Cdc42; Q is the spatial function representing the level of stimulation by the landmark cue and αQ represents the basal recruitment rate controlled by the landmark cue; 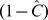 is the fraction of the cytoplasmic Cdc24; βC is the disassociation rate of C from the membrane to the cytoplasm;
is the fraction of the cytoplasmic Cdc24; βC is the disassociation rate of C from the membrane to the cytoplasm; 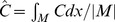 represents the average value of C over the membrane.
represents the average value of C over the membrane.
The dynamic of Bem1 complex is much faster than that of Cdc42. In the system of Cdc42, we obtain the particle density of Bem1 complex at every time t by solving the quasi-steady-state solution of the equation [4]. By assuming right hand side of [4] to be zero, the steady state equation of [4] can be written as follows:
| (5) |
By taking the average value of right hand side of [5] over the membrane (taking integration over the membrane and then dividing by the area of the membrane), we have
which leads to
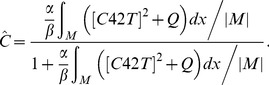 |
(6) |
By substituting [6] into [5], we can obtain C in term of [C42T]:
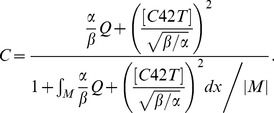 |
(7) |
By defining [cue] = αQ/β and 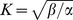 , C can be rewritten into the form.
, C can be rewritten into the form.
 |
(8) |
If we assume that the activation rate of Cdc42 is proportional to the quasi-steady-state solution of particle density of Bem1 complex and define [cue] = αQ/β, the form of the activation rate of Cdc42 will be
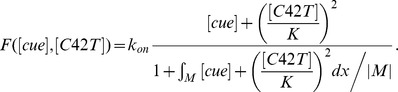 |
.
Parameter Estimation
For simplicity, we considered a diploid daughter cell as a 4 µm-diameter circle, since daughter cells are generally smaller than mother cells, which are typically 5×6 µm ellipsoids. For a yeast cell of radius 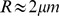 , the membrane diffusion coefficient of Cdc42 is estimated to be.
, the membrane diffusion coefficient of Cdc42 is estimated to be.
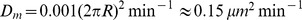 [29]. According to [29], we estimate an off-rate from membrane to cytoplasm to be
[29]. According to [29], we estimate an off-rate from membrane to cytoplasm to be 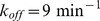 . Yeast cell polarization is mainly achieved by Bem1-mediated positive feedback and landmark cue is just an initial tracker for polarization so the level of landmark cue should be small comparing with feedback strength. Here we took the basal level of landmark cue to be C
0 = 0.1 and C
1, C
2 = 0.15–0.25 which were small comparing with feedback strength we observed in simulations. The recruitment rate was estimated to be
. Yeast cell polarization is mainly achieved by Bem1-mediated positive feedback and landmark cue is just an initial tracker for polarization so the level of landmark cue should be small comparing with feedback strength. Here we took the basal level of landmark cue to be C
0 = 0.1 and C
1, C
2 = 0.15–0.25 which were small comparing with feedback strength we observed in simulations. The recruitment rate was estimated to be 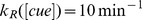 [29]. According to the definition of
[29]. According to the definition of 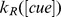 , we took
, we took 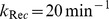 and
and 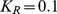 . For the activation rate coefficient of Cdc42, we took
. For the activation rate coefficient of Cdc42, we took 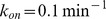 [30]. We assumed that the number of Cdc42-GTP on membrane is much smaller than total number of Cdc42, and thus we took inactivation rate coefficient
[30]. We assumed that the number of Cdc42-GTP on membrane is much smaller than total number of Cdc42, and thus we took inactivation rate coefficient 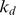 of Cdc42 to be between
of Cdc42 to be between 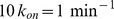 and
and 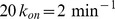 .
.
Numerical Method for Simulations
The simulations used a second-order central difference approximation for the diffusion terms, and the temporal discretization was carried out using a fourth order Adams-Moulton predictor-corrector method. FORTRAN 77 was used for the simulation shown in Figures 2 and 7 and plots were generated using MATLAB 7.
Strains, Plasmids and Genetic Methods
Standard methods of yeast genetics, DNA manipulation, and growth conditions were used [52], [53] unless indicated otherwise. Plasmids YIp211-GIC2-PBD-1.5tdTomato and YIp128-CDC3-GFP (kindly provided by E. Bi, University of Pennsylvania) were used to construct strains expressing Gic2-PBD-RFP and Cdc3-GFP, respectively, as previously described [34]. Plasmids pRS314-HO and YCp50-HO (from the Park lab collection), which carry the HO gene, were used to generate a/α diploids. See Table 3 for a list of strains used in this study.
Table 3. Yeast strains used in this study.
| Strain | Relevant Genotypea | Source/Comments | |
| YEF473* | a/α | his3- Δ 200/his3- Δ 200 leu2- Δ 1/leu2- Δ 1 lys2-801/lys2-801 trp1- Δ 63/trp1- Δ 63 ura3-52/ura3-52 | [54] |
| YEF1233* | a/α | rga1 Δ ::HIS3/rga1 Δ ::HIS3 | [34] |
| YHH415* | a/α | bud8- Δ 1::TRP1/bud8- Δ 1::TRP1 | [20] |
| HPY2353* | a/α | CDC3-GFP::LEU2/CDC3-GFP::LEU2 GIC2-PBD-RFP::URA3/GIC2-PBD-RFP::URA3 | This study |
| HPY2204* | a/α | rga1 Δ ::HIS3/rga1 Δ ::HIS3 CDC3-GFP::LEU2/CDC3-GFP::LEU2 GIC2-PBD-RFP::URA3/GIC2-PBD-RFP::URA3 | This study |
| HPY2370* | a/α | bud8- Δ 1::TRP1/bud8- Δ 1::TRP1 CDC3-GFP::LEU2/CDC3-GFP::LEU2 GIC2-PBD-RFP::URA3/GIC2-PBD-RFP::URA3 | This study |
| HPY2371* | a/α | rga1 Δ ::HIS3/rga1 Δ ::HIS3 bud8- Δ 1::TRP1/bud8- Δ 1::TRP1 CDC3-GFP::LEU2/CDC3-GFP::LEU2 GIC2-PBD-RFP::URA3/GIC2-PBD-RFP::URA3 | This study |
| HPY2385* | a/α | rga1 Δ ::HIS3/rga1 Δ ::HIS3 bud8- Δ 1::TRP1/bud8- Δ 1::TRP1 | This study |
| HPY1680@ | a/α | his3- Δ 1/his3- Δ 1 leu2 Δ 0/leu2 Δ 0 met15 Δ 0/met15 Δ 0 ura3 Δ 0/ura3 Δ 0 | Diploid of BY4741 (Open Biosystems) |
| HPY2205@ | a/α | rga1 Δ ::kanMX4/rga1 Δ ::kanMX4 | This study |
| HPY2246@ | a/α | rga2 Δ ::kanMX4/rga2 Δ ::kanMX4 | This study |
| HPY2384@ | a/α | bem2 Δ ::kanMX4/bem2 Δ ::kanMX4 | This study |
| HPY2426@ | a/α | bem3 Δ ::KanMX4/bem3 Δ ::KanMX4 | This study |
Strains marked with *are isogenic to YEF473 and strains marked with @are isogenic to HPY1680 except as indicated.
Determination of the Budding Pattern and Localization of Bud8
To determine budding patterns, cells were spotted on a YPD plate after a brief sonication and then the position of each bud was monitored under the dissecting microscope at 25oC. For time-lapse imaging by DIC microscopy, cells were grown similarly, spotted on a slab of YPD medium containing 1% agarose, and then imaged using a Nikon E800 microscope (Nikon, Tokyo, Japan) fitted with a 100X oil-immersion objective (NA = 1.30) with a Hamamatsu ORCA-2 CCD camera (Hamamatsu Photonics, Bridgewater, NJ) and Slidebook software (Intelligent Imaging Innovations, Denver, CO) at 25oC. Localization of Bud8 was examined as previously described [19] using YEpGFP-BUD8F [16].
3D Time-lapse Microscopy
To visualize GFP- and RFP-fusion proteins, a slab of SC-Ura was prepared as above using exponentially growing cells in SC-Ura media. Images were captured at 23–24oC every 2 min using a spinning disk confocal microscope (UltraView ERS, Perkin Elmer Life and Analytical Sciences, Waltham, MA) equipped with a 100×/1.4 NA objective lens (Nikon, Melville, NY), a 488-nm solid state laser and 568-nm argon ion laser, and a cooled charge-coupled device camera (ORCA-AG, Hamamatsu, Bridgewater, NJ). Maximum intensity projections of Z-sections (spaced at 0.4–0.5 µm) are generated using UltraView ERS software. All time-point images are shown in Movies S1, S2, S3, S4, S5, and the selected time-point images are shown in Figs. 1, 4, and 5.
Supporting Information
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α wild-type cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α rga1Δ cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α rga1 Δ cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α rga1 Δ bud8D cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α bud8 Δ cells.
(AVI)
Acknowledgments
We thank E. Bi for providing strains and plasmids; J.-Q. Wu and people in his lab for help with their confocal microscope; and P. Cullen and P. J. Kang for their thoughtful comments on the manuscripts.
Funding Statement
This work has been supported by the Mathematical Biosciences Institute and National Science Foundation (NSF) grant DMS 0931642 (to WCL and CSC), NSF grant DMS-1020625 (to CSC), and National Institutes of Health/National Institute of General Medical Sciences grant GM076375 (to HOP). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Etienne-Manneville S (2004) Cdc42 - the centre of polarity. J Cell Sci 117: 1291–1300. [DOI] [PubMed] [Google Scholar]
- 2. Park H-O, Bi E (2007) Central roles of small GTPases in the development of cell polarity in yeast and beyond.. Microbiol Mol Biol Rev 71: 48–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chant J, Herskowitz I (1991) Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65: 1203–1212. [DOI] [PubMed] [Google Scholar]
- 4. Chant J, Pringle JR (1995) Patterns of bud-site selection in the yeast Saccharomyces cerevisiae . J Cell Biol 129: 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Freifelder D (1960) Bud position in Saccharomyces cerevisiae . J Bacteriol 80: 567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hicks JB, Strathern JN, Herskowitz I (1977) Interconversion of yeast mating types III. Action of the homothallism (HO) gene in cells homozygous for the mating type locus. Genetics 85: 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bender A, Pringle JR (1989) Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1 . Proc Natl Acad Sci USA 86: 9976–9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chant J, Corrado K, Pringle JR, Herskowitz I (1991) Yeast BUD5, encoding a putative GDP-GTP exchange factor, is necessary for bud site selection and interacts with bud formation gene BEM1 . Cell 65: 1213–1224. [DOI] [PubMed] [Google Scholar]
- 9. Powers S, Gonzales E, Christensen T, Cubert J, Broek D (1991) Functional cloning of BUD5, a CDC25-related gene from S. cerevisiae that can suppress a dominant-negative RAS2 mutant. Cell 65: 1225–1231. [DOI] [PubMed] [Google Scholar]
- 10. Park H-O, Chant J, Herskowitz I (1993) BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature 365: 269–274. [DOI] [PubMed] [Google Scholar]
- 11. Bender A (1993) Genetic evidence for the roles of the bud-site-selection genes BUD5 and BUD2 in control of the Rsr1p (Bud1p) GTPase in yeast. Proc Natl Acad Sci, U S A 90: 9926–9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zahner JE, Harkins HA, Pringle JR (1996) Genetic analysis of the bipolar pattern of bud-site selection in the yeast Saccharomyces cerevisiae . Mol Cell Biol 16: 1857–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ni L, Snyder M (2001) A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae . Mol Biol Cell 12: 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sheu YJ, Barral Y, Snyder M (2000) Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol Cell Biol 20: 5235–5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang S, Ayscough KR, Drubin DG (1997) A role for the actin cytoskeleton of Saccharomyces cerevisiae in bipolar bud-site selection. J Cell Biol 136: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schenkman LR, Caruso C, Pagé N, Pringle JR (2002) The role of cell cycle-regulated expression in the localization of spatial landmark proteins in yeast. J Cell Biol 156: 829–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen T, Hiroko T, Chaudhuri A, Inose F, Lord M, et al. (2000) Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science 290: 1975–1978. [DOI] [PubMed] [Google Scholar]
- 18. Fujita A, Lord M, Hiroko T, Hiroko F, Chen T, et al. (2004) Rax1, a protein required for the establishment of the bipolar budding pattern in yeast. Gene 327: 161–169. [DOI] [PubMed] [Google Scholar]
- 19. Kang PJ, Angerman E, Nakashima K, Pringle JR, Park H-O (2004) Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol Biol Cell 15: 5145–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Harkins HA, Pagé N, Schenkman LR, De Virgilio C, Shaw S, et al. (2001) Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol Biol Cell 12: 2497–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wedlich-Soldner R, Altschler S, Wu L, Li R (2003) Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299: 1231–1235. [DOI] [PubMed] [Google Scholar]
- 22. Irazoqui JE, Gladfelter AS, Lew DJ (2003) Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol 5: 1062–1070. [DOI] [PubMed] [Google Scholar]
- 23. Wedlich-Soldner R, Wai SC, Schmidt T, Li R (2004) Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J Cell Biol 166: 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, et al. (2008) Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol 18: 1719–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ozbudak EM, Becskei A, van Oudenaarden A (2005) A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev Cell 9: 565–571. [DOI] [PubMed] [Google Scholar]
- 26. Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF (2007) Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell 129: 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Slaughter BD, Das A, Schwartz JW, Rubinstein B, Li R (2009) Dual modes of cdc42 recycling fine-tune polarized morphogenesis. Dev Cell 17: 823–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, et al. (2012) Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 149: 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Altschuler SJ, Angenent SB, Wang Y, Wu LF (2008) On the spontaneous emergence of cell polarity. Nature 454: 886–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goryachev AB, Pokhilko AV (2008) Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Letters 582: 1437–1443. [DOI] [PubMed] [Google Scholar]
- 31. Johnson JM, Jin M, Lew DJ (2011) Symmetry breaking and the establishment of cell polarity in budding yeast. Curr Opin Genet Dev 21: 740–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, et al. (2011) Modeling vesicle traffic reveals unexpected consequences for Cdc42p-mediated polarity establishment. Curr Biol 21: 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Arkowitz RA, Bassilana M (2011) Polarized growth in fungi: symmetry breaking and hyphal formation. Semin Cell Dev Biol 22: 806–815. [DOI] [PubMed] [Google Scholar]
- 34. Tong Z, Gao X-D, Howell AS, Bose I, Lew DJ, et al. (2007) Adjacent positioning of cellular structures enabled by a Cdc42 GTPase-activating protein mediated zone of inhibition. J Cell Biol 179: 1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Richman TJ, Sawyer MM, Johnson DI (2002) Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot Cell 1: 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Caviston JP, Longtine M, Pringle JR, Bi E (2003) The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol Biol Cell 14: 4051–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knaus M, Pelli-Gulli M-P, van Drogen F, Springer S, Jaquenoud M, et al. (2007) Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J 26: 4501–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sopko R, Huang D, Smith JC, Figeys D, Andrews BJ (2007) Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J 26: 4487–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozminski KG, Beven L, Angerman E, Tong AHY, Boone C, et al. (2003) Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol Biol Cell 14: 4958–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng Y, Hart MJ, Shinjo K, Evans T, Bender A, et al. (1993) Biochemical comparisons of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. Delineation of a limit Cdc42 GTPase-activating protein domain. J Biol Chem 268: 24629–24634. [PubMed] [Google Scholar]
- 41. Zheng Y, Cerione R, Bender A (1994) Control of the yeast bud-site assembly GTPase Cdc42: catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem 269: 2369–2372. [PubMed] [Google Scholar]
- 42. Marquitz AR, Harrison JC, Bose I, Zyla TR, McMillan JN, et al. (2002) The Rho-GAP Bem2p plays a GAP-independent role in the morphogenesis checkpoint. EMBO J 21: 4012–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bacon JS, Davidson ED, Jones D, Taylor IF (1966) The location of chitin in the yeast cell wall. Biochem J 101: 36C–38C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frydlová I, Malcová I, Vasicová P, Hasek J (2009) Deregulation of DSE1 gene expression results in aberrant budding within the birth scar and cell wall integrity pathway activation in Saccharomyces cerevisiae. Eukaryot Cell 8: 586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cullen PJ, Sprague GF Jr (2002) The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol Biol Cell 13: 2990–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Howell AS, Lew DJ (2012) Morphogenesis and the cell cycle. Genetics 190: 51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Smith GR, Givan SA, Cullen P, Sprague GF Jr (2002) GTPase-activating proteins for Cdc42. Eukaryotic Cell 1: 469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen GC, Zheng L, Chan CS (1996) The LIM domain-containing Dbm1 GTPase-activating protein is required for normal cellular morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol 16: 1376–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stevenson BJ, Ferguson B, De Virgilio C, Bi E, Pringle JR, et al. (1995) Mutation of RGA1, which encodes a putative GTPase-activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes & Dev 9: 2949–2963. [DOI] [PubMed] [Google Scholar]
- 50. Watanabe M, Watanabe D, Nogami S, Morishita S, Ohya Y (2009) Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr Genet 55: 365–380. [DOI] [PubMed] [Google Scholar]
- 51. Taheri N, Köhler T, Braus GH, Mösch H-U (2000) Asymmetrically localized Bud8p and Bud9p proteins control yeast cell polarity and development. EMBO J 19: 6686–6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, et al.. (1999) Current Protocols in Molecular Biology: John Wiley & Sons, Inc.
- 53.Guthrie C, Fink GR (1991) Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press. 933 p.
- 54. Bi E, Pringle JR (1996) ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae . Mol Cell Biol 16: 5264–5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α wild-type cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α rga1Δ cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α rga1 Δ cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α rga1 Δ bud8D cells.
(AVI)
Localization of Gic2-PBD-RFP (left) and Cdc3-GFP (right) in a/α bud8 Δ cells.
(AVI)