Abstract
Staphylococcus aureus is an important human pathogen in both hospital and the community that has demonstrated resistance to all currently available antibiotics over the last two decades. Multidrug-resistant isolates of methicillin-resistant S. aureus (MRSA) exhibiting decreased susceptibilities to glycopeptides has also emerged, representing a crucial challenge for antimicrobial therapy and infection control. The availability of complete whole-genome nucleotide sequence data of various strains of S. aureus presents an opportunity to explore novel compounds and their targets to address the challenges presented by antimicrobial drug resistance in this organism. Study compounds α-amyrin [3β-hydroxy-urs-12-en-3-ol (AM)], betulinic acid [3β-hydroxy-20(29)-lupaene-28-oic acid (BA)] and betulinaldehyde [3β-hydroxy-20(29)-lupen-28-al (BE)] belong to pentacyclic triterpenoids and were reported to exhibit antimicrobial activities against bacteria and fungi, including S. aureus. The MIC values of these compounds against a reference strain of methicillin-resistant S. aureus (MRSA) (ATCC 43300) ranged from 64 µg/ml to 512 µg/ml. However, the response mechanisms of S. aureus to these compounds are still poorly understood. The transcription profile of reference strain of MRSA treated with sub-inhibitory concentrations of the three compounds was determined using Affymetrix GeneChips. The findings showed that these compounds regulate multiple desirable targets in cell division, two-component system, ABC transporters, fatty acid biosynthesis, peptidoglycan biosynthesis, aminoacyl-tRNA synthetase, ribosome and β-lactam resistance pathways which could be further explored in the development of therapeutic agents for the treatment of S. aureus infections.
Introduction
Antibiotic resistance is a growing global threat to the effective treatment of infectious diseases. Over the past two decades resistance has been reported in many clinically important pathogens and to a broad range of antimicrobials in clinical use today [1], [2], [3]. This has been the driving force for the discovery of novel agents with antimicrobial activity that may potentially lead to therapeutic agents. The isolation of vancomycin, β-lactam, macrolide, tetracycline, aminoglycoside and quinolone-resistant strains of S. aureus provides a message that the availability of structurally novel antibacterials with multiple modes of action need to be part of the antibacterial armamentarium [4]. Over the past 30 years, only two new classes of antibiotics for the treatment of staphylococcal infections have been introduced: the oxazolidinones represented by linezolid, and the lipopeptide daptomycin [5]. However, reports of resistance to these antibiotics have also been reported [6]. Clearly, new approaches and innovative strategies are required to discover and identify agents that target essential bacterial targets in novel pathways, i.e. not targeted by currently used antibiotics, or novel targets in existing pathways. Promising targets for novel antibacterials against S. aureus include cell division, DNA replication and biosynthesis of fatty acid, peptidoglycan and protein [7].
To capture and exploit the wealth and diversity of genetic information of a variety of microorganisms, development of new technologies such as microarray provides a unique tool to understand the molecular basis of drug activity as it allows simultaneous analysis of the expression of all genes in the organism under any given condition. Transcriptional profiles generated by GeneChip analysis of bacteria have been used to investigate differential gene expression in response to antimicrobial agents [8], such as the response of S. aureus strain 8325-4 to antibiotics that act on cell wall such as oxacillin, D-cycloserine and bacitracin [9], and the effects of vancomycin on gene expression in S. aureus [10].
Plant-derived natural products have been reported to show anti-staphylococcal activities with micromolar MICs [11], [12], [13], [14], [15], [16]. In a preliminary study, the effects of 191 extracts towards different strains of bacteria were evaluated. Extracts from Callicarpa farinosa were shown to exhibit strong antimicrobial activities against MRSA [16]. Three pentacyclic triterpenoids, namely α-amyrin, betulinic acid and betulinaldehyde, have been investigated and found to have antibacterial activity against reference strains of methicillin-sensitive and methicillin-resistant S. aureus. Their activity was potentiated by currently used antistaphylococcal drugs [17]. In this study, the genome-wide transcription induced by the exposure of reference strain of S. aureus to pentacyclic triterpenoids was used to elucidate the mechanism of action of the compounds.
Materials and Methods
Bacterial Strains and Materials
The S. aureus strain used in this study was reference strain ATCC 43300, representing methicillin-resistant strains. The bacterial strain was maintained on Nutrient Agar (NA) (Difco, USA) slopes. Bacterial cultures used for microarray experiments were grown on cation-adjusted Mueller-Hinton broth (CAMHB II) (Difco, USA). Commercial sources of pentacyclic triterpenoids α-amyrin (AM) and betulinic acid (BA) were obtained from Sigma-Aldrich (USA), while betulinaldehyde (BE) was obtained from Advanced Technology and Industrial Co., Ltd (Hong Kong).
Growth of Staphylococcus aureus in the Presence of AM, BA and BE
In order to obtain good microarray results, the concentrations of the compounds need to be optimized at sub-inhibitory concentrations that do not affect the growth of the organism [18], [19], [20], [21]. The treatment time with AM, BA and BE was determined by exposure of the organism to the study compounds and bacterial growth curve analysis. The bacterial culture was grown in CAMHB and incubated at 37°C with shaking at 200 rpm, to an optical density of 0.3 at 600 nm; i.e. at mid-logarithmic phase as culture conditions at this phase were most constant and RNA levels were at the highest due to high metabolic rate. The optical density of the cultures was measured using a spectrophotometer (Nanophotometer, Germany). The cultures were then distributed into 100 ml aliquots. AM, BA and BE were added to the aliquots to obtain concentrations of 1/2× MIC (32 µg/ml for AM and BA; 256 µg/ml for BE) and 1× MIC (64 µg/ml for AM and BA; 512 µg/ml for BE) [17]. The cultures were then incubated, and cell growth was spectrophotometrically monitored at OD 600 nm for 5 hours at time intervals of 30 minutes. Untreated bacterial culture was used as control. The growth curve for each treatment and concentration was carried out in triplicates.
Total RNA Isolation and Purification
Three independent bacterial cultures for each treatment and control were prepared as biological replicates for total RNA isolation. After treatment of bacterial cells with the study compounds at concentrations and incubation times as determined from the growth curve, the cells were immediately stabilized with RNA Protect Bacteria Reagent (Qiagen, USA) to minimize RNA degradation before harvesting. Total RNA was isolated from treated and control bacterial cultures using the RNeasy Midikit (Qiagen) protocol. The cell wall of S. aureus was disrupted with Tris-EDTA (TE) buffer with 50 mg/ml lysozyme (Sigma Aldrich) containing proteinase K (Qiagen). DNA was removed using RNase-free DNase I (Qiagen). After purification, RNA concentration was determined by absorbance at 260 nm on Nanophotometer (1 absorbance unit is equivalent to 40 µg/ml RNA). RNA integrity was determined using Agilent RNA 6000 Nano chips (Agilent Technologies, USA). The procedure in preparing the assay, loading the chip and running the assay was carried out according to the manufacturer’s protocol at Malaysia Genome Institute, Bangi, Malaysia.
The total RNA analysis showed that the A260/A280 and A260/A230 readings of all the total RNA samples were in the range of 1.8 to 2.1 and a single peak at 260 nm indicating that the samples were free from contaminants such as proteins, phenol and salts, which could interfere with down-stream applications such as cDNA synthesis and microarray analysis. The RIN values were above 7.0, indicative of non-degraded RNA (supporting information Table S1).
cDNA Synthesis, Fragmentation and Terminal Labeling
cDNA was synthesized with a starting material of 10 µg of the total RNA, according to manufacturer’s protocol (prokaryotic array, Affymetrix, USA). The procedure was carried out in a Mastercycler Gradient thermal cycler (Eppendorf, Germany). The cDNA obtained was then purified using a MinElute PCR Purification Column Kit (Qiagen) and quantified using Nanophotometer at 260 nm absorbance. A minimum of 1.5 µg cDNA was then fragmented in a reaction mix containing DNase I and DNase running buffer (Promega, USA). The fragmented cDNA was then applied directly to the terminal labeling reaction using GeneChip DNA Labeling Reagent (Affymetrix). The fragmented and labeled cDNA (target) was then used for hybridization.
GeneChip Hybridization
The labeled fragmented cDNA from independent RNA preparations were hybridized onto a Genechip probe array masked with known genome of S. aureus N315 (methicillin-resistant strain), Mu50 (methicillin and vancomycin-resistant strain), COL (methicillin-resistant strain) and NCTC 8325 strains (methicillin-sensitive strain) (Affymetrix). The array contains probe sets of over 3300 S. aureus ORFs and over 4800 intergenic regions throughout the S. aureus genome. The labeled cDNA was hybridized, washed and scanned according to manufacturer’s protocol (prokaryotic array, Affymetrix). The data obtained was analyzed using the Affymetrix GeneChip Command Console software (AGCC).
Microarray Analysis
For gene expression analysis, data from the Affymetrix platform were processed with GeneSpring version 11 software. Data was normalized to minimize systematic non-biological differences to reveal true biological differences. Significance analysis using ANOVA test with multiple testing correction Benjamini Hochberg to correct false discovery rate and asymptotic p-value computation was performed to determine the change of expression of a particular gene due to the effect of different treatment on S. aureus. In addition, fold-change analysis was also performed and reported as the up- or down-regulated fold change. To select the differentially expressed genes, threshold values of ≥2 fold-change between pentacyclic triterpenoid-treated samples and control samples and significance level of <5% were used. From the statistical analysis, the gene ontology (GO) analysis was carried out. The pathways affected by the differentially expressed genes were analyzed using the KEGG database (http://www.genome.jp/kegg/pathway.html). To group genes with similar expression profiles together, hierarchical clustering analysis was used in which an entity tree and a condition tree were generated.
Validation of Gene Expression by Quantitative Real-time Quantitative PCR
To determine the validity of the microarray data, transcript level changes obtained with the microarray analysis were compared with those from quantitative real-time PCR (qPCR). Aliquots of cDNA preparations used in the microarray experiments were used for qPCR procedure. A list of genes and the primer sequences used for the real-time PCR analysis is shown in Table 1. The housekeeping gene adhE and gyrA were used as endogenous reference genes. The procedure was performed using the iCycler iQ5 Real Time PCR Detection System (Bio-Rad Laboratories, USA) with Quantitect SYBR Green PCR kit (Qiagen). For each gene, three biological replicates with three technical replicates each were used. Real-time cycler conditions were carried out according to manufacturer’s protocol: initial activation step at 95°C for 15 min, followed by 45 cycles of denaturation at 15 s and 94°C, annealing at 30 s and 53 to 60°C (as optimized for each target gene, data not shown), and extension at 30 s and 72°C. Melting curve analysis was also performed to verify PCR specificity. PCR efficiencies were derived from standard curve slopes in the iCycler software v3.1. Relative quantification based on the relative expression of the target gene compared to reference genes adhE and gyrA in treated and control samples was used to determine transcript level changes [22]. Paired student’s T test was performed to assess whether the means of a treated and control group (samples) is significantly different from each other using the SPSS software (PASW Statistic 18).
Table 1. Primers used in real-time PCR for validation of target genes.
| Genesymbol | Sequence accession number | Amplicon length | Primer sequence | |
| Forward | Reverse | |||
| ftsZ | 1123860 | 85 | AGCTGCAGAGGAATCTCGTGAACA | TCCGCCACCCATACCAGAAGTA |
| opp-1C | 1125181 | 81 | CGTGCGTGTGATGTTATGTTGGCA | TTCGGCACCCATTCCAAACAATGC |
| oppF | 1122975 | 81 | GGTGGGCAACGTCAGCGTGTAA | GGACACTGCCTCGTCGCAAACA |
| asnC | 1124125 | 120 | TCGGTGGATCTGAACGTGTGGA | GTGGCACACTACCATAACGACG |
| fabZ | 1124802 | 131 | TGAAGAAGGTCAACGTTGTGTGGC | CCGCACCTGTTTGAGCTAACGC |
| fmhB | 1124980 | 151 | AAGCGAGGTACGACAGTAGAACG | CATCTCCATCTTCATGCAACGCA |
| pbp2 | 1124121 | 90 | CGTTTCTACGAACATGGCGCAC | GGCACCTTCAGAACCAAATCCACCA |
| ccrA | 1122832 | 106 | CAGCTTGGCCGAACTTGAATCAGA | ACCAAAGGGCGCATGGGTTG |
| mecR1 | 1122813 | 112 | GTGCTCGTCTCCACGTTAATTCCA | GACTAACCGAAGAAGTCGTGTCAG |
| adhE | 1122918 | 105 | TTAGCGAAGTCGAACCGAACCCA | CTGAACCACCACCAAGTGCAATGA |
| gyrA | 1122777 | 80 | TATCCGCTTGTTGATGGCCAAGG | CGCGCTTCAGTATAACGCATTGC |
Results and Discussion
Growth Profile of Staphylococcus aureus in the Presence of AM, BA and BE
The effect of AM, BA and BE on the growth of reference strain of MRSA is as shown in Figure 1. The growth rate of the organism decreased at concentrations ½ MIC and MIC after 30 min for AM; and 45 min for BA and BE. After 300 min, the growth of the organism at ½ MIC was generally higher than the growth at MIC. These effects of the growth indicates that AM, BA and BE are bacteriostatic compounds as reported by Chung et al. [17]. Based on the growth curves, the organism was then treated with the three study compounds at ½ MIC (32 µg/ml for AM and BA; and 256 µg/ml for BE) and further incubated for 30 min (control and treatment with AM) and 45 min (treatment with BA and BE). Subinhibitory concentrations of the compounds were used in the microarray procedure to avoid secondary responses caused by high concentrations of the compounds.
Figure 1. Growth curves of reference strain of methicillin-resistant Staphylococcus aureus treated with α-amyrin, betulinic acid and betulinaldehyde.
Overview of Transcriptional Profiles of Staphylococcus aureus Treated with Pentacyclic Triterpenoids
From the data processed using GeneSpring version 11 software, an enormous number of genes were significantly differentially regulated (p<0.05, fold change ≥2) in response to treatment with AM, BA and BE as compared to control (Table 2). For example, a total of 1265 genes were significantly affected in the treatment of reference methicillin-resistant strains with BE. There is also a vast number of the genes which are uncharacterized, without specific functional category and pathways, and with unknown function (Tables S2, S3).
Table 2. Total number of differentially regulated genes in response to α-amyrin, betulinic acid and betulinaldehyde.
| Treatment | Up-regulated genes | Down-regulated genes | Total number of significantly regulated genes |
| α-amyrin | 77 | 102 | 179 |
| Betulinic acid | 178 | 69 | 247 |
| Betulinaldehyde | 523 | 742 | 1265 |
Overall, the changes in expression levels of most genes were moderate with ratio in the range of 2- to 3-fold (Tables 3, 4, 5, 6). There are 31 genes coding for ribosome which were highly down-regulated (more than 5-fold and up to 21-fold), indicating that ribosome assembly may play a prominent role in the response of S. aureus to BE. Four genes in the fatty acid biosynthesis and three genes in the peptidoglycan biosynthesis were down-regulated more than 3-fold while three genes in the ABC transporters were at least 3-fold up-regulated after treatment with BE. ABC transporters and ribosomes are significantly regulated by all three pentacyclic triterpenoids. Only BE affects cell division, while only BA targets genes in β-lactam resistance and DNA replication pathways.
Table 3. Selected genes which are differentially up-regulated in response to α-amyrin, betulinic acid and betulinaldehyde.
| N315 ORF | Gene symbol | Product | Fold changeδ | Functional category |
| (A) Treatment with α-amyrin | ||||
| ABC transporters | ||||
| SA2253 | opp-1C | Oligopeptide transporter permease | +2.1 | Transport and binding proteins |
| (B) Treatment with betulinic acid | ||||
| ABC transporters | ||||
| SA2253 | opp-1C | Oligopeptide transporter permease | +2.1 | Transport and binding proteins |
| β-lactam resistance | ||||
| SA0039 | mecR1 | Methicillin resistance protein | +2.1 | Transcription |
| DNA replication | ||||
| SA0057 | ccrB | Cassette chromosome recombinase B | +2.2 | DNA replication |
| SA0058 | ccrA | Cassette chromosome recombinase A | +2.4 | DNA replication |
| (C) Treatment with betulinaldehdye | ||||
| Two component system | ||||
| SAS066 | agrD | AgrD protein | +2.1 | Signal transduction mechanisms |
| Aminoacyl-tRNA biosynthesis | ||||
| SA0986 | pheT | Phenylalanine-tRNA synthetase beta chain | +3.0 | Translation |
| ABC transporters | ||||
| SA0109 | sirC | Lipoprotein (iron complex transport protein) | +2.6 | Inorganic ion transport |
| SA0110 | sirB | Lipoprotein (iron complex transport protein) | +5.8 | Inorganic ion transport |
| SA0198 | oppF | Oligopeptide transport ATP-binding protein | +3.8 | Inorganic ion transport |
| SA1214 | opp2B | Oligopeptide transporter permease | +3.5 | Inorganic ion transport |
Table 4. Selected genes which are differentially down-regulated in response to α-amyrin.
| N315 ORF | Gene symbol | Product | Fold changeδ | Functional category |
| Two-component system | ||||
| SA0661 | saeR | Response regulator | −3.9 | Regulatory function |
| SA0060 | saeS | Histidine protein kinase | −2.2 | Regulatory function |
| SA1881 | kdpA | Potassium-transporting ATPase subunit A | −2.6 | Inorganic ion transport |
| SA1936 | luxS | S-ribosylhomocysteinasae | −2.1 | Signal transduction |
| Aminoacyl-tRNA biosynthesis | ||||
| SA0488 | cysS | Cysteinyl-tRNA synthetase | −2.1 | Translation |
| SA1106 | proS | Prolyl-tRNA synthetase | −2.6 | Translation |
| SA1287 | asnC | Asparaginyl-tRNA synthetase | −2.4 | Translation |
| SA1506 | thrS | Threonyl-tRNA synthetase | −2.4 | Translation |
| Fatty acid biosynthesis | ||||
| SA1073 | fabD | Malonyl coA-acyl carrier protein transacylase | −2.5 | Fatty acid and phospholipid metabolism |
| Peptidoglycan biosynthesis | ||||
| SA2057 | fmhB | FmhB protein | −2.2 | Cell envelope |
| Ribosome | ||||
| SAS042 | rpmG | 50s ribosomal protein L33 | −2.0 | Transcription |
| SA0354 | rpsR | 30s ribosomal protein S18 | −2.1 | Transcription |
| SA0497 | rplJ | 50s ribosomal protein L10 | −2.5 | Transcription |
Table 5. Selected genes which are differentially down-regulated in response to betulinic acid.
| N315 ORF | Gene symbol | Product | Fold changeδ | Functional category |
| Two component system | ||||
| SA1883 | kdpE | KDP operon transcriptional regulatory protein | −2.1 | Signal transduction mechanisms |
| SA0068 | kdpA | Potassium-transporting ATPase subunit A | −2.1 | Signal transduction mechanisms |
| ABC transporters | ||||
| SA0198 | oppF | Oligopeptide transport ATP-binding protein | −2.2 | Inorganic ion transport |
| Ribosome | ||||
| SAS042 | rpmG | 50s ribosomal protein L33 | −2.1 | Transcription |
Table 6. Selected genes which are differentially down-regulated in response to betulinaldehyde.
| N315 ORF | Gene symbol | Product | Fold changeδ | Functional category |
| Cellular process | ||||
| SA1029 | ftsZ | Cell division protein FtsZ | −4.8 | Cell division |
| SA1028 | ftsA | Cell division protein FtsA | −5.2 | Cell division |
| Two-component system | ||||
| SA1844 | agrA | Accessory gene regulator A | −2.4 | Signal transduction mechanisms |
| SA1881 | kdpA | Potassium-transporting ATPase subunit A | −9.0 | Signal transduction mechanisms |
| SA1883 | kdpE | KDP operon transcriptional regulatory protein | −2.7 | Signal transduction mechanisms |
| Aminoacyl-tRNA biosynthesis | ||||
| SA0209 | serS | Seryl-tRNA synthetase | −2.7 | Translation |
| SA0475 | lysS | Lysyl-tRNA synthetase | −3.3 | Translation |
| SA1106 | proS | Propyl-tRNA synthase | −2.3 | Translation |
| SA1287 | asnC | Asparaginyl-tRNA synthase | −2.3 | Translation |
| SA1506 | thrS | Threonyl-tRNA synthase I | −2.6 | Translation |
| Fatty acid biosynthesis | ||||
| SA0843 | fabF | 3-oxoacyl synthase | −3.4 | Fatty acid and phospholipid metabolism |
| SA0889 | fabI | Enoyl-(acyl carrier protein) reductase | −2.7 | Fatty acid and phospholipid metabolism |
| SA1073 | fabD | Malonyl coA-acyl carrier protein transacylase | −3.0 | Fatty acid and phospholipid metabolism |
| SA1074 | fabG | 3-oxoacyl-{acyl-carrier protein} reductase | −3.9 | Fatty acid and phospholipid metabolism |
| SA1901 | fabZ | (3R)-hydroxymyristoyl ACP dehydratase | −3.7 | Fatty acid and phospholipid metabolism |
| Peptidoglycan biosynthesis | ||||
| SA1251 | murG | Undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase | −3.9 | Cell envelope |
| SA1283 | pbp2 | Penicillin binding protein 2 | −4.5 | Cell envelope |
| SA1561 | murC | UDP-N-acetylmuramate L-alanine ligase | −2.5 | Cell envelope |
| SA1902 | murA | UDP-N-acetylglucoasmine 1-carboxyvinyltransferase | −5.1 | Cell envelope |
| SA2057 | fmhB | FmhB protein | −2.2 | Cell envelope |
| Ribosome | ||||
| SA0503 | rpsL | 30S ribosomal protein S12 | −7.3 | Translation |
| SA2025 | rpsM | 30S ribosomal protein S13 | −6.1 | Translation |
| SA1081 | rpsP | 30S ribosomal protein S16 | −2.9 | Translation |
| SA2038 | rpsQ | 30S ribosomal protein S17 | −10.2 | Translation |
| SA0354 | rpsR | 30S ribosomal protein S18 | −4.8 | Translation |
| SA2043 | rpsS | 30S ribosomal protein S19 | −4.8 | Translation |
| SA1099 | rpsB | 30S ribosomal protein S2 | −9.9 | Translation |
| SA1414 | rpsT | 30S ribosomal protein S20 | −6.6 | Translation |
| SA1404 | rpsU | 30S ribosomal protein S21 | −12.9 | Translation |
| SAS052 | rpsD | 30S ribosomal protein S4 | −5.7 | Translation |
| SA0496 | rplA | 50S ribosomal protein L1 | −6.1 | Translation |
| SA2031 | rpsE | 30S ribosomal protein S5 | −8.3 | Translation |
| SA0352 | rpsF | 30S ribosomal protein S6 | −5.7 | Translation |
| SA0497 | rplJ | 50S ribosomal protein L10 | −6.5 | Translation |
| SA0495 | rplK | 50S ribosomal protein L11 | −6.2 | Translation |
| SA2017 | rplM | 50S ribosomal protein L13 | −9.0 | Translation |
| SA2037 | rplN | 50S ribosomal protein L14 | −9.2 | Translation |
| SA2029 | rplO | 50S ribosomal protein L15 | −6.3 | Translation |
| SA2040 | rplP | 50S ribosomal protein L16 | −7.4 | Translation |
| SA2022 | rplQ | 50S ribosomal protein L17 | −3.7 | Translation |
| SA2032 | rplR | 50S ribosomal protein L18 | −7.6 | Translation |
| SA1473 | rplU | 50S ribosomal protein L21 | −6.9 | Translation |
| SA2036 | rplX | 50S ribosomal protein L24 | −9.7 | Translation |
| SA0459 | rplY | 50S ribosomal protein L25 | −5.4 | Translation |
| SA1471 | rpmA | 50S ribosomal protein L27 | −6.3 | Translation |
| SA1067 | rpmB | 50S ribosomal protein L28 | −6.3 | Translation |
| SA2039 | rpmC | 50S ribosomal protein L29 | −9.4 | Translation |
| SA2047 | rplC | 50S ribosomal protein L3 | −4.9 | Translation |
| SA2030 | rpmD | 50S ribosomal protein L30 | −6.2 | Translation |
| SA1922 | rpmE | 50S ribosomal protein L31 type B | −4.8 | Translation |
| SAS033 | rpmF | 50S ribosomal protein L32 | −7.7 | Translation |
| SAS042 | rpmG | 50S ribosomal protein L33 1,2,3 | −8.4 | Translation |
| SAS093 | rpmH | 50S ribosomal protein L34 | −6.3 | Translation |
| SA1503 | rpmI | 50S ribosomal protein L35 | −6.0 | Translation |
| SAS078 | rpmJ | 50S ribosomal protein L36 | −21.0 | Translation |
| SA2035 | rplE | 50S ribosomal protein L5 | −6.3 | Translation |
| SA0498 | rplL | 50S ribosomal protein L7/L12 | −8.3 | Translation |
| SA0014 | rplI | 50S ribosomal protein L9 | −2.8 | Translation |
Differentially expressed genes in promising pathways such as cell division (ftsZ), ABC transporters (opp-1C), biosynthesis of fatty acid (fabZ), peptidoglycan (fmhB, pbp2) and protein (asnC), DNA replication (ccrA) and β-lactam resistance (mecR1) were selected to be validated by real-time PCR. The paired student’s T test analysis showed that all these selected genes were extremely statistically significantly different between bacterial cells treated with AM, BA or BE, and untreated cells. The difference in the expression of reference genes adhE and gyrA was considered not significantly different (Table S4).
A comparison of genes identified in the treatments of all three study compounds showed that genes rpmG and kdpA, which code for the protein of the large subunit 50S and potassium-transporting ATPase subunit A, respectively, are down-regulated. AM and BA showed up-regulation of opp-1C in the ABC transporter pathway, while kdpE in the two-component systems are similarly down-regulated in BA and BE. Treatment of the bacterial strain with AM and BE affected the highest number of similarly regulated genes, involving a total of seven genes in the two-component systems, fatty acid biosynthesis, peptidoglycan biosynthesis, aminoacyl-tRNA biosynthesis and ribosome pathways (Table 7). Generally, all three study compounds acted on more than one target/pathway that is unique to the microorganisms. This is a highly desirable property of an antibacterial agent and it could potentially delay the development of bacterial resistance to the compounds [23], [24].
Table 7. Similarly regulated genes in response to treatment with α-amyrin, betulinic acid and/or betulinaldehyde.
| N315 ORF | Gene symbol | Product | Pathway |
| Treatment with α-amyrin, betulinic acid and betulinaldehyde | |||
| SA1881 | kdpA | Potassium-transporting ATPase subunit A | Two-component system |
| SAS042 | rpmG | 50s ribosomal protein L33 1,2,3 | Ribosome |
| Treatment with α-amyrin and betulinic acid | |||
| SA1881 | kdpA | Potassium-transporting ATPase subunit A | Two-component system |
| SA2253 | opp-1C | Oligopeptide transporter permease | ABC transporter |
| SAS042 | rpmG | 50s ribosomal protein L33 1,2,3 | Ribosome |
| Treatment with α-amyrin and betulinaldehyde | |||
| SA1881 | kdpA | Potassium-transporting ATPase subunit A | Two-component system |
| SA1287 | asnC | Asparaginyl-tRNA synthase | Aminoacyl-tRNA biosynthesis |
| SA1073 | fabD | Malonyl coA-acyl carrier protein transacylase | Fatty acid biosynthesis |
| SA2057 | fmhB | FmhB protein | Peptidoglycan biosynthesis |
| SA0354 | rpsR | 30s ribosomal protein S18 | Ribosome |
| SA0497 | rplJ | 50s ribosomal protein L10 | Ribosome |
| SAS042 | rpmG | 50s ribosomal protein L33 1,2,3 | Ribosome |
| Treatment with betulinic acid and betulinaldehyde | |||
| SA1881 | kdpA | Potassium-transporting ATPase subunit A | Two-component system |
| SA1883 | kdpE | KDP operon transcriptional regulatory protein | Two-component system |
| SAS042 | rpmG | 50s ribosomal protein L33 1,2,3 | Ribosome |
Validation of Gene Expression by qPCR
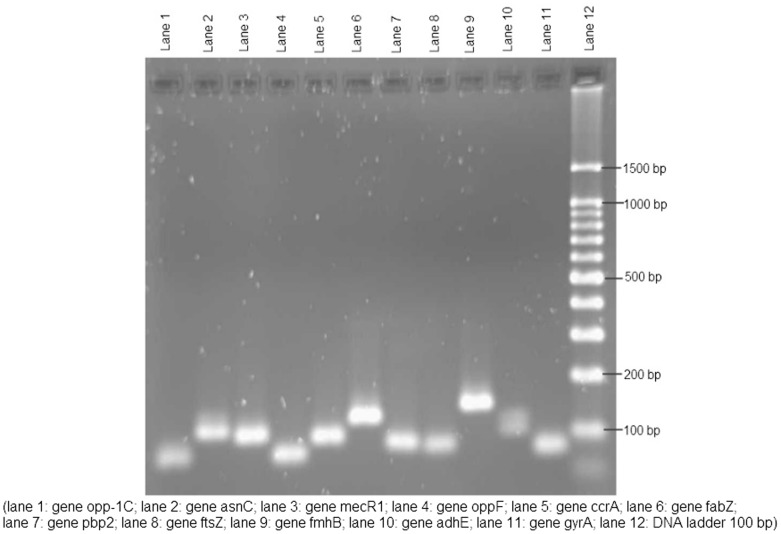
The target genes selected for validation represent significant pathways in S. aureus identified as promising targets in the literature, and two genes selected as reference genes because they were not differentially regulated by AM, BA and BE. The target genes were ftsZ (cell division), opp-1C and oppF (ABC transporters), asnC (aminoacyl-tRNA biosynthesis), fabZ (fatty acid biosynthesis), fmhB and pbp2 (peptidoglycan biosynthesis), ccrA (DNA replication), and mecR1 (β-lactam resistance), while the reference genes were adhE (metabolism of fatty acid and glycolysis) and gyrA (DNA replication). All the target and reference genes validated showed single peaks in melting curves, indicating the amplification of specific amplicons (data not shown). The high specificity of the reactions was further confirmed by the single band and size of the amplicons observed in the agarose gel electrophoresis (Figure 2). The expression of the target genes was quantified using the relative quantification method which measures the changes in the expression of the target genes in response to the different treatment normalized to the expression of reference gene(s). The expression of genes opp-1C, oppF, asnC and mecR1 was observed at greater fold-change by real-time PCR than with microarray analysis. For three other genes fabZ, fmhB and ccrA, the levels of gene expression did not differ markedly between the microarray data and real-time PCR analysis. Genes ftsZ and pbp2 showed higher gene expression level in microarray analysis. The comparison of the gene expression level by qPCR and microarray analysis is shown in Table 8.
Figure 2. Agarose gel electrophoresis of PCR product.
Table 8. Comparison of gene expression level by quantitative real-time PCR and microarray analysis.
| N315 ORF | Gene name# | Product | Fold change | |
| Real-time PCR | Microarray | |||
| SA1029 | ftsZ(3) | Cell division protein | −2.2 | −4.8 |
| SA2253 | opp-1C(1) | Oligopeptide transporter permease | +3.4 | +2.1 |
| SA0198 | oppF(3) | Oligopeptide transport ATP-binding protein | +3.4 | +2.2 |
| SA1287 | asnC(1) | Asparaginyl-tRNA synthetase | −7.4 | −2.4 |
| SA1901 | fabZ(3) | (3R)-hydroxymyristoyl ACP dehydratase | −3.1 | −3.7 |
| SA2057 | fmhB(1) | FmhB protein | −2.1 | −2.2 |
| SA1283 | pbp2(3) | Penicillin-binding protein 2 | −3.1 | −4.5 |
| SA0058 | ccrA(2) | Cassette chromosome recombinase A | +2.7 | +2.4 |
| SA0039 | mecR1(2) | Methicillin resistance protein | +5.9 | +2.1 |
Treatment of bacterial cell with pentacyclic triterpenoids:
α-amyrin,
betulinic acid,
betulinaldehyde.
Novel Pathways Targeted by AM, BA and BE
Cell division
BE inhibits genes encoding proteins FtsZ and FtsA in the cell division process of the bacterial cell. FtsZ is an essential division protein and forms a ring structure that recruits further cell division proteins. Cell division starts with the polymerization of FtsZ into the Z ring, which is the heart of the division apparatus and serves as a landing pad for the recruitment of other proteins to the division site. Suppression of FtsZ prevents cell division, leading to mass increase and eventually cell lysis. Even if inhibition of division does not result in cell death, it should prevent proliferation of the bacteria within the host and the spread of bacteria from infection sites, thus enhancing the efficiency of host-defense factors [25]. FtsA which tethers FtsZ to the membrane, recruits downstream divisome proteins and stabilizes Z rings at membrane. Thus, suppression of FtsZ and FtsA proteins inhibits growth and could exert bactericidal effect [26].
Two-component system
From the microarray analysis, AM, BA and BE have the potential to target the virulence factors of MRSA, which includes structures that promote adhesion, colonization and dissemination; the components responsible for cell communication and regulatory functions; the factors that facilitate evasion of the immune system; and the machineries for toxin production and excretion [27]. Expression of these factors is regulated by a range of global regulators comprising two-component regulatory systems which are sensitive to environmental signals [28], [29], [30]. The study compounds regulates three of the systems of the 16 two-component systems identified by genomic scans in S. aureus, i.e. Agr, Sae and Kdp.
The expression of gene agrD and inhibition of agrA, as observed in the treatment with BE, leads to the decreased production of capsule, toxins and proteases, thus decreasing the virulence of the organism. AM which inhibits saeR and saeS also decreases virulence as a result of reduction in the production in several exoproteins and cell-bound proteins involved in pathogenicity [31]. With the inhibition of kdpA and kdpE as observed in the treatment with BA and BE, the transcription of genes encoding cell wall proteins and polysaccharides, which are beneficial to colonization, are repressed [32]. As a result, the virulence of the organism is reduced with the exposure to each of the compounds.
ABC transporters
The up-regulation of oligopermease transporter oppC by AM and BA, and oppF by BE may increase the efficient transport of peptides for the salvage of essential amino acids from the environment of the bacterial cell. The uptake of these peptides could be to compensate for the halt in protein synthesis due to the inhibition of the aminoacyl-tRNA synthetase and ribosomes. The up-regulation of gene sirB as observed in the treatment with BE, indicates there could be an uptake of iron (III)-staphylobactin to satisfy its requirement for iron, which is an essential micronutrient for bacteria and serves as the catalytic center of enzymes involved in critical cellular processes such as DNA synthesis and electron transport [33]. This may represent an attempt by the organism to acquire iron for successful establishment of infection, as a response to the environmental stress caused by BE.
Novel Targets in Existing Pathways Regulated by AM, BA and BE
Fatty acid biosynthesis
BE down-regulates a group of genes in the type II fatty acid synthesis (FAS) pathway, which include genes encoding for enzymes that are involved in the assembly of important cellular components in the bacteria, such as phospholipids, lipoproteins, lipopolysaccharides, myolic acids and the cell envelope [34]. The inhibition of enzymes encoded by fabF, fabI, fabD, fabG and fabZ may cause modification of the permeability of the phospholipid bilayer and destabilization of the cell membrane. As a result, processes such as passive transport of hydrophobic molecules, active transport of solutes and protein-protein interactions could be affected [35]. Thus, AM and BE may be able to enter the cells relatively easier via passive transport. However, due to the high partition coefficients of AM and BE at log P values of 11.01 and 9.07, respectively (Chemspider database, http://www.chemspider.com/Search.aspx), the entry of these compounds may still not increase significantly, thus the high observed MIC values at 64 and 512 µg/ml as compared to current anti-staphylococcal agents such as methicillin and vancomycin.
Peptidoglycan biosynthesis
Closely linked to cell division is peptidoglycan synthesis. Coordinated synthesis of the peptidoglycan is crucial for the integrity of the bacterial cell, attachment sites for virulence factors and resistance to external stress conditions [36]. BE inhibits three mur genes (murA, murC and murG), fmhB and pbp2 which are involved in the synthesis initiation phase, modification, and polymerization and cross-linking of lipid II, respectively. The suppression of the synthesis of lipid II which is the substrate for Pbp2 and transpeptidation reactions results in the weakening of the peptidoglycan, leading to cell lysis and death. The majority of approved anti-staphylococcal drugs that target the peptidoglycan biosynthesis act on extracellular steps and resistance to these drugs are on the rise. An attractive alternative is to exploit the intracellular steps, involving the mur and fem genes. There are currently no known antibiotics or synthetic chemicals of therapeutic usefulness that inhibits the Mur enzymes, except for MurA which is inhibited by fosfomycin [37].
Aminoacyl-tRNA biosynthesis
The suppression of six aminoacyl-tRNA synthetase, namely cysS, proS, asnC, thrS, serS and lysS, by AM and BE causes the accumulation of the deacylated form of tRNA. The shortage of corresponding aminoacyl-tRNA of a required amino acid prompts the stringent response with increased levels of guanosine tetraphosphates (ppGpp) and guanosine pentaphosphates (pppGpp). RNA polymerase is inhibited, thus directly reducing the production of RNA. This leads to subsequent inhibition of all key downstream cellular processes such as the biosynthesis of protein and peptidoglycan [38]. By perturbing several metabolically important processes, the inhibition of aminoacyl-tRNA synthetase leads to the cessation of bacterial growth and the attenuation of virulence in vivo [39].
Ribosome
The assembly of the ribosome begins with the transcription of rRNA [40] and proceeds through the synthesis of ribosomal proteins [41]. In comparison to the other study compounds, BE inhibits the expression of the highest number of genes in the ribosome, with 13 and 23 of the 30S and 50S subunits, respectively. The compounds could bind to the precursor forms of rRNA and ribosomal proteins and stall the normal assembly sequence of the ribosome. As a result, continued translation could be reduced, leading to inhibition of protein synthesis and prevention of growth of the bacteria. The interference with the assembly of the ribosomal subunit from its constituent rRNA and proteins could be a new target for inhibition of antibacterial agents.
β-lactam resistance
MecA, the main determinant for methicillin resistance, is inducible and modulated by a signal-transduction system encoded by genes from the SCCmec [42], which comprises a sensor/signal transducer MecR1, and a transcriptional repressor, methicillin repressor MecI [43]. Treatment with BA causes the expression of gene mecR1 that results in the production of fully functional MecR1 and MecI that appear to repress the production of pbp2a [44], [45]. The observation indicates that BA could be modified and developed into a more potent inhibitor of penicillin-binding protein (PBP) that can target MRSA. In addition, BA has the potential to restore the in vitro activity of β-lactams and glycopeptides against MRSA, as shown in the strong synergistic interactions with methicillin and vancomycin [17].
Conclusions
Microbial genomics has allowed the identification of a greater number of desirable and essential antibacterial targets for further evaluation. From the transcriptional profile, AM, BA and BE regulate multiple desirable targets in essential pathways which could be further explored in the development of therapeutic agents for the treatment of S. aureus infections. In addition, the structure of the study compounds differ from the currently used anti-staphylococcal agents, and thus cross-resistance is unlikely and development of resistance to these novel compounds will be delayed. While the compounds have the ability to affect multiple and essential targets, the relatively high MICs in S. aureus suggests that the permeability of the compounds into the bacterial cell may not be optimum and compound modifications may be required to improve the permeability. A more detailed examination of these gene responses is also needed to further characterize the antimicrobial action of each of the regulated pathways.
Supporting Information
Quantification and quality assessment of total RNA isolated from reference strain of methicillin-resistant Staphylococcus aureus treated with α-amyrin, betulinic acid and betulinaldehyde at ½× MIC and control.
(DOCX)
Genes differentially expressed in response to α-amyrin, betulinic acid and betulinaldehyde without identified pathways.
(DOCX)
Genes differentially expressed in response to α-amyrin, betulinic acid and betulinaldehyde with identified pathways.
(DOCX)
Significance analysis of target and reference genes validated with quantitative real-time PCR.
(DOCX)
Acknowledgments
The authors wish to acknowledge Malaysia Genome Institute (Microarray) for providing the Bioanalyzer analysis of the total RNA and GeneSpring software for the statistical analysis of the microarray data. The authors also thank the following representatives for their technical support: C Chan (Qiagen) for the valuable insights and guidance in the extraction of pure and intact total RNA, LH Teng and ET Siah (Affymetrix) for their assistance with the DNA microarray analysis, and C Lee (BioRad Laboratories) for the real-time PCR analysis.
Funding Statement
This project was partly funded by the Ministry of Science, Technology and Innovation, Malaysia (eScience 02-02-10-SF0056) (March 2010 to February 2012) and Monash University (Seed Grant 5140029-000-00) (April 2009 to March 2010) under Parasakthi Navaratnam. Both the funding bodies provided financial support for the conduct of the research and are not involved in the study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Levy SB, Marshall B (2004) Antibacterial resistance worldwide: causes, challenges and responses. Nat Med 12 (Supplement): S122–S129. [DOI] [PubMed] [Google Scholar]
- 2.Livermore DM (2003) Bacterial resistance: origins, epidemiology and impact. Clin Infect Dis 36 (Supplement): S11–S23. [DOI] [PubMed] [Google Scholar]
- 3. Barrett CT, Barrett JF (2003) Antibacterials: are the new entries enough to deal with the emerging resistance problems? Curr Opin Biotechnol 14: 621–626. [DOI] [PubMed] [Google Scholar]
- 4. Monaghan RL, Barrett JF (2006) Antibacterial drug discovery: then, now and the genomics future. Biochem Pharmacol 71: 901–909. [DOI] [PubMed] [Google Scholar]
- 5.IDSA Report (2004) Bad bugs, no drugs as antibiotic discovery stagnates, a public health crises brews. Available: http://www.idsociety.org.
- 6. Tsiodras S, Gold HS, Sakoulas G, Eliopoulos GM, Wennersten C, et al. (2001) Linezolid resistance in a clinical isolate of Staphylococcus aureus . The Lancet 358: 207–208. [DOI] [PubMed] [Google Scholar]
- 7. Ohlsen K, Lorenz U (2007) Novel targets for antibiotics in Staphylococcus aureus . Future Microbiol 2: 655–666. [DOI] [PubMed] [Google Scholar]
- 8. Qiu J, Zhou D, Qin L, Han Y, Wang X, et al. (2006) Microarray expression profiling of Yersinia pestis in response to chloramphenicol. FEMS Microbiol Lett 263: 26–31. [DOI] [PubMed] [Google Scholar]
- 9. Utaida S, Dunman PM, Macapagal D, Murphy E, Projan SJ, et al. (2003) Genome-wide transcriptional profiling of the responses of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149: 2719–2732. [DOI] [PubMed] [Google Scholar]
- 10. McAleese F, Wu SW, Sieradzki K, Dunman P, Murphy E, et al. (2006) Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate S. aureus-type resistance to vancomycin. J Bacteriol 188: 1120–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mallavadhani UV, Mahapatra A, Jamil K, Reddy S (2004) Antimicrobial activity of some pentacyclic triterpenes and their synthesized 3-O-lipophilic chain. Biol Pharm Bull 27: 1576–1579. [DOI] [PubMed] [Google Scholar]
- 12. Singh B, Singh S (2003) Antimicrobial activity of terpenoids from Trichodesma amplexicaule Roth. Phytother Res 17: 814–816. [DOI] [PubMed] [Google Scholar]
- 13. Chandramu C, Manohar RD, Krupadanam DG, Dashavantha RV (2003) Isolation, characterization and biological activity of betulinic acid and ursolic acid from Vitex negundo L. Phytother Res. 17: 129–134. [DOI] [PubMed] [Google Scholar]
- 14. Schuhly W, Heilmann J, Calis I, Sticher O (1999) New triterpenoids with antibacterial activity from Zizyphus joazeiro . Planta Med 65: 740–743. [DOI] [PubMed] [Google Scholar]
- 15. Gibbons S (2004) Antistaphylococcal plant natural products. Nat Prod Rep 21: 263–277. [DOI] [PubMed] [Google Scholar]
- 16. Chung PY, Chung LY, Ngeow YF, Goh SH, Imiyabir Z (2004) Antimicrobial activities of Malaysian plant species. Pharm Biol 42: 292–300. [Google Scholar]
- 17. Chung PY, Navaratnam P, Chung LY (2011) Synergistic antimicrobial activity between pentacyclic triterpenoids and antibiotics against Staphylococcus aureus strains. Ann Clin Microb Antimicrob 10: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng H, Xiang H, Zhang J, Liu G, Guo N, et al.. (2009) Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cryptotanshione. J Biomed Biotech ID 617509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang D, Yu L, Xiang H, Fan J, He L, et al. (2008) Global transcriptional profiles of Staphylococcus aureus treated with berberine chloride. FEMS Microbiol Lett 279: 217–225. [DOI] [PubMed] [Google Scholar]
- 20. Yu L, Xiang H, Fan J, Wang D, Yang F, et al. (2008) Global transcriptional response of Staphylococcus aureus to rhein, a natural plant product. J Biotech 135: 304–308. [DOI] [PubMed] [Google Scholar]
- 21. Hutter B, Schaab C, Albrecht S, Borgmann M, Brunner NA, et al. (2004) Prediction of mechanisms of action of antibacterial compounds by gene expression profiling. Antimicrob Agents Chemother 48: 2838–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.BioRad Laboratories (2006) Real-time PCR applications guide. [Google Scholar]
- 23. Lange RP, Locher HH, Wyss PC, Then RL (2007) The targets of currently used antibacterial agents: lessons for drug discovery. Curr Pharm Des 13: 3140–3154. [DOI] [PubMed] [Google Scholar]
- 24. Silver LL (2007) Multi-targeting by monotherapeutic antibacterials. Nat Rev Drug Disc 6: 41–55. [DOI] [PubMed] [Google Scholar]
- 25. Vollmer W (2008) Targeting the bacterial Z-ring. Chem Biol 15: 93–94. [DOI] [PubMed] [Google Scholar]
- 26. Lock RL, Harry EJ (2008) Cell-division inhibitors: new insights for future antibiotics. Nat Rev Drug Disc 7: 324–338. [DOI] [PubMed] [Google Scholar]
- 27. Brotz-Oesterhelt H, Sass P (2010) Post-genomic strategies in antibacterial drug discovery. Future Microbiol 5: 1553–1579. [DOI] [PubMed] [Google Scholar]
- 28. Bronner S, Monteil H, Prevost G (2004) Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 28: 183–200. [DOI] [PubMed] [Google Scholar]
- 29. Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ (2004) Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus . FEMS Immunol Med Microbiol 40: 1–9. [DOI] [PubMed] [Google Scholar]
- 30. Novick RP (2003) Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48: 1429–1449. [DOI] [PubMed] [Google Scholar]
- 31. Giraudo AT, Calzolari AA, Cataldi AA, Bogni C, Nagel R (1999) The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol Lett 177: 15–22. [DOI] [PubMed] [Google Scholar]
- 32. Xue T, You Y, Hong D, Sun H, Sun B (2011) The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect Immun 79: 2154–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Speziali CD, Dale SE, Henderson JA, Vines ED, Heinrichs DE (2006) Requirement of Staphylococcus aureus ATP-binding cassette-ATPase FhuC for iron-restricted growth and evidence that it functions with more than one iron transporter. J Bacteriol 188: 2048–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park HS, Yoon YM, Jung SJ, Kim CM, Kim JM, et al. (2007) Antistaphylococcal activities of CG400549, a new bacterial enoyl-acyl carrier protein reductase (FabI) inhibitor. J Antimicrob Chemother 60: 568–574. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y, Rock CO (2008) Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6: 222–233. [DOI] [PubMed] [Google Scholar]
- 36. Macheboeuf P, Contretas-Martel C, Job V, Diderberg O, Dessen A (2006) Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol Rev 30: 673–691. [DOI] [PubMed] [Google Scholar]
- 37. El Zoeiby A, Sanschagrin F, Levesque RC (2003) Structure and function of the Mur enzymes: development of novel inhibitors. Mol Microbiol 47: 1–12. [DOI] [PubMed] [Google Scholar]
- 38. Cassels R, Olivia B, Knowles D (1995) Occurrence of the regulatory nucleotides ppGpp and pppGpp following induction of the stringent response in staphylococci. J Bacteriol 177: 5161–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tao J, Schimmel P (2000) Inhibitors of amino-acyl tRNA synthetase as novel anti-infectives. Expert Opin Investig Drugs 9: 1767–1775. [DOI] [PubMed] [Google Scholar]
- 40. Maguire BA (2009) Inhibition of bacterial ribosome assembly: a suitable drug target? Microbiol Mol Biol Rev 73: 22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Champney WS (2006) The other target for ribosomal antibiotics: inhibition of bacterial ribosomal subunit formation. Infect Disord – Drug Targets 6: 377–390. [DOI] [PubMed] [Google Scholar]
- 42. Zhang HZ, Hackbarth CJ, Chansky KM, Chambers HF (2001) A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science 291: 1962–1965. [DOI] [PubMed] [Google Scholar]
- 43. Weller TMA (1999) The distribution of mecA, mecR1 and mecI and sequence analysis of mecI and the mec promoter region in staphylococci expressing resistance to methicillin. J Antimicrob Chemother 43: 15–22. [DOI] [PubMed] [Google Scholar]
- 44. Kuwahara-Arai K, Kondo N, Hori S, Tateda-Suuzuki E, Hiramatsu K (1996) Suppression of methicillin resistance in mecA-containing pre-methicillin-resistant Staphylococcus aureus is caused by the mecI-mediated repression of PBP2′ production. Antimicrob Agents Chemother 40: 2680–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ryffel C, Kayser FH, Berger-Bachi B (1992) Correlation between regulation of mecA transcription and expression of methicillin resistance Staphylococcus aureus . Antimicrob Agents Chemother 36: 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification and quality assessment of total RNA isolated from reference strain of methicillin-resistant Staphylococcus aureus treated with α-amyrin, betulinic acid and betulinaldehyde at ½× MIC and control.
(DOCX)
Genes differentially expressed in response to α-amyrin, betulinic acid and betulinaldehyde without identified pathways.
(DOCX)
Genes differentially expressed in response to α-amyrin, betulinic acid and betulinaldehyde with identified pathways.
(DOCX)
Significance analysis of target and reference genes validated with quantitative real-time PCR.
(DOCX)