Abstract
Background
Due to the possible involvement of Glutathione S-transferase Mu-1 (GSTM1) and Glutathione S-transferase theta-1 (GSTT1) in the detoxification of environmental carcinogens, environmental toxins, and oxidative stress products, genetic polymorphisms of these two genes may play important roles in the susceptibility of human being to hepatocellular carcinoma. However, the existing research results are not conclusive.
Methods
A systematic literature search using databases (PubMed, Scopus, Embase, Chinese Biomedical Database, Chinese National Knowledge Infrastructure, Wanfang Data, etc.) for the eligible studies meeting the inclusion criteria including case-control studies or cohort studies is evaluated using an updated systematic meta-analysis.
Results
Significant increase in the risk of HCC in the Chinese population is found in GSTM1 null genotype (OR = 1.47, 95% CI: 1.21 to 1.79, P<0.001) and GSTT1 null genotype (OR = 1.38, 95% CI: 1.14 to 1.65, P<0.001). Analysis using the random-effects model found an increased risk of HCC in GSTM1-GSTT1 dual null population (OR = 1.79, 95% CI: 1.26 to 2.53, P<0.001). In addition, subgroup analyses showed a significant increase in the association of GST genetic polymorphisms (GSTM1, GSTT1, and GSTM1-GSTT1) with HCC in southeast and central China mainland. However, available data collected by this study fail to show an association between GST genetic polymorphisms and HCC in people from the Taiwan region (for GSTM1: OR = 0.78, 95% CI: 0.60 to 1.01, P = 0.06; for GSTT1: OR = 0.94, 95% CI: 0.78 to 1.14, P = 0.546; for GSTM1-GSTT1: OR = 1.04, 95% CI: 0.81 to 1.32, P = 0.77). Sensitivity analysis and publication bias diagnostics confirmed the reliability and stability of this meta-analysis.
Conclusions
Our results indicate that both GSTM1 and GSTT1 null genotypes are associated with an increased HCC risk in Chinese population. Peoples with dual null genotypes of GSTM1-GSTT1 are more susceptible to developing HCC. In conclusion, GST genetic polymorphisms play vital roles in the development of HCC in the Chinese population.
Introduction
Due to a high mortality, hepatocellular carcinoma (HCC) is one of the most serious health problems worldwide [1]–[2], which is consisted of approximately 80% of all primary tumors of liver [3]. Incidence rates in males and females are listed sixth and ninth as the most common cancers, respectively. Incidence rate of HCC has been increasing for several years while overall cancer incidence rate has been decreasing in recent years [4]–[5]. Environment and genetic factors are believed to be the pathogenesis of HCC [6]–[8]. Furthermore, previous studies indicated that racial and ethnic variations in the same geographic location could cause result bias in meta-analysis [9]–[11]. In Asia, people are at higher risk of developing HCC because of chronic infection with hepatitis B virus (HBV) [12]–[13]. In Europe, not only hepatitis C virus (HCV) and cirrhosis, but alcohol and tobacco smoking are also clearly able to accelerate HCC development [2]. Due to its substantial morbidity and mortality, HCC has been a hot research topic in China in recent years.
The Glutathione S-transferases (GSTs) family is an important phase II isoenzyme which can detoxify environmental carcinogens and toxins, oxidative stress products, and modulate the induction of other enzymes and proteins in the cell at the same time [14]–[16]. Enzymes of GSTs family are composed of many cytosolic, mitochondrial, and MAPEG proteins. Human GSTs can be divided into eight main classes including alpha, mu, pi, theta, sigma, kappa, omega and zeta [17]. GSTM1 and GSTT1 (encoding the Mu and Theta, respectively) both play important roles in human carcinogenesis. Epidemiologic investigations related to genetic association including case-control and cohort studies suggested the association between GST genetic polymorphisms and HCC risk. However, some of these studies with sparse data, unreasonable and highly underpowered designs, and differential in research methodology could all inevitably influence the robustness of their results. Meta-analysis can avoid these weaknesses by selecting all eligible studies and reducing random error. To identify the association of GST genetic polymorphisms with the susceptibility to hepatocellular carcinoma in the Chinese population, an updated systematic meta-analysis was performed in this study by using a full reference search (from January 1996 to October 2012) and a careful reinvestigation strategy.
Methods
1. Literature and Research Strategy
A computerized literature search was carried out in Embase, PubMed, Scopus, Chinese Biomedical Database (CBM), CochraneLibrary, Chinese National Knowledge Infrastructure (CNKI), and Wanfang Data (the latest research was retrospected to October 2012) to collect articles with case-control or cohort studies related to the association of GSTM1 and/or GSTT1 polymorphisms with the susceptibility of HCC in China. Meanwhile, reference lists of the relevant articles were also collected. Search was performed through websites of http://www.baidu.com and http://scholar.google.cn to identify additional eligible studies. MeSH terms (“glutathione S-transferase” or “GST” or “GSTM1” or “GSTT1”) and (“hepatocellular carcinoma” or “liver cancer” or “HCC”) and (“China” or “Chinese” or “Taiwan”) were used in PubMed. These keyword retrieval strategies were also used in other databases. When there was more than one article published in a same case series, the latest and/or the comprehensive one would be adopted only. Eligible research articles not captured by above research strategies would be further searched by bibliographies.
2. Inclusion and Exclusion Criteria
Inclusion criteria are: (1) case-control and cohort studies, in which individuals or samples used for evaluation of the association between GST genetic variances and HCC risk included these owning either with a balance match or not; (2) in the Chinese population; (3) the articles provided raw data including odds ratio (OR) with 95% confidence interval (CI) and respective variance, or the relevant information could be calculated.
Exclusion criteria are: (1) raw data not available for retrieval; (2) multiple articles based on a same population and published by a same research team, only the latest and/or the largest population study was adopted, others would be excluded; (3) meeting abstract, case reports, editorials, review articles and other meta-analysis were exclusive.
3. Data Extraction and Synthesis
To decide inclusively or exclusively, articles were identified by two independent reviewers using a standardized data extraction form designed by our group. Data with discrepancies in identification were discussed. If consensus was not achieved, the decision was made by a third reviewer. Both title and abstract from all potential included articles were screened to identify their relevance. Additionally, if title and abstract were ambiguous, full articles were also investigated. The following information was collected from each study: first author, year of publication, geographical location, study time, pathologic diagnosis, source of control, characteristic of cases and controls, and genotype frequency of null GSTM1, GSTT1 and null of both genotypes in cases and controls.
4. Statistical Analysis
(1) The pooled OR and 95% CI were determined by Z test with P<0.05 considered statistically significant; (2) Statistical heterogeneity among studies was assessed with the Q and I2 statistics [18]. The Q test and I2 were claimed to test the variation which was due to heterogeneity or by random error [19]. When P value of heterogeneity tests was no more than 0.1 (P≤0.1), we used random effects model. When P value of heterogeneity test was more than 0.1 (P>0.1), we used fixed effects model [20]; (3) Sensitivity analysis was also tested by removing one study at a time to calculate the overall homogeneity and effect size; (4) Publication bias was investigated with Beggar’s funnel plot, in which the standard error of log OR of each study was plotted against its OR [21]; (5) Publication bias was further assessed by the method of Egger’s linear regression test which could assess the relationship between effect size and variance differs between large and small studies [22]; (6) In this meta-analysis, subgroup analyses were used to better investigate possible reasons of between-study heterogeneity [23]. The subgroups are as following: geographical location (southeast and central China mainland, and Taiwan region), number of case (<100 vs. ≥100), source of control (population-based vs. hospital-based); (7) All analyses were performed using the software State version12.0 (StataCorp LP,College Station,Texas,USA), Review Manager 5.0 (Cochrane collaboration, http://www.cc-ims.net/RevMan/relnotes.htm). All the P values were two sided.
Results
1. Study Selection and Study Characteristics
We ultimately identified a total of 27 articles reporting the relationship between GST genetic polymorphisms and HCC risk by both Chinese and English database [24]–[50] (Figure 1). According to the inclusive and exclusive criteria, all articles were retrieved and carefully reviewed to assess the eligibility. The characteristics of the studies including 26 articles of GSTM1 (3712 cases and 6024 controls), 21 articles of GSTT1 (3378 cases and 5400 controls) and 12 articles of both GSTM1 and GSTT1 (1562 cases and 2537 controls) are shown in Table 1.
Figure 1. Flow chart of study selection.
Table 1. Characteristics of the studies related with the effects of GSTs genetic polymorphisms and HCC risk.
| No. | Study (ref.) | Region | Study time | Pathologic diagnosis | Source of controls | Case group | Control group | Null GSTM1/Group number | Null GSTT1/Group number | Dual Null/Group number | Overlapped (ref.) | |||||
| case | control | case | control | case | control | |||||||||||
| *1 | Hsieh LL 1996([24]) | Taiwan | 1990–1992 | ALL | NA | 46 male caseswith HBsAg (+) | 88 male controls with HBsAg (+) matchedon age | 25/46 | 47/88 | |||||||
| ^2 | Bian JC 1996([25]) | Zhejiang,etc. | NA | ALL | Population | 65 cases | 106 healthy controls | 44/65 | 50/106 | |||||||
| ^3 | Hu Y 1997([26]) | Jiangsu | NA | NA | Population | 45 cases | 147 healthy controls without consanguineous relationship | 37/45 | 75/147 | |||||||
| ^4 | Dong CH 1997([27]) | Hebei, etc. | NA | NA | Hospital | 110 cases | 112 controls | 62/110 | 50/112 | 63/110 | 42/112 | 36/110 | 20/112 | [58] | ||
| *5 | Dong CH 1998([28]) | Jiangsu | 1996 | NA | Population | 64 cases | 64 healthy controls, matched on age and sex | 29/56 | 24/58 | 33/56 | 23/58 | 21/56 | 9/58 | |||
| *6 | Yu MW 1999([29]) | Taiwan | 1988–1996 | PARTIAL | Population | 84 cases (81HBsAg (+)) | 375 controls (153 HBsAg (−) and 222 HBsAg (+) ), matched on age etc | 42/84 | 216/375 | 41/83 | 181/375 | [59]–[61] | ||||
| *7 | Bian JC 2000([30]) | Jiangsu, etc. | NA | ALL | Population | 63 cases(47male) | 88 healthy controls (67male), without consanguineous relationship | 36/63 | 37/88 | 8/63 | 33/88 | 1/63 | 16/88 | [62] | ||
| *8 | Ma Y 2001([31]) | Guangxi | NA | ALL | Population | 120 cases | 100 healthy controls without any tumors, matched on age and sex | 71/120 | 52/100 | |||||||
| ^9 | Wu HL 2000([32]) | Hunan | 1997–1999 | ALL | Population | 54 cases(46 male) | 136 healthy controls | 38/54 | 62/136 | |||||||
| ^10 | Zhu WC 2001([33]) | Guangdong | NA | ALL | Population | 52 cases | 100 healthy controls equally comparable in sex, age, birthplace and ethnicity | 34/52 | 41/100 | |||||||
| *11 | Sun CA 2001([34]) | Taiwan | 1991–1997 | PARTIAL | Population | 79 cases withHBsAg (+) | 149 controls with HBsAg (+), matched on age, sex, residential township etc | 26/69 | 77/128 | 30/67 | 77/128 | [63] | ||||
| ^12 | Liu CZ 2002([35]) | Shanghai, etc. | NA | ALL | Population | 84 cases | 144 healthy controls, equally comparable in age and birthplace, but not in sex | 56/84 | 69/144 | 34/84 | 36/144 | 23/84 | 19/144 | |||
| ^13 | Liu ZG 2003([36]) | Guangxi | NA | ALL | Population | 51 cases | 53 healthy controls without any tumors,equally comparable in age and sex | 28/51 | 18/53 | |||||||
| *14 | Yu MW 2003([37]) | Taiwan | 1997–2001 | PARTIAL | Population | 577 caseswith HBsAg (+) | 389 controls with HBsAg (+), matched on age and sex | 322/577 | 231/389 | 298/577 | 199/389 | 171/577 | 116/389 | [51] | ||
| *15 | McGlynn KA 2003([38]) | Jiangsu | 1992–1993 | PARTIAL | Population | 231 cases (73%HBsAg (+)) | 256 controls matched on age, sex and township of residence | OR (95% CI) = 0.83 (0.57, 1.21)§ | OR(95% CI) = 0.88 (0.59, 1.31)§ | |||||||
| ^16 | Li SP 2004([39]) | Jiangsu | 1998–2002 | NA | Population | 207 cases | 207 healthy controls, matched on sex, age and residence | 122/207 | 118/207 | 108/207 | 97/207 | |||||
| ^17 | He SJ 2004([40]) | Guangxi | 2001–2002 | ALL | Population | 105 HCC cases | 151 healthy controls equally comparable in age, sex, ethnicity | 68/105 | 77/151 | 43/105 | 50/151 | 30/105 | 31/151 | [64], [65], [66] | ||
| ^18 | Guo HY 2005([41]) | Henan | 1999–2002 | PARTIAL | Population | 95 HCC cases | 103 healthy controls equallycomparable in age, sex, residence | 67/95 | 52/103 | 58/95 | 45/103 | 39/95 | 21/103 | |||
| ^19 | Ma DL 2005([42]) | Guangxi | 2003–2004 | ALL | Population | 62 cases withHBsAg (+) | 73 controls with HBsAg (+), without any tumor,equally comparable in age and sex | 37/62 | 29/73 | 35/62 | 21/73 | |||||
| *20 | Long XD 2005([43]) | Guangxi | 2002–2003 | ALL | Hospital | 140 cases | 536 controls without any tumor,equally comparable in sex, age, ethnicity | 92/140 | 254/536 | 82/140 | 234/536 | 60/140 | 127/536 | [67] | ||
| *21 | Deng ZL 2005([44]) | Guangxi | 1998–2002 | ALL | Population | 181 cases | 360 controls without any tumor | 117/181 | 172/360 | 108/181 | 154/360 | 38.2% | 18.5% | [68], [69] | ||
| *22 | Long XD 2006([45]) | Guangxi | 2004–2005 | ALL | Population | 257 cases | 649 controls without clinical evidence of liver disease,matched on age, sex, ethnicityand HBV infection | 179/257 | 312/649 | 146/257 | 297/649 | |||||
| ^23 | Yang ZG 2009([46]) | Guangxi | 2002–2008 | ALL | Population | 100 cases | 60 healthy controls withouthepatitis virus infection, tumorsand AFP negative, equally comparable in age and sex | 59/100 | 41/60 | 33/100 | 11/60 | 22/100 | 2/60 | |||
| *24 | Kao CC 2010([47]) | Taiwan | 2006–2008 | ALL | Population | 102 cases | 386 healthy controls, matched onethnicity, sex and residential area | 54/102 | 211/386 | 51/102 | 200/386 | 31/102 | 104/386 | |||
| *25 | Wei YP 2012([48]) | Guangxi | NA | ALL | Hospital | 181 cases (78.5%HBsAg (+)) | 641 controls (9.8%HBsAg (+))without cancer disease, matched on age and sex | 118/181 | 305/641 | 104/181 | 276/641 | [70] | ||||
| ^26 | Tang YT 2012([49]) | Guangxi | 2008–2010 | ALL | Population | 150 malecases | 150 male healthy controls, equallycomparable in age | 76/150 | 77/150 | 63/150 | 68/150 | 30/150 | 32/150 | |||
| *27 | Ling CG 2012([50]) | NA | 2005–2007 | ALL | Population | 476 cases (54.7%HBsAg (+), 13.4%Anti-HCV (+)) | 481 controls (43.6%HBsAg (+), 2.5%Anti-HCV (+)), withoutmalignancy diseases etc., equally comparable in age and sex | 244/476 | 211/481 | 120/476 | 94/481 | |||||
ALL: HCC cases were confirmed by pathologic diagnosis; PARTIAL: part of HCC cases were confirmed by pathologic diagnosis; NA: relative data were not available in original studies;
Articles published in English;
Articles published in Chinese.
McGlynn et al. did not show genotype frequency of cases and controls, but presented OR with 95% CI;
Southeast regions in China mainland include Hebei, Shanghai, Jiangsu, Zhejiang, Anhui, Jiangxi, and Guangxi. Central regions in China mainland include Hunan and Henan.
2. Meta-analysis Results
2.1. GSTM1 null genotype with HCC risk
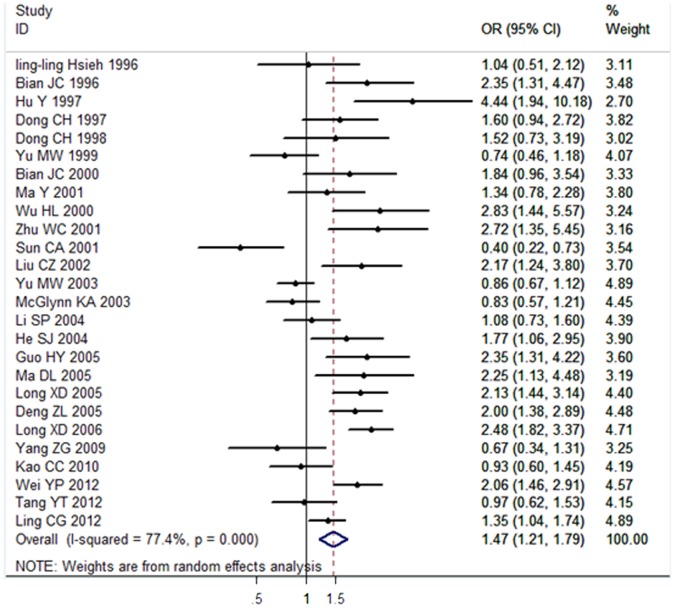
26 articles [24–35, and 37–50] including 3712 cases and 6024 controls were investigated in this study to evaluate the association between GSTM1 null genotype and HCC susceptibility. 12 articles were published in Chinese and 14 articles in English. Results obtained from a random-effects model showed a significant association between the GSTM1 null genotype and HCC risk in the Chinese population (OR = 1.47, 95% CI: 1.21 to 1.79, P<0.001). The forest plot was showed in Figure 2.
Figure 2. Association between GSTM1 null genotype and HCC risk analyzed by forest plot of meta-analysis.
The forest plots of pooled OR with 95% CI (Null genotype vs. Present genotype; OR = 1.47, 95% CI: 1.21 to 1.79; Random-effects model, P<0.001).
2.2. GSTT1 null genotype with HCC risk
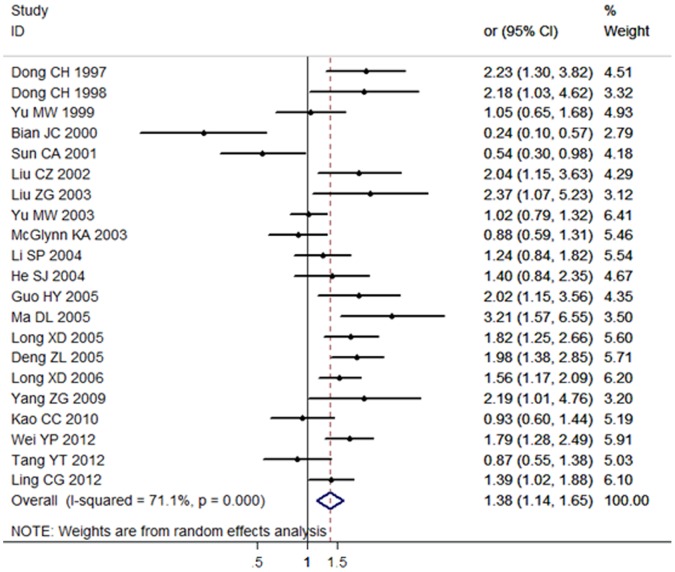
21 articles including 3378 cases and 5400 controls were used for the investigation of the association between GSTT1 null genotype and HCC susceptibility. 9 articles were published in Chinese and 12 articles were published in English. Results showed that the GSTM1 null genotype was significantly associated with HCC risk demonstrated by random-effects model in the Chinese population (OR = 1.38, 95% CI: 1.14 to 1.65, P<0.001). The forest plot was shown in Figure 3.
Figure 3. Association between GSTT1 null genotype and HCC risk analyzed by forest plot of meta-analysis.
The forest plots of pooled OR with 95% CI (Null genotype vs. Present genotype; OR = 1.38, 95% CI: 1.14 to 1.65; Random-effects model, P<0.001).
2.3. Dual-null genotype of GSTM1-GSTT1 with HCC risk
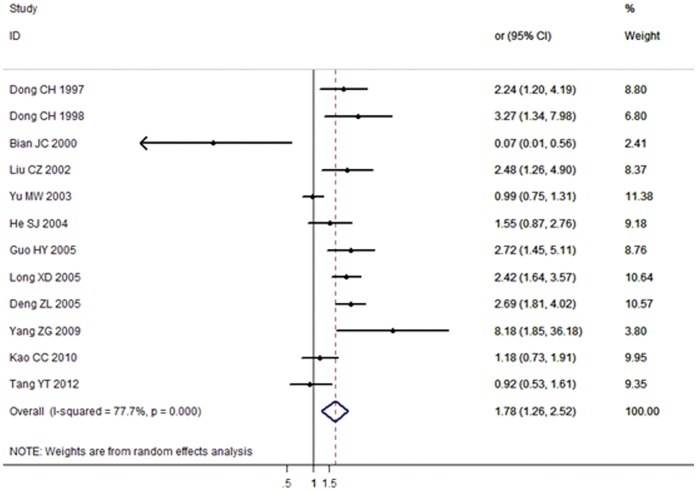
12 articles (6 articles in Chinese and 6 articles in English) including 1763 cases and 2537 controls were used to evaluate the relationship between GSTM1-GSTT1 null genotype and HCC susceptibility. Results indicated that dual-null genotype of GSTM1-GSTT1 also had a significant association with HCC risk in the Chinese population (OR = 1.79, 95% CI: 1.26 to 2.53, P<0.001). The forest plot was shown in Figure 4.
Figure 4. Association between GSTM1-GSTT1 dual-null genotype and HCC risk analyzed by forest plot of meta-analysis.
The forest plots of pooled OR with 95% CI (Dual-null genotype vs. Present genotype; OR = 1.79, 95% CI: 1.26 to 2.53; Random-effects model, P<0.001).
3. Subgroup Analysis
The substantial between-study heterogeneity of the three above analyses were observed (P values for GSTM1, GSTT1, and the interaction of GSTM1-GSTT1 were all less than 0.001, I2 values were 77.4%, 71.1%, and 77.7%, respectively). In this meta-analysis, subgroup analyses contained geographical location (southeast regions in China mainland, central regions in China mainland, and Taiwan region), case number (<100 vs. ≥100), source of control (population-based vs. hospital-based). The between-study heterogeneity showed that the major source of heterogeneity came from China mainland population. Association between GST genetic polymorphisms and HCC risk increase was significant in subgroup analyses of both southeast and central regions in China mainland population, but no significant in Taiwan population. Other subgroup analyses results were shown in Table 2, Table 3, and Table 4.
Table 2. Subgroup analysis of the association between GSTM1 null genotype and HCC risk.
| Polymorphism | Null vs. Present | No. of studies (cases/controls) | Odds ratio | M | Heterogeneity | PE | ||
| OR [95% CI] | POR | I2 (%) | PH | |||||
| GSTM1 | All studies | 26(3712/6024) | 1.47[1.21,1.79] | <0.001 | R | 77.4% | <0.001 | 0.367 |
| subgroup analyses by geographical location | ||||||||
| Southeast regions in mainland China | 18(2209/3938) | 1.69[1.38,2.07] | <0.001 | R | 67.0% | <0.001 | 0.805 | |
| Central regions in mainland China | 2(149/239) | 2.55[1.64,3.97] | <0.001 | F | 0.0% | 0.680 | @ | |
| Taiwan province | 5(878/1366) | 0.78[0.60,1.01] | 0.06 | F | 38.1% | 0.164 | 0.555 | |
| subgroup analyses by number of case | ||||||||
| <100 | 12(775/1546) | 1.59[1.33,1.90] | <0.001 | R | 77.8% | <0.001 | 0.031 | |
| ≥100 | 14(2937/4478) | 1.36[1.23,1.50] | <0.001 | R | 78.4% | <0.001 | 0.859 | |
| subgroup analyses by source of control | ||||||||
| population-based | 21(3133/4261) | 1.47[1.17,1.84] | <0.001 | R | 79.4% | <0.001 | 0.238 | |
| hospital-based | 4(533/1675) | 1.62[1.11,2.37] | 0.012 | R | 69.1% | 0.021 | 0.472 | |
M: model of meta-analysis; R: random-effects model; F: fixed-effects model. PH: P value of heterogeneity test. PE: P value of Egger’s test. POR: P<0.001 replace P = 0.000 and P less than 0.001. @: P values could not be calculated.
Table 3. Subgroup analysis of the association between GSTT1 null genotype and HCC risk.
| Polymorphism | Null vs. Present | No. of studies (cases/controls) | Odds ratio | M | Heterogeneity | PE | ||
| OR [95% CI] | POR | I2 (%) | PH | |||||
| GSTT1 | All studies | 21(3378/5400) | 1.38[1.14,1.65] | <0.001 | R | 71.1% | <0.001 | 0.795 |
| subgroup analyses by geographical location | ||||||||
| Southeast regions in mainland China | 16(2454/4019) | 1.51[1.35,1.69] | <0.001 | R | 67.1% | <0.001 | 0.952 | |
| Central regions in mainland China | 1(95/103) | 2.02[1.15,3.56] | 0.020 | F | @ | @ | @ | |
| Taiwan province | 4(829/1278) | 0.94[0.78,1.14] | 0.546 | F | 24.4% | 0.265 | 0.315 | |
| subgroup analyses by number of case | ||||||||
| <100 | 8(561/1022) | 1.34[0.78,2.28] | 0.258 | R | 81.8% | <0.001 | 0.961 | |
| ≥100 | 13(2817/4378) | 1.38[1.16,1.64] | 0.002 | R | 61.0% | <0.001 | 0.560 | |
| subgroup analyses by source of control | ||||||||
| population-based | 17(2845/3725) | 1.32[1.06,1.64] | <0.001 | R | 72.1% | <0.001 | 0.746 | |
| hospital-based | 4(533/1675) | 1.60[1.14,2.26] | 0.007 | R | 63.6% | 0.041 | 0.929 | |
M: model of meta-analysis; R: random-effects model; F: fixed-effects model. PH: P value of heterogeneity test. PE: P value of Egger’s test. POR: P<0.001 replace the P = 0.000 and the P less than 0.001. @: P values could not be calculated.
Table 4. Subgroup analysis of the association between GSTM1-GSTT1 null genotype and HCC risk.
| Polymorphism | Null vs. Present | No. of studies (cases/controls) | Odds ratio | M | Heterogeneity | PE | ||||
| OR [95% CI] | POR | I2 (%) | PH | |||||||
| GSTM1-GSTT1 | All studies | 12(1763/2537) | 1.78[1.26,2.52] | <0.001 | R | 77.7% | <0.001 | 0.535 | ||
| subgroup analyses by geographical location | ||||||||||
| Southeast regions in mainland China | 9(989/1659) | 1.98[1.32,2.95] | <0.001 | R | 70.3% | <0.001 | 0.497 | |||
| Central regions in mainland China | 1(95/103) | 2.72[1.45,5.11] | 0.002 | F | @ | @ | @ | |||
| Taiwan province | 2(679/775) | 1.04[0.81,1.32] | 0.770 | F | 0.0% | 0.536 | @ | |||
| subgroup analyses by number of case | ||||||||||
| <100 | 4(298/393) | 1.73[0.70,4.28] | 0.235 | R | 75.9% | 0.001 | 0.115 | |||
| ≥100 | 8(1465/2144) | 1.70[1.17,2.48] | 0.006 | R | 78.8% | 0.001 | 0.263 | |||
| subgroup analyses by source of control | ||||||||||
| population-based | 9(1411/1503) | 1.75[1.09,2.80] | 0.020 | R | 81.0% | 0.001 | 0.531 | |||
| hospital-based | 3(352/1034) | 1.86[1.16,2.97] | 0.010 | R | 63.8% | 0.063 | 0.856 | |||
M: model of meta-analysis; R: random-effects model; F: fixed-effects model. PH: P value of heterogeneity test. PE: P value of Egger’s test. POR: P<0.001 replace the P = 0.000 and the P less than 0.001. @: P values could not be calculated.
4. Sensitivity and Heterogeneity Analysis
Sensitivity analysis was performed by sequential excluding one article each time. The significance of all ORs was not changed. We used Galbraith plot to omit some possible major sources of heterogeneous articles. The results were showed in Figure 5. In Figure 5A, we found more than 6 articles (No. 3, 6, 11, 13, 14, and 21) spotted by Galbraith plot. However, it might cause some biases by excluding those articles as the sources of heterogeneity. So we didn’t reduce the obvious between-study heterogeneity in the analyses on the GSTM1 polymorphisms. In Figure 5B, 3 articles (No. 4, 5 and 13) were obviously spotted as the outliers and the possible sources of heterogeneity in the analysis pooled of total available studies, but another 3 articles (No. 8, 9 and 15) the outliers were not reduced because it could cause some biases. After adjustment, the association between GSTT1 polymorphisms and HCC risk was increased (OR = 1.45, 95% CI: 1.24 to 1.69, P<0.001, random-effects model). Galbraith plots (Figure 5C) spotted 5 articles (No. 3, 5, 8, 9 and 10) as the possible sources of heterogeneity, but only 3 articles (No. 3, 5 and 9) were omitted for the obvious between-study heterogeneity in the analyses on the GSTM1-GSTT1 polymorphisms. The adjusted OR and 95% CI between GSTM1/GSTM1-GSTT1 polymorphisms and HCC risk was significantly increased although heterogeneity (I2 GSTT1 = 56.1%, PGSTT1 = 0.002; I2 GSTM1-GSTT1 = 59.0%, PGSTM1-GSTT1 = 0.012) still existed. These results were shown in Table 5 and Table 6.
Figure 5. Galbraith plot of association between GST polymorphisms and HCC risk.
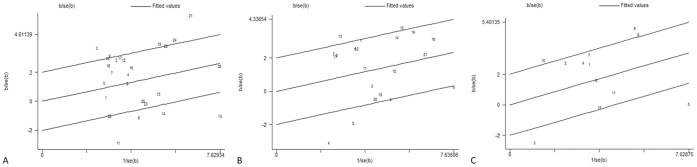
Each figure represents a unique article in this meta-analysis. The figures outside the three lines are spotted as the outliers and the possible sources of heterogeneity in the analysis pooled of total available studies. (A) Galbraith plot identifies the outliers from 26 studies about GSTM1 polymorphisms and HCC risk. (B) Galbraith plot identifies the outliers from 21 studies about GSTT1 polymorphisms and HCC risk. (C) Galbraith plot identifies the outliers from 12 studies about GSTM1-GSTT1 polymorphisms and HCC risk.
Table 5. Subgroup analysis of $the adjusted association between GSTT1 null genotype and HCC risk.
| Polymorphism | Null VS. Present | No. of studies(cases/controls) | Odds ratio | M | Heterogeneity | PE | ||
| OR [95% CI] | POR | I2 (%) | PH | |||||
| GSTT1 | All studies | 18(3186/5111) | 1.45[1.24,1.69] | <0.001 | R | 56.1% | 0.002 | 0.142 |
Table 6. Subgroup analysis of $the adjusted association between GSTM1-GSTT1 null genotype and HCC risk.
| Polymorphism | Null vs. Present | No. of studies (cases/controls) | Odds ratio | M | Heterogeneity | PE | ||
| OR [95% CI] | POR | I2 (%) | PH | |||||
| GSTM1-GSTT1 | All studies | 9(942/1674) | 1.98[1.43, 2.74] | <0.001 | R | 59.0% | 0.012 | 0.236 |
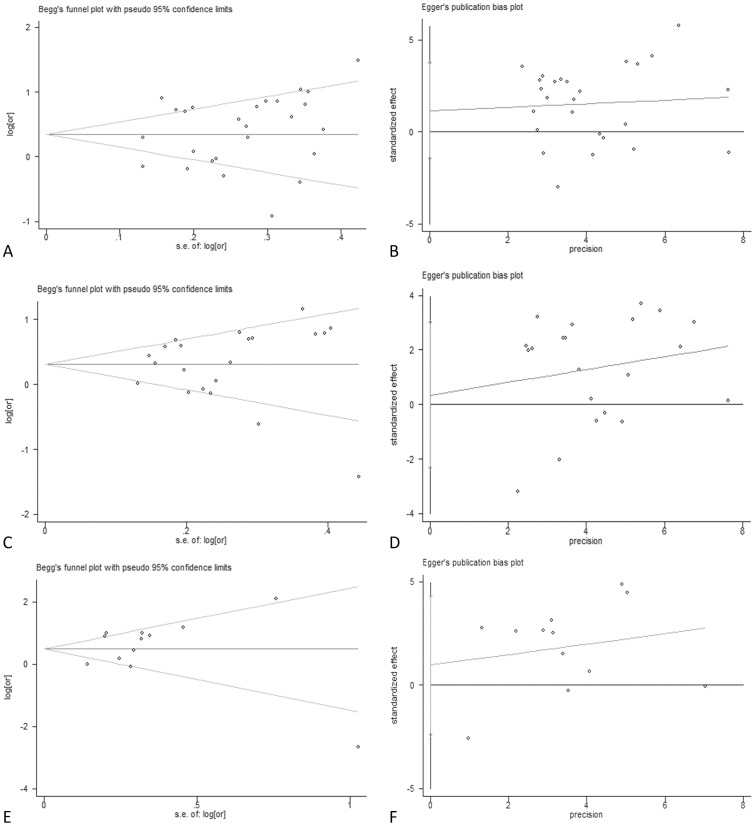
5. Potential Publication Bias
Beggar’s funnel plots and Egger’s publication bias plots were used to assess the potential publication bias for GSTM1, GSTT1, and dual-null genotype of GSTM1-GSTT1 (Figure 6). No publication bias was detected by Egger’s test (PE = 0.367 for GSTM1, PE = 0.795 for GSTT1 and PE = 0.64 for dual-null genotype of GSTM1-GSTT1).
Figure 6. Beggar’s test and Egger’s test of GST polymorphisms and HCC risk.
Beggar’s funnel plot is used to detect potential publication bias in which a symmetric funnel shape means no publication bias. Egger’s linear regression test is used to quantify the potential presence of publication bias. Both Beggar’s test and Egger’s test show that no publication bias has been found from 26 inclusive studies about the association between GSTM1 polymorphisms and HCC risk (A and B), 21 inclusive studies about the association between GSTT1 polymorphisms and HCC risk (C and D), and 12 inclusive studies about the association between dual-null genotype of GSTM1-GSTT1 and HCC risk polymorphisms and HCC risk (E and F).
Discussion
The association between GST genetic polymorphisms and HCC risk are inconsistent according to the present research results. This may be caused by several reasons. Improper matching or insufficient case and control numbers used in the studies are all possible reasons. One meta-analysis [10] published in 2009 with the association between GST genetic polymorphisms and HCC risk didn’t cover all conclusive articles published in Chinese and English databases. In this meta-analysis paper, overlapped data was found in two adopted studies [37], [51] (two different articles with different case and control numbers written by the same research group). Another two adopted studies [52], [53] in this meta-analysis didn’t match properly for the cases (HBV carried) and controls (HBV negative). The other meta-analysis [11] published in 2012 about Asian population included a study [38] with unclear case and control numbers. In addition, some more studies [47]–[50] related with the association between GST genetic polymorphisms and HCC risk have emerged since these two meta-analysis papers were published.
To evaluate the association of GST genetic polymorphisms and susceptibility to HCC in the Chinese population, we performed an updated systematic meta-analysis. In this study, 27 articles (3781 patients and 6104 controls) were selected from Chinese and English databases. 26 studies (3712 cases and 6024 controls) out of the 27 articles were used for investigation of the relationship between GSTM1 null genotype and HCC susceptibility. 21 studies (3378 cases and 5400 controls) out of the 27 articles were used to evaluate the relationship between GSTT1 null genotype and HCC susceptibility. 12 studies (1763 cases and 2537 controls) were applied for evaluation for the GSTM1-GSTT1 gene. Random-effects model of meta-analysis shows significant associations of polymorphisms of GSTM1 null gene (OR = 1.47, 95% CI: 1.21 to 1.79, P<0.001), GSTT1 null gene (OR = 1.38, 95% CI: 1.14 to 1.65, P<0.001), and GSTM1-GSTT1 dual null gene (OR = 1.79, 95% CI: 1.26 to 2.53, P<0.001), respectively, with HCC risk in the Chinese population. Subgroup analyses on GSTM1 null gene indicate that geographical location (China mainland, but not in Taiwan region), case numbers and source of controls are significantly associated with HCC risk. Results of subgroup analyses on GSTT1 null gene and GSTM1-GSTT1 dual null gene indicate that geographical location (China mainland, but not in Taiwan region), case numbers (≥100, but not <100) and source of controls are also significantly associated with HCC risk. Reasons for inconsistent in conclusions between China mainland and Taiwan region may be caused by environmental factors. Moreover, limited investigative numbers of the case-control/followed up studies from Taiwan region may result in difficulty for getting stable risk estimation, though these investigations own low between-study heterogeneity. In addition, studies with case number less than 100 may have effects on drawing a proper estimation for the association between GST genetic polymorphisms and HCC risk. Therefore, further well design case-control/followed-up studies, especially with a larger case number, are necessary to provide better evidences for the evaluation. Heterogeneity analysis is a key part of meta-analysis. Q statistic test (Cochran’s Q statistic) and I2 statistic test are commonly used to test and quantify the between-study heterogeneity. The major source of heterogeneity in the China mainland population detected in the subgroup analysis might come from the environmental difference which could affect their sensitivity to particular genomic variants. In this meta-analysis, Galbraith plot was performed for identifying the articles with possible heterogeneity. However, in the analyses on the GSTM1 polymorphism and HCC risk, we kept several articles with obvious between-study heterogeneity because too many articles omitting could cause some biases. For the association of GSTT1 null gene and HCC risk, we deleted 3 articles [30], [34], [42] which were obviously spotted as the outliers with major source of between-heterogeneity, and same procedures were done for GSTM1-GSTT1 gene (3 article deletion [30], [37], [44]). Regretfully, the between-heterogeneity didn’t decrease significantly even if the adjustment was done in both GSTT1 and GSTM1-GSTT1 genetic polymorphisms (I2 GSTT1 = 56.1%, PGSTT1 = 0.002; I2 GSTM1-GSTT1 = 59.0%, PGSTM1-GSTT1 = 0.012). Therefore, we applied the random-effects model to evaluate the pooled OR for GSTT1 and GSTM1-GSTT1 genes, respectively. After the above adjustments, the associations were increased between GSTT1 and GSTM1-GSTT1 polymorphisms and HCC risk (ORGSTT1 = 1.45, 95% CI: 1.24 to 1.69; ORGSTM1-GSTT1 = 1.98, 95% CI: 1.43 to 2.74). In this study, Beggar’s funnel plots and Egger’s linear regression test were applied to assess the potential publication bias. No publication bias was detected (PE = 0.367 for GSTM1, PE = 0.795 for GSTT1 and PE = 0.64 for dual-null genotype of GSTM1-GSTT1, Egger’s linear regression test).
Research evidences suggest that GST genetic polymorphisms are associated with the susceptibility to several carcinomas. Takahiko Katoh et al. [54] showed the GSTM1 null genotype might be associated with susceptibility to gastric adenocarcinoma and distal colorectal adenocarcinoma in Japanese population. Wang J et al. [55] found that the combination of GSTM1 null and GSTP1 Val was significantly associated with an elevated lung adenocarcinoma risk (OR = 2.4, 95% CI: 1.1 to 5.1). Helzlsouer K J et al. [56] considered that genetic variability in members of the GST gene family might be associated with an increased susceptibility to breast cancer (OR = 3.77, 95% CI: 1.10 to 12.88). Compared to the control group value of 41.8%, Zhong S et al. [57] found a significant excess of 56.1% GSTM1 gene null individuals in colorectal cancer group. Our meta-analysis results demonstrate that there is an association between GST genetic polymorphisms and susceptibility to HCC in the Chinese population. Thus, further epidemiological and molecular biological studies are necessary to clarify the role of GST genetic polymorphisms in HCC and other carcinomas.
Nevertheless, there were several limitations to this meta-analysis. (1) Observational studies were susceptible to various biases such as selection bias. Due to some studies without clear explanation for the pathologic diagnostic results of all/part subjects (Table 1), therefore, some selection bias might be unavoidable. (2) In some studies, participants in control groups stemmed from hospital-based population might not fully represent the population-based controls, which could distort the results (Table 1). (3) The conclusions draw from subgroup analysis might be limited due to a low statistic power from the small sample size. (4) Each study had its own inclusive criteria. For example, some studies selected from HbsAg positive population, while others selected the common people or healthy population. Due to these reasons, some bias might bring influence on the results. (5) Not only genetic polymorphisms but other factors such as alcohol consumption, AFB1 status, and chronic infection of HBV/HCV might also play vital roles in the development of HCC. Owning to the lack of sufficient data, gene-environment interactive functions were not evaluated in this meta-analysis, which might also have an influence on the precision of the conclusion.
In summary, our results suggest GST genetic polymorphisms are associated with the increased risk of HCC in the Chinese population. To further evaluate gene-to-gene and gene-to-environment combined effects on GST genetic polymorphisms and HCC,both large scale multicenter epidemiological studies in total population and/or selected population with different environmental background are urgently needed.
Acknowledgments
The excellent assistance of Linda Bowman, Dechuan Kong and Jian Chen in the preparation of this article is greatly appreciated. This work was partly supported by K.C. Wong Magna Fund in Ningbo University.
Funding Statement
Funding provided by grant number: the National Nature Science Foundation of China (Grant No. 81273111), the Foundations of Innovative Research Team of Educational Commission of Zhejiang Province (T200907), the Nature Science Foundation of Ningbo city (Grant No. 2012A610185), the Ningbo Scientific Projects (2012C5019 and SZX11073), the Scientific Innovation Team Project of Ningbo (No. 2011B82014), Innovative Research Team of Ningbo (2009B21002). This work was partly supported by K. C. Wong Magna Fund in Ningbo University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cabibbo G, Maida M, Genco C, Antonucci M, Cammà C (2012) Causes of and Prevention Strategies for Hepatocellular Carcinoma; 2012. Elsevier. pp. 374–383. [DOI] [PubMed]
- 2.Aghemo A, Colombo M (2012) Hepatocellular carcinoma in chronic hepatitis C: from bench to bedside; Springer. pp. 1–10. [DOI] [PubMed]
- 3. French SW, Lee J, Zhong J, Morgan TR, Buslon V, et al. (2012) Alcoholic liver disease-Hepatocellular carcinoma transformation. Journal of Gastrointestinal Oncology 3: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jemal A, Siegel R, Ward E, Hao Y, Xu J, et al. (2009) Cancer statistics, 2009. CA: a cancer journal for clinicians 59: 225–249. [DOI] [PubMed] [Google Scholar]
- 5. Jemal A (2010) Center MM, DeSantis C, Ward EM (2010) Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiology Biomarkers & Prevention 19: 1893–1907. [DOI] [PubMed] [Google Scholar]
- 6.Annick Buendia M. Genetics of hepatocellular carcinoma; 2000. Elsevier. 185–200. [DOI] [PubMed]
- 7. Thorgeirsson SS, Grisham JW (2002) Molecular pathogenesis of human hepatocellular carcinoma. Nature genetics 31: 339–346. [DOI] [PubMed] [Google Scholar]
- 8. Farazi PA, DePinho RA (2006) Hepatocellular carcinoma pathogenesis: from genes to environment. Nature Reviews Cancer 6: 674–687. [DOI] [PubMed] [Google Scholar]
- 9. Wang B, Huang G, Wang D, Li A, Xu Z, et al. (2010) Null genotypes of GSTM1 and GSTT1 contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. Journal of Hepatology 53: 508–518. [DOI] [PubMed] [Google Scholar]
- 10. Yu L, Wang CY, Xi B, Sun L, Wang RQ, et al. (2011) GST polymorphisms are associated with hepatocellular carcinoma risk in Chinese population. World Journal of Gastroenterology: WJG 17: 3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J, Ma L, Peng NF, Wang SJ, Li LQ (2012) A meta-analysis of the relationship between glutathione S-transferases gene polymorphism and hepatocellular carcinoma in Asian population. Molecular Biology Reports: 1–11. [DOI] [PubMed]
- 12. Beasley RP, Lin CC, Hwang LY, Chien CS (1981) Hepatocellular carcinoma and hepatitis B virus. The Lancet 318: 1129–1133. [DOI] [PubMed] [Google Scholar]
- 13. McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, et al. (1995) Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proceedings of the National Academy of Sciences 92: 2384–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Udomsinprasert R, Pongjaroenkit S, Wongsantichon J, Oakley AJ, Prapanthadara L, et al. (2005) Identification, characterization and structure of a new Delta class glutathione transferase isoenzyme. Biochemical Journal 388: 763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Atkinson HJ, Babbitt PC (2009) Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry 48: 11108–11116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayes JD, Flanagan JU, Jowsey IR (2005) Glutathione transferases. Annu Rev Pharmacol Toxicol 45: 51–88. [DOI] [PubMed] [Google Scholar]
- 17. Strange RC, Spiteri MA, Ramachandran S, Fryer AA (2001) Glutathione-S-transferase family of enzymes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 482: 21–26. [DOI] [PubMed] [Google Scholar]
- 18. Higgins J, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Statistics in Medicine 21: 1539–1558. [DOI] [PubMed] [Google Scholar]
- 19. Huedo-Medina TB, Sánchez-Meca J, Marín-Martínez F, Botella J (2006) Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychological methods 11: 193. [DOI] [PubMed] [Google Scholar]
- 20. Hedges LV, Vevea JL (1998) Fixed-and random-effects models in meta-analysis. Psychological methods 3: 486. [Google Scholar]
- 21.Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics: 1088–1101. [PubMed]
- 22. Egger M, Smith GD, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higgins J, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsieh LL, Huang RC, Yu MW, Chen CJ, Liaw YF (1996) L-myc, GST Ml genetic polymorphism and hepatocellular carcinoma risk among chronic hepatitis B carriers. Cancer Letters 103: 171–176. [DOI] [PubMed] [Google Scholar]
- 25. Bian JC, Wang JB, Wu Y (1996) Relationship between GSTM1 NULL genotype and genetic susceptibility to primary hepatocellular carcinoma. Chinese Journal of Medical Genetics 13: 353–353. [Google Scholar]
- 26. Hu Y, Sheng FM (1997) Association between GSTM1 gene polymorphism of primary hepatocellular carcinoma and mutation of p53 codon 249. Chinese Journal of Medical Genetics 14: 76–78. [Google Scholar]
- 27. Dong CH, Yu SC, Chen GC, Zhao DM, Fu YP (1997) Polymorphisms of GSTT1 and M1 Genotypes and Their Effects on Elevated Aflatoxin Exposure and Increased Risk of Hepatocellular Carcinoma. Zhong Liu Fang Zhi Yan Jiu 24: 327–329. [Google Scholar]
- 28. Dong CH, Yu SZ, Chen GC, Zhao DM, Hu Y (1998) Association of polymorphisms of glutathione S-transferase M1 and T1 genotypes with elevated aflatoxin and increased risk of primary liver cancer. World Chinese Journal of Digestology 6: 463–466. [Google Scholar]
- 29. Yu MW, Chiu YH, Chiang YC, Chen CH, Lee TH, et al. (1999) Plasma carotenoids, glutathione S-transferase M1 and T1 genetic polymorphisms, and risk of hepatocellular carcinoma: Independent and interactive effects. American Journal of Epidemiology 149: 621–629. [DOI] [PubMed] [Google Scholar]
- 30. Bian JC, Shen FM, Shen L, Wang TR, Wang XH, et al. (2000) Susceptibility to hepatocellular carcinoma associated with null genotypes of GSTM1 and GSTT1. World Journal of Gastroenterology 6: 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma Y, Deng ZL, Wei YP (2001) Study of the deletion mutation of glutathione S transferase M1 gene and its role in susceptibility to hepatocellular carcinoma. Chinese Journal of Cancer Research 13: 176–178. [Google Scholar]
- 32. Wu HL, Cheng ML, Zhang RI (2000) Relationship Between GSTM1 Gene Polymorphism and Genetic Susceptibility to Primary Hepatocellular Carcinoma. The Practical Journal of Cancer 15: 463–465. [Google Scholar]
- 33. Zhu WC, Cheng Q, Luo CL, Chu XW, Wu M (2001) Relationship Study Between Gene Polymorphism of CYP1A1,GSTM1 and Genetic Susceptibility of Primary Hepatocellar Carcinoma. China J Cancer Prev Treat 8: 572–574. [Google Scholar]
- 34. Sun CA, Wang LY, Chen CJ, Lu SN, You SL, et al. (2001) Genetic polymorphisms of glutathione S-transferases M1 and T1 associated with susceptibility to aflatoxin-related hepatocarcinogenesis among chronic hepatitis B carriers: A nested case-control study in Taiwan. Carcinogenesis 22: 1289–1294. [DOI] [PubMed] [Google Scholar]
- 35. Liu CZ, Bian JC (2002) Genetic Polymorphism of Glutathione S-transferase M 1,T1,P1on Susceptibility Hepatocellular Carcinoma. China Public Health 18: 935–936. [Google Scholar]
- 36. Liu ZG, Wei YP, Ma Y, ZL D (2003) Population with GSTT1 gene deletion and the relationship to hepatocellular carcinoma from guangxi. Journal of Guangxi Medical University 2: 002. [Google Scholar]
- 37. Yu MW, Yang SY, Pan IJ, Lin CL, Liu CJ, et al. (2003) Polymorphisms in XRCC1 and glutathione S-transferase genes and hepatitis B-related hepatocellular carcinoma. Journal of the National Cancer Institute 95: 1485–1488. [DOI] [PubMed] [Google Scholar]
- 38. McGlynn KA, Hunter K, LeVoyer T, Roush J, Wise P, et al. (2003) Susceptibility to aflatoxin B1-related primary hepatocellular carcinoma in mice and humans. Cancer Research 63: 4594–4601. [PubMed] [Google Scholar]
- 39. Li SP, Wu JZ, Ding JH, Gao CM, Cao HX, et al. (2004) Impact of Genetic Polymorphism of Glutathione S-transferase T1,M1 on the Risk of Primary Hepatocellular Carcinoma in Alcohol Drinkers. The Practical Journal of Cancer 19: 229–232. [Google Scholar]
- 40. He SJ, Tan JR, Gu YY, Zhong WG, Su SG (2004) Relationship between Polymorphisms of Phase II Metabolic Genes and the Susceptibility to Hepatocellular Carcinoma in Guangxi. The Practical Journal of Cancer 19: 460–462. [Google Scholar]
- 41. Guo HY, Bian JC, Jiang F, Wang QM, Zhang ZM, et al. (2005) The null genotypes of GSTM1 and GSTT1 and the genetic susceptibility of primary liver cancer in Luoyang,China. Tumor Jan 25: 58–61. [Google Scholar]
- 42. Ma DL, Chen YX, Li Y, Zhao HT, Xie XM, et al. (2005) Glutathione-S-transferase M1 and T1 polymorphisms and susceptibility to liver cancer in hepatitis B surface antigen positive population. Medical Journal of the Chinese People’s Armed Police 27: 656–657. [Google Scholar]
- 43. Long XD, Ma Y, Wei YP, Deng ZL (2005) Study on the detoxication gene gstM1-gstT1-null and susceptibility to aflatoxin B1 related hepatocellular carcinoma in Guangxi. Chinese Journal of Epidemiology 26: 777–781. [PubMed] [Google Scholar]
- 44. Deng ZL, Wei YP, Ma Y (2005) Polymorphism of glutathione S-transferase mu 1 and theta 1 genes and hepatocellular carcinoma in southern Guangxi, China. World Journal of Gastroenterology 11: 272–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Long XD, Ma Y, Wei YP, Deng ZL (2006) The polymorphisms of GSTM1, GSTT1, HYL1*2, and XRCC1, and aflatoxin B1-related hepatocellular carcinoma in Guangxi population, China. Hepatology Research 36: 48–55. [DOI] [PubMed] [Google Scholar]
- 46. Yang ZG, Xie YA, Kuang ZP, Luo XL, Zhang WM, et al. (2009) Relationship between genetic polymorphisms of glutathione-s-transferase M1,T1 genes and susceptibility to hepatocellular carcinoma in population of Fusui District of Guangxi Zhuang Autonomous Region. Chinese Journal of Cancer Prevention and Treatment 16: 970–973. [Google Scholar]
- 47. Kao CC, Chen MUK, Kuo WUH, Chen TY, Su SC, et al. (2010) Influence of glutathione-S-transferase theta (GSTT1) and mu (GSTM1) gene polymorphisms on the susceptibility of hepatocellular carcinoma in Taiwan. Journal of Surgical Oncology 102: 301–307. [DOI] [PubMed] [Google Scholar]
- 48. Wei Y, Long X, Liu Z, Ma Y, Deng Z (2012) Genetic polymorphism of glutathione-S-transferase M1 and T1 in associated with carcinogenesis of hepatocellular carcinoma and nasopharyngeal carcinoma. Chinese-German Journal of Clinical Oncology 11: 138–141. [Google Scholar]
- 49.Tang YT, Li XP, Liu TQ, Yang JR, Luo JQ, et al.. (2012) A study of genetic polymorphisms of Glutathione S-transferase in patients with hepatocellular carcinoma. Chin J Lab Diagn 16.
- 50. Li CG, Zhao ZM, Hu MG, et al. (2012) Predictive Role of Glutathione-S-transferase Gene Polymorphisms in Risk and Prognosis of Hepatocellular Carcinoma. Asian Pacific Journal of Cancer Prevention 13: 3247–3252. [DOI] [PubMed] [Google Scholar]
- 51. Chen CC, Yang SY, Liu CJ, Lin CL, Liaw YF, et al. (2005) Association of cytokine and DNA repair gene polymorphisms with hepatitis B-related hepatocellular carcinoma. International Journal of Epidemiology 34: 1310–1318. [DOI] [PubMed] [Google Scholar]
- 52. Zhang YC, Deng CS, Zhu YQ (2005) Study on genetic polymorphisms of xenobiotica metabolizing enzymes in hepatitis B virus- associated hepatic disease. Wenzhou Yi Xue Yuan Xue Bao 35: 464–467. [Google Scholar]
- 53. Zhu MH, Chen XH, Zhou LF (2005) Association of genetic polymorphisms in glutathione S-transferases M1 with hepatitis beta-related hepatocellular carcinoma. Journal of Zhejiang University (Medical Sciences) 34: 126–130. [DOI] [PubMed] [Google Scholar]
- 54. Katoh T, Nagata N, Kuroda Y, Itoh H, Kawahara A, et al. (1996) Glutathione S-transferase M1 (GSTM1) and T1 (GSTT1) genetic polymorphism and susceptibility to gastric and colorectal adenocarcinoma. Carcinogenesis 17: 1855–1859. [DOI] [PubMed] [Google Scholar]
- 55. Wang J, Deng Y, Cheng J, Ding J, Tokudome S (2003) GST genetic polymorphisms and lung adenocarcinoma susceptibility in a Chinese population. Cancer Letters 201: 185. [DOI] [PubMed] [Google Scholar]
- 56. Helzlsouer KJ, Huang HY, Hoffman S, Alberg AJ, Comstock GW, et al. (1998) Association between glutathione S-transferase M1, P1, and T1 genetic polymorphisms and development of breast cancer. Journal of the National Cancer Institute 90: 512–518. [DOI] [PubMed] [Google Scholar]
- 57. Zhong S, Wyllie A, Barnes D, Wolf C, Spurr N (1993) Relationship between the GSTM1 genetic polymorphism and susceptibility to bladder, breast and colon cancer. Carcinogenesis 14: 1821–1824. [DOI] [PubMed] [Google Scholar]
- 58. Deng CH, Zi XL (1997) Relationship between Deletion of Slutathion S-trausferase Gene and Susceptibility to Primary Hepatocellular Carcinoma. Chinese Journal of Public Health 16: 141–142. [Google Scholar]
- 59. Yu MW, Gladek-Yarborough A, Chiamprasert S, Santella RM, Liaw YF, et al. (1995) Cytochrome P450 2E1 and glutathione S-transferase M1 polymorphisms and susceptibility to hepatocellular carcinoma. Gastroenterology 109: 1266–1273. [DOI] [PubMed] [Google Scholar]
- 60. Chen CJ, Yu MW, Liaw YF, Wang LW, Chiamprasert S, et al. (1996) Chronic hepatitis B carriers with null genotypes of glutathione S-transferase M1 and T1 polymorphisms who are exposed to aflatoxin are at increased risk of hepatocellular carcinoma. American Journal of Human Genetics 59: 128. [PMC free article] [PubMed] [Google Scholar]
- 61. Yu MW, Lien JP, Chiu YH, Santella RM, Liaw YF, et al. (1997) Effect of aflatoxin metabolism and DNA adduct formation on hepatocellular carcinoma among chronic hepatitis B carriers in Taiwan. Journal of Hepatology 27: 320–330. [DOI] [PubMed] [Google Scholar]
- 62. Bian J, Shen F, Wang J, Chen G, Zhang B, et al. (1999) The mutation of deletion for glutathione S-transferase M1 gene in the tissue of hepatocellular carcinoma. Chinese Journal of Medical Genetics 16: 171–173. [PubMed] [Google Scholar]
- 63. Sun C, Wu D, Wang L, Chen C, You S, et al. (2002) Determinants of formation of aflatoxin-albumin adducts: a seven-township study in Taiwan. British Journal of Cancer 87: 966–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. He S (2005) the analysis of GSTM1,GSTT1 gene polymorphism in HCC. Joural of Guangxi Medical University 22: 875–877. [Google Scholar]
- 65. He SJ, Gu YH, M ZH (2007) Effect of interaction between smoking, alcohol drinking and polymorphisms of phase II metabolic genes on the susceptibility of hepatocellular carcinoma. Basic Medical Sciences and Clinics 27: 666–669. [Google Scholar]
- 66.He SJ, Gu YH, M ZH (2008) The association between GSTM1 gene polymorphism and wine,tobacco habit and susceptibility in primary liver cancer. Journal of Guangxi Medical Unversity 25.
- 67. Long X, Ma Y, Wei Y, Deng ZL (2005) A study about the association of detoxication gene GSTM1 polymorphism and the susceptibility to aflatoxin B1-related hepatocellular carcinoma. Chinese Journal of Hepatology 13: 668. [PubMed] [Google Scholar]
- 68. Deng Z, Wei Y, Ma Y (2001) Glutathione-S-transferase M1 genotype in patients with hepatocellular carcinoma. Chinese Journal of Oncology 23: 477–479. [PubMed] [Google Scholar]
- 69. Wei YP, Ma Y, Deng ZL (2004) Genetic polymorphisms of Glutathione S-transferase M1 and T1 and the risk of hepatocellular carcinoma. Tumor 23: 464–466. [Google Scholar]
- 70. Wei YP, Long XD, Liu CZ, Ma Y, Deng ZL (2010) Genetic polymorphism of glutathione-S-transferase M1 and T1 in associated with carcinogenesis of hepatocellular carcinoma and nasopharyngeal carcinoma. Cancer Research On Prevention And Treatment 37: 1162–1165. [Google Scholar]