Abstract
Background
The 677C>T polymorphism of methylenetetrahydrofolate reductase (MTHFR) gene is considered to have a significant effect on colorectal cancer susceptibility, but the results are inconsistent. In order to investigate the association between the MTHFR 677C>T polymorphism and the risk of colorectal cancer, a meta-analysis was held based on 71 published studies.
Methods
Eligible studies were identified through searching the MEDLINE, EMBASE, PubMed, Web of Science, Chinese Biomedical Literature database (CBM) and CNKI database. Odds ratios (OR) and 95% confidence intervals (CIs) were used to assess the association. The statistical heterogeneity across studies was examined with x2-based Q-test. Begg's and Egger's test were also carried out to evaluate publication bias. Sensitive and subgroup analysis were also held in this meta-analysis.
Results
Overall, 71 publications including 31,572 cases and 44,066 controls were identified. The MTHFR 677 C>T variant genotypes are significantly associated with increased risk of colorectal cancer. In the stratified analysis by ethnicity, significantly increased risks were also found among Caucasians for CC vs TT (OR = 1.076; 95%CI = 1.008–1.150; I2 = 52.3%), CT vs TT (OR = 1.102; 95%CI = 1.032–1.177; I2 = 51.4%) and dominant model (OR = 1.086; 95%CI = 1.021–1.156; I2 = 53.6%). Asians for CC vs TT (OR = 1.226; 95%CI = 1.116–1.346; I2 = 55.3%), CT vs TT (OR = 1.180; 95%CI = 1.079–1.291; I2 = 36.2%), recessive (OR = 1.069; 95%CI = 1.003-1.140; I2 = 30.9%) and dominant model (OR = 1.198; 95%CI = 1.101-1.303; I2 = 52.4%), and Mixed populations for CT vs TT (OR = 1.142; 95%CI = 1.005-1.296; I2 = 0.0%). However, no associations were found in Africans for all genetic models.
Conclusion
This meta-analysis suggests that the MTHFR 677C>T polymorphism increases the risk for developing colorectal cancer, while there is no association among Africans found in subgroup analysis by ethnicity.
Introduction
Colorectal cancer (CRC) is a worldwide public health problem,which is the third most commonly diagnosed cancer in males and the second in females with over 1.2 million new CRC patients and 608,700 deaths occurred in the world [1], [2]. Previous studies demonstrated that colorectal carcinogenesis is a complicated multi-step progress involving changes of many oncogenes and tumor suppressor genes induced by the interaction of many factors. Simultaneously, other factors such as alcohol, low methionine, low folate diets, heavy drink, smoking and environmental carcinogenic agents are assumed the possible risk factors. Whereas, not all individuals exposed to the above exogenous risk factors will develop CRC, which suggested that individual susceptibility factors may play an important role in the cancer development.
Methylenetetrahydrofolate reductase (MTHFR) located on chromosome 1 at 1p36.3, is an important enzymes involved in the folate metabolic pathway. It reduces 5,10-methylenetetra-hydrofolate which protects cells from DNA damage induced by uridylate misincorporation to 5-methyltetrahydrofolate, a carbon donor for the homocysteine/methionine conversion. In addition, 5-methyltetrahydrofolate is the major circulating form of folate and is important for DNA synthesis, repair, and methylation [3], [4]. Folate, in its 5- methyl form, participates in single-carbon transfers that occur as part of the synthesis of nucleotides, the synthesis of S-adenosyl-methionine, the remethylation of homocysteine to methionine, the methylation of DNA, proteins, neurotransmitters and phospholipids. DNA aberrant methylation might also increase the risk of CRC [5]. It has been considered as one of the molecular mechanisms by which gene expression is regulated [6].
Reduced activity of the MTHFR, due to C677T polymorphisms, increases the pool of 5,10-methy-lene-THF that reduces misincorporation of uracil into DNA, which might lead to double-strand breaks during uracil excision repair processes, and increase chromosomal aberrations risk [7]. C677T (rs1801133) is a single nucleotide polymorphism (SNP) in exon 4 at the folate binding site of the MTHFR gene which influences the general balance between DNA synthesis, repair, and methylation processes. Individuals with the MTHFR 677TT genotype have been shown to have only 30% of the in vitro MTHFR enzyme activity compared with the wild type, whereas those with the heterozygous CT genotype have been found to have 60% of wild-type MTHFR enzyme activity [8]. Up to 15% of individuals are homozygous 677TT for the variant, which is associated with higher plasma homocysteine levels and reduced plasma folate levels [9]. Considering the functional effects of the polymorphisms of these enzymes, it is expected that these gene polymorphisms may be associated with the CRC risk.
Previous epidemiological studies have been conducted trying to clarify the association between the MTHFR C677T polymorphism and CRC susceptibility, but obtain controversial results. To date, we performed a meta-analysis from all eligible studies and provided more accurate estimate the association between the MTHFR C677T polymorphism and the risk of CRC.
Materials and Methods
Literature and search strategy
We considered all studies that examined the association of the MFTHR 677C>T polymorphism with CRC. The sources included MEDLINE, EMBASE, PubMed, Web of Science, Chinese Biomedical Literature database (CBM) and CNKI database (last search was update on July 2012). The search strategy to identify all possible studies involved uses the combinations of the following key words: “methylenetetrahydrofolate reductase”, “MTHFR”, “MTHFR C677T”, “SNPs”, “rs1801133”; “polymorphism”, “genotype”; “colorectal”, “colon”, “rectal”; “cancer”, “carcinoma”, “adenocarcinoma”. No language restrictions were imposed. The reference lists of review articles, clinical trials, and meta-analyses, were also hand-searched for the collection of other relevant studies, and two authors conducted all searches independently. Non-familial case-control studies were eligible if they determined the distribution for this polymorphism in unrelated patients with CRC and in a concurrent control group of healthy subjects using molecular methods for genotyping. We did not include abstracts or unpublished reports. When overlapping data of the same patient population were included in more than one publication, only the most recent or complete study was used in this meta-analysis.
Eligibility criteria
The studies accepted for this meta-analysis had to meet the following criteria: (1) utilized platinum-based regimens for patients with pathologically proven colorectal cancer; (2) controls were consisted with normal persons; (3) only cohort studies and case-control studies included in this meta-analysis; (4)evaluation of the MTHFR C677T polymorphisms and colorectal cancer risks; (5) The paper should clearly describe the sources of cases and controls; (6) The authors must offer the size of the sample, OR and their 95% CI or the information that can help infer the results in the papers.
Accordingly, major reasons for exclusion of studies were: (1) not designed as case/control or cohort studies; (2) reviews and repeated research studies; (3) not offering the sources of cases and controls and other essential information; (4) control population including malignant tumor patients; (5) duplicated publications.
Quality assessment
The Newcastle – Ottawa Quality Assessment Scale for cohort studies was used to assess the quality of the studies [10], [11]. This scale is composed of eight items that assess patient selection, study comparability and outcome. The scale was recommended by the Cochrane Non-Randomized Studies Methods Working Group [12]. Two investigators performed quality assessment independently. Disagreement was resolved by consensus.
Data extraction
Information was carefully extracted from all eligible publications independently by two authors according to the inclusion and exclusion criteria listed above. Disagreement was resolved by discussion between the two authors. The following data were collected from each study: first author's surname, year of publication, ethnicity, and numbers of cases and controls with the CC, CT and TT genotypes, and Hardy – Weinberg equilibrium (HWE) in controls, respectively. Different ethnicity descents were categorized as Caucasian, Asian, African, and Mixed population. When studies included subjects of more than one ethnicity and were able to separate, data were extracted separately for each ethnic group. We did not define any minimum number of patients to include a study in our meta-analysis.
Statistical analysis
We used the crude odds ratios (ORs) with 95% confidence intervals (CIs) to assess the strength of this association according to the method of Woolf [13]. Subgroup analyses were done by ethnicity. Both fixed-effects model using the Mantel – Haenszel method [14] and random-effects model using the DerSimonian and Laird method [15] were used to pool the results. The fixed-effects model was used when there was no heterogeneity of the results of studies; otherwise, the random-effects model was used.
Heterogeneity assumption was checked by the Chi-square-based Q-test [16]. A P-value greater than 0.10 for the Q-test indicates a lack of heterogeneity among studies. Simultaneously, it was also assessed using the I2 statistic, which takes values between 0% and 100% with higher values denoting greater degree of heterogeneity (I2 = 0–25%: no heterogeneity; I2 = 25–50%: moderate heterogeneity; I2 = 50–75%: large heterogeneity; I2 = 75–100%: extreme heterogeneity) [17]. The significance of the pooled OR was determined by the Z-test, and P<0.05 was considered as statistically significant. One-way sensitivity analyses were performed to assess the stability of the results. An estimate of potential publication bias was carried out by the funnel plot, Begg's and Egger's test. The significance of the intercept was determined by the t-test suggested by Egger (P<0.05 was considered representative of statistically significant publication bias) [18]. Hardy – Weinberg equilibrium in the control group was tested by the Chi-square test, and P-value of <0.05 was considered significant. All of the calculations were performed using STATA (version 12.0; Stata Corporation, College Station, TX), using two-sided P-values.
Results
Study characteristics
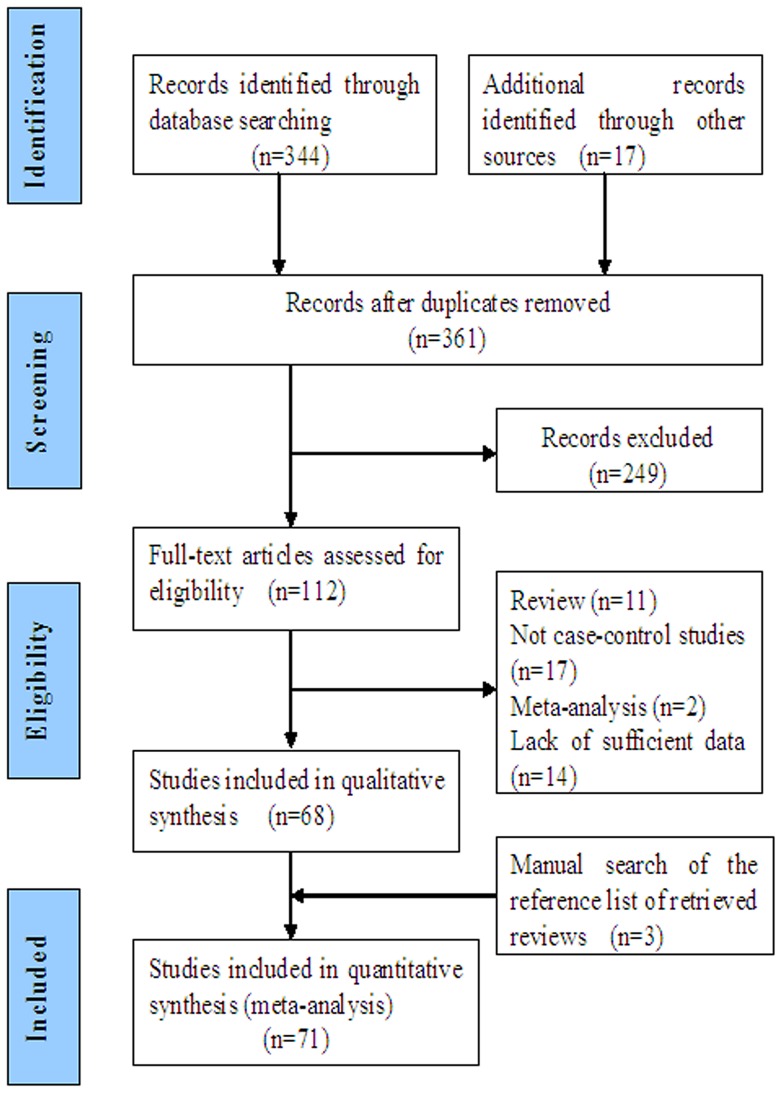
Studies relevant to the searching words were retrieved originally. The flow diagram of the literature search strategy is shown in Figure 1 . 71 publications involving 31,572 CRC cases and 44,066 controls were ultimately analyzed, addressing the association between MTHFR C677T polymorphism and CRC susceptibility was preliminarily eligible [19]–[89]. The main characteristics of these studies included in this meta-analysis were presented in Table S1 . All the cases were histologically confirmed. Controls were mainly healthy populations. Therefore, each study was considered separately for pooling subgroup analysis. There were a total of 71 studies, including 77 independent case-control or cohort researches, which included 38 groups of Caucasians, 27 groups of Asians, 8 groups of Mixed populations and 4 groups of Africans. The distribution of genotypes in the controls of all studies was in agreement with Hardy – Weinberg equilibrium except 13 researches [20], [21], [24], [40], [42], [43], [46], [49], [58], [75], [81], [84], [88].
Figure 1. Flow diagram summarizing the search strategy.
Meta-analysis results
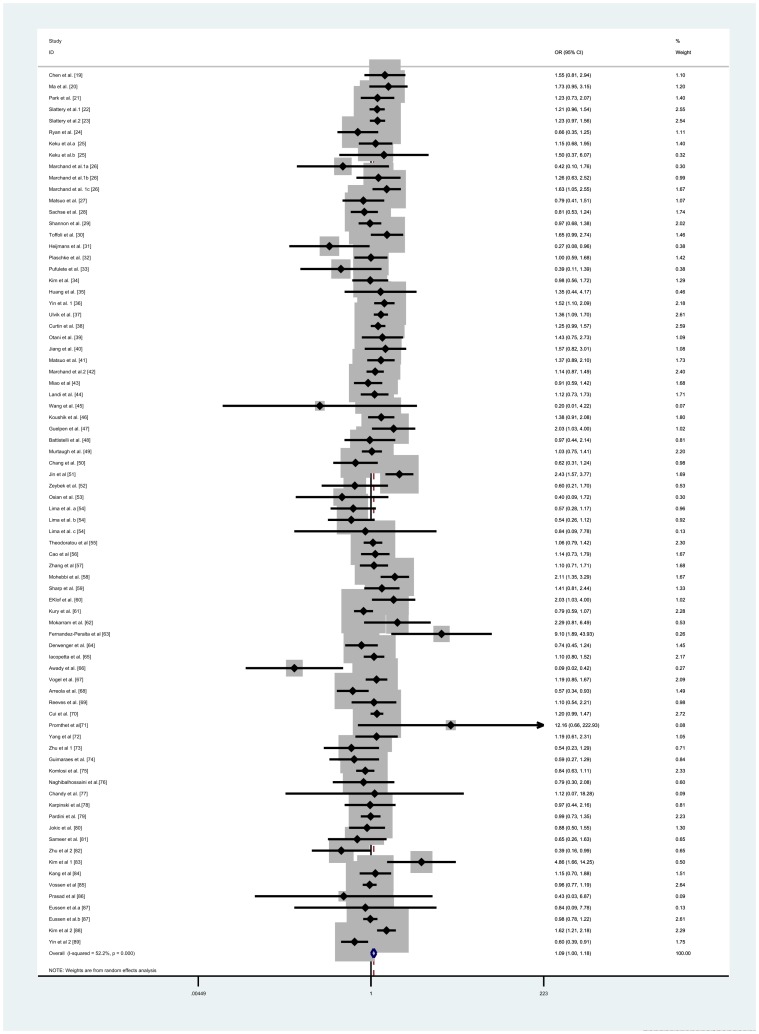
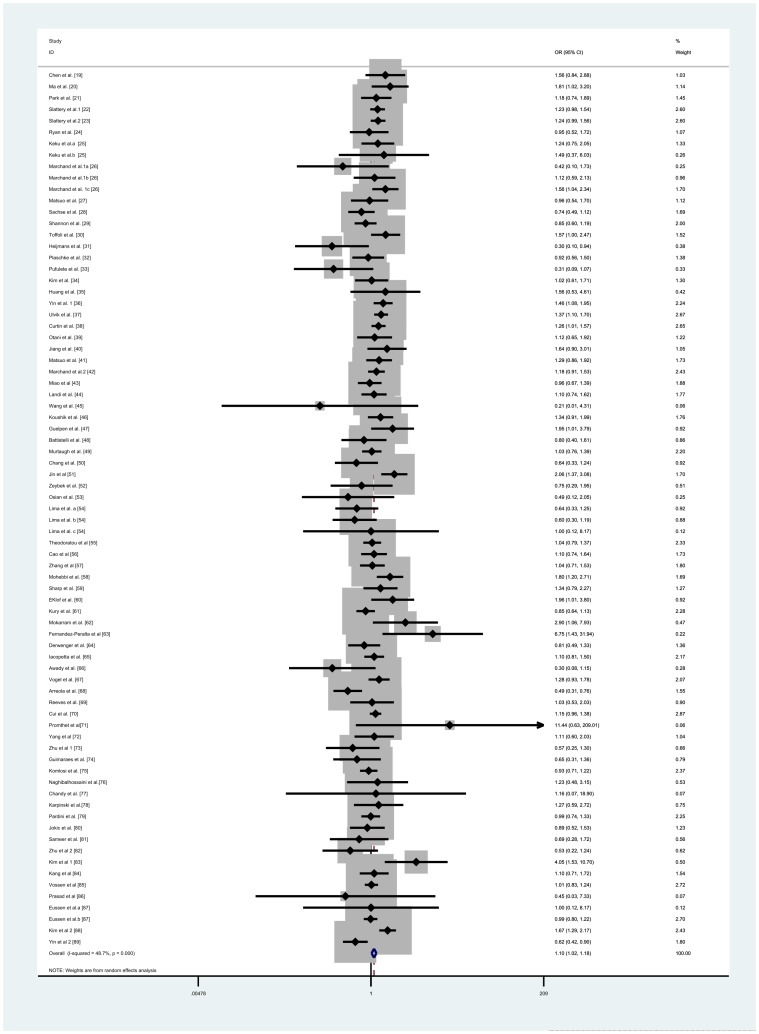
The main results of this meta-analysis were listed in Table 1 . Overall, significantly elevated CRC risk were associated with some genetic models when all the eligible studies were pooled into the meta-analysis (OR = 1.089; 95%CI = 1.002–1.183; I2 = 52.2% for CC vs TT, Figure 2 ; OR = 1.113; 95%CI = 1.037–1.195; I2 = 38.5% for CT vs TT, Figure S1 ; OR = 1.097; 95%CI = 1.019–1.182; I2 = 48.7% for dominant model, Figure 3 ). In contrast, there were no associations were found in other genetic models. Simultaneously, the C-allele genotype was not associated with an increased CRC risk compared with the T-allele genotype (OR = 1.014; 95%CI = 0.973–1.056; I2 = 60.9%, Figure S2 ).
Table 1. Meta-analysis on the association between 677 C>T MTHFR polymorphism and colorectal cancer susceptibility.
| Variables | Study number | Statistic | Test of heterogeneity | Test of Association | |||
| Model | Q | P | I2 | OR(95% CL) | P | ||
| CC vs TT | |||||||
| Total | 77 | Random | 159.08 | 0.000 | 52.2% | 1.089 (1.002 1.183) | 0.044 |
| Caucasian | 38 | Random | 77.51 | 0.000 | 52.3% | 1.076(1.008 1.150) | 0.029 |
| Asian | 27 | Random | 58.18 | 0.000 | 55.3% | 1.226(1.116 1.346) | 0.000 |
| Mixed population | 8 | Fixed | 7.70 | 0.360 | 9.0% | 1.112(0.980 1.262) | 0.098 |
| African | 4 | Fixed | 7.61 | 0.055 | 60.6% | 1.119(1.065 1.176) | 0.141 |
| CC vs CT | |||||||
| Total | 77 | Random | 125.60 | 0.000 | 39.5% | 0.972(0.928 1.019) | 0.241 |
| Caucasian | 38 | Random | 82.78 | 0.000 | 55.3% | 0.961(0.923 1.002) | 0.060 |
| Asian | 27 | Fixed | 21.48 | 0.717 | 0.0% | 1.026 (0.958 1.098) | 0.462 |
| Mixed population | 8 | Fixed | 2.59 | 0.920 | 0.0% | 0.975 (0.901 1.055) | 0.531 |
| African | 4 | Random | 14.13 | 0.003 | 78.8% | 0.710 (0.498 1.010) | 0.057 |
| CT vs TT | |||||||
| Total | 77 | Random | 123.62 | 0.000 | 38.5% | 1.113(1.037 1.195) | 0.003 |
| Caucasian | 38 | Random | 76.06 | 0.000 | 51.4% | 1.102(1.032 1.177) | 0.004 |
| Asian | 27 | Random | 40.75 | 0.033 | 36.2% | 1.180(1.079 1.291) | 0.000 |
| Mixed population | 8 | Fixed | 4.87 | 0.676 | 0.0% | 1.142(1.005 1.296) | 0.041 |
| African | 4 | Fixed | 0.41 | 0.938 | 0.0% | 1.089(0.476 2.494) | 0.840 |
| CC vs CT+TT | |||||||
| Total | 77 | Random | 153.54 | 0.000 | 50.5% | 0.994(0.946 1.044) | 0.806 |
| Caucasian | 38 | Random | 88.16 | 0.000 | 58.0% | 0.982(0.944 1.021) | 0.356 |
| Asian | 27 | Fixed | 37.63 | 0.065 | 30.9% | 1.069(1.003 1.140) | 0.041 |
| Mixed population | 8 | Fixed | 4.34 | 0.740 | 0.0% | 1.002(0.930 1.080) | 0.960 |
| African | 4 | Random | 16.22 | 0.001 | 81.5% | 0.714(0.508 1.002) | 0.051 |
| CC+CT vs TT | |||||||
| Total | 77 | Random | 148.27 | 0.000 | 48.7% | 1.097(1.019 1.182) | 0.015 |
| Caucasian | 38 | Random | 79.69 | 0.000 | 53.6% | 1.086(1.021 1.156) | 0.009 |
| Asian | 27 | Random | 54.62 | 0.001 | 52.4% | 1.198(1.101 1.303) | 0.000 |
| Mixed population | 8 | Fixed | 6.75 | 0.455 | 0.0% | 1.126(0.999 1.270) | 0.052 |
| African | 4 | Fixed | 2.85 | 0.415 | 0.0% | 1.123(1.073 1.177) | 0.469 |
| C-allele vs T-allele | |||||||
| Total | 77 | Random | 194.58 | 0.000 | 60.9% | 1.014(0.973 1.056) | 0.508 |
| Caucasian | 38 | Random | 96.87 | 0.000 | 61.8% | 1.000(0.948 1.056) | 0.986 |
| Asian | 27 | Random | 63.92 | 0.000 | 59.3% | 1.054 (0.974 1.140) | 0.193 |
| Mixed population | 8 | Fixed | 7.09 | 0.420 | 1.2% | 1.028(0.970 1.088) | 0.351 |
| African | 4 | Random | 14.68 | 0.002 | 79.6% | 0.664(0.315 1.400) | 0.282 |
Note: There are 71 studies which some one includes two or three researches in one work, including 77 case-control or cohort researches.
Figure 2. Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism (for CC vs TT).
Figure 3. Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism at dominant model (CC + CT vs TT).
In the stratified analysis by ethnicity, significantly increased risks were found among Caucasians for CC vs TT (OR = 1.076; 95%CI = 1.008–1.150; I2 = 52.3%, Figure S3 ), CT vs TT (OR = 1.102; 95%CI = 1.032–1.177; I2 = 51.4%, Figure S4 ) and dominant model (OR = 1.086; 95%CI = 1.021 −1.156; I2 = 53.6%, Figure S5 ), ( Table 1 ). Simultaneously, significantly increased risks were also found among Asians for CC vs TT (OR = 1.226; 95%CI = 1.116–1.346; I2 = 55.3%, Figure S3 ), CT vs TT (OR = 1.180; 95%CI = 1.079–1.291; I2 = 36.2%, Figure S4 ), recessive (OR = 1.069; 95%CI = 1.003–1.140; I2 = 30.9%, Figure S6 ) and dominant model (OR = 1.198; 95%CI = 1.101–1.303; I2 = 52.4%, Figure S5 ), ( Table 1 ). For Mixed populations, significantly increased risks were found for CT vs TT (OR = 1.142; 95%CI = 1.005–1.296; I2 = 0.0%, Figure S4 ) ( Table 1 ). However, no significant associations were found in Africans for all genetic models ( Table 1 ).
There was significant heterogeneity for all genetic model comparisons among worldwide populations. After assessing the source of heterogeneity for all genetic model comparison by subgroup analysis on ethnicity the heterogeneity was partly decreased or removed. However, there was still significant heterogeneity among different descent population studies. When we deleted the study on evaluating different descent populations for departure from HWE, the heterogeneity was completely removed.
Sensitivity analysis
In order to compare the difference and evaluate the sensitivity of the meta-analyses, we conducted one-way sensitivity analysis to evaluate the stability of the meta-analysis. The statistical significance of the results was not altered when any single study was omitted, confirming the stability of the results. Hence, results of the sensitivity analysis suggest that the data in this meta-analysis are relatively stable and credible (Date was not shown).
Publication bias
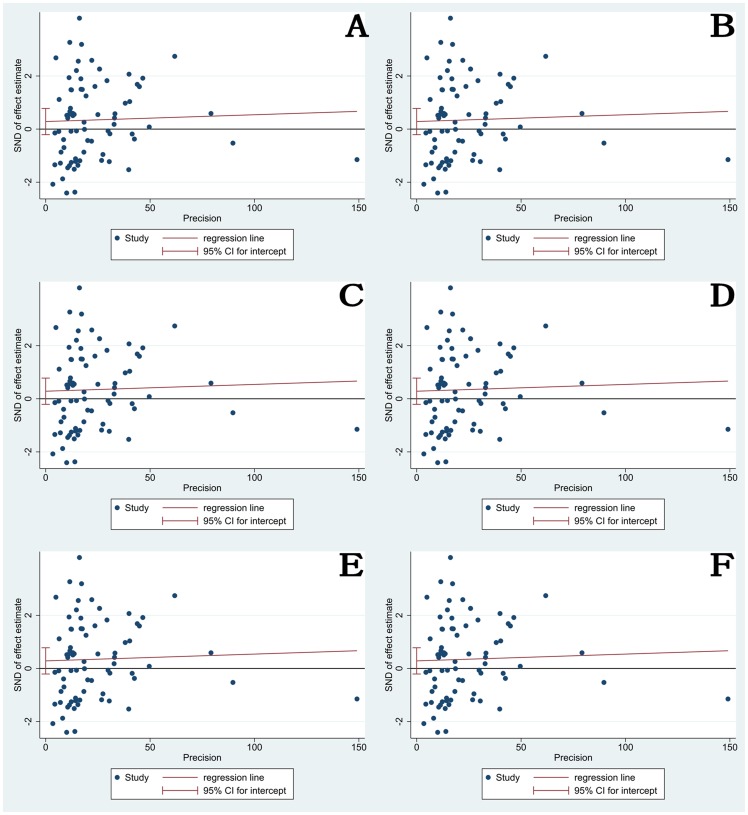
Begg's funnel plot and Egger's test were performed to assess the publication bias of the literature. The shapes of the funnel plots did not reveal any evidence of obvious asymmetry in all comparison models. Furthermore, Egger's test was used to provide statistical evidence for funnel plot symmetry ( Figure 4 ). The results still did not suggest any evidence of publication bias.
Figure 4. Eegger's funnel plot with pseudo 95% confidence limit under all different genetic models of 677C>T genotype(A: CC vs TT; B: CC vs CT; C: CT vs TT; D: CC vs CT+TT; E: CC+CT vs TT; F: C-allele vs T-allele).
Discussion
It is well recognized that there is a range of individual susceptibility to the CRC even with identical environmental exposure. Single nucleotide polymorphism (SNP) is the most common form of human genetic variation, and may contribute to individual's susceptibility to cancer, however, the underlying molecular mechanism is unknown. Therefore, genetic susceptibility to cancer has been a research focus in scientific community. Some variants, especially those in the promoter regions of genes, may affect either the expression or activity levels of enzymes [90]–[93] and therefore may be mechanistically associated with cancer risk. Previous studies investigating the association between MTHFR C677T polymorphism and CRC risk have provided inconsistent results. These inconsistent results are possibly because most of those studies involved no more than a few hundred CRC cases, which is too few to assess any genetic effects reliably, which resulted in a small effect of the polymorphism on CRC risk and relatively low statistical power of the published studies. Hence, a meta-analysis was needed to provide a quantitative approach for combining the results of various studies with the same topic, and for estimating and explaining their diversity.
Meta-analysis has been considered as an important tool to more precisely define the effect of selected genetic polymorphisms on risk of disease and to identify potentially important sources of between-study heterogeneity. Previous meta-analysis included only 8 case-control studies in the analysis of Asian population, which was too little to confirm the association between MTHFR C677T polymorphism and CRC risk [94]. Yang et al included 21 studies to research the associations in his work but only from Asian descent [95]. In order to provide the most comprehensive assessment of the association between MTHFR C677T polymorphism and CRC risk in worldwide populations, we did an updated meta-analysis of all available studies. At last, we performed this updated meta-analysis on the association between MTHFR C677T polymorphism and CRC risk by critically reviewing 71 individual studies including 31,572 cases and 44,066 controls.
In the present meta-analysis, we found that the variant genotypes of the MTHFR C677T polymorphisms were significantly associated with CRC risk. Simultaneously, subgroup analyses by ethnicity further identified this association. We found that the variant genotype of the MTHFR C677T polymorphism, in Caucasian population, was associated with significant increase in CRC risk. The same results were detected among Asian, and Mixed populations. Although the MTHFR C677T polymorphism may be associated with DNA repair activity, no significant associations of some variant genotypes with CRC risk were found in Caucasian, Asian and Mixed populations, suggesting the influence of the genetic variant may be masked by the presence of other as-yet unidentified causal genes involved in CRC. Moreover, there is no significant association was found among African descent populations.
Possible limitations of this meta-analysis should be acknowledged. Firstly, publication bias may have occurred because only published researches were included in this study. Though Funnel plots were performed to access the publication bias in this meta-analysis and the outcome suggested that publication bias was not evident in the present study, but the publication bias in the present analysis was still not negligible. Secondly, misclassification bias was possible. For example, most studies could not exclude latent cancer cases in the control group. The controls in some studies were selected from non-cancer patients, while the controls in other several studies were just selected from asymptomatic individuals. Thirdly, in the subgroup analyses by ethnicity, only four studies in African populations included in our meta-analysis, which means relatively limited study number made it impossible to perform ethnic subgroup analysis. Thus, additional studies are warranted to evaluate the effect of this functional polymorphism on CRC risk in different ethnicities, especially in African descent populations. Finally, gene-environmental interactions were not fully addressed in this work for the lack of sufficient data. As we know, aside from genetic factor, smoking, diet habit, smoking, drinking status and some environmental risk factors are major risk factors for CRC. However we didn't perform subgroup analyses based on environmental explosion owing to the limited reported information on such associations in the included studies. In addition, our analysis did not consider the possibility of gene-gene or SNP-SNP interactions or the possibility of linkage disequilibrium between polymorphisms. Considering that, a more precise analysis should be conducted adjusted by all the factors stated above.
Despite of those limitations, this meta-analysis provided evidence of the association between the MTHFR C677T polymorphisms and CRC risk, supporting the hypothesis that MTHFR C677T polymorphism is contributed to overall CRC risk. In subgroup analysis, the same results were found in Caucasian, Asian and Mixed populations, but not in African descent men. In order to verify our findings, well-designed studies are needed to further evaluate the association between the MTHFR C677T polymorphism and CRC risk.
Supporting Information
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism (for CT vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism at additive model ( C-allele vs T-allele ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations ( CC vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations ( CT vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations at dominant model ( CC + CT vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations at recessive model ( CC vs CT + TT ).
(TIF)
Main characters of studies included in this meta-analysis.
(DOC)
Funding Statement
These authors have no support or funding to report.
References
- 1. Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics. CA Cancer J Clin 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 3. Rozen R (1997) Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR). Thromb Haemost 78(1): 523–526. [PubMed] [Google Scholar]
- 4. Ames BN (2001) DNA damage from micronutrient deficiencies is likely to be a major cause of cancer. Mutat Res 475: 7–20. [DOI] [PubMed] [Google Scholar]
- 5. Giovannucci E, Rimm EB, Ascherio A, Stampfer MJ, Colditz GA (1995) Alcohol, low-methionine-low-folate diets, and risk of colon cancer in men. J Natl Cancer Inst 87: 265–273. [DOI] [PubMed] [Google Scholar]
- 6. Jacob RA, Gretz DM, Taylor PC, James SJ, Pogribny IP, et al. (1998) Moderate folate depletion increases plasma homocysteine and decreases lymphocyte DNA methylation in postmenopausal women. J. Nutr 128: 1204–1212. [DOI] [PubMed] [Google Scholar]
- 7. Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, et al. (2002) A common mutation in the 5,10-methylenetetrahydrofo-late reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA 99(8): 5606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, et al. (1995) A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10(1): 111–113. [DOI] [PubMed] [Google Scholar]
- 9. Deloughery TG, Evans A, Sadeghi A, McWilliams J, Henner WD, et al. (1996) Common mutation in methylenetetrahydrofolate reductase. Correlation with homocysteine metabolism and late-onset vascular disease 94: 3074–3078. [DOI] [PubMed] [Google Scholar]
- 10.Wells GA, Shea B, O'Connell D, Peterson J, Welch V, et al.. (2003) The Newcastle–Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. Available: http://www.ohri.ca/programs/clinical epidemiology/oxford.htm. Accessed 2009 Sept 25.
- 11. Schoenleber SJ, Kurtz DM, Talwalkar JA, Roberts LR, Gores GJ (2009) Prognostic role of vascular endothelial growth factor in hepatocellular carcinoma: systematic review and meta-analysis. Br J Cancer 100: 1385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.The Cochrane Collaborative Review Group on HIV Infection and AIDS. Editorial Policy: Inclusion and Appraisal of Experimental and Non-experimental (Obser-vational) Studies; 2009. Available: http://www.igh.org/Cochrane. Accessed 2009 Sept 29.
- 13. Woolf B (1955) On estimating the relation between blood group and disease. Ann Hum Genet 19: 251–3. [DOI] [PubMed] [Google Scholar]
- 14. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–48. [PubMed] [Google Scholar]
- 15. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177–88. [DOI] [PubMed] [Google Scholar]
- 16. Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10: 101–129. [Google Scholar]
- 17. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Egger M, Smith DG, Schneider M, Minder C (1997) Bias in meta analysis detected by a simple, graphical test. BMJ 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen J, Giovannucci E, Kelsey K, Rimm EB, Stampfer MJ, et al. (1996) A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res 56(21): 4862–4864. [PubMed] [Google Scholar]
- 20. Ma J, Stampfer MJ, Giovannucci E, Artigas C, Hunter DJ, et al. (1997) Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res 57(6): 1098–1102. [PubMed] [Google Scholar]
- 21. Park KS, Mok JW, Kim JC (1999) The 677C>T mutation in 5, 10-methylenetetrahydrofolate reductase and colorectal cancer risk. Genet Test 3: 233–236. [DOI] [PubMed] [Google Scholar]
- 22. Slattery ML, Potter JD, Samowitz W, Schaffer D, Leppert M (1999) Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol Biomarkers Prev 8(6): 513–518. [PubMed] [Google Scholar]
- 23. Slattery ML, Edwards SL, Samowitz W, Potter J (2000) Associations between family history of cancer and genes coding for metabolizing enzymes (United States). Cancer Causes Control 11: 799–803. [DOI] [PubMed] [Google Scholar]
- 24. Ryan BM, Molloy AM, McManus R, Arfin Q, Kelleher D, et al. (2001) The methylenetetrahydrofolate reductase (MTHFR) gene in colorectal cancer: role in tumor development and significance of allelic loss in tumor progression. Int J Gastrointest Cancer 30(3): 105–111. [DOI] [PubMed] [Google Scholar]
- 25. Keku T, Millikan R, Worley K, Winkel S, Eaton A, et al. (2002) 5,10-Methylenetetrahydrofolate reductase codon 677 and 1298 polymorphisms and colon cancer in African Americans and whites. Cancer Epidemiol Biomarkers Prev 11(12): 1611–1621. [PubMed] [Google Scholar]
- 26. Le Marchand L, Donlon T, Hankin JH, Kolonel LN, Wilkens LR, et al. (2002) B-vitamin intake, metabolic genes, and colorectal cancer risk (United States). Cancer Causes Control 13(3): 239–248. [DOI] [PubMed] [Google Scholar]
- 27. Matsuo K, Hamajima N, Hirai T, Kato T, Inoue M, et al. (2002) Methionine synthase reductase gene A66G polymorphism is associated with risk of colorectal cancer. Asian Pac J Cancer Prev 3: 353–359. [PubMed] [Google Scholar]
- 28. Sachse C, Smith G, Wilkie MJ, Barrett JH, Waxman R, et al. (2002) A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis 23(11): 1839–1849. [DOI] [PubMed] [Google Scholar]
- 29. Shannon B, Gnanasampanthan S, Beilby J, Iacopetta B (2002) A polymorphism in the methylenetetrahydrofolate reductase gene predisposes to colorectal cancers with microsatellite instability. Gut 50(4): 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toffoli G, Gafà R, Russo A, Lanza G, Dolcetti R, et al. (2003) Methylenetetrahydrofolate reductase 677C→T polymorphism and risk of proximal colon cancer in north Italy. Clinical cancer research 9(2): 743–748. [PubMed] [Google Scholar]
- 31. Heijmans BT, Boer J, Suchiman HED, Cornelisse CJ, Westendorp RGJ, et al. (2003) A common variant of the methylenetetrahydrofolate reductase gene (1p36) is associated with an increased risk of cancer. Cancer research 63(6): 1249. [PubMed] [Google Scholar]
- 32. Plaschke J, Schwanebeck U, Pistorius S, Saeger HD, Schackert HK (2003) Methylenetetrahydrofolate reductase polymorphisms and risk of sporadic and hereditary colorectal cancer with or without microsatellite instability. Cancer Lett 191(2): 179–185. [DOI] [PubMed] [Google Scholar]
- 33. Pufulete M, Al-Ghnaniem R, Leather AJ, Appleby P, Gout S, et al. (2003) Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology 124(5): 1240–1248. [DOI] [PubMed] [Google Scholar]
- 34.Kim DH, Ahn YO, Lee BH, Tsuji E, Kiyohara C, et al.. (2004) Methylenetetrahydrofolate reductase polymorphism, alcohol intake, and risks of colon and rectal cancers in Korea. Cancer Lett, 216: 199–205. [DOI] [PubMed] [Google Scholar]
- 35.Huang P, Zhou ZY, Ma HT (2003) MTHFR polymorphisms and colorectal cancer susceptibility in Chongging people. Acta Academiae Med Militaris Tertiae, 25: 1710–1715. [Google Scholar]
- 36. Yin G, Kono S, Toyomura K, Hagiwara T, Nagano J, et al. (2004) Methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and colorectal cancer, the Fukuoka Colorectal Cancer Study. Cancer Sci 95: 908–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ulvik A, Vollset SE, Hansen S, Gislefoss R, Jellum E, et al. (2004) Colorectal cancer and the methylenetetrahydrofolate reductase 677C>T and methionine synthase 2756A>G polymorphisms: a study of 2,168 case-control pairs from the JANUS cohort. Cancer Epidemiol Biomarkers Prev 13(12): 2175–2180. [PubMed] [Google Scholar]
- 38. Curtin K, Bigler J, Slattery ML, Caan B, Potter JD, et al. (2004) MTHFR C677T and A1298C polymorphisms: diet, estrogen, and risk of colon cancer. Cancer Epidemiol Biomarkers Prev 13: 285–292. [DOI] [PubMed] [Google Scholar]
- 39. Otani T, Iwasaki M, Hanaoka T, Kobayashi M, Ishihara J, et al. (2005) Folate, vitamin B6, vitamin B12, and vitamin B2 intake, genetic polymorphisms of related enzymes, and risk of colorectal cancer in a hospital-based case-control study in Japan. Nutr Cancer 53(1): 42–50. [DOI] [PubMed] [Google Scholar]
- 40. Jiang Q, Chen K, Ma X, Li Q, Yu W, et al. (2005) Diets, polymorphisms of methylenetetrahydrofolate reductase, and the susceptibility of colon cancer and rectal cancer. Cancer Detect Prev 29: 146–154. [DOI] [PubMed] [Google Scholar]
- 41. Matsuo K, Ito H, Wakai K, Hirose K, Saito T, et al. (2005) One-carbon metabolism related gene polymorphisms interact with alcohol drinking to influence the risk of colorectal cancer in Japan. Carcinogenesis 26: 2164–2171. [DOI] [PubMed] [Google Scholar]
- 42. Le Marchand L, Wilkens LR, Kolonel LN, Henderson BE (2005) The MTHFR C677T polymorphism and colorectal cancer: the multiethnic cohort study. Cancer Epidemiology Biomarkers & Prevention 14(5): 1198–1203. [DOI] [PubMed] [Google Scholar]
- 43. Miao XP, Yang S, Tan W, Zhang XM, Ye YJ, et al. (2005) Association between genetic variations in methylenetetrahydrofolate reductase and risk of colorectal cancer in a Chinese population. Zhonghua Yu Fang Yi Xue Za Zhi 39: 409–411. [PubMed] [Google Scholar]
- 44. Landi S, Gemignani F, Moreno V, Gioia-Patricola L, Chabrier A, et al. (2005) A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet Genomics 15: 535–546. [DOI] [PubMed] [Google Scholar]
- 45. Wang J, Gajalakshmi V, Jiang J, Kuriki K, Suzuki S, et al. (2006) Associations between 5,10-methylenetetrahy-drofolate reductase codon 677 and 1298 genetic polymorphisms and environmental factors with reference to susceptibility to colorectal cancer: a case–control study in an Indian population. Int J Cancer 118: 991–997. [DOI] [PubMed] [Google Scholar]
- 46. Koushik A, Kraft P, Fuchs CS, Hankinson SE, Willett WC, et al. (2006) Nonsynonymous polymorphisms in genes in the one-carbon metabolism pathway and associations with colorectal cancer. Cancer Epidemiol Biomarkers Prev 15(12): 2408–2417. [DOI] [PubMed] [Google Scholar]
- 47. Van Guelpen B, Hultdin J, Johansson I, Hallmans G, Stenling R, et al. (2006) Low folate levels may protect against colorectal cancer. Gut 55(10): 1461–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Battistelli S, Vittoria A, Stefanoni M, Bing C, Roviello F (2006) Total plasma homocysteine and methylenetetrahydrofolate reductase C677T polymorphism in patients with colorectal carcinoma. World J Gastroenterol 12: 6128–6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Murtaugh MA, Curtin K, Sweeney C, Wolff RK, Holubkov R, et al. (2007) Dietary intake of folate and co-factors in folate metabolism, MTHFR polymorphisms, and reduced rectal cancer. Cancer Causes Control 18: 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chang SC, Lin PC, Lin JK, Yang SH, Wang HS, et al. (2007) Role of MTHFR polymorphisms and folate levels in different phenotypes of sporadic colorectal cancers. Int J Colorectal Dis 22: 483–489. [DOI] [PubMed] [Google Scholar]
- 51. Jin XX, Zhu ZZ, Wang AZ, Hangruo Jia (2007) Association of methylenetetrahydrofoIate reductase C677T polymorphism with genetic susceptibility to colorectal cancer. Shijie Huaren Xiao Hua Za Zhi 15: 2754–2757. [Google Scholar]
- 52. Zeybek U, Yaylim I, Yilmaz H, Agachan B, Ergen A, et al. (2007) Methylenetetrahydrofolate reductase C677T polymorphism in patients with gastric and colorectal cancer. Cell Biochem Funct 25(4): 419–422. [DOI] [PubMed] [Google Scholar]
- 53. Osian G, Procopciuc L, Vlad L (2007) MTHFR polymorphisms as prognostic factors in sporadic colorectal cancer. J Gastrointest Liver Dis 16: 251–256. [PubMed] [Google Scholar]
- 54. Lima CS, Nascimento H, Bonadia LC, Teori MT, Coy CS, et al. (2007) Polymorphisms in methylenetetrahydrofolate reductase gene (MTHFR) and the age of onset of sporadic colorectal adenocarcinoma. Int J Colorectal Dis 22(7): 757–63. [DOI] [PubMed] [Google Scholar]
- 55. Theodoratou E, Farrington SM, Tenesa A, McNeill G, Cetnarskyj R, et al. (2008) Dietary vitamin B6 intake and the risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev 17(1): 171–182. [DOI] [PubMed] [Google Scholar]
- 56. Cao HX, Gao CM, Takezaki T, Wu JZ, Ding JH, et al. (2008) Genetic polymorphisms of methylenetetrahydrofolate reductase and susceptibility to colorectal cancer. Asian Pac J Cancer Prev 9: 203–208. [PubMed] [Google Scholar]
- 57. Zhang YL, Yuan XY, Zhang C, Yang Y, Pan YM, et al. (2008) Relationship between polymorphisms of thymidylate synthase and methylenetetrahydrofolate reductase and susceptibility in Liaoning Benxi colorectal cancer pafients. Linchuang Zhong Liu Xue Za Zhi 13: 769–774. [Google Scholar]
- 58. Mohebbi SR, Khatami F, Ghiasi S, Derakhshan F, Atarian H, et al. (2008) Reverse association between MTHFR polymorphism (C677T) with sporadic colorectal cancer. Gastroenterology and Hepatology from bed to bench 1(2): 57–63. [Google Scholar]
- 59. Sharp L, Little J, Brockton NT, Cotton SC, Masson LF, et al. (2008) Polymorphisms in the methylenetetrahydrofolate reductase (MTHFR) gene, intakes of folate and related B vitamins and colorectal cancer: a case-control study in a population with relatively low folate intake. Br J Nutr 99(2): 379–389. [DOI] [PubMed] [Google Scholar]
- 60. Eklof V, Van Guelpen B, Hultdin J, Johansson I, Hallmans G, et al. (2008) The reduced folate carrier (RFC1) 80G [A and folate hydrolase 1 (FOLH1) 1561C [T polymorphisms and the risk of colorectal cancer: a nested case-referent study. Scand J Clin Lab Invest 68: 393–401. [DOI] [PubMed] [Google Scholar]
- 61. Kury S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, et al. (2008) Low-penetrance alleles predisposing to sporadic colorectal cancers: a French case-controlled genetic association study. BMC Cancer 8: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mokarram P, Naghibalhossaini F, Saberi Firoozi M, Hosseini SV, Izadpanah A, et al. (2008) Methylenetetrahydrofolate reductase C677T genotype affects promoter methylation of tumor-specific genes in sporadic colorectal cancer through an interaction with folate/vitamin B12 status. World J Gastroenterol 14: 3662–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fernandez-Peralta AM, Daimiel L, Nejda N, Iglesias D, Medina Arana V, et al. (2010) Association of polymorphisms MTHFR C677T and A1298C with risk of colorectal cancer, genetic and epigenetic characteristic of tumors, and response to chemotherapy. Int J Colorectal Dis 25(2): 141–151. [DOI] [PubMed] [Google Scholar]
- 64. Derwinger K, Wettergren Y, Odin E, Carlsson G, Gustavsson B (2009) A study of the MTHFR gene polymorphism C677T in colorectal cancer. Clin Colorectal Cancer 8: 43–48. [DOI] [PubMed] [Google Scholar]
- 65. Iacopetta B, Heyworth J, Girschik J, Grieu F, Clayforth C, et al. (2009) The MTHFR C677T and DeltaDNMT3B C-149T polymorphisms confer different risks for right- and left-sided colorectal cancer. Int J Cancer 125: 84–90. [DOI] [PubMed] [Google Scholar]
- 66. El Awady MK, Karim AM, Hanna LS, El Husseiny LA, El Sahar M, et al. (2009) Methylenetetrahydrofolate reductase gene polymorphisms and the risk of colorectal carcinoma in a sample of Egyptian individuals. Cancer Biomark 5(6): 233–40. [DOI] [PubMed] [Google Scholar]
- 67. de Vogel S, Wouters KA, Gottschalk RW, van Schooten FJ, de Goeij AF, et al. (2009) Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiol Biomarkers Prev 18: 3086–3096. [DOI] [PubMed] [Google Scholar]
- 68. Gallegos-Arreola MP, Garcia-Ortiz JE, Figuera LE, Puebla-Perez AM, Morgan-Villela G, et al. (2009) Association of the 677C>T polymorphism in the MTHFR gene with colorectal cancer in Mexican patients. Cancer Genomics Proteomics 6: 183–188. [PubMed] [Google Scholar]
- 69. Reeves SG, Meldrum C, Groombridge C, Spigelman AD, Suchy J, et al. (2009) MTHFR 677 C>T and 1298 A>C polymorphisms and the age of onset of colorectal cancer in hereditary nonpolyposis colorectal cancer. Eur J Hum Genet 17: 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cui LH, Shin MH, Kweon SS, Kim HN, Song HR, et al. (2010) Methylenetetrahydrofolate reductase C677T polymorphism in patients with gastric and colorectal cancer in a Korean population. BMC Cancer 10: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Promthet SS, Pientong C, Ekalaksananan T, Wiangnon S, Poomphakwaen K, et al. (2010) Risk factors for colon cancer in Northeastern Thailand: interaction of MTHFR codon 677 and 1298 genotypes with environmental factors. J Epidemiol 20: 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang XX, Li FX, Yi JP, Bai J, Fu S (2010) Association of MTHFR C677T polymorphism and susceptibility of lung cancer, gastric cancer and colorectal cancer risk. Guangdong Yi Xue 31: 2375–2378. [Google Scholar]
- 73. Zhu F, Wang YM, Zhang QM (2010) A case-control study of plasma homocysteine, serum folate, the polymorphism of methylenetetrahydrofolate reductase in colorectai cancer. Dongnan Da Xue Xue Bao 29: 88–92. [Google Scholar]
- 74. Guimaraes JL, Ayrizono Mde L, Coy CS, Lima CS (2011) Gene polymorphisms involved in folate and methionine metabolism and increased risk of sporadic colorectal adenocarcinoma. Tumour Biol 32(5): 853–861. [DOI] [PubMed] [Google Scholar]
- 75. Komlosi V, Hitre E, Pap E, Adleff V, Reti A, et al. (2010) SHMT1 1420 and MTHFR 677 variants are associated with rectal but not colon cancer. BMC Cancer 10: 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Naghibalhossaini F, Mokarram P, Khalili I, Vasei M, Hosseini SV, et al. (2010) MTHFR C677T and A1298C variant genotypes and the risk of microsatellite instability among Iranian colorectal cancer patients. Cancer Genet Cytogenet 197: 142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chandy S, Sadananda Adiga MN, Ramachandra N, Krishnamoorthy S, Ramaswamy G, et al. (2010) Association of methylenetetrahydrofolate reductase gene polymorphisms colorectal cancer in India. Indian J Med Res 131: 659–664. [PubMed] [Google Scholar]
- 78. Karpinski P, Myszka A, Ramsey D, Misiak B, Gil J, et al. (2010) Polymorphisms in methyl-group metabolism genes and risk of sporadic colorectal cancer with relation to the CpG island methylator phenotype. Cancer Epidemiol 34: 338–344. [DOI] [PubMed] [Google Scholar]
- 79. Jokic M, Brcic-Kostic K, Stefulj J, Catela Ivkovic T, Bozo L, et al. (2011) Association of MTHFR, MTR, MTRR, RFC1, and DHFR gene polymorphisms with susceptibility to sporadic colon cancer. DNA Cell Biol 30(10): 771–776. [DOI] [PubMed] [Google Scholar]
- 80. Pardini B, Kumar R, Naccarati A, Prasad RB, Forsti A, et al. (2011) MTHFR and MTRR genotype and haplotype analysis and colorectal cancer susceptibility in a case-control study from the Czech Republic. Mutat Res 721(1): 74–80. [DOI] [PubMed] [Google Scholar]
- 81. Sameer AS, Shah ZA, Nissar S, Mudassar S, Siddiqi MA (2011) Risk of colorectal cancer associated with the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism in the Kashmiri population. Genet Mol Res 10(2): 1200–1210. [DOI] [PubMed] [Google Scholar]
- 82. Zhu Q, Jin Z, Yuan Y, Lu Q, Ge D, et al. (2011) Impact of MTHFR gene C677T polymorphism on Bcl-2 gene methylation and protein expression in colorectal cancer. Scand J Gastroenterol 46: 436–445. [DOI] [PubMed] [Google Scholar]
- 83. Kim JW, Park HM, Choi YK, Chong SY, Oh D, et al. (2011) Polymorphisms in genes involved in folate metabolism and plasma DNA methylation in colorectal cancer patients. Oncol Rep 25: 167–172. [PubMed] [Google Scholar]
- 84. Kang BS, Ahn DH, Kim NK, Kim JW (2011) Relationship between metabolic syndrome and MTHFR polymorphism in colorectal cancer. J Korean Soc Coloproctol 27: 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Vossen CY, Hoffmeister M, Chang-Claude JC, Rosendaal FR, Brenner H (2011) Clotting factor gene polymorphisms and colorectal cancer risk. J Clin Oncol 29: 1722–1727. [DOI] [PubMed] [Google Scholar]
- 86. Prasad VV, Wilkhoo H (2011) Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Onkologie 34: 422–426. [DOI] [PubMed] [Google Scholar]
- 87. Eussen SJ, Vollset SE, Igland J, Meyer K, Fredriksen A, et al. (2010) Plasma folate, related genetic variants, and colorectal cancer risk in EPIC. Cancer Epidemiol Biomarkers Prev 19: 1328–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719–748. [PubMed] [Google Scholar]
- 89. Yin G, Ming H, Zheng X, Xuan Y, Liang J, et al. (2012) Methylenetetrahydrofolate reductase C677T gene polymorphism and colorectal cancer risk: A case-control study. Oncol Lett 4(2): 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Skoog T, van't Hooft FM, Kallin B, Jovinge S, Boquist S, et al. (1999) A common functional polymorphism (C3A substitution at position-863) in the promoter region of the tumor necrosis factor (TNF-) gene associated with reduced circulating levels of TNF-. Hum. Mol Genet 8: 1443–1449. [DOI] [PubMed] [Google Scholar]
- 91. Momparler RL, Bovenzi V (2000) DNA methylation and cancer. J Cell Physiol 183: 145–154. [DOI] [PubMed] [Google Scholar]
- 92. Shen H, Wang L, Spitz MR, Hong WK, Mao L, et al. (2002) A novel polymorphism in human cytosine DNA-methyltransferase-3B promoter is associated with an increased risk of lung cancer. Cancer Res 62: 4992–4995. [PubMed] [Google Scholar]
- 93. Skoog T, van't Hooft FM, Kallin B, Jovinge S, Boquist S, et al. (1999) A common functional polymorphism (C>A substitution at position -863) in the promoter region of the tumor necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet 8: 1443–1449. [DOI] [PubMed] [Google Scholar]
- 94. Taioli E, Garza M, Ahn Y, Bishop D, Bost J, et al. (2009) Meta-and pooled analyses of the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and colorectal cancer: a HuGE-GSEC review. American journal of epidemiology 170(10): 1207–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yang Z, Zhang XF, Liu HX, Hao YS, Zhao CL (2012) MTHFR C677T polymorphism and colorectal cancer risk in Asians, a meta-analysis of 21 studies. Asian Pac J Cancer Prev 13(4): 1203–1208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism (for CT vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism at additive model ( C-allele vs T-allele ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations ( CC vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations ( CT vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations at dominant model ( CC + CT vs TT ).
(TIF)
Forest plot of colorectal cancer susceptibility associated with MTHFR 677C>T polymorphism in different descent populations at recessive model ( CC vs CT + TT ).
(TIF)
Main characters of studies included in this meta-analysis.
(DOC)