Abstract
Purpose
To evaluate the prognostic value of fibroblast growth factor receptor 4 (FGFR4) protein expression in patients with advanced-stage, high-grade serous ovarian cancer, delineate the functional role of FGFR4 in ovarian cancer progression, and evaluate the feasibility of targeting FGFR4 in serous ovarian cancer treatment.
Experimental Design
Immunolocalization of FGFR4 was performed on 183 ovarian tumor samples. The collected FGFR4 expression data were correlated with overall survival using Kaplan-Meier and Cox regression analyses. The effects of FGFR4 silencing on ovarian cancer cell growth, survival, invasiveness, apoptosis and FGF1-mediated signaling pathway activation were evaluated by transfecting cells with FGFR4-specific small interfering RNAs (siRNAs). An orthotopic mouse model was used to evaluate the effect of injection of FGFR4-specific siRNAs and FGFR4 trap protein encapsulated in nanoliposomes on ovarian tumor growth in vivo.
Results
Overexpression of FGFR4 protein was significantly associated with decreased overall survival durations. FGFR4 silencing significantly decreased the proliferation, survival, and invasiveness and increased apoptosis of ovarian cancer cells. Also, downregulation of FGFR4 significantly abrogated the mitogen-activated protein kinase (MAPK), nuclear factor-κB (NFκB), and WNT signaling pathways, which are activated by FGF1. Targeting FGFR4 with the FGFR4-specific siRNAs and FGFR4 trap protein significantly decreased ovarian tumor growth in vivo.
Conclusions
FGFR4 is a prognostic marker for advanced-stage, high-grade serous ovarian carcinoma. Silencing FGFR4 and inhibiting ligand-receptor binding significantly decrease ovarian tumor growth both in vitro and in vivo, suggesting that targeting ovarian cancer cells with high levels of FGFR4 protein expression is a new therapeutic modality for this disease and will improve survival of it.
Keywords: serous ovarian carcinoma, FGFR4, FGF1, nanoliposomes, FGF trap
Introduction
High-grade, late-stage metastatic serous carcinoma accounts for the majority of annual epithelial ovarian cancer deaths in the United States (1). More than 15,000 deaths per year result from ovarian cancer, making it the most lethal gynecologic malignancy. Researchers have made minimal advances in extending overall survival durations over the past 3 decades. Currently, the cornerstone of treatment of this cancer is surgery with the aim of reducing the tumor burden to microscopic disease (2). This is usually followed by adjuvant combination chemotherapy with platinum and taxane, which produces initial complete responses in 80% of patients and disease-free periods ranging from 12 to 65 months (3, 4). However, abdominal and pelvic recurrence rates approach 80%, and response to further chemotherapy is limited (5). Attempts at using biologic agents to improve outcomes of this disease, including trap proteins, small interfering RNA (siRNA) encapsulated in nanoparticles, and humanized antibodies, are ongoing (6–8). Thus far, though, only the anti-vascular endothelial growth factor antibody bevacizumab has demonstrated clinical activity as reported in a phase 3 clinical trial (7). Poly(ADP-ribose) polymerase inhibitors also have demonstrated promising activity in pre-phase 3 trials (9). Prognostic and predictive biomarkers that can stratify patients for treatment are still lacking.
Using a 60-mer 22K oligonucleotide-based array comparative genome hybridization platform with DNA isolated from microdissected tumor tissue samples, Birrer et al. (10) demonstrated that a gain frequency of 5q31-35.3 in ovarian cancer cells was a negative prognostic indicator for advanced-stage, high-grade serous ovarian cancer, with an overall prevalence of 25%. Further studies showed that fibroblast growth factor 1 (FGF1) in 5q31-35.3 may be one of the genes that drive ovarian cancer progression (10). FGFR4, one of the key receptors for FGF1, is located on the same chromosomal segment and has exhibited preferential binding to FGF1 (11, 12). An alteration in FGFR4 expression would suggest further activation of the FGF ligand and receptor axis, which would subsequently impact ovarian cancer progression. It would also suggest targeting the FGF ligand and receptor axis can be a new regimen in ovarian cancer treatment.
In the present retrospective study, we evaluated the clinical significance of FGFR4 expression in ovarian cancer patients and delineated the receptor’s signaling pathways in conferring an aggressive phenotype to ovarian cancer cells. Furthermore, we evaluated the feasibility of targeting FGFR4 with siRNA delivered in nanoparticles and an FGFR4 trap protein in the treatment of ovarian cancer
Materials and Methods
Patient samples
183 paraffin-embedded high-grade, International Federation of Gynecology and Obstetrics stage IIIB-IV (advanced-stage) serous ovarian tumor samples were used in this study. They were obtained from the ovarian cancer repository at The University of Texas MD Anderson Cancer Center. The samples were collected from patients undergoing primary cytoreductive surgery for ovarian cancer from 1995 to 2006. After surgery, patients received platinum-based combination chemotherapy. Optimal surgical cytoreduction was defined by a residual tumor no more than 1 cm in diameter. The overall survival duration was measured from the date of diagnosis to the date of death or censored at the date of the last follow-up examination. Clinical data, including age, cytoreduction status (optimal vs. suboptimal), and overall survival were abstracted from the patients’ medical records. 10 normal ovarian and 10 normal fallopian tube tissue samples were collected from patients with benign gynecologic diseases for use as controls. All samples and clinical data were collected with the approval of the MD Anderson Institutional Review Board.
Evaluation of FGFR4 overexpression by immunohistochemistry
Immunolocalization of FGFR4 was performed on 183 serous carcinoma, 10 normal ovarian tissue, and 10 normal fallopian tube tissue samples. Slides were stained with a commercially available anti-FGFR4 antibody (1:250; sc-124, Santa Cruz Biotechnology) (Supplementary Table S1). FGFR4 protein expression was visualized by 3,3'-diaminobenzidine (DAB). Normal rabbit IgG applied to high-grade serous carcinoma samples with high levels of FGFR4 expression was used as a negative control. Digital photomicrographs of representative areas of each slide were taken at ×20 magnification. Quantitative FGFR4 staining intensity was determined and localization measurements were obtained using the Image-Pro Plus software (version 5.1; MediaCybernetics). Briefly, areas of interest were drawn around tumor regions. Color segmentation was then used to isolate DAB from background staining. A luminescence grayscale filter was then used, with each pixel carrying an intensity value from 0 (black, maximum DAB saturation) to 255 (white, no DAB). The FGFR4 staining intensity score was calculated by dividing the sum of the intensity values in a tumor area of interest by the number of pixels. This score was used to group the patients according to low and high FGFR4 expression using the median score as the cutoff point. Survival analyses using both Kaplan-Meier modeling (with log-rank significance testing) and a Cox proportional hazards model were performed to determine the effect of FGFR4 expression levels on overall survival and risk of death. A P value <0.05 was considered significant.
Effects of FGFR4 silencing on ovarian cancer cell proliferation, invasion potential, survival, and apoptosis
Endogenous FGFR4 expression was first evaluated in six ovarian cancer cell lines (A2780, SKOV3, OVCA3, OVCA5, OVCA432, and OVCA433) and a normal ovarian surface epithelial (OSE) cell line using western blot analysis. FGFR4 was knocked down in ovarian cancer cells by using two validated commercially available FGFR4-specific siRNAs (Hs_FGFR4_5 and Hs_FGFR4_6) and a nontarget scrambled siRNA sequence as a control (Qiagen Sciences). Transfection of ovarian cancer cells with the siRNAs was performed using siRNA duplexes at a final concentration of 10 nM and Lipofectamine RNAiMAX reagent (Life Technologies). Validation of successful FGFR4 knockdown was performed at the protein and mRNA levels using western blot analysis and quantitative real-time PCR analysis, respectively (Supplementary Fig. S1A).
The effect of FGFR4 expression on ovarian cell survival was determined by incubating high-grade serous ovarian carcinoma cell lines transfected with siRNAs in medium containing 2% fetal bovine serum for 3 days, after which the medium was removed, and water-soluble tetrazolium salt (WST-1) reagent (Roche Applied Science) diluted in serum-free media was added to the cells. After 2 hours of incubation, the assay was read using a FLUOstar Omega plate reader (BMG LABTECH). All data were normalized to respective controls.
A real-time cell proliferation assay was performed using the xCELLigence system (Roche Applied Science). This system uses measurement of electrical impedance, created by cells attached to the microelectrode integrated cell culture plated. to measure cell proliferation in real time (13). Cells were plated at 10,000 cells per well in serum-free Opti-MEM (Life Technologies) and allowed to attach for 12 hours. A unit-less parameter termed the cell index was derived and used to represent the cell number based on the measured relative change in electrical impedance that occurred in the presence and absence of cells in the wells (14). The cell index was normalized to the baseline reading at time point 0 following attachment and transfection. Cell proliferation as measured according to the cell index was observed for 3.5 days.
The invasion potential of three high-grade serous ovarian carcinoma cell lines (SKOV3, OVCA3 and OVCA433) was assessed using the Oris cell invasion assay (Platypus Technologies) as described by the manufacturer (15). In brief, cells were seeded at a density of 30,000 per well in serum-free Opti-MEM on a 96-well plate previously coated with a collagen I layer and with silicon stoppers covering the central areas of the wells to create a cell-free gap. The cells were then transfected with Hs_FGFR4_5 and Hs_FGFR4_6 and with the nontarget scrambled siRNA sequence for 24 hours, after which fresh culture medium containing 2% fetal bovine serum was added to the cells. 12 hours after the addition of fresh medium, the stoppers were removed, and cells were immediately overlaid with a thick layer of the same matrix. Cells were then allowed to invade into the collagen matrix at the central area of the wells for another 48 hours. Cells were then stained with calcein AM (Life Technologies). The invasion potential of the cells was assessed by measuring the fluorescence of labeled cancer cells invaded into the center of the collagen matrix. with a FLUOstar Omega plate reader. Photomicrographs were taken under a fluorescent microscope for each of the wells.
Apoptosis assays were performed with two high-grade serous ovarian carcinoma cell lines (SKOV3 and OVCA432) harvested after transfection with Hs_FGFR4_5 and Hs_FGFR4_6 and with the nontarget scrambled siRNA sequence for 72 hours using the Cell Death Detection cell ELISA system (Roche Diagnostics), which quantified the relative number of apoptotic cancer cells by colorimetrically measuring cell-free nucleosomes (16).
Effects of FGFR4 overexpression on ovarian cancer cell proliferation
To determine the effect of FGFR4 overexpression on cancer cell proliferation, FGFR4 was overexpressed in ovarian cancer cell lines OVCA429 and OVCA5, which have low endogenous FGFR4 expression levels. The full-length FGFR4 gene was delivered into the cells via transient transduction using FGFR4 lentiviral particles (GeneCopoeia). 48 hours after transduction, cells were harvested, and overexpression of FGFR4 in transduced cancer cells was confirmed. A cell proliferation assay was performed with FGFR4-overexpressing cells or control mock-transduced cells seeded on a 96-well plate at 2500 cells per well. 24 hours after cell seeding, cell proliferation was assayed by incubating cells with WST-1 for 1 hour, and the absorbance at 450 nm was measured.
Effect of FGFR4 silencing on FGF1-mediated cell signaling
OVCA432 cells stably transduced with different reporter response elements from the Cignal reporter system (SABiosciences) were transfected with Hs_FGFR4_5 and Hs_FGFR4_6 and with the nontarget scrambled siRNA sequence. They were then plated in 96-well plates at a density of 30,000 cells per well and allowed to attach to the plate for 24 hours. After cell attachment, cell culture medium was replaced with fresh medium containing 10 ng/mL FGF1 and incubated for 6 hours. Next, cells were lysed with cell lysis buffer and assayed according to the manufacturer’s instructions (Promega) using a FLUOstar Omega plate reader. Data were normalized to the respective negative control groups.
Effect of FGFR4 silencing on transcriptome profiles in ovarian cancer cells
Total RNAs were isolated from SKOV3 and OVCA432 cells transiently transfected with Hs_FGFR4_5 and Hs_FGFR4_6 and with the nontarget scrambled siRNA sequence for 36 hours. The isolated RNA samples were then subjected to whole-genome transcriptome profiling using the GeneChip Human Genome U133 Plus 2.0 array (Affymetrix). Differentially expressed genes (>twofold) common to both FGFR4-specific siRNAs and both cell lines were selected for further analysis using the Ingenuity Pathway Analysis program (Ingenuity Systems). Upregulation of two of the identified genes CXCR4 and BNIP3, was validated in OVCA432, OVCA433, and SKOV3 cells using TaqMan gene expression assays (Life Technologies).
To identify the key canonical pathways most relevant to the set of differential expressed genes identified in the FGFR4-silenced ovarian cancer cells, canonical pathway analysis was performed using the Ingenuity Pathway Analysis program. Differentially expressed genes with expression levels that differed more than twofold from those in both OVCA432 and SKOV3 cells were input into the program for analysis. P values <0.05 for key pathways were considered significant, and association of a specific pathway with the uploaded data set was unlikely to result from chance.
Targeting FGFR4 with FGFR4 trap proteins and siRNA in an orthotopic mouse model
The FGFR4 fusion trap protein FTP-091 (FGFR4mut:Fc:R4-Trap; FGFR4:Fc) generated by Five Prime Therapeutics was used for targeting FGFR4 in the in vivo study. FGFR4mut:Fc is a chimeric fusion protein consisting of the three Ig-like extracellular domains of FGFR4 (D1-3) with the acid box linker between D1 and D2 replaced with the corresponding acid box region of FGFR1. The Biacore profile for the affinity of FGFR4:Fc to different FGF ligands is shown in Supplementary Table S2. Intraperitonal tumor growth in a murine orthotopic model was used as described previously (17). Briefly, a firefly luciferase-expressing serous ovarian cancer cell line (OVCA432-Luc) was generated by transfecting the paternal cell line with commercially available lentiviral particles (GenTarget) and selected for luciferase expressing cells using puromycin. Mice received intraperitoneal injections of 1 × 106 OVCA432-Luc cells and were subsequently placed in experimental and control groups of 12 mice each. The experimental group received 20 mg/kg FTP-091 twice weekly, whereas the control group received pooled human IgGs (Sigma-Aldrich) at the same dose and schedule. Weekly bioluminescent imaging, performed on the IVIS 100 imaging system (Caliper Life Sciences, Hopkinton, MA), was used to follow intraperitoneal tumor nodule development for 4 weeks, after which the animals were euthanized by CO2 inhalation followed by cervical dislocation. All visible tumors were resected and weighed. Ex vivo bioluminescent imaging of all of the tumors in the mice was performed, and the signal intensity of ex vivo tumors was quantified using the Living Image software (Caliper Life Sciences).
In addition, an in vivo siRNA silencing study was performed. Specifically, 2 × 106 OVCA432-Luc cells suspended in phosphate-buffered saline (PBS) were intraperitoneally injected into 30 nude mice. The mice were then randomized and placed in three groups for injection of dioleoyl phosphatidylcholine (DOPC) nanoliposomes containing a control siRNA (control-siRNA-DOPC), Hs_FGFR4_5 (siRNA-Hs_FGFR4_5-DOPC), or Hs_FGFR4_6 (siRNA-Hs_FGFR4_6-DOPC). DOPC nanoliposomes were prepared as described previously (18). Briefly, DOPC and siRNA were mixed and lyophilized. The lyophilized preparations were hydrated in PBS at a concentration of 5 µg/100 µL before administration. The DOPC nanoliposomes were determined to have a mean diameter of 30 nm. Injection of nanoliposomes began 1 week after the initial cancer cell injection. Each mouse was intraperitoneally injected with 5 µg of the corresponding siRNA-DOPC reconstituted in 200 µL of Ca++ and Mg++ free PBS. Intraperitoneal injections were performed twice a week for 7 weeks, after which the mice were euthanized by CO2 inhalation followed by cervical dislocation and submitted to necropsy. The mice and their tumors were weighed, and resected tumors were processed for histologic evaluation.
Immunostaining of tissue sections from the orthotopic mouse model
Ki-67 staining was performed to visualize proliferating ovarian cancer cells using a polyclonal anti-human Ki-67 antibody (1:200; Life Technologies) with paraffin-embedded ovarian tumor sections obtained in the FGFR4 trap protein and in vivo siRNA silencing studies. Slides were incubated with the antibody and staining was visualized by incubating with fast red chromogen. For each tissue section, the Ki-67-positive cancer cells in five random fields under a ×20 objective were counted, and the average number of Ki-67-positive cells per unit tumor area was calculated.
To validate the differential expression of the candidate genes identified from our microarray study, immunolocalization of the two genes with upregulated expression identified in the FGFR4-knockdown cancer cells, CXCR4 and BNIP3, was performed using polyclonal anti-human CXCR4 (1:200; LifeSpan BioSciences) and polyclonal anti-human BNIP3 (1:200; Sigma-Aldrich) antibodies with tumor tissue sections obtained in the in vivo FGFR4 trap protein study. The staining intensity in each section in both treatment groups was quantified and presented in a box plot.
Statistical analysis
The SPSS software (version 17; IBM Corporation) was used to perform all statistical tests. All in vitro experiments were repeated independently in triplicate. Data were pooled with mean and standard error of the mean values calculated for all end points where appropriate. The two-tailed Student t-test was used to test the significance of differences in sample means for data with normally distributed means. Alternatively, the Mann-Whitney U test was used for the analysis of nonparametric data. Kaplan-Meier survival curves were generated and compared using a two-sided log-rank statistic. The Cox proportional hazards model was used for the multivariate analysis of patient survival. The Pearson correlation coefficient was used to test linear associations. A P value <0.05 was considered significant.
Results
FGFR4 overexpression is associated with overall survival
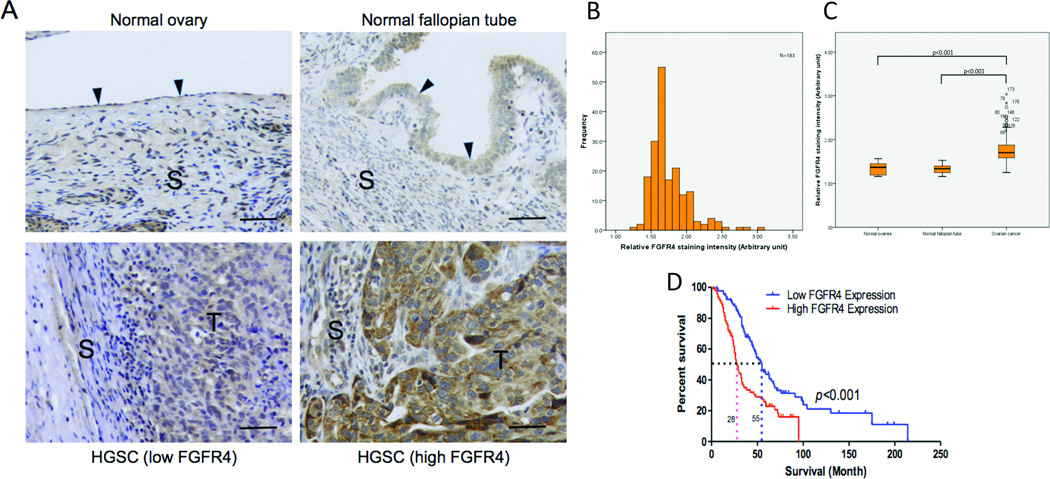
Previous studies showed that overexpression of FGF1, which is located on chromosome 5q31-35, was significantly associated with poor survival (10). Because FGFR4 is one of the key receptors for FGF1, we evaluated the association between FGFR4 expression and outcomes in high-grade serous ovarian carcinoma patients. Staining of both normal ovarian tissue and ovarian tumor samples demonstrated cell surface expression of FGFR4. Immunolocalization of FGFR4 in the 183 high-grade serous ovarian carcinoma samples showed that FGFR4 expression was significantly higher in the tumor samples than in the normal ovarian and fallopian tube tissue samples, which are postulated to be the tissues of origin for the majority of high-grade serous ovarian carcinomas (P < 0.01) (Fig. 1A–C) (19). Furthermore, using the median staining intensity as a cutoff, Kaplan-Meier analysis showed a significant correlation between high FGFR4 protein expression and poor overall survival (log-rank test; P < 0.001) (Fig. 1D). We further confirmed the prognostic significance of FGFR4 expression using Cox regression analysis after adjusting for age and debulking status (hazard ratio, 2.1; 95% CI:1.4–4.3; P < 0.01)
Figure 1.
Overexpression of FGFR4 protein is associated with overall survival. (A) Immunolocalization of FGFR4 demonstrating low FGFR4 protein expression in normal ovarian and fallopian tube tissue samples and low and high FGFR4 expression in an advanced-stage, high-grade serous ovarian carcinoma sample. S, stroma; T, tumor cells; bar = 10 µg. (B) Histogram showing the distribution of FGFR4 signaling intensity in the 183 high-grade serous ovarian carcinoma samples used in the survival analysis. (C) Box plot showing significantly higher FGFR4 protein expression in high-grade serous ovarian tumor samples than in normal ovarian and fallopian tube tissue samples. The bottom of the box indicates the 25th percentile, the top indicates the 75th percentile, and the whiskers represent 95% confidence intervals. (D) Kaplan-Meier analysis of the 183 study patients with advanced-stage, high-grade serous ovarian carcinoma showing a significant correlation between FGFR4 protein expression and overall survival with use of the median FGFR4 staining intensity as the cutoff (log rank test; P < 0.001). The median survival in the high FGFR4 expression group was 24 months, whereas that in the low expression group was 55 months. Correlation of FGFR4 protein expression with survival was maintained after stratification according to age and debulking status.
FGFR4 silencing decreases ovarian cancer cell survival, proliferation, and invasion potential in vitro
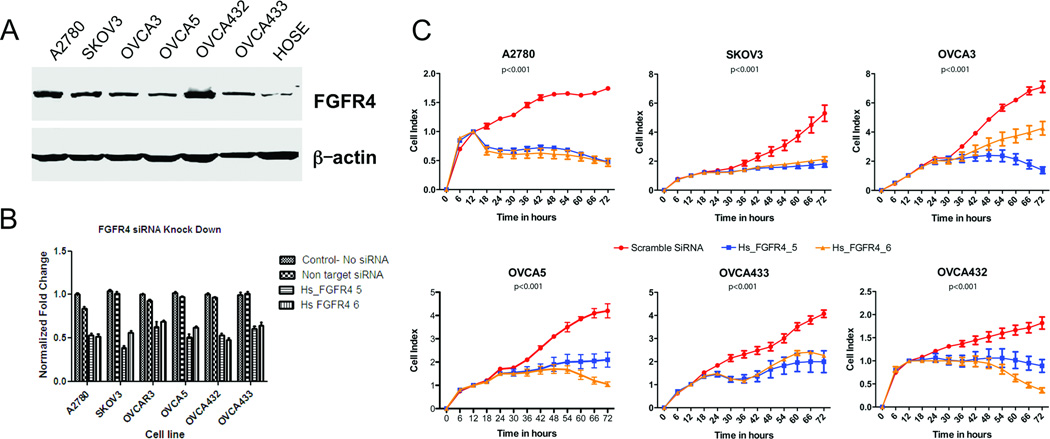
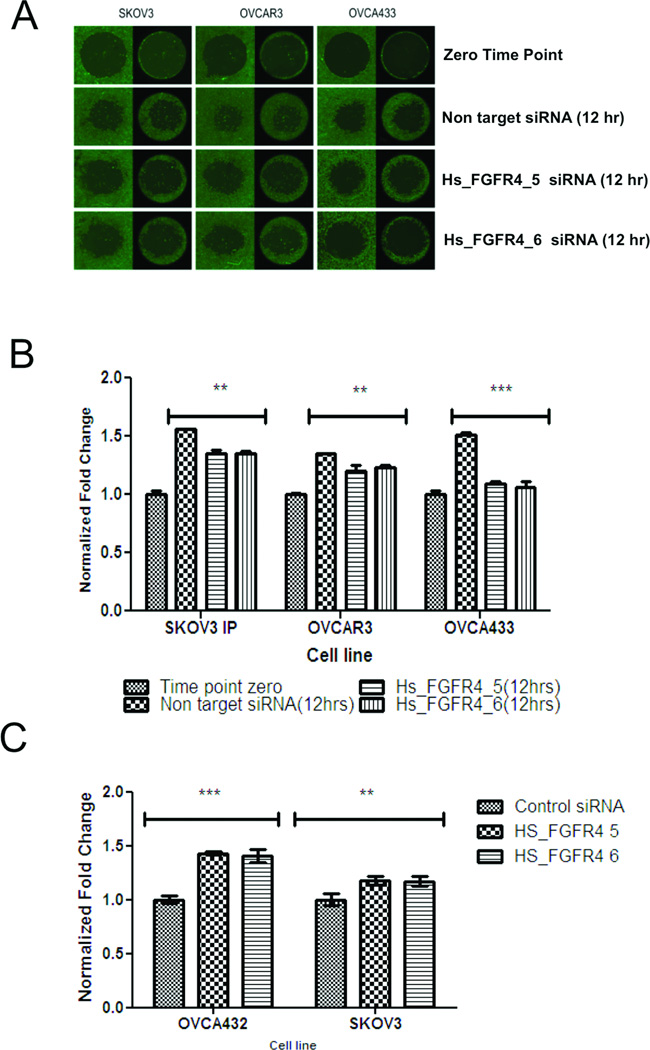
The association between FGFR4 expression and survival suggests that FGFR4 expression plays a key role in ovarian cancer progression. To determine the effect of FGFR4 expression on ovarian cancer cells, we first evaluated FGFR4 overexpression using Western blot analysis with the ovarian cancer cell lines A2780, SKOV3, OVCA3, OVCA5, OVCA432, and OVCA433 and compared it with that in HOSE cells. All six ovarian cancer cell lines exhibited higher FGFR4 expression (Fig. 2A). We confirmed successful FGFR4 silencing in all of the cell lines by the FGFR4-specific siRNAs Hs_FGFR4_5 and Hs_FGFR4_6 using quantitative PCR and Western blot analyses (Supplementary Fig. S1B and C). Cell survival analysis showed that transfection with both FGFR4-targeting siRNAs in all six cell lines significantly decreased cell survival in serum-reduced media more so than did transfection with the nontarget scrambled siRNA sequence and untransfected control (Fig. 2B). Furthermore, cell proliferation assays showed that all six ovarian cancer cell lines transfected with FGFR4-specific siRNAs demonstrated significant decreases in proliferation after 18–24 hours (Fig. 2C). Moreover, invasion assays with three cell lines that exhibited invasion potential in vitro demonstrated that the number of FGFR4-specific siRNA-transfected cells invading into the central zone of the Oris assay was significantly lower than that of the nontarget siRNA transfected control cells (Fig. 3A and B). Finally, the number of SKOV3 and OVCA432 cells transfected with Hs_FGFR4_5 and Hs_FGFR4_6 that were apoptotic was significantly higher than that of the cells transfected with the nontarget scrambled siRNA sequence (Fig. 3C).
Figure 2.
FGFR4 silencing in vitro decreases ovarian cancer cell survival and proliferation. (A) Western blot analysis showing higher levels of FGFR4 expression in the ovarian cancer cell lines A2780, SKOV3, OVCA3, OVCA5, OVCA432, and OVCA433 than in the normal ovarian epithelial cell. Fifty micrograms of protein was loaded in each lane. The same blot was probed with an anti-β-actin antibody for the normalization of protein loading and transfer. (B) Bar charts showing the survival of high-grade serous ovarian cancer cell lines after transfection with Hs_FGFR4_5 and Hs_FGFR4_6 and with the nontarget scrambled siRNA and incubation for 72 hours in serum-reduced media. Cells transfected with the FGFR4-specific siRNAs had significantly decreased survival than did those transfected with the control siRNA as measured using WST-1 assays (P < 0.001). (C) Real-time cell proliferation assay using electric impedence as a measure of ovarian cancer cell proliferation. Electrical impedance was normalized according to background measurement at time point 0. Results demonstrated a significantly lower rate of proliferation of high-grade serous ovarian carcinoma cells transiently transfected with Hs_FGFR4_5 and Hs_FGFR4_6 than of cells transfected with the nontarget scrambled siRNA (P < 0.001).
Figure 3.
FGFR4 silencing decreases ovarian cancer cell invasion potential in vitro. (A) Fluorescent micrographs showing SKOV3, OVCA3, and OVCA433 cells transiently transfected with Hs_FGFR4_5 and Hs_FGFR4_6 and the nontarget scrambled siRNA invading into the central zone of the Oris assay at 0 and 12 hours. Cells were then stained with calcein AM for visualization. (B) Bar charts showing significantly lower invasion potential of SKOV3, OVCA3, and OVCA433 cells after transfection with FGFR4-specific siRNAs than of control cells. *P = 0.024; **P = 0.003; ***P = 0.001. Error bars represent standard errors of the mean. (C) Bar chart showing significantly higher apoptosis rates in response to serum starvation as measured according to increased number of cell-free nucleosomes in SKOV3 and OVCA432 cells after transfection with FGFR4-specific siRNAs than in the control cells. **P < 0.01; ***P < 0.001. Error bars represent standard errors of the mean.
FGFR4 overexpression promotes ovarian cancer cell proliferation
Our results showed that FGFR4 overexpression in OVCA429 and OVCA5 cells, which have low levels of endogenous FGFR4 expression, promoted cell proliferation. The rate of proliferation of both cell lines was more than 50% higher than that of their corresponding control cells at 24 hours after initial cell seeding (Supplementary Fig. S2A and B), suggesting that overexpression of FGFR4 led to an aggressive phenotype.
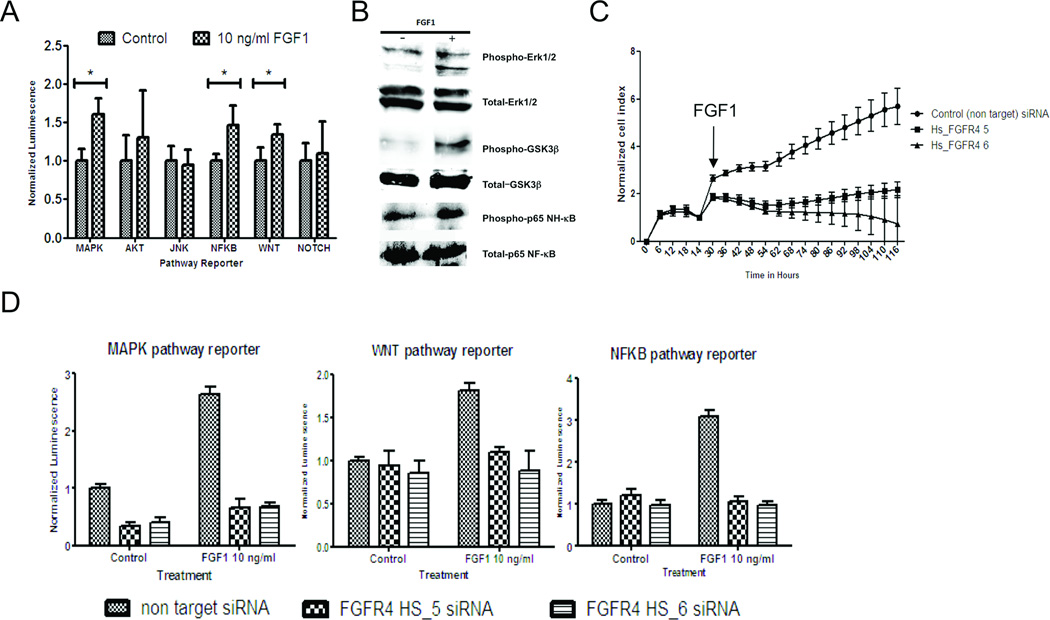
FGFR4 silencing abrogates the effect of FGF1 on ovarian caner cell growth and downstream signaling pathway activation
Researchers have shown that FGFR4 preferentially binds to FGF1 (11, 12), suggesting that FGF1-activated downstream signaling pathways are mediated by FGFR4. To confirm this in ovarian cancer cells, we first treated OVCA432 cells with exogenous FGF1 (1–10 ng/mL), which produced a significant increase in cell proliferation rates (Supplementary Fig. S2C). The cells also demonstrated a significant increase in reporter activity, which reflected activation of the mitogen-activated protein kinase (MAPK), NF-κB, and WNT signaling pathways (Fig. 4A). In addition, Western blot analysis demonstrated a significant increase in phosphorylated form of extracellular signal-regulated kinase (ERK) 1/2 and glycogen synthase kinase 3β (GSK-3β) (Fig. 4B). However, when we treated OVCA432 cells transfected with FGFR4-specific siRNAs with FGF1 (10 ng/mL), their growth rates were significantly lower than those of cells transfected with the nontarget scrambled siRNA (P < 0.001) (Fig. 4C). Furthermore, the reporter activities of the MAPK, NF-κB, and WNT signaling pathways activated by FGF1 were abrogated (Fig. 4D).
Figure 4.
FGFR4 silencing abrogates the effect of FGF1 on ovarian cancer cell growth and downstream signaling pathway activation. (A) Bar chart showing a significant increase in reporter activity in FGF1-treated OVCA432 cells stably transfected with the Cignal lenti reporter system for the MAPK, NF-κB, and WNT signaling pathways. *P < 0.001. (B) Western blot analysis demonstrating significantly higher level of phosphorylated ERK1/2, GSK-3β, and NF-κB protein in OVCA432 cells treated with FGF1 at 10 ng/mL for 1 hour (+) versus control cells (−). (C) Real-time cell proliferation assay results showing a significantly lower proliferation rate in OVCA432 cells transfected with Hs_FGFR4_5 and Hs_FGFR4_6 than in those transfected with the nontarget scrambled siRNA under treatment with FGF1 at 10 ng/mL (P < 0.001). (D) Bar charts showing the effect of FGF1 on reporter activity, which was significantly lower in ovarian cancer cells transfected with Hs_FGFR4_5 and Hs_FGFR4_6 than in those transfected with the scrambled siRNA (P < 0.001).
FGFR4 silencing alters transcriptome profiles in ovarian cancer cells
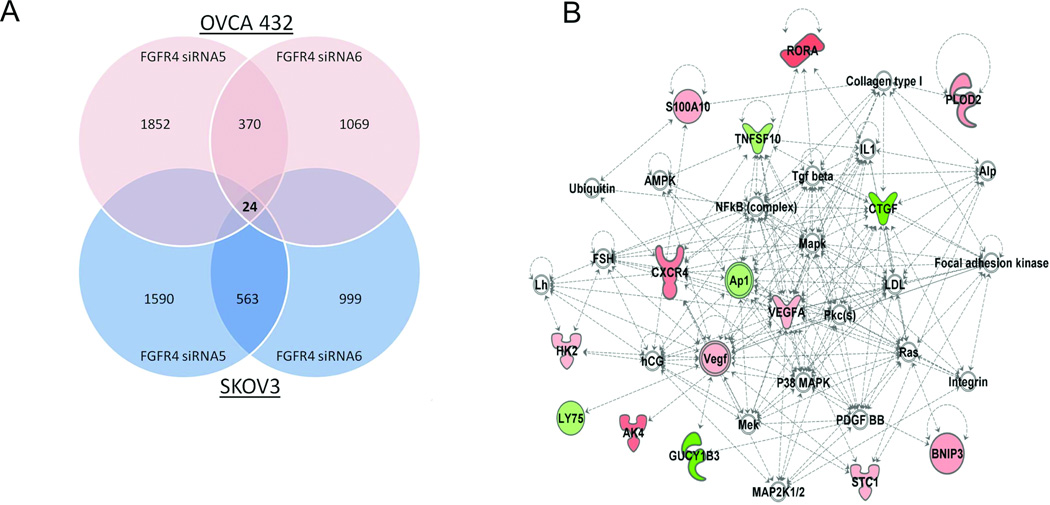
To further delineate the underlying molecular mechanism of FGFR4’s control of the malignant phenotypes of ovarian cancer cells, we performed transcriptome profiling analysis on SKOV3 and OVCA432 cells transiently transfected with Hs_FGFR4_5 and Hs_FGFR4_6 and with the nontarget scrambled siRNA sequences. A total of 24 genes were identified to be altered by over twofold in expression, which are common for both OVCA432, and SKOV3 cells transiently transfected with the siRNAs than in those transfected with the scrambled siRNA sequence (Fig. 5A; Supplementary Table S3; Gene Expression Omnibus, GES34828). Network functions analysis showed that of these 24 genes, 13 were associated with free radical scavenging, cell death, cellular growth, and proliferation (Table 1). Two of these genes were CXCR4 and BNIP3 (Fig. 5B) We further validated upregulation of the expression of CXCR4, which regulates cell proliferation and apoptosis upon binding of its ligand CXCL12 in T cells and cancer cells (20, 21), and of BNIP3, which induces apoptosis (22), using quantitative -PCR analysis of three ovarian cancer cell lines in which FGFR4 was silenced by siRNA (Supplementary Fig. S3A and B). In addition, pathway analyses showed that several canonical signaling pathways, including ERK/MAPK (P = 0.03), NF-κB (P = 0.05), and tumor necrosis factor receptor 1 (TNFR1) (P = 0.04), were significantly associated with the differentially expressed gene signature identified in the transcriptome profiling study. Identification of the MAPK and NF-κB pathways in the array data was consistent with the results of the reporter assay.
Figure 5.
Transcriptome analysis on FGFR4 silenced ovarian cancer cell lines. (A) Venn diagram showing the number of differentially expressed genes (>twofold) common to both OVCA432 and SKOV3 cells transiently transfected with Hs_FGFR4_5 and Hs_FGFR4_6 for 36 hours when compared with those transfected with the nontarget scrambled siRNA. (B) The most significant network associated with FGFR4 silencing. Genes with upregulated and downregulated expression are shown in red and green, respectively.
Table 1.
Networks involved by the differentially expressed genes in FGFR4 silenced ovarian cancer cells. Function networks involved by the differentially expressed genes (>twofold) common to OVCA432 and SKOV3 cells. Network scores were calculated by the Ingenuity Pathway Analysis software program using the Fisher exact test according to the number of network-eligible molecules in a specific network, molecules analyzed in the submitted data set, and molecules in the Ingenuity Pathway Analysis database. The network score equals −log (Fisher exact test result): the higher the score, the lower the chance of getting the corresponding network when randomly picking molecules in the Ingenuity Pathway Analysis database.
| Top Networks | |||
|---|---|---|---|
| ID | Associated Network Functions | Score | Focus Molecules |
| 1 | Free Radical Scavenging, Cell Death, Cellular Growth and Proliferation | 34 | 13 |
| 2 | Cell Death, Free Radical Scavenging, Lipid Metabolism | 13 | 6 |
| 3 | Amino Acid Metabolism, Small Molecule Biochemistry, Cellular Growth and Proliferation | 2 | 1 |
| 4 | Gene Expression, Cell-To-Cell Signaling and Interaction, Molecular Transport | 2 | 1 |
Targeting FGFR4 suppresses ovarian tumor growth in vivo
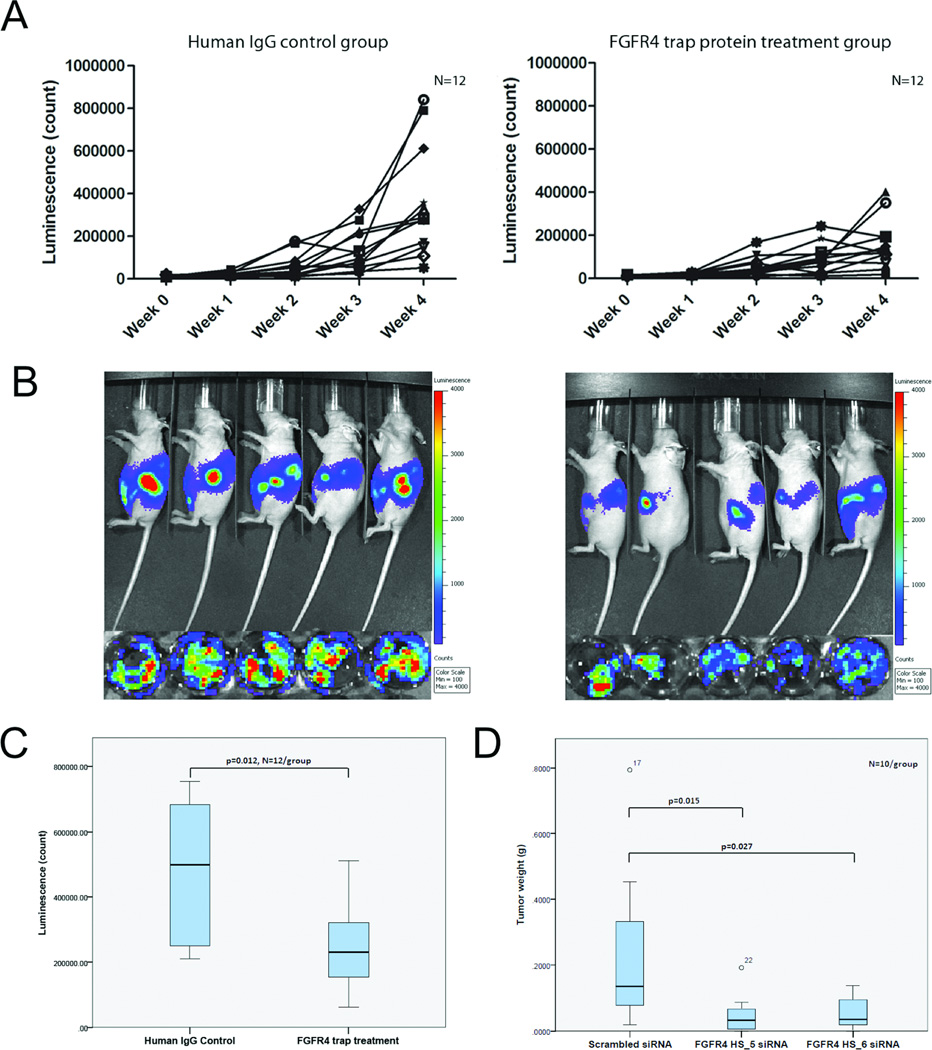
The antitumor effect of silencing of FGFR4 in vitro suggests that targeting FGFR4 is a new strategy for treatment of ovarian cancer. We therefore examined the effect of blocking the binding of ligands to FGFR4 using the novel FGFR4 fusion trap protein FTP-091 (FGFR4mut:Fc:R4-Trap; FGFR4:Fc) and downregulating FGFR4 expression by injection of DOPC nanoliposomes containing FGFR4-specific siRNAs in a murine xenograft OVCA432 ovarian tumor model. The results showed that mice injected with 20 mg/kg FTP-091 had significantly lower luciferase activity in ex vivo tumors than did those given control human IgG (Fig. 6A–C), illustrating a decrease in the number of viable tumor cells in mice given the trap protein. Furthermore, mice injected with FGFR4 HS_5 and FGFR4 HS_6 in DOPC nanoliposomes had significantly smaller tumors than did those injected with scrambled siRNA (Fig. 6D). Histologic examination of the resected tumor tissue collected from the xenografts showed that the tumors had histologies consistent with serous ovarian carcinoma. Ki-67 staining of tumor sections obtained in the FGFR4 trap protein and siRNA studies demonstrated significantly fewer Ki-67–positive cancer cells per unit tumor area in the FGFR4 trap protein-injected mice and in the FGFR4-knockdown mice (via injection of siRNAs in DOPC nanoliposomes) than in their corresponding controls (Supplementary Fig. S4A and B), suggesting that targeting FGFR4 in vivo can suppress ovarian tumor growth via inhibition of cancer cell proliferation. Furthermore, immunolocalization of CXCR4 and BNIP3 protein in tumor samples collected in the FGFR4 trap protein study demonstrated significantly lower expression of both proteins in the FGFR4 trap protein-treated mice than in their controls, suggesting that blockage of FGFR4 signaling with the trap fusion protein upregulated the expression of the CXCR4 and BNIP3 genes (Supplementary Fig. S4C and D), which was consistent with our findings of transcriptome profiling of FGFR4-silenced ovarian cancer cells in vitro.
Figure 6.
Targeting FGFR4 decreases ovarian cancer cell growth in vitro. (A) Bioluminescence signals in 12 mice given 20 mg/kg FGFR4:Fc and 12 mice given control IgG from week 0 to week 4. (B) Bioluminescent images of mice given FGFR4:Fc and those given human IgG at week 4 and their corresponding ex vivo tumor samples. (C) Box plot showing significantly lower bioluminescence signals in the 12 mice given FGFR4:Fc than in those given control IgG in week 4 (P = 0.012). The bottom of the box indicates the 25th percentile, the top indicates the 75th percentile, and whiskers indicate 95% confidence intervals. (D) Box plot showing significantly lower tumor weights in OVCA432 tumor-bearing mice given siRNA-Hs_FGFR4_5-DOPC and siRNA-Hs_FGFR4_6-DOPC than in those given control-siRNA-DOPC (P = 0.015 and P = 0.027, respectively). Values greater than one and a half box lengths from either end of the box are denoted by circles and identified as outliers. All values, including the outliers in all experimental groups, were included in statistical analyses.
Discussion
In a previous study, we demonstrated decreased survival in patients with high-grade serous carcinomas harboring amplification of 5q31-35.3. Specifically, we observed that overexpression of FGF1, located in this amplicon, was a poor prognostic indicator for these tumors (10). One of the key receptors for FGF1 is FGFR4, which is located on the same amplicon and preferentially binds to FGF1 (11, 12). In the present study, we demonstrated that FGFR4 overexpression is associated with a more aggressive high-grade serous carcinoma phenotype in vitro and in vivo, suggesting that FGF axis activation through overexpression of both FGFRs and FGF ligands may represent a targetable autocrine signaling loop associated with poor overall survival in ovarian cancer patients.
Birrer and colleagues observed a 25% gain frequency of chromosome segment 5q31-35.3 harboring the FGFR4 gene in ovarian cancer cases (10). Their data also demonstrated a significant correlation between FGFR4 copy number and overall survival. However, recently reported data from The Cancer Genome Atlas (TCGA) demonstrated a 6% amplification/overexpression rate for FGFR4 with no significant correlation with survival in ovarian cancer cases (23). The discrepancy between these two sets of data may result from several factors. First, the TCGA tumor samples were bulk tissue samples with stromal cell contamination rates ranging from 5% to 50%. Tissue samples with high levels of stromal contamination will show significantly low levels of FGFR4 gene amplification and/or expression, as the FGFR4 gene and mRNA copies are diluted by the stromal DNA and mRNA. This may impact survival correlation studies. In comparison, in our analyses, we used DNA and RNA extracted from microdissected ovarian tumor cells, which had minimal stromal DNA and RNA contamination. Second, the TCGA samples were collected from multiple institutions, whereas we collected our samples at a single institution, which may have given us a more homogenous patient population. Third, we found a significant correlation between FGFR4 protein expression and survival. Whereas FGFR4 amplification may not correlate with survival as indicated in the TCGA data set, FGFR4 protein expression may correlate with it, as gene amplification and mRNA expression may not correlate with protein expression owing to mechanisms that regulate mRNA and/or protein expression. However, researchers have not immunohistochemically evaluated FGFR4 protein expression in the TCGA data set.
FGFR4 differs from the other three members of the FGFR family in genomic structure, ligand binding, and signal transduction (24). FGFR activation, either by activating mutations (25–29) or overexpression (30–35), occurs in multiple solid tumors. FGFR2 and FGFR3 mutations are common in endometrial cancer (36) and bladder cancer (31). In comparison, FGFR1 and FGFR4 mutations are not common in carcinoma cells; instead, overexpression of FGFR1 and FGFR4 is more prevalent (37). To exclude the presence of several rare activating mutations of FGFR4, we sequenced DNA isolated from microdissected high-grade serous carcinoma samples. We did not identify any mutations in either the kinase or intermembrane domain except for the Gly388Arg polymorphism, which has exhibited no effect on cancer prognosis (38), in 6 of 43 (14%) high-grade serous carcinoma samples. Our data suggest that, similar to breast cancer (39), FGFR overexpression is the main mechanism implicated in high-grade serous ovarian carcinoma.
Unlike other FGFR family members, FGFR4 preferentially binds to acidic FGF (FGF1) (11, 12). Our prior work demonstrated that poor survival and related phenotypic changes induced by FGF1 in high-grade serous carcinoma parallel those in FGFR4. Hence, overexpression of both FGF1 and FGFR4 may provide an autocrine loop that drives high-grade serous ovarian carcinoma growth. This may result from activation of the MAPK/ERK signaling pathway by FGF1 as described previously (40) and confirmed by the pathway reporter and Western blot analysis data in the present study. In addition to activation of the MAPK/ERK pathway, we observed activation of the proliferative NF-κB and WNT signaling pathways in FGF1-treated cells, which our pathway reporter and Western blot analysis confirmed. In addition, our canonical pathway analysis of transcriptome profiling data demonstrated a significant correlation between these pathways and the differentially expressed genes identified by manipulating the level of FGFR4 expression in ovarian cancer cells. These data suggest that multiple pathways can be activated by FGF1, most likely mediated via FGFR4, because the effect of FGF1 on pathways activation can be abrogated by downregulation of FGFR4 expression. Furthermore, our data suggest that the effects of FGFR4 on ovarian cancer cell proliferation and survival result from upregulation of expression of CXCR4, which can regulate cell proliferation and apoptosis upon binding of its ligand CXCL12 in T cells and cancer cells (20, 21), and from upregulation of expression of BNIP3, which is known to induce apoptosis (22). We confirmed upregulation of these two proteins in the ovarian tumor samples obtained from mice given the FGF trap protein. However, the molecular mechanisms involved by these two proteins in mediating the effect of the FGFR4 signaling pathway on ovarian cancer cell growth remain to be elucidated.
In the absence of activating mutations of FGFR4, downregulation of FGFR4 expression and prevention of binding of ligands to FGFR4 may be effective in the treatment of serous ovarian cancer. Using two strategies for targeting FGFR4 with an orthotopic mouse model of high-grade serous carcinoma, we demonstrated that decreasing FGFR4 expression leads to a decrease in tumor growth. Although the use of FGFR4 siRNAs in vitro can completely silence FGFR4 expression, leading to decreased proliferation and survival of ovarian cancer cell lines, ovarian tumors, although small, still developed in mice given FGFR4-specific siRNAs delivered in DOPC nanoliposomes. In addition, the tumors had FGFR4 expression, suggesting that the nanoliposomes may not efficiently deliver the siRNAs to all tumor cells. In addition to the use of siRNAs, our data demonstrated that use of the soluble fusion protein FPT-091 containing the extracellular domain of FGFR4 and an IgG Fc fragment may have significant clinical applications. Replacement of the FGFR4 acid box region with the corresponding FGFR1 region improved the bioavailability and pharmacokinetics of the fusion protein in vivo. The chimeric FGFR4/FGFR1 extracellular domain in FGFR4mut:Fc is fused to the Fc fragment of IgG1 and can bind to several members of the FGF protein family, including FGF1. The ability to sequester several FGF ligands and inhibit the pathway upstream of the FGF receptor enables the trap protein to inhibit all of the possible effects of overexpression of FGFs and FGFR4. Given the high degree of genetic heterogeneity of high-grade serous carcinomas, activation of multiple pathways and/or targets, such as Notch3 (41) and NAC1 (42), may be involved in the pathogenesis of different ovarian cancer patients. A challenge remains in identifying ovarian cancer patients who may benefit from targeting a specific pathway or pathways, possibly via individual tumor expression profiling.
In conclusion, the present study demonstrated that overexpression of FGFR4 is an indicator of poor prognosis for high-grade serous ovarian carcinoma. It also identified mechanisms by which activation of FGFR4 by ligands such as FGF1 may lead to an aggressive cancer phenotype. Successful targeting of FGF axis actication in our orthotopic mouse model suggests that this approach is feasible in clinical trials.
Supplementary Material
Translational Relevance.
High grade serous carcinoma accounts for the majority of epithelial ovarian cancer. Prognostic or predictive biomarkers that can stratify patients for treatment are lacking. This study seeks to validate the prognostic value of FGFR4 in high grade serous carcinoma overexpression, to delineate the functional role of FGFR4 in ovarian cancer progression and to evaluate the feasibility of targeting FGFR4 in serous ovarian cancer treatment. The results of this study demonstrated that FGFR4 protein is a prognostic marker for advanced stage high grade serous carcinoma, and targeting ovarian cancer expressing high levels of the protein with FGFR4 specific siRNAs and FGFR4 trap protein suppressed ovarian tumor growth in a xenograft mouse model. These studies provide evidences that silencing FGFR4, or inhibiting ligand-receptor binding can significantly inhibit ovarian cancer progression, suggesting that targeting ovarian cancer expressing high levels of the protein may be a new modality in the treatment of the disease and improve survival.
Acknowledgments
Grant Support
This study was supported in part by grants R01 CA133057 (S.C.M), RC4 CA156551 (M.J.B.), and U54 CA151668 (A.K.S.); the Gilder Foundation (A.K.S.); and MD Anderson Ovarian Cancer SPORE grant P50 CA083639 (R.C.B. and S.C.M.) from the National Institutes of Health. This research is supported in part by the MD Anderson Cancer Center Support Grant CA016672.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.NIH consensus conference. Ovarian cancer. Screening, treatment, and follow-up. NIH Consensus Development Panel on Ovarian Cancer. JAMA. 1995;273:491–497. [PubMed] [Google Scholar]
- 3.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 4.McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, et al. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med. 1996;334:1–6. doi: 10.1056/NEJM199601043340101. [DOI] [PubMed] [Google Scholar]
- 5.Markman M, Rothman R, Hakes T, Reichman B, Hoskins W, Rubin S, et al. Second-line platinum therapy in patients with ovarian cancer previously treated with cisplatin. J Clin Oncol. 1991;9:389–393. doi: 10.1200/JCO.1991.9.3.389. [DOI] [PubMed] [Google Scholar]
- 6.Yap TA, Carden CP, Kaye SB. Beyond chemotherapy: targeted therapies in ovarian cancer. Nat Rev Cancer. 2009;9:167–181. doi: 10.1038/nrc2583. [DOI] [PubMed] [Google Scholar]
- 7.Burger RA. Overview of anti-angiogenic agents in development for ovarian cancer. Gynecol Oncol. 2011;121:230–238. doi: 10.1016/j.ygyno.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 8.Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- 9.Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, et al. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 28:2512–2519. doi: 10.1200/JCO.2009.26.9589. [DOI] [PubMed] [Google Scholar]
- 10.Birrer MJ, Johnson ME, Hao K, Wong KK, Park DC, Bell A, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J Clin Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 11.Kan M, Wu X, Wang F, McKeehan WL. Specificity for fibroblast growth factors determined by heparan sulfate in a binary complex with the receptor kinase. J Biol Chem. 1999;274:15947–15952. doi: 10.1074/jbc.274.22.15947. [DOI] [PubMed] [Google Scholar]
- 12.Partanen J, Makela TP, Eerola E, Korhonen J, Hirvonen H, Claesson-Welsh L, et al. FGFR-4, a novel acidic fibroblast growth factor receptor with a distinct expression pattern. EMBO J. 1991;10:1347–1354. doi: 10.1002/j.1460-2075.1991.tb07654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irelan JT, Wu MJ, Morgan J, Ke N, Xi B, Wang X, et al. Rapid and quantitative assessment of cell quality, identity, and functionality for cell-based assays using real-time cellular analysis. J Biomol Screen. 2011;16:313–322. doi: 10.1177/1087057110397359. [DOI] [PubMed] [Google Scholar]
- 14.Xing D, Orsulic S. A genetically defined mouse ovarian carcinoma model for the molecular characterization of pathway-targeted therapy and tumor resistance. Proc Natl Acad Sci U S A. 2005;102:6936–6941. doi: 10.1073/pnas.0502256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carragher NO, Frame MC. Modelling distinct modes of tumour invasion and metastasis. Drug Discovery Today: Disease Models. 2011;8:103–112. [Google Scholar]
- 16.Holdenrieder S, Stieber P, Chan LY, Geiger S, Kremer A, Nagel D, et al. Cell-free DNA in serum and plasma: comparison of ELISA and quantitative PCR. Clin Chem. 2005;51:1544–1546. doi: 10.1373/clinchem.2005.049320. [DOI] [PubMed] [Google Scholar]
- 17.Liao S, Liu J, Lin P, Shi T, Jain RK, Xu L. TGF-beta blockade controls ascites by preventing abnormalization of lymphatic vessels in orthotopic human ovarian carcinoma models. Clin Cancer Res. 2011;17:1415–1424. doi: 10.1158/1078-0432.CCR-10-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray MJ, Van Buren G, Dallas NA, Xia L, Wang X, Yang AD, et al. Therapeutic targeting of neuropilin-2 on colorectal carcinoma cells implanted in the murine liver. J Natl Cancer Inst. 2008;100:109–120. doi: 10.1093/jnci/djm279. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn E, Kurman RJ, Shih IM. Ovarian cancer is an imported disease: fact or fiction? Curr Obstet Gynecol Rep. 2012;1:1–9. doi: 10.1007/s13669-011-0004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colamussi ML, Secchiero P, Gonelli A, Marchisio M, Zauli G, Capitani S. Stromal derived factor-1 alpha (SDF-1 alpha) induces CD4+ T cell apoptosis via the functional up-regulation of the Fas (CD95)/Fas ligand (CD95L) pathway. J Leukoc Biol. 2001;69:263–270. [PubMed] [Google Scholar]
- 21.Drury LJ, Wendt MK, Dwinell MB. CXCL12 chemokine expression and secretion regulates colorectal carcinoma cell anoikis through Bim-mediated intrinsic apoptosis. PLoS One. 2010;5:e12895. doi: 10.1371/journal.pone.0012895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray R, Chen G, Vande Velde C, Cizeau J, Park JH, Reed JC, et al. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J Biol Chem. 2000;275:1439–1448. doi: 10.1074/jbc.275.2.1439. [DOI] [PubMed] [Google Scholar]
- 23.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vainikka S, Partanen J, Bellosta P, Coulier F, Birnbaum D, Basilico C, et al. Fibroblast growth factor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1992;11:4273–4280. doi: 10.1002/j.1460-2075.1992.tb05526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Behrens C, Lin HY, Lee JJ, Raso MG, Hong WK, Wistuba II, et al. Immunohistochemical expression of basic fibroblast growth factor and fibroblast growth factor receptors 1 and 2 in the pathogenesis of lung cancer. Clin Cancer Res. 2008;14:6014–6022. doi: 10.1158/1078-0432.CCR-08-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allerstorfer S, Sonvilla G, Fischer H, Spiegl-Kreinecker S, Gauglhofer C, Setinek U, et al. FGF5 as an oncogenic factor in human glioblastoma multiforme: autocrine and paracrine activities. Oncogene. 2008;27:4180–4190. doi: 10.1038/onc.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baird K, Davis S, Antonescu CR, Harper UL, Walker RL, Chen Y, et al. Gene expression profiling of human sarcomas: insights into sarcoma biology. Cancer Res. 2005;65:9226–9235. doi: 10.1158/0008-5472.CAN-05-1699. [DOI] [PubMed] [Google Scholar]
- 29.Toyokawa T, Yashiro M, Hirakawa K. Co-expression of keratinocyte growth factor and K-sam is an independent prognostic factor in gastric carcinoma. Oncol Rep. 2009;21:875–880. doi: 10.3892/or_00000297. [DOI] [PubMed] [Google Scholar]
- 30.Tomlinson DC, Hurst CD, Knowles MA. Knockdown by shRNA identifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007;26:5889–5899. doi: 10.1038/sj.onc.1210399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Rhijn BW, van Tilborg AA, Lurkin I, Bonaventure J, de Vries A, Thiery JP, et al. Novel fibroblast growth factor receptor 3 (FGFR3) mutations in bladder cancer previously identified in non-lethal skeletal disorders. Eur J Hum Genet. 2002;10:819–824. doi: 10.1038/sj.ejhg.5200883. [DOI] [PubMed] [Google Scholar]
- 32.van Rhijn BW, Montironi R, Zwarthoff EC, Jobsis AC, van der Kwast TH. Frequent FGFR3 mutations in urothelial papilloma. J Pathol. 2002;198:245–251. doi: 10.1002/path.1202. [DOI] [PubMed] [Google Scholar]
- 33.Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, et al. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590–592. doi: 10.1038/ng1571. [DOI] [PubMed] [Google Scholar]
- 34.Jang JH, Shin KH, Park JG. Mutations in fibroblast growth factor receptor 2 and fibroblast growth factor receptor 3 genes associated with human gastric and colorectal cancers. Cancer Res. 2001;61:3541–3543. [PubMed] [Google Scholar]
- 35.Dutt A, Salvesen HB, Chen TH, Ramos AH, Onofrio RC, Hatton C, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc Natl Acad Sci U S A. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollock PM, Gartside MG, Dejeza LC, Powell MA, Mallon MA, Davies H, et al. Frequent activating FGFR2 mutations in endometrial carcinomas parallel germline mutations associated with craniosynostosis and skeletal dysplasia syndromes. Oncogene. 2007;26:7158–7162. doi: 10.1038/sj.onc.1210529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haugsten EM, Wiedlocha A, Olsnes S, Wesche J. Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res. 2010;8:1439–1452. doi: 10.1158/1541-7786.MCR-10-0168. [DOI] [PubMed] [Google Scholar]
- 38.Spinola M, Leoni VP, Tanuma J, Pettinicchio A, Frattini M, Signoroni S, et al. FGFR4 Gly388Arg polymorphism and prognosis of breast and colorectal cancer. Oncol Rep. 2005;14:415–419. [PubMed] [Google Scholar]
- 39.Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rouge C, et al. Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res. 2005;3:655–667. doi: 10.1158/1541-7786.MCR-05-0128. [DOI] [PubMed] [Google Scholar]
- 40.Luo Y, Ye S, Kan M, McKeehan WL. Control of fibroblast growth factor (FGF) 7- and FGF1-induced mitogenesis and downstream signaling by distinct heparin octasaccharide motifs. J Biol Chem. 2006;281:21052–21061. doi: 10.1074/jbc.M601559200. [DOI] [PubMed] [Google Scholar]
- 41.Shih Ie M, Wang TL. Notch signaling, gamma-secretase inhibitors, and cancer therapy. Cancer Res. 2007;67:1879–1882. doi: 10.1158/0008-5472.CAN-06-3958. [DOI] [PubMed] [Google Scholar]
- 42.Yap KL, Fraley SI, Thiaville MM, Jinawath N, Nakayama K, Wang J, et al. NAC1 is an actin-binding protein that is essential for effective cytokinesis in cancer cells. Cancer Res. 2012;72:4085–4096. doi: 10.1158/0008-5472.CAN-12-0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.