Abstract
Radical damage to DNA has been implicated in cell death, cellular dysfunction and cancer. A recently developed method for detecting DNA radicals uses the nitrone spin trap DMPO (5,5-dimethyl-1-pyrroline N-oxide) to trap radicals. The trapped radicals then decay into stable nitrone adducts detectable with anti-DMPO antibodies and quantifiable by ELISA or dot blot assays. However, the sequences of DNA that are damaged are likely to be as important as the total level of damage. Therefore, we have developed immunoblotting methods for detection of DNA nitrone adducts on electrophoretically separated DNA, comparable to Western blotting for proteins. These new techniques not only allow the assessment of relative radical adduct levels, but can reveal specific DNA fragments, and ultimately nucleotides, as radical targets. Moreover, we have determined that denaturation of samples into single-stranded DNA enhances the detection of DNA-DMPO adducts in our new blotting methods and also ELISA assays.
Keywords: Free radicals, oxidatively generated damage, DNA, ELISA, immuno-spin trapping, immunoblotting
Introduction
Biologically relevant reactive oxygen species (ROS) include radicals such as superoxide radical anion, hydroxyl radical (•OH) and peroxyl radical (ROO•) and non-radicals such as hydrogen peroxide (H2O2) and hypochlorous acid. ROS have been implicated in DNA damage induced by drugs [1–3], environmental hazards such as arsenic [4, 5] and ionizing radiation [1], and endogenous processes [6, 7]. Unrepaired DNA damage can lead to cell death, cellular dysfunction and cancer [6, 8].
Electron spin resonance (ESR) is used in in vitro studies of a wide range of biological radicals [9–12] but lacks the sensitivity to detect DNA radicals in intact cells. ESR spin trapping involves the use of a “spin trap” that reacts with the free radical to form a more stable radical adduct. While spin trapping increases the effective lifetime of radicals, thereby enhancing the sensitivity of ESR, it is still not generally applicable in intact cell studies. A more recent advance, termed immuno-spin trapping, enhances the sensitivity of radical detection by orders of magnitude through combining the specificity of spin trapping with the sensitivity of immunological techniques.
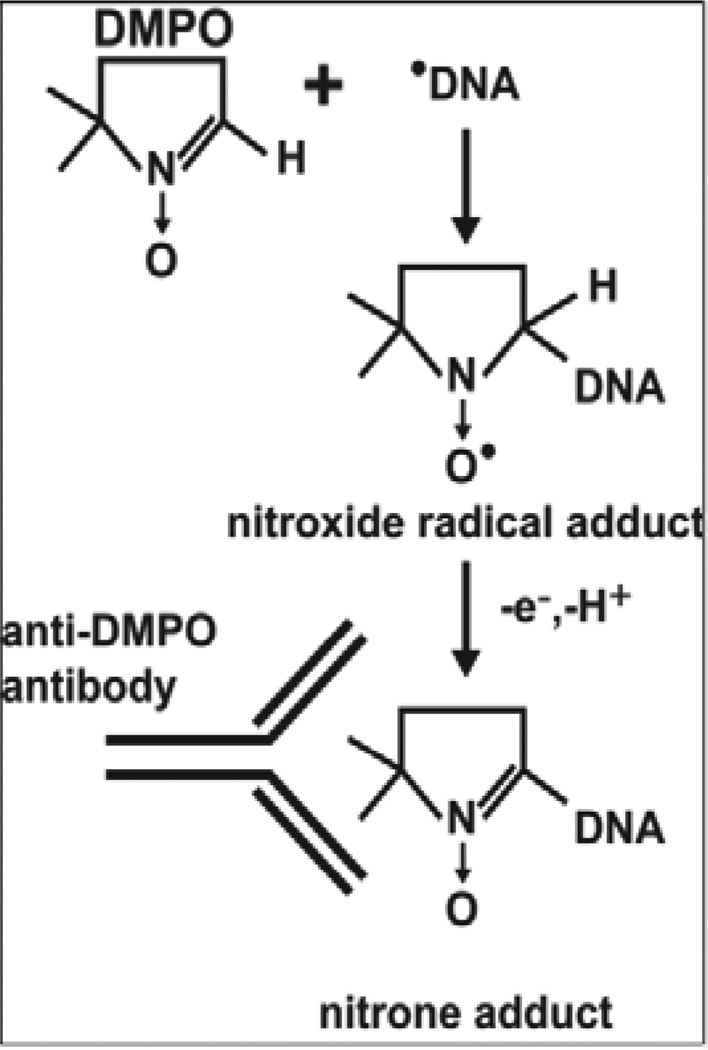
Immuno-spin trapping (Scheme 1) comprises two parts: 1) A spin-trapping reaction between a radical and the spin trap DMPO and 2) Immunological detection of the DMPO nitrone adducts (hereafter referred to as DMPO adducts) using an anti-DMPO antibody that recognizes DMPO covalently attached to a macromolecule, such as DNA or protein, at the site of the radical [13, 14]. DMPO freely permeates cell membranes and animal organs [15, 16], and is non-toxic at concentrations necessary for effective radical trapping. When added to in vitro systems, cell cultures or animals where radicals are being generated, DMPO reacts with radicals to form DMPO nitroxide radical adducts, which decay to far longer-lived, ESR-silent nitrone adducts recognized by the anti-DMPO antibody [13, 17].
Scheme 1.
Reaction of the DMPO spin trap with a DNA radical to form a DNA-DMPO nitrone adduct, which is detectable using an anti-DMPO antibody.
Immuno-spin trapping was first used to study protein radicals [13] but has now been used successfully in DNA radical studies [4, 11, 14, 17]. One disadvantage of immuno-spin trapping is that the chemical structure of the free radical is not identified in this process but mass spectrometry has been used to identify the structure of a DMPO adduct formed on adenine [18]. Analyses of physiological DNA oxidation products, such as 8-oxo-7,8-dihydro-2’-deoxyguanosine (also referred to as 8-hydroxy-2’-deoxyguanosine), by ELISA has been limited due to cross-reactivity of the antibodies with unoxidized 2'-deoxyguanosine [19], which is in relatively high abundance relative to 8-oxo-7,8-dihydro-2’-deoxyguanosine. Analysis of 8-oxo-7,8-dihydro-2’-deoxyguanosine by mass spectrometry is complicated by artifactual oxidation, which can occur easily during DNA extraction and sample work-up, leading to discrepancies in the measurement which can vary by as much as 1000-fold depending on which procedure is used [20–22] but this has become less variable with improvements in sample preparation to minimize spurious oxidation [23–26]. In immuno-spin trapping, by contrast, after spin trap reactions are complete, sample processing decreases DMPO concentration to below 1 mM, a level too low to trap radicals [12]. Moreover, anti-DMPO antibodies do not cross-react with DNA [14, 17], which is a problem that ELISA measurements of 8-oxo-7,8-dihydro-2’-deoxyguanosine have. Relative levels of DNA-DMPO adducts can be measured by ELISA or dot blot [14, 17], and differences can then be observed between treatments or over time and can be correlated with a functional effect [4, 11]. However, the genes that are damaged are likely to be as important as the total level of damage. The ability to identify specific genes prone to radical damage under specific physiological or developmental regimes would allow connections to be drawn between mutated DNA and health outcomes. Therefore, to more precisely analyze the extent and location of radical-mediated damage throughout the genome, there is a need to extend immuno-spin trapping to detection of DMPO adducts on DNA, analogous to Western blotting.
To develop this method, we used an in vitro system consisting of DNA, copper (II) and H2O2 to generate DNA radicals in the presence of DMPO. Under these conditions, no assignable ESR spectrum has been obtained [18]. H2O2, a non-radical oxidant, does not react with DNA but can react with iron and copper through Fenton-type reactions to produce •OH that can react with DNA at a diffusion-limited rate [9, 27–29]. Copper ions bind preferentially to the N7 of guanine and to a lesser extent the N7 of adenine [30–32]. Hydroxyl radical scavengers are relatively ineffective at inhibiting Cu-mediated damage, suggesting that scavengers in bulk solution cannot effectively compete when hydroxyl radical is formed at the damage site [14, 27, 29]. A less likely alternative is that the DNA radical damage may be due to a species closely related to the hydroxyl radical that does not react with hydroxyl radical scavengers.
Although the copper-Fenton system is an in vitro model of DNA damage, it may have physiological relevance. Wilson’s disease, for example, is due to a mutation which blocks copper efflux from the liver, resulting in copper accumulation and liver cirrhosis. The bulky DNA lesions detected in liver DNA extracted from Wilson’s disease patients are similar to the bulky DNA lesions formed in vitro by a copper-Fenton system [33]. Moreover, penicillamine and triethylenetetramine, drugs used to treat patients with Wilson’s disease, can chelate copper and inhibit radical formation as measured by ESR [9].
This work was undertaken to expand the utility of DNA immuno-spin trapping through development of a blotting technique for spin-trapped DNA comparable to Western blotting. Examination of a number of standard techniques for nucleic acid transfer allowed us to identify reproducible methods for immunoblotting of both high and low molecular weight DNA.
Material and methods
Materials
Nitrocellulose and AG® 501-X8 resin were from BioRad. DMPO was from Dojindo Molecular Technologies Inc. Immobilon-FL PVDF membrane was from Millipore™. The LumiGLO peroxidase chemiluminescent substrate kit was from KPL, Inc. Reacti-Bind™ DNA coating solution, stabilized goat anti-mouse IgG (H+L) conjugated to HRP and rabbit anti-chicken IgY (H+L) conjugated to HRP were from Pierce Scientific. Donkey anti-chicken IgG-800 (H+L) IRDye 800CW, donkey anti-mouse IgG-800 (H+L) IRDye 800CW, and 10X Orange loading dye were obtained from LI-COR Biotechnology. Low IgG fetal bovine serum, Iscove’s modified Dulbecco’s media, 6% (wt/vol) DNA retardation polyacrylamide gels, Novex® TBE running buffer and SYTO® 60 red fluorescent nucleic acid stain were from Invitrogen Life Technologies. Calf thymus DNA, copper (II) chloride, casein, diethylenetriaminepentaacetic acid (DTPA), polydeoxyguanylic acid.polydeoxycytidylic acid sodium salt (poly(dG).poly(dC)), poly(deoxyguanylic-deoxycytidylic) acid sodium salt (poly(dG-dC).poly(dG-dC)), polydeoxyadenylic acid.polythymidylic acid sodium salt (poly(dA).(dT)), poly(deoxyadenylic-thymidylic) acid sodium salt (poly(dA-dT).poly(dA-dT)), glyoxal trimer dihydrate and all other chemicals were from Sigma Chemical Co. The chicken polyclonal antibody to anti-5,5-dimethyl-2-(8-octonoic acid)-1-pyrrolone-N-oxide conjugated to BSA (anti-DMPO adduct) was made by Aves Labs, Inc. in a way similar to that described previously for polyclonal rabbit anti-DMPO adduct antibody [13]. The hybridoma clone N1664A that produces mouse anti-DMPO adduct monoclonal antibody was grown in a humidified incubator in 5% CO2 at 37 °C in 10% (vol/vol) low IgG fetal bovine serum, 90% (vol/vol) Iscove’s modified Dulbecco’s media supplemented with 100 U mL−1 penicillin and 0.1 mg mL−1 streptomycin. The monoclonal antibody was purified by protein G chromatography in-house, but this antibody can be obtained commercially.
Preparation of DNA radicals and spin-trapping with DMPO
DNA was incubated at 37 °C with copper(II) chloride, hydrogen peroxide and DMPO in PBS (2 mM potassium phosphate, 8 mM sodium phosphate, 2.7 mM potassium chloride, and 137 mM sodium chloride, pH 7.4) with DMPO being added last. After 1 h, DTPA was added to a final concentration of 1 mM to terminate the reaction. The DNA was precipitated with 1/10th vol. 3 M sodium acetate, pH 5.2, and 2 vol. ice-cold ethanol and incubated for 10 min at RT because the DMPO precipitated in this mixture if incubated at 4 °C. The DNA was centrifuged at 13,000 rpm for 15 min at RT, washed with 70% (vol/vol) ethanol and redissolved in TE (10 mM Tris, 1 mM EDTA, pH 8.0).
DNA electrophoresis
DNA was denatured immediately before electrophoresis by adding deionized formamide to a final concentration of 60% (vol/vol), with 1/10th vol. 10X orange loading dye and 1 µL 5 µM SYTO® 60 (for sample volumes ranging from 10 to 30 µL). The samples were denatured by heating for 5 min at 65 °C, followed by immediate chilling on ice for 5 min before loading onto the gel. DNA to be run under native conditions was mixed with 1/5th vol. 10X orange loading dye and 1 µL 5 µM SYTO® 60 and incubated for 5 min at RT. DNA (5 µg/lane) was electrophoresed on either 1% (wt/vol) agarose gels in TAE (40 mM Tris-acetate, 1 mM EDTA) for 45 min at 90 V or on 6% (wt/vol) DNA retardation polyacrylamide gels in 0.5X TBE (44.5 mM Tris, 44.5 mM borate, 1 mM EDTA) for 90 min at 100 V. The DNA stained with SYTO® 60 was visualized by scanning the gels using the 700 nm channel on the Odyssey Infrared Imaging System (LI-COR Biotechnology).
DNA transfer to membranes
Agarose gels were equilibrated in 20X SSC with two 20-min incubations at RT with constant gentle agitation and the DNA transferred to a nitrocellulose membrane by downward capillary transfer [34] overnight in 20X SSC (3 M sodium chloride, 300 mM sodium citrate) [34]. A Stratalinker® UV crosslinker 2400 (Agilent Technologies) was used for UV (254 nm) irradiation of the nitrocellulose membrane to crosslink the DNA using an exposure of 120,000 microjoules/cm2. PVDF membranes were pre-wet in methanol for several seconds and then equilibrated in 0.5X TBE for 15 min; the DNA from polyacrylamide gels was then transferred onto the PVDF membrane by electroblotting for 20 min at 15 V on a Trans-Blot® SD Semi-Dry Transfer Cell (BioRad). Exposing PVDF membranes to UV crosslinking did not appear to improve binding of the DNA to the membrane as the UV crosslinked membranes gave the same DMPO signal as membranes which had not been subjected to UV crosslinking.
DNA immunoblot
The membrane was blocked with 1% (wt/vol) casein in PBS for 1 h, followed by incubation for 1 h with 5 µg mL−1 monoclonal anti-DMPO nitrone adduct antibody or 10 µg mL−1 polyclonal anti-DMPO nitrone adduct antibody in blocking solution, and followed by incubation for 1 h with a 1:15,000 dilution of IRDye 800CW donkey anti-mouse IgG (H+L) or 1:15,000 dilution of IRDye 800CW donkey anti-chicken IgG (H+L), respectively, in blocking solution. As the chicken polyclonal anti-DMPO antibody bound nonspecifically to residual agarose on the membrane the polyclonal antibody was pre-incubated with 1% (w/v) agarose in blocker for 2 h at room temperature and then the mixture centrifuged to remove the agarose prior to use. The membrane was washed after each of the antibody steps with three washes in PBS of 5 min each. The membranes were dried prior to being scanned on the Odyssey Infrared Imaging System.
ELISA
For the DNA binding step, we tested a 2 h incubation (data not shown) and an overnight incubation at room temperature (manufacturer’s instructions). The overnight incubation gave better detection (approximately two-fold) of DMPO adducts than the 2 h incubation, suggesting that extending the incubation time allowed more DNA to bind to the ELISA plate and that the DMPO adducts were stable in this time frame.
DNA (0.5 µg) was applied to white ELISA plates and Reacti-Bind® DNA coating solution added to 200 µL. The DNA was allowed to bind to the ELISA plate at RT overnight in the dark with gentle agitation. The plate was washed with PBS and in experiments where the DNA was glyoxalated, the DNA was subsequently treated with 66% (vol/vol) dimethyl sulfoxide, 1 M glyoxal and 1.5 mM sodium phosphate [35] at 37 °C for 1 h. The plate was washed with PBS and blocked with 1% (wt/vol) casein in PBS, pH 7.4. The DMPO nitrone adducts were detected with 5 µg mL−1 mouse monoclonal anti-DMPO antibody and 1:100 goat anti-mouse IgG-HRP or with 10 µg mL−1 chicken anti-DMPO nitrone IgY and 1:20,000 rabbit anti-chicken IgY (H+L)-HRP (each 1-h incubations at 37 °C). The plate was washed three times with PBS between incubations. The LumiGLO substrate was added and incubated at RT for 5 min prior to the luminescence being measured on a GENios platereader (Tecan). The statistical analyses were performed using GraphPad Prism version 5.03, GraphPad Software, La Jolla, California, USA.
Results
Agarose gel electrophoresis and detection of DMPO adducts
To develop an immunoblotting method to assess the extent of radical-mediated damage throughout the genome, an in vitro oxidizing system consisting of calf thymus DNA, Cu2+ and H2O2 was used both with and without the radical spin trap DMPO. High molecular weight DNA is routinely resolved on agarose gels (Fig. 1a) for transfer to nitrocellulose. The standard technique of denaturing double-stranded DNA (dsDNA) in-gel with 0.5 M NaOH and 1.5 M NaCl prior to transfer [34] degraded the DMPO adducts (data not shown). Because nylon membranes can bind dsDNA and nitrocellulose binds only single stranded DNA (ssDNA) [34], nylon membranes were tried to avoid in-gel denaturation [34]. However, even with extensive blocking, nylon membranes gave a very high background and were deemed unsuitable (data not shown).
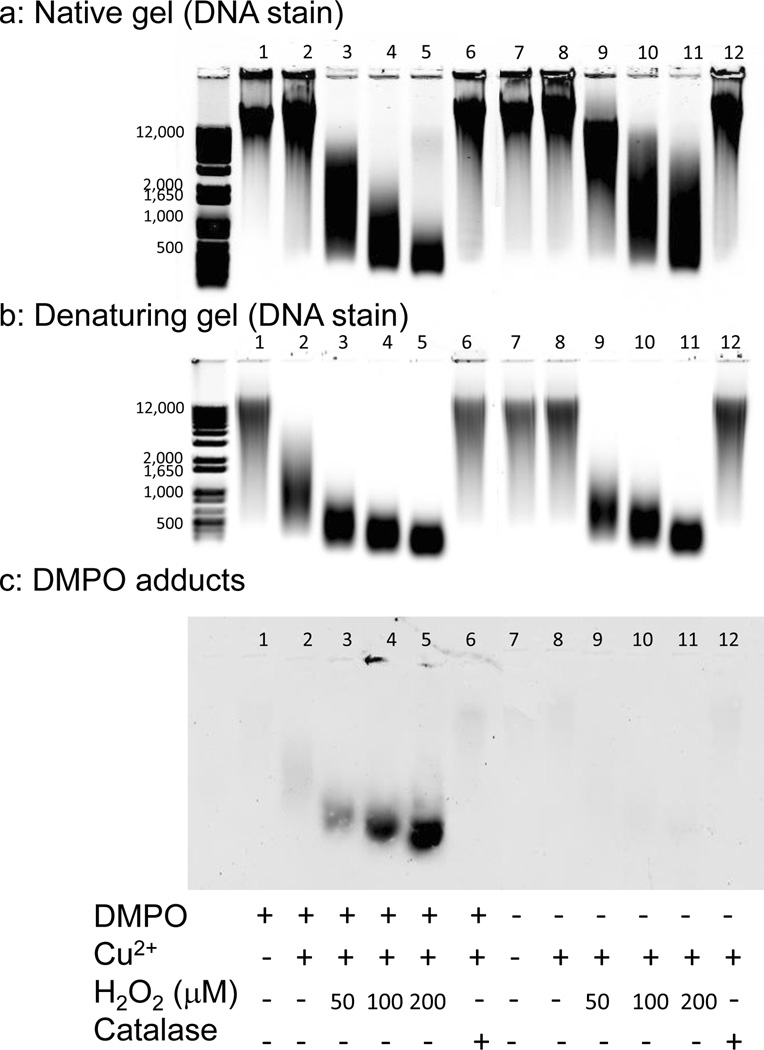
Figure 1.
Agarose gel electrophoresis of DNA fragmentation under native and denaturing conditions and detection of DMPO adducts on DNA oxidized by Cu2+ and H2O2 in the presence and absence of DMPO. DNA (5 µg/lane) was electrophoresed on a 1% (wt/vol) agarose gel in TAE and stained with SYTO® 60 either under native conditions (panel a) or having been denatured in hot formamide prior to electrophoresis (panel b). Denatured DNA was transferred to a nitrocellulose membrane by capillary transfer and the DMPO adducts were detected using a monoclonal anti-DMPO adduct antibody (panel c). The DNA (250 µg mL−1) was treated with 50 µM Cu2+, 0–200 µM H2O2, 100 mM DMPO and 1 U mL−1 catalase as indicated. Results are representative of at least three independent experiments.
Instead, the DNA was denatured prior to gel loading using hot formamide (Fig. 1b), a simplified yet equally effective alternative to the more common 3-(N-morpholino)propane-sulfonic acid and formaldehyde agarose gel protocol [36]. Agarose gels run under denaturing conditions were used for immunoblot experiments (Fig. 1c).
Treatment with Cu2+ and H2O2 extensively fragmented the DNA, independent of the presence or absence of DMPO, with fragmentation proportional to H2O2 concentration (Fig. 1a and b). DNA run under denaturing conditions (Fig. 1b), however, appears more fragmented because ssDNA reveals both single- and double-stranded breaks while dsDNA shows only double-stranded breaks. When DNA was incubated with DMPO and Cu2+ fragmentation due to single-stranded breaks was visible only in the denaturing gel (lane 2). The addition of catalase, which catalyzes the decomposition of H2O2 to water and oxygen, to reactions with DNA, DMPO and Cu2+ prevented this fragmentation completely (Fig. 1b, compare lane 6 to lane 2). Individual treatment with DMPO (Fig. 1b, lane 1) or Cu2+ (Fig. 1b, lane 8) did not cause single-stranded breaks.
DNA fragmenting in the presence of DMPO and Cu2+ was attributed to H2O2 generation by DMPO and Cu2+ [37, 38], followed by reaction of this H2O2 with DNA-bound Cu2+. The sample containing DNA, Cu2+ and DMPO (Fig. 1b, lane 2) was less fragmented than the sample containing DNA, Cu2+ and 50 µM H2O2 (Fig. 1b, lane 9), suggesting that less than 50 µM H2O2 was generated.
Denatured DNA was transferred from agarose to nitrocellulose by capillary action and DMPO adducts detected with a monoclonal anti-DMPO antibody. The antibody recognized only DNA oxidized in the presence of DMPO, and the DMPO adducts increased with H2O2 concentration in the presence of Cu2+ (Fig. 1c). In contrast, few DMPO adducts were detected in the DNA sample incubated with DMPO and Cu2+ (Fig. 1c, lane 2) and, even though DNA single strand breaks had formed (Fig. 1b, lane 2), these breaks were attributed to artifactual H2O2 generation as they were inhibited by catalase (Fig. 1b, compare lane 6 with lane 2).
Polyacrylamide gel electrophoresis and immunoblot
Polyacrylamide gels are suitable for separation of low molecular weight DNA. Electroblotting is necessary for efficient transfer of DNA out of polyacrylamide, but low ionic strength buffers must be used because high ionic strength buffers generate too much heat. Therefore, nitrocellulose could not be used for electroblotting because high ionic strength buffers are needed for efficient retention of DNA [34]. PVDF was investigated as an alternative because it has been used in capillary transfer with low ionic strength buffers and binds both dsDNA and ssDNA [39, 40].
DNA was run under native and denaturing conditions (formamide treatment prior to electrophoresis) on polyacrylamide gels; the patterns of DNA fragmentation (Fig. 2a) were similar to those seen on the agarose gels (Fig. 1a and b) except that there was better resolution of lower molecular weight DNA. DNA was transferred from the polyacrylamide gel to PVDF membrane, and a variety of methods were investigated for fixation of the DNA. DMPO adducts were undetectable if DNA was fixed to the PVDF membrane by baking at 80 °C (data not shown), a commonly used technique for permanently immobilizing DNA to membranes. DMPO adducts could be detected if the DNA was fixed by UV crosslinking if the membrane was kept wet (data not shown). However, the same level of immunodetection could be achieved without UV crosslinking, and this step was omitted (Fig. 2b).
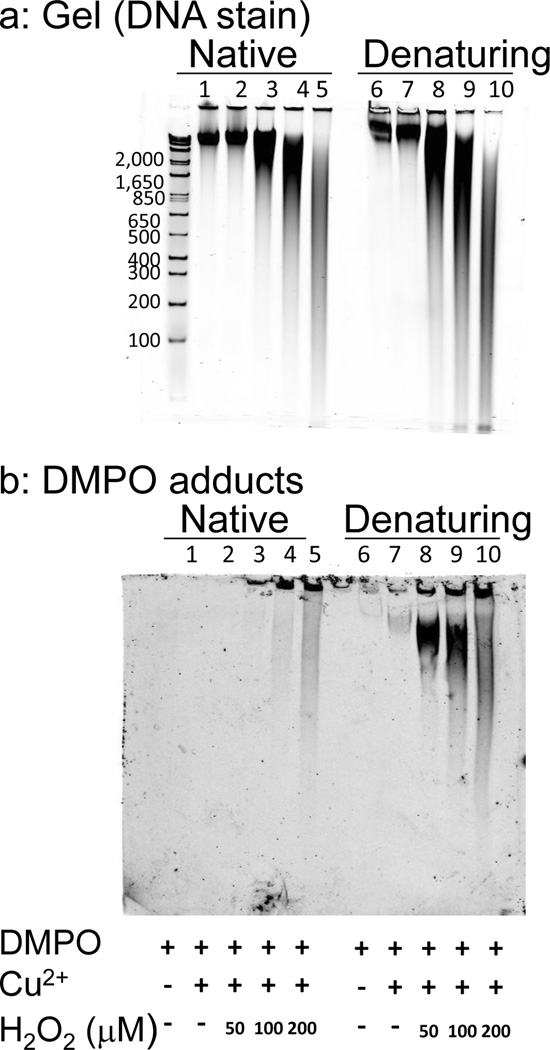
Figure 2.
Polyacrylamide gel electrophoresis of DNA fragmentation and detection of DMPO adducts on DNA oxidized by Cu2+, H2O2 and DMPO. DNA was run under native conditions (lanes 1–5) or denaturing conditions after hot formamide treatment (lanes 6–10). DNA (5 µg/lane) was electrophoresed on 6% (wt/vol) polyacrylamide gel in 0.5X TBE (panel a). The DNA was transferred to a PVDF membrane by electroblotting, and the DMPO adducts were detected using a monoclonal anti-DMPO adduct (panel b). The DNA (250 µg mL−1) was treated with 50 µM Cu2+, 0–200 µM H2O2 and 100 mM DMPO as indicated. Results are representative of at least three independent experiments.
Similar to immunodetection of DMPO adducts on nitrocellulose (Fig. 1c), DMPO adducts were detected on PVDF in samples containing DNA, Cu2+, H2O2 and DMPO, and the levels of DMPO adducts increased with H2O2 concentration (Fig. 2b). There was better detection of DMPO adducts in the denatured samples, perhaps due to better accessibility of nitrone adducts to the DMPO antibodies.
Glyoxal denatured DNA ELISA
We examined the possibility that ELISA detection of DMPO adducts could be improved by converting dsDNA to ssDNA (Fig. 2b). DNA samples bound to an ELISA plate were subsequently treated with or without dimethyl sulfoxide and glyoxal. Dimethyl sulfoxide denatures DNA, and glyoxal reacts with the bases, in particular guanine, to produce stable glyoxalated derivatives unable to form hydrogen bonds with cytosine, thus preventing DNA renaturation [35].
Monoclonal anti-DMPO recognized only DNA oxidized in the presence of DMPO, and the DMPO adducts increased with H2O2 concentration in the presence of Cu2+ (Fig. 3), consistent with previous results using rabbit polyclonal anti-DMPO [14, 17]. Glyoxal treatment of DNA improved the detection of DMPO adducts, with a greater effect at lower H2O2 concentrations. In particular, DMPO adducts were virtually undetectable in the sample containing DNA, Cu2+, 50 µM H2O2 and DMPO without glyoxal treatment but with glyoxal treatment, DMPO adducts were easily detectable. Glyoxal treatment of DNA lacking DMPO adducts gave no signal, indicating that monoclonal anti-DMPO does not recognize glyoxalated DNA (Fig. 3), but polyclonal chicken anti-DMPO did crossreact with glyoxal-treated DNA (data not shown).
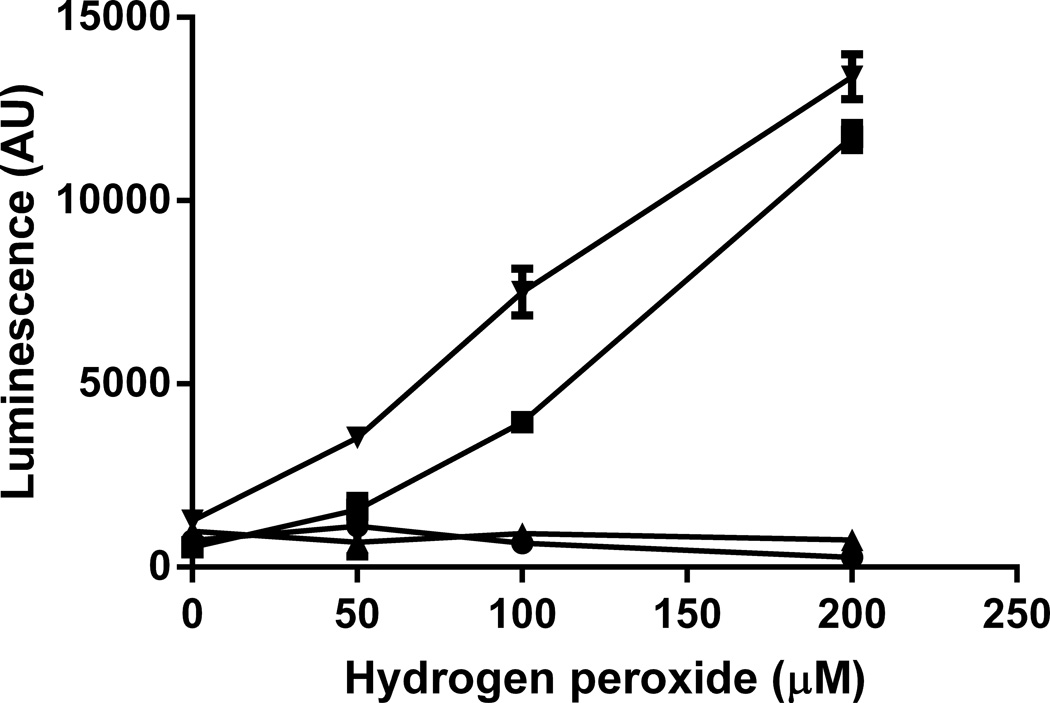
Figure 3.
Detection of DMPO adducts on DNA oxidized with Cu2+ and H2O2 with and without DMPO and with and without DMSO- and glyoxal-denaturation of DNA. DNA was oxidized with 50 µM Cu2+ and varying H2O2 concentrations in the presence (▼,■) and absence (▲,●) of DMPO. DNA (0.5 µg/well) was bound to an ELISA plate overnight in a Reacti-Bind DNA coating solution (Pierce). The DNA was denatured by incubating in 66% (vol/vol) DMSO, 1 M glyoxal and 1.5 mM sodium phosphate (▼,▲) at 37 °C for 1 h, and control samples were incubated in PBS (■,●). Subsequently, the denaturant was washed away and the DMPO adducts were detected with a mouse monoclonal antibody. Results are means ± SD of triplicate measurements and are representative of three independent experiments. Two-way ANOVA with Bonferroni post-tests showed there was no significant difference between 0 DMPO samples with or without glyoxal treatment. In the absence of glyoxal treatment, there was a significant difference between 100 mM DMPO samples and 0 mM DMPO samples at 100 and 200 uM H2O2 (p<0.05). There was a significant difference between 100 mM DMPO samples with glyoxal treatment and 0 DMPO samples at 50, 100 and 200 uM H2O2 (p<0.05).
DMPO adduct formation on polynucleotides of defined composition
Oxidation of DNA by Cu2+ and H2O2 produces a number of oxidation products [28], many of which are likely to have DNA radical precursors trappable by DMPO, yet only one DNA-DMPO adduct has been characterized to date: Bhattacharjee et al. recently described the identification of an adenine radical trapped by DMPO on calf thymus DNA oxidized with copper and H2O2 [18]. To investigate whether other radicals are formed, polynucleotides with defined base compositions were oxidized with Cu2+ and H2O2 in the presence of DMPO. Polynucleotides used were poly(dG).poly(dC) and poly(dA).poly(dT), where each strand of the dsDNA is a homopolymer; and poly(dG-dC).poly(dG-dC) and poly(dA-dT).poly(dA-dT), where each strand of the dsDNA is a heteropolymer of alternating bases.
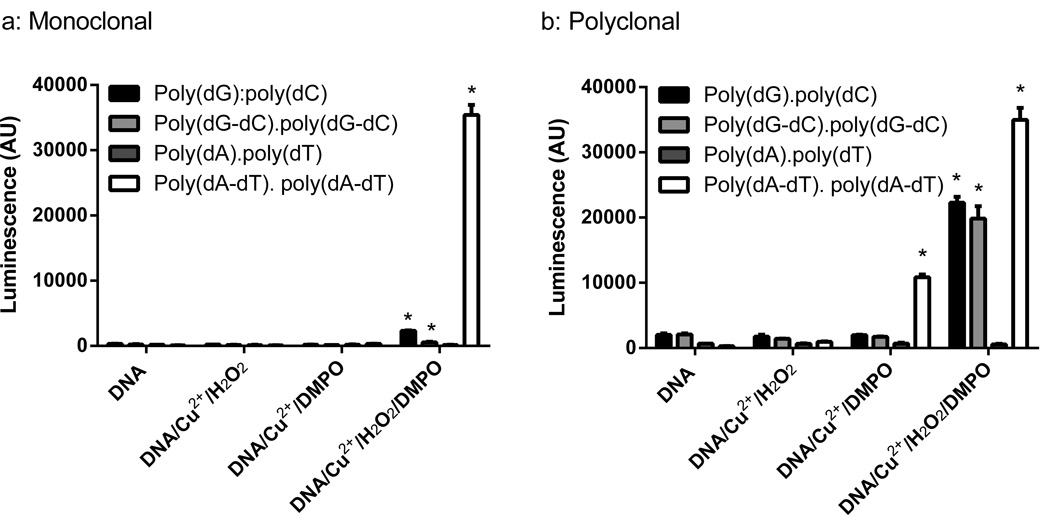
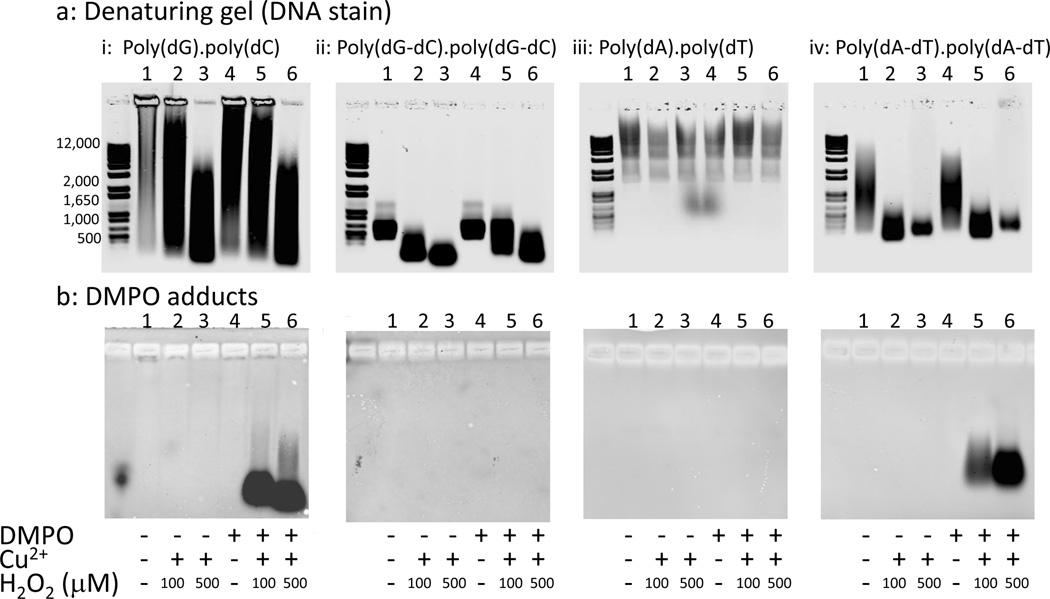
ELISA analysis of these samples used both monoclonal anti-DMPO (Fig. 4a) and chicken polyclonal anti-DMPO (Fig. 4b). Immunoblotting analysis of these samples on nitrocellulose membranes used chicken polyclonal anti-DMPO (Fig. 5b). DMPO adducts were detectable on poly(dA-dT).poly(dA-dT) treated with Cu2+ and H2O2 in the presence of DMPO using either monoclonal or polyclonal anti-DMPO (Figs. 4 and 5b (iv)). DMPO adducts were also detected in poly(dA-dT).poly(dA-dT) treated with DMPO and Cu2+ in the absence of H2O2 (Fig. 4), which is likely due to H2O2 formation from Cu2+ and DMPO [37, 38]. Detection of DMPO adduct(s) on poly(dA-dT).poly(dA-dT) is consistent with the previous characterization of an adenine radical [18]. However, no DMPO adducts were detected on poly(dA).poly(dT) treated with Cu2+, H2O2 and DMPO using either monoclonal or polyclonal anti-DMPO (Figs. 4a and 5b (iii)). Cu2+ and H2O2 did not fragment poly(dA).poly(dT) (Fig. 5a (iii)), whereas extensive fragmentation was seen with poly(dA-dT).poly(dA-dT) (Fig. 5 a (iv)).
Figure 4.
ELISA for DMPO adduct detection on polynucleotides oxidized by the Cu2+ and H2O2 in the presence and absence of DMPO. Polynucleotides were: poly(dG).poly(dC), poly(dG-dC). poly(dG-dC), poly(dA).poly(dT), and poly(dA-dT).poly(dA-dT). Polynucleotides (100 µg mL−1), 20 µM Cu2+, 500 µM H2O2and 100 mM DMPO in PBS were incubated for 1 h at 37 °C. DNA (0.5 µg/well) was bound to an ELISA plate overnight in a Reacti-Bind DNA coating solution (Pierce). DMPO adducts were detected with a mouse monoclonal anti-DMPO antibody (panel a) or a chicken polyclonal anti-DMPO antibody (panel b). Results are means ± SD of triplicate measurements and representative of at least three independent experiments. One-way ANOVA with Dunnett’s Multiple Comparison Test showed the treatments that produced a significant increase over the untreated polynucleotides as indicated (* p <0.05).
Figure 5.
Agarose gel electrophoresis of polynucleotide fragmentation and detection of DMPO adducts on polynucleotides oxidized by Cu2+, H2O2 and DMPO. The polynucleotides were run under denaturing conditions after hot formamide treatment (panel a). Denatured polynucleotides (1.2 µg/lane) were transferred to a nitrocellulose membrane by capillary transfer and DMPO adducts detected using a chicken polyclonal anti-DMPO antibody (panel b). The polynucleotides (100 µg mL−1) were treated with 20 µM Cu2+, 0–500 µM H2O2 and 100 mM DMPO as indicated. This experiment is representative of at least three independent experiments.
By ELISA, polyclonal anti-DMPO detected DMPO adducts on poly(dG).poly(dC) and poly(dG-dC). poly(dG-dC) treated with Cu2+, H2O2 and DMPO (Fig. 4b), but these adducts were poorly detected with monoclonal anti-DMPO (Fig. 4a). This suggests that there are multiple DMPO-containing epitopes, and the DMPO adduct epitope(s) formed on guanine- and cytosine-containing polynucleotides are distinct from those formed on poly(dA-dT).poly(dA-dT). When the sample was immunoblotted with chicken polyclonal anti-DMPO, DMPO adducts were detected on poly(dG).poly(dC) (Fig. 5b (i)) but not on poly(dG-dC).poly(dG-dC) (Fig 5b, (ii)); however, fragmentation was observed in both cases (Fig. 5 a (i and ii)). The inability to detect DMPO adducts on poly(dG-dC).poly(dG-dC) treated with Cu2+, H2O2 and DMPO was ascribed to poor retention of this polynucleotide on nitrocellulose, which has a lower limit of 500 nucleotides for efficient retention [34]. Detection of DMPO adducts on poly(dG-dC).poly(dG-dC) was also unsuccessful on PVDF but was successful for poly(dG).poly(dC) and poly(dA-dT).poly(dA-dT), suggesting that the PVDF has a similar cutoff for efficient retention of polynucleotides (data not shown). Detection of DMPO adducts on polynucleotides lacking adenine indicates that other nucleotides can form radical adducts trappable by DMPO. The poor detection of non-adenine DMPO adducts by monoclonal anti-DMPO may, in part, explain the detection of only an adenine DMPO adduct by Bhattacharjee et al. [18], because an immunoprecipitation step using monoclonal anti-DMPO was used to enrich for DMPO adducts prior to identification by mass spectrometry.
Discussion
To date, DNA immuno-spin trapping has emphasized quantification of radical adducts via ELISA [4, 14, 17]. While information on relative DNA radical accumulation is important, identification of specific radical lesions should provide the means to connect these lesions with specific physiological conditions such as cancer and aging. Here we describe the development of DNA-DMPO immunoblots comparable to protein Western blots. Using a Fenton-like chemistry model system for widespread generation of hydroxyl radicals, we investigated and adapted a variety of standard molecular biology methods. We have developed blotting systems for both high (Fig. 1c) and low molecular weight DNA (Fig. 2b) that detect a genome-wide distribution of the DNA-DMPO adducts expected from the chemistry used to generate the radicals. Immunodetection of DMPO adducts was enhanced following DNA denaturation by either of two different methods: 1) hot formamide (Figs. 1, 2 and 5), which disrupts hydrogen-bonding, or 2) glyoxal and DMSO denaturation, in which hydrogen-bonding is disrupted by the glyoxalation of guanine bases (Fig. 3). This enhancement of the standard protocol of using dsDNA to quantify DNA DMPO adducts [4, 14, 17] will facilitate the detection of lower levels of these adducts.
One of the foremost advantages of immuno-spin trapping is that DMPO spin-trap chemistry has been extensively characterized, with DMPO cited in PubMed over 1,000 times. In general, DMPO suffers few artifactual reactions of biological importance. The most significant exception is the nucleophilic addition of water to DMPO, which can be catalyzed by cupric [37, 38] and ferric ions [38, 41]. The nucleophilic addition of water to DMPO and the subsequent auto-oxidation to DMPO/hydroxyl radical produces a false positive with ESR spectroscopy [37, 38, 41] but this adduct cannot be detected by immuno-spin trapping, which detects only DMPO adducts on macromolecules [13, 14]. However, in this study artifactual formation of H2O2 resulting from reactions subsequent to the nucleophilic addition of water to DMPO was detected in samples containing DNA, DMPO and Cu2+, resulting in single-strand breakage (Fig. 1b, lane 2 and Fig. 2a, lane 8) and a low level of DMPO adducts (Fig. 1c, lane 2; Fig. 2b, lane 8 and Fig. 4b) that were formed in the absence of added H2O2.
This generation of H2O2 from free copper and/or iron ions in the presence of DMPO is a theoretical limitation in the use of DMPO to identify areas of oxidatively damaged DNA. However, such an artifact is likely to occur only in the presence of unchelated copper or iron, as artifactual generation of H2O2 is totally prevented if the copper or iron is chelated by chelating agents, ligands or proteins even if the metal is able to redox cycle in the chelated form [37, 38]. Phosphate and citrate buffers completely suppress H2O2 formation from iron and DMPO but not copper and DMPO [38]. In vivo, most copper and iron is in proteins such as ceruloplasmin for copper and hemoglobin for iron. In vivo, even copper unassociated with proteins is likely to be chelated by ligands such as reduced glutathione, present intracellularly at millimolar concentrations, and free histidine as well as histidine residues of proteins [10, 37]. These ligands prevent DMPO from interacting with the metal center, precluding generation of H2O2 in cells and tissues. The intracellular environment has such an overcapacity for the chelation of copper that intracellular free copper concentration is estimated to be less than one free copper per cell [42]. Therefore, the artifactual generation of H2O2 from copper and DMPO seen in this in vitro study is highly unlikely to be a problem in the use of immuno-spin trapping in cell cultures or animals.
Both monoclonal and polyclonal anti-DMPO detected DMPO adducts on ELISA plates and nitrocellulose or PVDF membranes (Figs. 1–5) without cross-reacting with DNA oxidized in the absence of DMPO. The use of DMPO as a tag and the high signal to low background achieved by detecting DMPO adducts with an anti-DMPO antibody is an advantage over antibodies raised against physiological oxidation products such as 8-oxo-7,8-dihydro-2’-deoxyguanosine, which have problems of specificity [19]. Unlike antibodies raised against oxidized nucleic acid constituents, anti-DMPO adduct antibodies are raised against a DMPO derivative conjugated to bovine serum albumin or ovalbumin [13]. In addition, the concentration of unattached DMPO is below 1 mM during DNA extraction, precluding the possibility of artifactual DMPO adduct formation (Fig. S1).
Both monoclonal and chicken polyclonal anti-DMPO detected DMPO adducts on poly(dA-dT). poly(dA-dT) (Figs. 4 and 5b (iv), monoclonal blots not shown) but not on poly(dA).poly(dT) after reaction with Cu2+ and H2O2 in the presence of DMPO (Fig. 4 and Fig. 5b (iii)), (monoclonal blots not shown). This detection of DMPO adduct(s) on poly(dA-dT).poly(dA-dT) is consistent with the previous characterization of an adenine radical trapped by DMPO on DNA [18]. The susceptibility of poly(dA-dT).poly(dA-dT) to oxidation may be significant, as repeating adenine and thymine sequences are found in TATA-boxes in the promoter region of many eukaryote genes [43]. Mutations in these regions can lead to changes in levels of gene expression or the emergence or disappearance of a transcription start site and have been associated with a number of human pathologies [43].
The absence of DMPO adducts on oxidized poly(dA).poly(dT) was initially surprising as this polynucleotide differs from poly(dA-dT).poly(dA-dT) only by different sequencing of the bases. However, poly(dA).poly(dT) also did not fragment with Cu2+ and H2O2 treatment (Fig. 5a (iii)), while with poly(dA-dT).poly(dA-dT) both DMPO adduct formation (Fig. 5b (iv)) and DNA fragmentation were observed (Fig. 5a (iv)). Because the sequence of bases determines DNA three-dimensional structure, this suggests that Cu2+ may be unable to bind to the poly(dA).poly(dT) and is thus only able to react in bulk solution, forming reactive intermediates far less damaging to DNA [29]. Poly(dA).poly(dT) has unusual structural properties and is more rigid than generic sequence DNA [44]. The binding of the intercalating fluorescent dye propridium iodide to poly(dA).poly(dT) is anomalous and far weaker than binding to poly(dA-dT).poly(dA-dT) and generic sequence DNA [45]. Poly(dA).poly(dT) also bound far less of the DNA stain Syto 60 than the other polynucleotides (Fig. 5a). The rigidity of poly(dA).poly(dT) is also evidenced by the inability of nucleosomes to form over sufficiently long stretches of poly(dA).poly(dT), whereas nucleosomes will form over poly(dA-dT).poly(dA-dT) and generic sequence DNA [44], and poly(dA).poly(dT) stretches have been proposed to be a major determinant of the nucleosome organization in eukaryotes.
In the ELISA experiments, the polyclonal anti-DMPO detected DMPO adducts formed on the two guanine- and cytosine-containing polynucleotides oxidized in the presence of DMPO (Fig. 4a), whereas with monoclonal anti-DMPO it detected these DMPO adducts poorly (Fig. 4b). Detection of DMPO adducts on polynucleotides without adenine indicates that radicals are being formed and trapped by DMPO other than the already characterized adenine radical [18]. DMPO adducts formed on poly(dG).poly(dC) and poly(dG-dC).poly(dG-dC) are likely to include guanine radicals, as copper binds preferentially to guanine bases [30–32] and 8-oxo-7,8-dihydro-2’-deoxyguanosine is the major product of the oxidation of DNA with Cu2+ and H2O2 in the absence of spin-trap [28]. The difference in the detection of DMPO adducts between monoclonal and polyclonal antibodies suggests that polyclonal anti-DMPO contains antibodies against multiple epitopes. Because anti-DMPO antibodies are not raised against nucleic acid constituents, the ability of polyclonal anti-DMPO to recognize DMPO adducts on guanine- and cytosine-containing polynucleotides must be due to the formation of DMPO covalently bound to these polynucleotides in a conformation different from the one recognized by monoclonal anti-DMPO. This raises the possibility that new monoclonal anti-DMPO antibodies could be raised against a putative different conformation of DMPO adducts. This could be particularly informative if certain DMPO conformations are more likely to arise in the oxidation of certain sequences.
The immuno-spin trapping techniques developed here allow for separation of genomic DNA based on size and detection of DMPO adducts on DNA immobilized on nitrocellulose and PVDF membranes. We expect to apply this technique to cell culture and animal models of oxidative stress by converting the genomic DNA to smaller fragments by restriction enzyme digestion or sonication. Although the degree of radical-mediated DNA damage is likely to be far lower in vivo than can be achieved using an in vitro copper-Fenton system, DNA radicals have been detected in cells [46][47][4][11] and in animals [48] by immuno-spin trapping using confocal microscopy and an older ELISA method. Here, we have shown that denaturing the DNA improves the detection of DNA-DMPO adducts by ELISA and that DMPO adducts undetectable by the older ELISA method can be detected with the improved method (Fig. 3).The detection of DMPO adducts on DNA fixed to a membrane is also enhanced by denaturation (Fig. 2). The detection of DNA-DMPO adducts on denatured DNA in the immunoblotting techniques (Fig.1 and 2) is comparable to that seen with the improved ELISA method (Fig. 3). The use of these immunoblotting techniques would allow the visualization of the extent of oxidatively generated damage throughout the genome in response to a particular oxidative stress, as free radical intermediates have been strongly implicated in oxidatively-generated damage. The next step, immunoprecipitation of DMPO-tagged DNA sequences, would lead the way to identification of specific sequences damaged by defined events or physiological processes. In this way, both the level of DNA damage, measured by DMPO adduct ELISA, and the specific DNA sequences could then be correlated with a functional effect.
Supplementary Material
Highlights.
► Radicals formed on DNA can be detected using anti-DMPO antibodies. ► We report new techniques for DMPO adduct detection on DNA blots. ► DNA denaturation improves DMPO adduct detection in blotting procedures and ELISA. ► Different DNA sequences showed differing susceptibilities to radical formation.
Acknowledgements
We are grateful to A.G. Motten, J. Corbett and M. Mason for valuable help in the preparation of the manuscript.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Environmental Health Sciences 52 (Z01 ES050139-13). Funding for open access charge: Intramural Research Program of the National Institutes of Health and the National Institute of Environmental Health Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 2.Kizek R, Adam V, Hrabeta J, Eckschlager T, Smutny S, Burda JV, Frei E, Stiborova M. Anthracyclines and ellipticines as DNA-damaging anticancer drugs: recent advances. Pharmacol. Ther. 2012;133:26–39. doi: 10.1016/j.pharmthera.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Sinha BK. Free radicals in anticancer drug pharmacology. Chem. Biol. Interact. 1989;69:293–317. doi: 10.1016/0009-2797(89)90117-8. [DOI] [PubMed] [Google Scholar]
- 4.Kojima C, Ramirez DC, Tokar EJ, Himeno S, Drobna Z, Styblo M, Mason RP, Waalkes MP. Requirement of arsenic biomethylation for oxidative DNA damage. J. Natl. Cancer Inst. 2009;101:1670–1681. doi: 10.1093/jnci/djp414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flora SJ. Arsenic-induced oxidative stress and its reversibility. Free Radic. Biol. Med. 2011;51:257–281. doi: 10.1016/j.freeradbiomed.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Ames BN. Endogenous DNA damage as related to cancer and aging. Mutat. Res. 1989;214:41–46. doi: 10.1016/0027-5107(89)90196-6. [DOI] [PubMed] [Google Scholar]
- 7.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev.Biochem. 2008:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 9.Gunther MR, Hanna PM, Mason RP, Cohen MS. Hydroxyl radical formation from cuprous ion and hydrogen peroxide: a spin-trapping study. Arch. Biochem. Biophys. 1995;316:515–522. doi: 10.1006/abbi.1995.1068. [DOI] [PubMed] [Google Scholar]
- 10.Milne L, Nicotera P, Orrenius S, Burkitt MJ. Effects of glutathione and chelating agents on copper-mediated DNA oxidation: pro-oxidant and antioxidant properties of glutathione. Arch. Biochem. Biophys. 1993;304:102–109. doi: 10.1006/abbi.1993.1327. [DOI] [PubMed] [Google Scholar]
- 11.Ogusucu R, Rettori D, Netto LES, Augusto O. Superoxide dismutase 1-mediated production of ethanol- and DNA-derived radicals in yeasts challenged with hydrogen peroxide: molecular insights into the genome instability of peroxiredoxin-null strains. J. Biol. Chem. 2009;284:5546–5556. doi: 10.1074/jbc.M805526200. [DOI] [PubMed] [Google Scholar]
- 12.Ranguelova K, Chatterjee S, Ehrenshaft M, Ramirez DC, Summers FA, Kadiiska MB, Mason RP. Protein radical formation resulting from eosinophil peroxidase-catalyzed oxidation of sulfite. J. Biol. Chem. 2010;285:24195–24205. doi: 10.1074/jbc.M109.069054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic. Biol. Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 14.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping of DNA radicals. Nat. Methods. 2006;3:123–127. doi: 10.1038/nmeth852. [DOI] [PubMed] [Google Scholar]
- 15.Khan N, Wilmot CM, Rosen GM, Demidenko E, Sun J, Joseph J, O'Hara J, Kalyanaraman B, Swartz HM. Spin traps: in vitro toxicity and stability of radical adducts. Free Radic. Biol. Med. 2003;34:1473–1481. doi: 10.1016/s0891-5849(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 16.Liu KJ, Miyake M, Panz T, Swartz H. Evaluation of DEPMPO as a spin trapping agent in biological systems. Free Radic. Biol. Med. 1999;26:714–721. doi: 10.1016/s0891-5849(98)00251-2. [DOI] [PubMed] [Google Scholar]
- 17.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping analyses of DNA radicals. Nat Protoc. 2007;2:512–522. doi: 10.1038/nprot.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharjee S, Deterding LJ, Chatterjee S, Jiang J, Ehrenshaft M, Lardinois O, Ramirez DC, Tomer KB, Mason RP. Site-specific radical formation in DNA induced by Cu(II)-H2O2 oxidizing system, using ESR, immuno-spin trapping, LC-MS, and MS/MS. Free Radic. Biol. Med. 2011;50:1536–1545. doi: 10.1016/j.freeradbiomed.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin B, Whyatt RM, Perera FP, Randall MC, Cooper TB, Santella RM. Determination of 8-hydroxydeoxyguanosine by an immunoaffinity chromatography-monoclonal antibody-based ELISA. Free Radic. Biol. Med. 1995;18:1023–1032. doi: 10.1016/0891-5849(95)00003-g. [DOI] [PubMed] [Google Scholar]
- 20.ESCODD. Comparison of different methods of measuring 8-oxoguanine as a marker of oxidative DNA damage. ESCODD (European Standards Committee on Oxidative DNA Damage) Free Radic. Res. 2000;32:333–341. doi: 10.1080/10715760000300331. [DOI] [PubMed] [Google Scholar]
- 21.Lunec J. ESCODD: European Standards Committee on Oxidative DNA Damage. Free Radic. Res. 1998;29:601–608. doi: 10.1080/10715769800300651. [DOI] [PubMed] [Google Scholar]
- 22.Möller L, Hofer T, Zeisig M. Methodological considerations and factors affecting 8-hydroxy-2'- deoxyguanosine analysis. Free Radic. Res. 1998;29:511–524. doi: 10.1080/10715769800300561. [DOI] [PubMed] [Google Scholar]
- 23.Cadet J, Douki T, Ravanat JL. Measurement of oxidatively generated base damage in cellular DNA. Mutat. Res. 2011;711:3–12. doi: 10.1016/j.mrfmmm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Gedik CM, Collins A. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 25.Collins AR, Cadet J, Moller L, Poulsen HE, Vina J. Are we sure we know how to measure 8- oxo-7,8-dihydroguanine in DNA from human cells? Arch. Biochem. Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 26.Chao MR, Yen CC, Hu CW. Prevention of artifactual oxidation in determination of cellular 8- oxo-7,8-dihydro-2'-deoxyguanosine by isotope-dilution LC-MS/MS with automated solid-phase extraction. Free Radic. Biol. Med. 2008;44:464–473. doi: 10.1016/j.freeradbiomed.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Chevion M. A site-specific mechanism for free radical induced biological damage: the essential role of redox-active transition metals. Free Radic. Biol. Med. 1988;5:27–37. doi: 10.1016/0891-5849(88)90059-7. [DOI] [PubMed] [Google Scholar]
- 28.Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Copper-ion-dependent damage to the bases in DNA in the presence of hydrogen peroxide. Biochem. J. 1991;273:601–604. doi: 10.1042/bj2730601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoewe R, Prutz WA. Copper-catalyzed DNA damage by ascorbate and hydrogen peroxide: kinetics and yield. Free Radic. Biol. Med. 1987;3:97–105. doi: 10.1016/s0891-5849(87)80003-5. [DOI] [PubMed] [Google Scholar]
- 30.Gao YG, Sriram M, Wang AHJ. Crystallographic studies of metal ion-DNA interactions: different binding modes of cobalt(II), coppeRII) and barium(II) to N7 of guanines in Z-DNA and a drug-DNA complex. Nucleic Acids Res. 1993;21:4093–4101. doi: 10.1093/nar/21.17.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geierstanger BH, Kagawa TF, Chen SL, Quigley GJ, Ho PS. Base-specific binding of coppeRII) to Z-DNA. The 1.3-Å single crystal structure of d(m5CGUAm5CG) in the presence of CuCl2. J. Biol. Chem. 1991;266:20185–20191. doi: 10.2210/pdb1d40/pdb. [DOI] [PubMed] [Google Scholar]
- 32.Kagawa TF, Geierstanger BH, Wang AHJ, Ho PS. Covalent modification of guanine bases in double-stranded DNA. The 1.2-Å Z-DNA structure of d(CGCGCG) in the presence of CuCl2. J. Biol. Chem. 1991;266:20175–20184. doi: 10.2210/pdb1d39/pdb. [DOI] [PubMed] [Google Scholar]
- 33.Carmichael PL, Hewer A, Osborne MR, Strain AJ, Phillips DH. Detection of bulky DNA lesions in the liver of patients with Wilson's disease and primary haemochromatosis. Mutat. Res. 1995;326:235–243. doi: 10.1016/0027-5107(94)00177-7. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn KM, DeRisi JL, Brown PO, Sarnow P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell. Biol. 2001;21:916–927. doi: 10.1128/MCB.21.3.916-927.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thurston CF, Perry CR, Pollard JW. In: Electrophoresis of RNA denatured with glyoxal or formaldehyde. Clifton NJ, editor. Humana Press; 1988. c1988. [DOI] [PubMed] [Google Scholar]
- 36.Masek T, Vopalensky V, Suchomelova P, Pospisek M. Denaturing RNA electrophoresis in TAE agarose gels. Anal. Biochem. 2005;336:46–50. doi: 10.1016/j.ab.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Hanna PM, Mason RP. Direct evidence for inhibition of free radical formation from Cu(I) and hydrogen peroxide by glutathione and other potential ligands using the EPR spin-trapping technique. Arch. Biochem. Biophys. 1992;295:205–213. doi: 10.1016/0003-9861(92)90507-s. [DOI] [PubMed] [Google Scholar]
- 38.Hanna PM, Chamulitrat W, Mason RP. When are metal ion-dependent hydroxyl and alkoxyl radical adducts of 5,5-dimethyl-1-pyrroline N-oxide artifacts? Arch. Biochem. Biophys. 1992;296:640–644. doi: 10.1016/0003-9861(92)90620-c. [DOI] [PubMed] [Google Scholar]
- 39.Williams DL. The use of a PVDF membrane in the rapid immobilization of genomic DNA for dotblot hybridization analysis. Biotechniques. 1990;8:14–15. [PubMed] [Google Scholar]
- 40.Hicks DA, Vecoli C. The use of PVDF membrane in the alkaline transfer of DNA. Biotechniques. 1987;5:206. [Google Scholar]
- 41.Makino K, Hagiwara T, Hagi A, Nishi M, Murakami A. Cautionary note for DMPO spin trapping in the presence of iron ion. Biochem. Biophys. Res. Commun. 1990;172:1073–1080. doi: 10.1016/0006-291x(90)91556-8. [DOI] [PubMed] [Google Scholar]
- 42.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O'Halloran TV. Undetectable intracellular free copper: The requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 43.Savinkova LK, Ponomarenko MP, Ponomarenko PM, Drachkova IA, Lysova MV, Arshinova TV, Kolchanov NA. TATA box polymorphisms in human gene promoters and associated hereditary pathologies. Biochemistry (Mosc) 2009;74:117–129. doi: 10.1134/s0006297909020011. [DOI] [PubMed] [Google Scholar]
- 44.Segal E, Widom J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson WD, Wang YH, Krishnamoorthy CR, Smith JC. Poly(dA).poly(dT) exists in an unusual conformation under physiological conditions: propidium binding to poly(dA).poly(dT) and poly[d(AT)]. poly[d(A-T)] Biochemistry. 1985;24:3991–3999. doi: 10.1021/bi00336a029. [DOI] [PubMed] [Google Scholar]
- 46.Bhattacharjee S, Chatterjee S, Jiang J, Sinha BK, Mason RP. Detection and imaging of the free radical DNA in cells - site-specific radical formation induced by Fenton chemistry and its repair in cellular DNA as seen by electron spin resonance, immuno-spin trapping and confocal microscopy. Nucleic Acids Res. 2012;40:5477–5486. doi: 10.1093/nar/gks180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomez-Mejiba SE, Zhai Z, Gimenez MS, Ashby MT, Chilakapati J, Kitchin K, Mason RP, Ramirez DC. Myeloperoxidase-induced genomic DNA-centered radicals. J. Biol. Chem. 2010;285:20062–20071. doi: 10.1074/jbc.M109.086579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dogan S, Ozlem Elpek G, Kirimlioglu Konuk E, Demir N, Aslan M. Measurement of intracellular biomolecular oxidation in liver ischemia-reperfusion injury via immuno-spin trapping. Free Radic. Biol. Med. 2012;53:406–414. doi: 10.1016/j.freeradbiomed.2012.05.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.