Abstract
The RNA-binding protein Musashi1 (Msi1) is one of two mammalian homologues of Drosophila Musashi, which is required for the asymmetric cell division of sensory organ precursor cells. In the mouse central nervous system (CNS) Msi1 is preferentially expressed in mitotically active progenitor cells in the ventricular zone (VZ) of the neural tube during embryonic development and in the subventricular zone (SVZ) of the postnatal brain. Previous studies showed that cells in the SVZ can contribute to long-term neurogenesis in the olfactory bulb (OB) but it remains unclear whether Msi1-expressing cells have self-renewing potential and can contribute to neurogenesis in the adult. Here we describe the generation of Msi1-CreERT2 knock-in mice and show by cell lineage tracing that Msi1-CreERT2-expressing cells mark neural stem cells (NSCs) in both the embryonic and adult brain. Msi1-CreERT2 mice thus represent a new tool in our arsenal for genetically manipulating NSCs, which will be essential for understanding the molecular mechanisms underlying neural development.
Keywords: Msi1, NSCs, SVZ, Olfactory epithelium, Knock-in mice
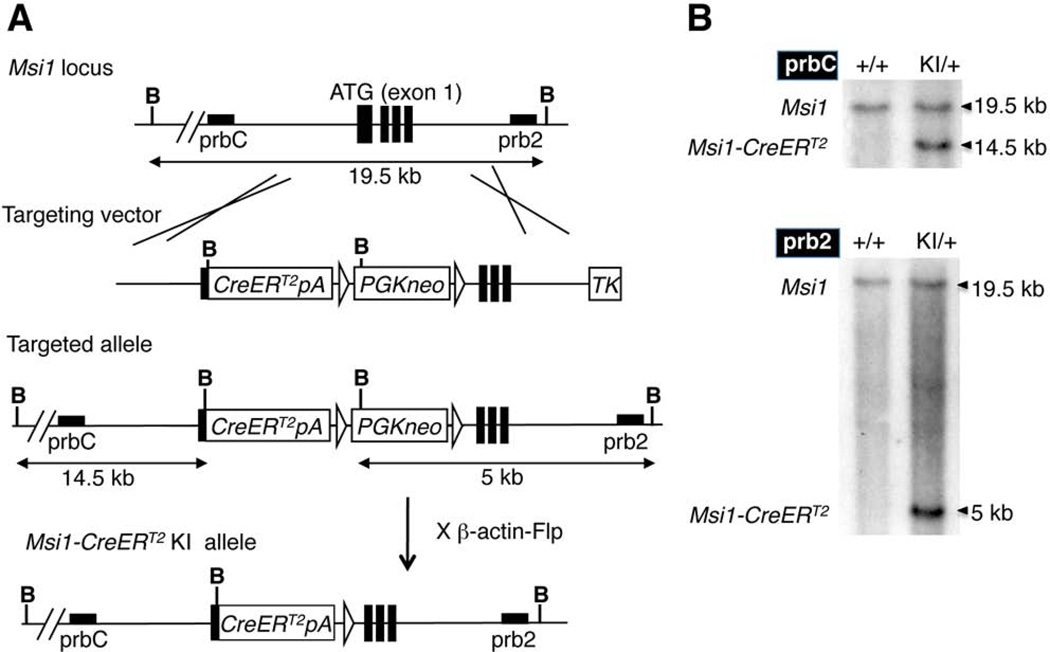
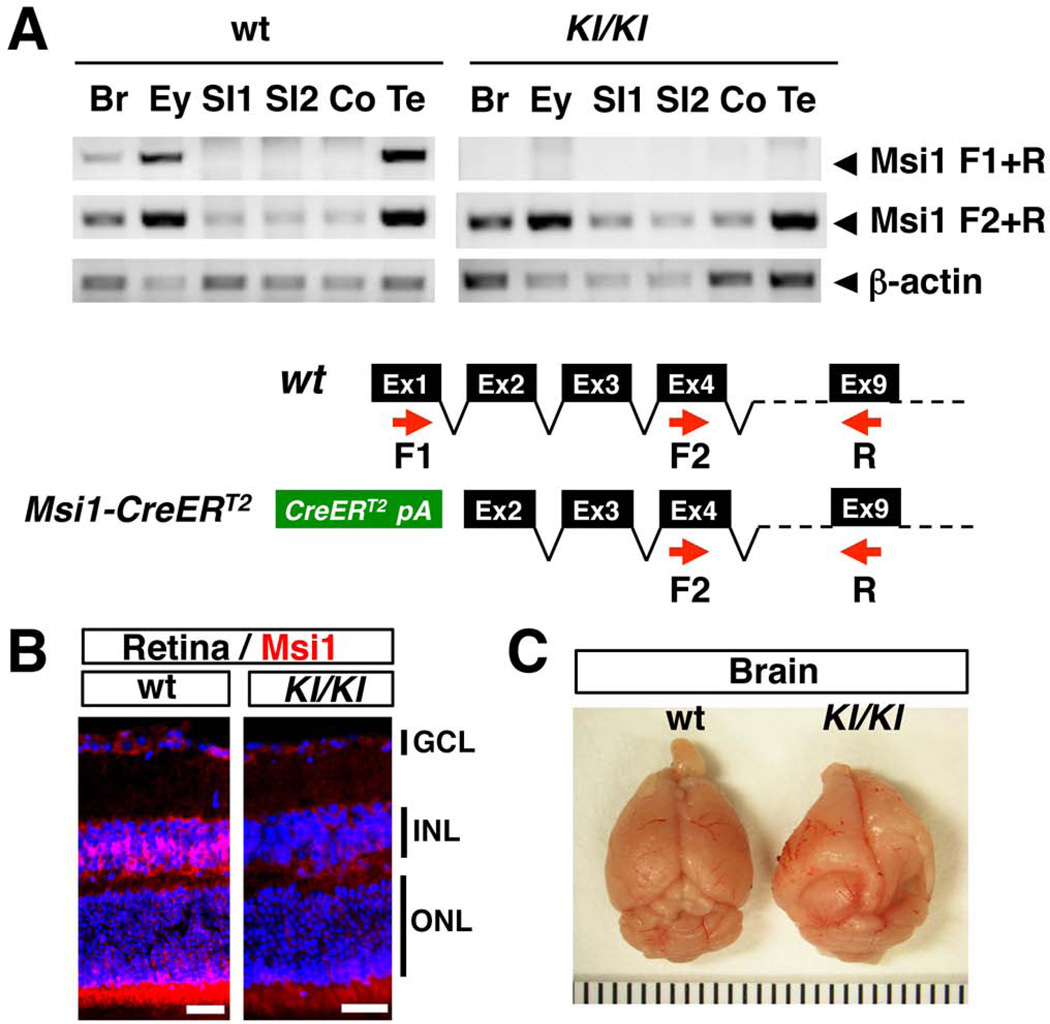
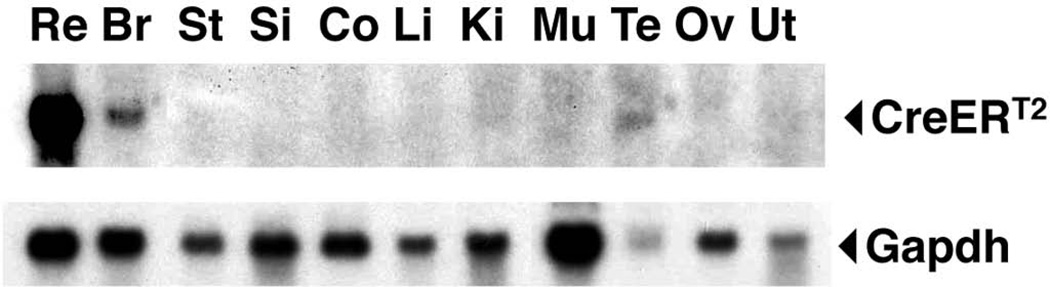
In order to generate the Msi1-CreERT2 knock-in mice, a targeting vector was created by inserting a CreERT2 expression cassette downstream of the first ATG in Msi1 exon 1 using recombineering, along with a PGKneo selection cassette (Copeland et al., 2001) (Fig. 1A). CreERT2 is an inducible form of the Cre recombinase that can be activated by tamoxifen administration (Feil et al., 1997). The targeting cassette was subsequently electroporated into ES cells, and the cells screened for homologous recombination by Southern blotting (Fig. 1A, 1B). Correctly targeted cells were subsequently injected into mouse blastocysts to generate Msi1-CreERT2 knock-in mice, and the selection cassette was then removed by crossing the mice to β-actin-Flp mice. RT-PCR confirmed that the knock-in allele produces no Msi1 transcript containing exon 1 (Fig 2A, see Msi1 F1+R). We examined the coding potential of the shorter transcript in the eye by using an antibody that recognizes an epitope coded by the exon 10 (LAPGYTYQFP) (Kaneko et al., 2000). Msi1 expression was detected in Muller glia and photoreceptors in the wild type mouse retina as previously reported (Nickerson et al., 2011) (Fig. 2B). However, the signal was not detected in the retina of homozygous Msi1-CreERT2 knock-in mice (Fig. 2B), indicating that the shorter transcript is not translated into an N-terminal truncated Msi1 protein in the eye. While heterozygous Msi1-CreERT2 mice were healthy and fertile, 50% of homozygous Msi1-CreERT2 mice (6 of 12) developed hydrocephalus (Fig. 2C), a phenotype described previously for the Msi1 mutant mice that lack exons 1–4 (Sakakibara et al., 2002), indicating that the inactivation of a transcript containing exon 1 is responsible for the phenotype. Northern blot analysis showed that the 1.4 kb Msi1-CreERT2 transcript was expressed in retina, brain and testis (Fig. 3), consistent with the known expression pattern of Msi1 (Sakakibara et al., 1996).
FIG. 1. Generation of Msi1-CreERT2 knock-in mice.
(A) A CreERT2-PGKneo expression cassette was inserted into the first ATG located in exon 1 of Msi1. The pBlight-Msi1-CreERT2-PGKneo targeting vector was used for homologous recombination to generate the Msi1-CreERT2 targeted allele. The PGKneo cassette was subsequently excised by flip-mediated recombination. B; BamHI, prbC (5’ probe), prb2 (3’ probe). (B) Southern blot analysis to screen ES cells for correctly targeted alleles. For the knock-in (KI) allele, BamHI digestion produces 14.5 kb and 5 kb bands for prbC and prb2, respectively. For the wild type (+) allele, BamHI digestion produces 19.5 kb bands for both probes.
FIG. 2. Expression of Msi1-CreERT2 in homozygous knock-in mice.
(A) RT-PCR analysis of Msi1 mRNA expression in wild type (wt) and Msi1-CreERT2 homozygous knock-in mice (KI/KI). Red arrows, PCR primers; Br, brain; Ey, eye; SI, small intestine; Co, colon; Te, testis. (B) Msi1 expression was detected in Muller glial cells in the INL and photoreceptors in the ONL of the wild type (wt) retina, but not in the retina of homozygous knock-in mice (KI/KI). GCL, ganglion cell layer; INL, inner nuclear layer; ONL, outer nuclear layer. The nucleus is counterstained with DAPI. Bars represent 50 µm. (C) Hydrocephalus was often observed in Msi1-CreERT2 homozygous knock-in mice (KI/KI).
FIG. 3. Tissue distribution of CreERT2 transcripts in Msi1-CreERT2 knock-in mice.
The expression levels of CreERT2 mRNA in adult tissues were determined by Northern blot analysis. The membrane was then reprobed with Gapdh as a loading control. Re, retina; Br, brain; St, stomach; Si, small intestine; Co, colon; Li, liver; Ki, kidney; Mu, muscle; Te, testis; Ov, ovary; Ut, uterus
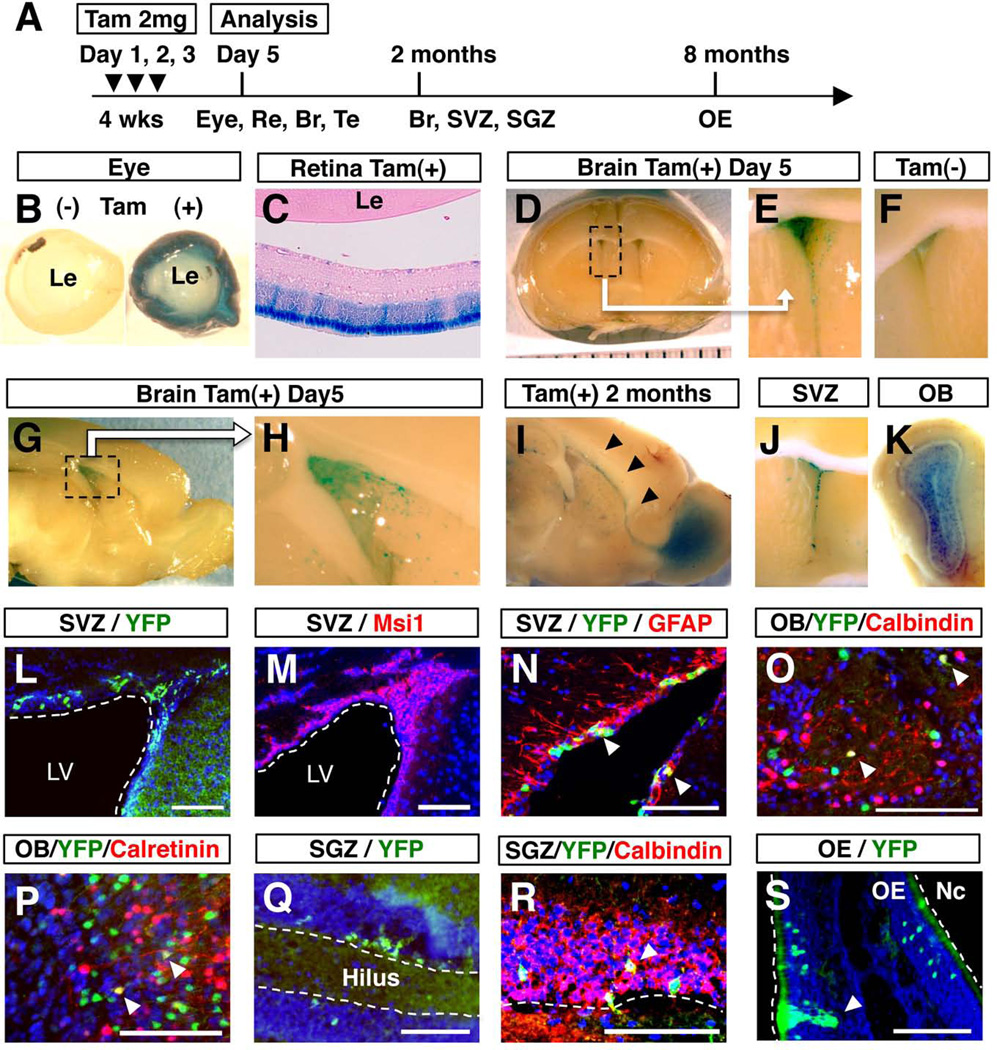
To more thoroughly characterize Msi1-CreERT2 expression, we crossed Msi1-CreERT2 mice with Rosa26-lsl-LacZ reporter (R26R3) mice (Soriano, 1999). In Msi1-CreERT2; R26R3 mice, LacZ expression is blocked by the presence of the transcriptional floxed-stop (lsl) cassette. However, once Msi1-CreERT2 is activated by tamoxifen, the stop cassette is removed by Cre recombination and LacZ expression can now be visualized by X-Gal staining. We focused on the eye, brain, olfactory bulb and testis since these are all known sites of Msi1 expression. Msi1-CreERT2; R26R3 mice were treated with tamoxifen at 4 weeks of age (Fig. 4A) and euthanized 5 days later. Whole mount staining of the eye showed that the entire retina was stained with X-Gal (Fig. 4B), while paraffin sections showed strong expression in the photoreceptor cell layer (Fig. 4C) where Msi1 expression has previously been observed (Susaki et al., 2009). In the brain, a small number of LacZ+ cells were observed in the wall of the lateral ventricle but not in the rostral migratory stream (RMS) and the OB (Fig. 4D, 4E, 4G and 4H), indicating the presence of Msi1-CreERT2-expressing cells in the SVZ. LacZ expression was tamoxifen dependent since no LacZ+ cells were detected in the absence of tamoxifen (Fig 4F).
FIG. 4. Lineage tracing of Msi1-CreERT2-expressing cells.
Msi1-CreERT2; R26R3 or Msi1-CreERT2; Rosa26YFP mice were treated for three consecutive days, with 2 mg of tamoxifen or corn oil, beginning at 4 weeks of age (A). X-Gal staining of the eye (B), retina (C) and brain (D–H) 5 days after tamoxifen or corn oil treatment. X-Gal staining of sagittal (I) and coronal (J, K) sections of the brain at 2 months after tamoxifen treatment. LacZ+ cells were observed in the RMS (I, arrow heads). YFP expression in the SVZ (L, N), OB (O, P) and SGZ (Q, R) at 2 months after tamoxifen treatment, or 8 months after tamoxifen treatment for the OE (S). Endogenous Msi1 expression was observed in the SVZ (M). In the SVZ, YFP+ cells and Msi1+ cells showed a similar pattern of localization (L, M). YFP+ cells in the SVZ expressed GFAP, a marker for NSCs (N, arrow heads). YFP+ cells in the OB expressed markers for interneurons, Calbindin (O, arrow heads) and Calretinin (P, arrow heads). YFP+ cells in the SGZ expressed Calbindin, a marker for dentate granule neuron (R, arrow head). In the OE, YFP+ cells with morphological characteristics of sustentacular cells were observed (S, arrow head). Br, brain; Le, lens; LV, lateral ventricle; Nc, nasal cavity; Re, retina; Tam, tamoxifen; Te, testis; Bars: 100 µm.
Next, we traced the lineage of Msi1-CreERT2-expressing cells by performing X-Gal staining 60-days after tamoxifen treatment. LacZ+ cells were found in the OB, in addition to the RMS and SVZ (Fig. 4I–K), demonstrating that recombination took place in NSCs in the SVZ rather than transit-amplifying cells. Lineage-tracing experiments have shown that labeled transit-amplifying cells and their progeny complete migration to the OB within one month, with no labeled cells remaining in the RMS and SVZ after that time (Kim et al., 2007).
Green fluorescent protein and its variants are useful in lineage tracing, because they allow detection of labeled cells without chemical substrates, and the morphology of labeled cells can be more easily analyzed in tissue sections. Thus, we next generated a second reporter line, Msi1-CreERT2; Rosa26YFP. Analysis of frozen sections of Msi1-CreERT2; Rosa26YFP mice at 2 months after tamoxifen treatment showed that YFP+ cells were present in the SVZ (Fig. 4L) where endogenous Msi1 is expressed (Figure 4M), indicating that Msi1-CreERT2 expression faithfully recapitulates endogenous Msi1 expression in the SVZ. Immunohistochemical analysis showed that YFP+ cells in the SVZ expressed GFAP, a marker for adult NSCs (Ming and Song, 2011) (Fig. 4N). NSCs in the SVZ give rise to different subtypes of interneurons in the OB (Ming and Song, 2011). Immunohistochemical analysis showed the presence of YFP-expressing cells that express markers for interneurons, such as Calbindin and Calretinin (Ming and Song, 2011) (Fig. 4O and 4P). Together, these data demonstrate that Msi1-CreERT2 is expressed in NSCs of the SVZ.
In the subgranular zone (SGZ), putative multipotent neural stem or progenitor cells, which will give rise to dentate granule cells in the hippocampus, have also been identified (Bonaguidi et al., 2011; Zhao et al., 2008). We detected clusters of YFP+ cells in the dentate gyrus in the hippocampus of Msi1-CreERT2; Rosa26YFP mice at 2 months after tamoxifen addition (Fig. 4Q). YFP+ cells express a marker for dentate granule cell, Calbindin (Ming and Song, 2011) (Fig. 4R), suggesting that Msi1-CreERT2 is expressed in the precursor cells for dentate granule cells.
The olfactory neuroepithelium (OE) undergoes continual neurogenesis throughout life. Msi1-CreERT2; Rosa26YFP mice were treated with tamoxifen at 4 weeks and the OE was analyzed 8 months later. Clusters of YFP+ cells were occasionally observed in the OE. Most of YFP+ cells appeared to be olfactory receptor neurons (ORNs) as judged by their position in the OE and their cellular morphology (Fig. 4O). It is possible that these clusters of YFP+ cells were derived from a single Msi1-CreERT2-expressing stem or progenitor cell, because YFP+ cells were usually visible as cell clusters or single cells. We also detected YFP+ cells whose morphology and position seemed to correspond to sustentacular cells (Fig. 4O). Genetic experiments have reported that ORNs and sustentacular cells may share a common progenitor (Beites et al., 2005). Our data suggest that Msi1-CreERT2 is expressed in the common progenitor cells of the OE in adult mice.
Northern blot analysis showed the expression of CreERT2 in the testis, however we could not detect any cells that express reporter genes in either Msi1-CreERT2; R26R3 or Msi1-CreERT2; Rosa26YFP mice at five days after tamoxifen injection (Fig. 4A), raising a possibility that the expression level of CreERT2 might not be strong enough to induce recombination in the testis.
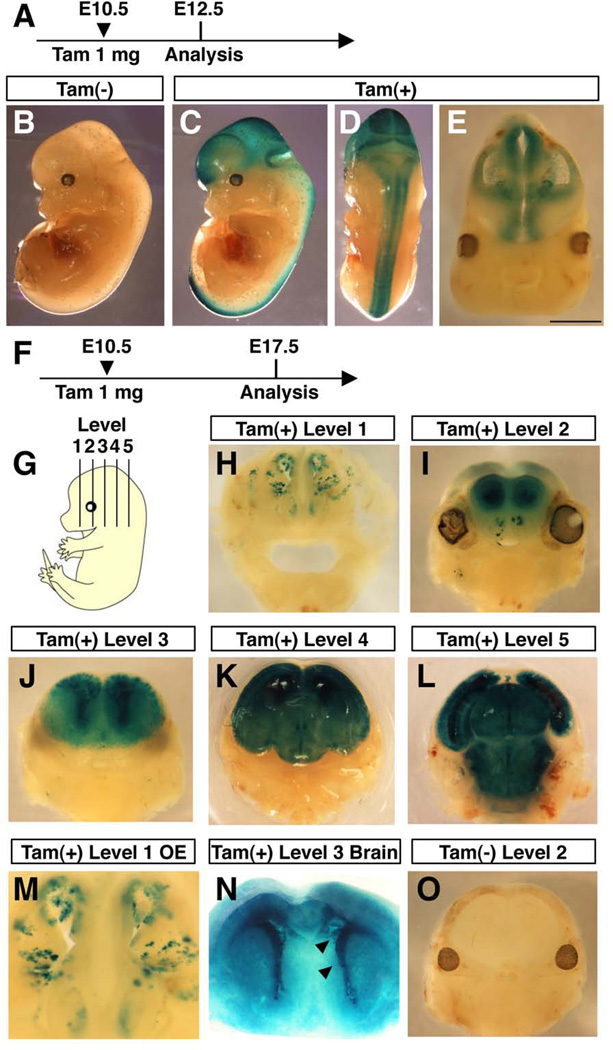
Next, we examined Msi1-CreERT2 expression during embryonic development. Pregnant Msi1-CreERT2; R26R3 mice were treated with tamoxifen or oil at E10.5 and the embryos stained for LacZ at E12.5 (Fig. 5A). No LacZ staining was observed in oil-treated embryos at E12.5 (Fig. 5B). LacZ+ cells were, however, observed within the entire nervous system at E12.5 in tamoxifen-treated embryos (Fig. 5C and 5D), indicating that Msi1-CreERT2 is expressed in the entire nervous system at E10.5. Coronal sections of embryonic brains showed strong X-Gal staining in the entire neuroepithelium (Fig. 5E), indicating that Msi1-CreERT2 is expressed in either NSCs or differentiating precursor cells in the developing cortex.
FIG. 5. Lineage tracing of Msi1-CreERT2-expressing cells in the embryonic neural system.
Time course of tamoxifen injection and X-Gal staining (A and F). Pregnant mice were treated with 1 mg of tamoxifen (C–E, H–N) or corn oil (B and O) at E10.5. Msi1-CreERT2; R26R3 embryos were stained with X-Gal in whole mount at E12.5 (A–E), or embryonic brains were sectioned at various levels using a vibratome (G), and stained with X-Gal at E17.5 (F–O). Strong X-Gal staining was observed in cells lining the lateral ventricle (N, arrowheads).
During neural development, the cortical layer is formed by an inside-out pattern of migration of differentiating cells, confining mitotic NSCs to the ventricular surface of the developing cortex, whereas differentiated cells occupy the peripheral region (Nadarajah and Parnavelas, 2002). To determine whether Msi1-CreERT2-expressing cells are NSCs or differentiating precursor cells in the developing cortex, we induced recombination in Msi1-CreERT2; R26R3 embryos at E10.5 and then examined the brain at E17.5. No LacZ+ cells were seen in oil-treated embryos (Fig. 5O), but in tamoxifen treated embryos, LacZ was expressed within the entire cortex in the forebrain, cerebellum and brain stem (Fig. 5I–L), showing that Msi1-CreERT2-expressing cells can give rise to differentiated cells in these regions. Importantly, intense LacZ staining was observed in the ventricular surface of the striatum (Fig. 5J and 5N), suggesting that Msi1-CreERT2-expressing cells have permanently labeled self-renewing stem cells in the VZ. These data indicate that Msi1-CreERT2 is also expressed in NSCs during embryonic development. Forni and colleagues showed that the activation of nestin-CreERT2 transgene induces brain abnormalities in a dose-dependent manner (Forni et al., 2006). However, the administration of 1 mg of tamoxifen did not induce any brain defects in the Msi1-CreERT2 line (Fig 5I–L and 5O), suggesting that the expression level of CreERT2 is not as strong as the nestin-CreERT2 line (Forni et al., 2006; Imayoshi et al., 2006).
In the adult OE we showed that Msi1-CreERT2 was likely expressed in progenitor cells for ORNs and sustentacular cells. Next we examined whether Msi1-CreERT2 is expressed in embryonic OE progenitor cells. During embryonic development of the OE, expansion of stem cells takes place by E12.5. Around E13.5-E14.5, the OE becomes organized into its mature pattern and established neurogenesis is initiated (Beites et al., 2005). We treated pregnant Msi1-CreERT2; R26R3 reporter mice with tamoxifen or oil at E10.5 and analyzed LacZ staining in embryos at E17.5. Clusters of LacZ+ cells were observed in the entire OE (Figure 5M), showing that Msi1-CreERT2 is expressed in stem or progenitor cells of the OE as early as E10.5.
Inducible recombination in NSCs has also been reported using Nestin-CreERT2 transgenes (Chen et al., 2009; Imayoshi et al., 2006; Lagace et al., 2007). Although we could not determine the efficiency of recombination due to the poor quality of CreERT2 immunostaining, X-Gal staining of the adult SVZ and the embryonic cortex in Msi1-CreERT2; R26R3 mice showed comparable patterns with those reported in Nestin-CreERT2 transgenic mice (Chen et al., 2009; Imayoshi et al., 2006). However, one striking difference was expression in the OE. The Nestin regulatory element directed reporter expression to a subpopulation of embryonic OE progenitor cells that are located in the dorsal medial zone (Murdoch and Roskams, 2008). In contrast, we did not detect regional restriction of CreERT2 expression in the OE. Tamoxifen administration of Msi1-CreERT2; Rosa26YFP adult mice induced clusters of LacZ+ cells in ORN and sustentacular cells in the OE, suggesting that CreERT2 is expressed in the common progenitor for these lineages. Moreover, tamoxifen treatment in the embryos as early as E10.5 induced clusters of LacZ+ cells in the OE, indicating that CreERT2 is expressed in expanding stem cells in the OE. Detailed lineage tracing of CreERT2 expressing cells will be needed to reveal the features of neurogenesis in the OE.
Msi1 is also reported to be a marker for intestinal stem cells (Potten et al., 2003), however by Northern blotting or lineage tracing, we failed to detect Msi1-CreERT2 expression in the intestine (data not shown). Msi1 is known to have several transcriptional variants (Susaki et al., 2009) (Ensemble Genome Browser, http://www.ensembl.org/). In Msi1-CreERT2 knock-in mice, Msi1-CreERT2 expression initiates from the first ATG in exon 1. The Msi1 transcript initiating from this ATG was not detected in the intestine (Fig. 2A, see Msi1 F1+R), whereas a shorter transcript lacking exon 1 is expressed in the intestine (Fig. 2A, see Msi1 F2+R), suggesting that the alternative transcript is abundant in the intestine. Our Msi1-CreERT2 knock-in mice should therefore represent a useful new tool for manipulating NSCs, which is not confounded by expression in the intestine.
Methods
Generation of Msi1-CreERT2 knock-in mice
A mouse BAC clone (RP23-268H8) containing the entire Msi1 genomic locus was obtained from the Children’s Hospital Oakland Research Institute (CHORI) and electroporated into the recombineering strain SW105 (Warming et al., 2005). The entire 10.0 kb Msi1 genomic locus in addition to 5.0 kb of upstream and 5.0 kb of downstream sequence, was then subcloned into pBlight (Warming et al., 2006) by recombineering. A CreERT2 fragment was subsequently excised from pCAG-CreERT2 (Addgene) and inserted into a plasmid derivative of pL451 (Liu et al., 2003) that has a Neomycin selection cassette flanked between two FRT sites. Sequences of the first exon located downstream of the first ATG were then replaced by the CreERT2-polyA, FRT-PGKneo-FRT cassette using recombineering. The Msi1 targeting vector was then linearized and transfected into B6/129 F1 hybrid embryonic stem (ES) cells (Southon and Tessarollo, 2009). G418 and FIAU double-resistant ES cell clones were selected and analyzed for homologous recombination using Southern blot hybridization of BamHI-digested genomic DNA. Probes were PCR amplified with the following primers. Probe C F: 5’-ccc agg act cca ccc atgc-3’, R: 5’-agg caa ctc att aac aaa ag-3’; Probe2 F: 5’-cct ctt gac tct agg cca tc-3’. R: 5’-cag cag gac ctg gca gcc tt-3’. Two independently targeted ES cell clones were injected into C57BL/6 blastocysts to produce chimeric mice (Reid and Tessarollo, 2009). The chimeras were then mated to C57BL/6 mice and germline transmission in agouti offspring confirmed by PCR. The neomycin selection cassette was later excised in vivo by crossing the mice with b-actin-FLPe mice (The Jackson Laboratory). Msi1-CreERT2 heterozygous mice were then intercrossed to obtain homozygous animals. Homozygous mice showing signs of hydrocephalus (an enlarged and domed head) were sent to necropsy, and the brains fixed in 4% PFA in PBS. All animal experiments were performed with institutional animal care and use committee (IACUC) approval. The mice will be made available to the scientific community upon request.
Reporter strains and tamoxifen treatment
The R26R3 reporter strain was obtained from Philippe Soriano (Mount Sinai School of Medicine, New York, NY) (Soriano, 1999), while the Rosa26YFP reporter strain (Srinivas et al., 2001) was purchased from the Jackson Laboratory. Tamoxifen (Sigma) was prepared as a 20 mg/ml stock solution in corn oil (Sigma). At 4 weeks of age, Msi1-CreERT2;R26R3 or Msi1-CreERT2;Rosa26YFP mice were injected intraperitoneally with 2 mg of tamoxifen for 3 consecutive days. Tissues were collected at 5 days, 2 months or 8 months after tamoxifen treatment. For embryonic induction, 1 mg of tamoxifen was injected intraperitoneally into pregnant mothers at E10.5, and embryos subsequently collected at E12.5 or E17.5.
Northern blotting and RT-PCR
Total RNA was isolated using Trizol (Invitrogen). Northern blotting was performed according to standard protocols. Ten µg of total RNA was run on a 1% agarose gel in MOPS buffer (0.4M of 3-morpholinopropanesulfonic acid, 0.1M of sodium acetate and 20mM of EDTA). The coding regions of Cre and Gapdh were used as probes. Superscript III (Invitrogen) was used to synthesize cDNA. PCR was performed using the following primers. Msi1 F1: 5’-CAC GAC CCC TGC AAG ATG-3’, F2: 5’-TCG TCA CTT TCA TGG ACC AG-3’, R: 5’-TGA AGG CAT CCA TTC CGT AG-3’; b-actin F: 5’-TAG ACT TCG AGC AGG AGA TG-3’, R: 5’-ACT CAT CGT ACT CCT GCT TG-3’.
X-Gal staining and histology
For whole-mount X-Gal staining, embryos and adult tissues were fixed in 1% formaldehyde, 0.2% glutaraldehyde, 0.02% IGEPAL CA-630 (Sigma) in phosphate buffered saline (PBS) for 10 min at room temperature (RT), rinsed in PBS three times, and stained at 37°C overnight in X-Gal staining buffer (1 mg/ml X-Gal, 2 mM MgCl2, 0.02% IGEPAL CA-630, 1 mM K3Fe(CN)6, 1 mM K4Fe(CN)6 in PBS). For X-Gal staining of tissue sections, embryos and adult brains were fixed and sectioned using a vibratome (100 mm). Then, free-floating sections were stained in X-Gal staining buffer as described above. For retinal sections, X-Gal stained eyes were fixed with 4% paraformaldehyde (PFA) in PBS, washed and embedded in paraffin wax prior to sectioning. Sections were counterstained with eosin. For visualization of YFP+ cells, brains were fixed with 4% PFA in PBS overnight at 4°C, washed in PBS, cryoprotected in 25% sucrose in PBS and embedded in Tissue-Tek (Sakura). Frozen sections were made using a cryostat (Leica CM3050 S) at 30 mm thickness. Sections were counter stained with 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma) and analyzed using an upright microscope (Zeiss AxioImager Z1). Antibodies used for immunohistochemical analysis were rabbit polyclonal anti-Musashi1 (Millipore, AB5977), rat monoclonal anti-Musashi1 (MBL, D270-3) for the detection of an epitope coded by the exon 10 (LAPGYTYQFP) and Alexa594-conjugated goat polyclonal anti-rabbit IgG (Molecular Probes).
Acknowledgments
We thank Keith Rogers, Susan M. Rogers and Vanessa Shiyun Tay for technical support. This work was supported by the Agency for Science, Technology and Research (A-STAR), Singapore, and the Cancer Prevention Research Institute of Texas. N.A.J. and N.G.C. are both CPRIT Scholars in Cancer Research. LT was supported by the Intramural Research Program of the NCI, Center for Cancer Research, NIH.
Footnotes
The authors declare no conflict of interests.
References
- Beites CL, Kawauchi S, Crocker CE, Calof AL. Identification and molecular regulation of neural stem cells in the olfactory epithelium. Experimental cell research. 2005;306:309–316. doi: 10.1016/j.yexcr.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kwon CH, Lin L, Li Y, Parada LF. Inducible site-specific recombination in neural stem/progenitor cells. Genesis. 2009;47:122–131. doi: 10.1002/dvg.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nature reviews. Genetics. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochemical and biophysical research communications. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Forni PE, Scuoppo C, Imayoshi I, Taulli R, Dastru W, Sala V, Betz UA, Muzzi P, Martinuzzi D, Vercelli AE, Kageyama R, Ponzetto C. High levels of Cre expression in neuronal progenitors cause defects in brain development leading to microencephaly and hydrocephaly. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:9593–9602. doi: 10.1523/JNEUROSCI.2815-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Ohtsuka T, Metzger D, Chambon P, Kageyama R. Temporal regulation of Cre recombinase activity in neural stem cells. Genesis. 2006;44:233–238. doi: 10.1002/dvg.20212. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, Sawamoto K, Ogawa Y, Toyama Y, Miyata T, Okano H. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Developmental neuroscience. 2000;22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Leung CT, Reed RR, Johnson JE. In Vivo Analysis of Ascl1 Defined Progenitors Reveals Distinct Developmental Dynamics during Adult Neurogenesis and Gliogenesis. JNeurosci. 2007;27:12764–12774. doi: 10.1523/JNEUROSCI.3178-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagace DC, Whitman MC, Noonan MA, Ables JL, DeCarolis NA, Arguello AA, Donovan MH, Fischer SJ, Farnbauch LA, Beech RD, DiLeone RJ, Greer CA, Mandyam CD, Eisch AJ. Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:12623–12629. doi: 10.1523/JNEUROSCI.3812-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome research. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70:687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch B, Roskams AJ. A novel embryonic nestin-expressing radial glia-like progenitor gives rise to zonally restricted olfactory and vomeronasal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:4271–4282. doi: 10.1523/JNEUROSCI.5566-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nature reviews. Neuroscience. 2002;3:423–432. doi: 10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- Nickerson PE, Myers T, Clarke DB, Chow RL. Changes in Musashi-1 subcellular localization correlate with cell cycle exit during postnatal retinal development. Experimental eye research. 2011;92:344–352. doi: 10.1016/j.exer.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Potten CS, Booth C, Tudor GL, Booth D, Brady G, Hurley P, Ashton G, Clarke R, Sakakibara S, Okano H. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation; research in biological diversity. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- Reid SW, Tessarollo L. Isolation, microinjection and transfer of mouse blastocysts. Methods Mol Biol. 2009;530:269–285. doi: 10.1007/978-1-59745-471-1_14. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, Nakajima K, Yasutomi D, Nagata T, Kurihara Y, Uesugi S, Miyata T, Ogawa M, Mikoshiba K, Okano H. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Developmental biology. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- Sakakibara S, Nakamura Y, Yoshida T, Shibata S, Koike M, Takano H, Ueda S, Uchiyama Y, Noda T, Okano H. RNA-binding protein Musahi familiy: Roles for CNS stem cells and a subpopulation of ependymal cells revealed by targeted disruption and antisense ablation. PNAS. 2002;99:15194–15199. doi: 10.1073/pnas.232087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Southon E, Tessarollo L. Manipulating mouse embryonic stem cells. Methods Mol Biol. 2009;530:165–185. doi: 10.1007/978-1-59745-471-1_9. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC developmental biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki K, Kaneko J, Yamano Y, Nakamura K, Inami W, Yoshikawa T, Ozawa Y, Shibata S, Matsuzaki O, Okano H, Chiba C. Musashi-1, an RNA-binding protein, is indispensable for survival of photoreceptors. Experimental eye research. 2009;88:347–355. doi: 10.1016/j.exer.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic acids research. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warming S, Rachel RA, Jenkins NA, Copeland NG. Zfp423 is required for normal cerebellar development. Molecular and cellular biology. 2006;26:6913–6922. doi: 10.1128/MCB.02255-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]