CONSPECTUS
The nitration of protein tyrosine residues to 3-nitrotyrosine represents an oxidative postranslational modification that unveils the disruption of nitric oxide (•NO) signaling and metabolism towards pro-oxidant processes. Indeed, excess levels of reactive oxygen species in the presence of •NO or •NO-derived metabolites lead to the formation of nitrating species such as peroxynitrite. Thus, protein 3-nitrotyrosine has been established as a biomarker of cell, tissue and systemic “nitroxidative stress”. Moreover, tyrosine nitration modifies key properties of the amino acid (i.e. phenol group pKa, redox potential, hydrophobicity and volume). Thus, the incorporation of a nitro group (−NO2) to protein tyrosines can lead to profound structural and functional changes, some of which contribute to altered cell and tissue homeostasis.
In this Account, I describe our current efforts to define 1) biologically-relevant mechanisms of protein tyrosine nitration and 2) how this modification can cause changes in protein structure and function at the molecular level. First, the relevance of protein tyrosine nitration via free radical-mediated reactions (in both peroxynitrite-dependent or independent pathways) involving the intermediacy of tyrosyl radical (Tyr•) will be underscored. This feature of the nitration process becomes critical as Tyr• can take variable fates, including the formation of 3-nitrotyrosine. Fast kinetic techniques, electron paramagnetic resonance (EPR) studies, bioanalytical methods and kinetic simulations have altogether assisted to characterize and fingerprint the reactions of tyrosine with peroxynitrite and one-electron oxidants and its further evolution to 3-nitrotyrosine. Recent findings show that nitration of tyrosines in proteins associated to biomembranes is linked to the lipid peroxidation process via a connecting reaction that involves the one-electron oxidation of tyrosine by lipid peroxyl radicals (LOO•).
Second, immunochemical and proteomic-based studies indicate that protein tyrosine nitration is a selective process in vitro and in vivo, preferentially directed to a subset of proteins, and within those proteins, typically one or two tyrosine residues are site-specifically modified. The nature and site(s) of formation of the proximal oxidizing/nitrating species, the physico-chemical characteristics of the local microenvironment and also structural features of the protein account for part of this selectivity. Then, how this relatively subtle chemical modification in one tyrosine residue can sometimes cause dramatic changes in protein activity has remained elusive. Herein, I will analyze recent structural biology data of two pure and homogenously nitrated mitochondrial proteins (i.e. cytochrome c and MnSOD) to illustrate regio-selectivity and structural effects of tyrosine nitration, and subsequent impact in protein loss- or even gain-of-function.
INTRODUCTION
With the emergence of peroxynitrite* in the early nineties as a biologically-relevant oxidant arising from the diffusion-controlled reaction of nitric oxide (•NO) and O2•˜, additional oxidative post-translational modifications in proteins were recognized: indeed, peroxynitrite can evolve into oxidizing and nitrating species that cause nitration of amino acids, most notably, tyrosine1 (and references therein). Later, it was found that nitrating species such as nitrogen dioxide (•NO2) could be also formed by peroxynitrite-independent pathways, most notably by the hemeperoxidase/hydrogen peroxide (H2O2)-dependent oxidation of nitrite (NO2−)1†. The main mechanism of protein tyrosine nitration in vivo relies in free radical reactions (Figure 1). The nitration of protein tyrosine residues to 3-nitrotyrosine uncovers the disruption of nitric oxide (•NO) signaling and metabolism towards pro-oxidant processes, in a condition also defined as “nitroxidative stress”.
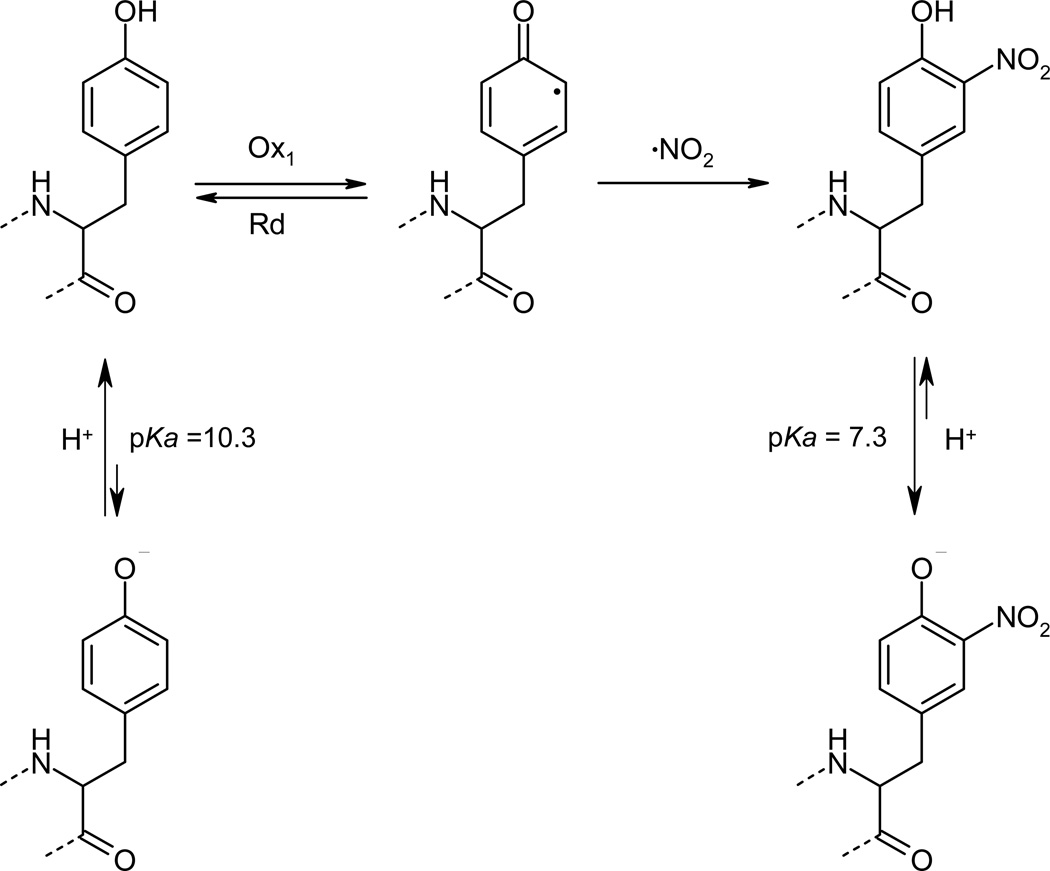
Figure 1. Tyrosine, tyrosyl radical and 3-nitrotyrosine.
The one-electron oxidation of tyrosine yields Tyr· that upon reaction with •NO2 evolves to 3-nitrotyrosine. To note, while the phenolic −OH group in free tyrosine is almost 100 % protonated at physiological pH, nitration causes a drop of three pH units in the pKa value resulting in a significant fraction (ca. 50 %) of the corresponding phenolate. Tyr· can be reduced back to tyrosine by reductants (Rd) in competition with the nitration reaction.
Tyrosine nitrated proteins have been detected in a large variety of disease conditions and are also associated to the aging process2. While immunochemical-based studies using antibodies against protein 3-nitrotyrosine have been useful for detection and cellular/tissue distribution of nitrated proteins, proteomic-based analyses have been fundamental to identify preferential protein targets of nitration and bioanalytical methods have assisted to quantitate the extents of nitration. The collected information indicates that a) protein tyrosine nitration occurs under basal physiological conditions, and is several-fold enhanced under circumstances that lead to augmented rates of oxidants and •NO formation (e.g. inflammation), b) the distribution of tyrosine nitrated proteins is largely dependent on the sites of formation of nitrating species, c) protein tyrosine nitration is preferentially directed to a subset of proteins, and within those proteins, typically one or at the most two specific tyrosine residues are nitrated, part of which depends on the nitration mechanism.
In spite of being now a well-established biomarker of nitroxidative stress, a fundamental question has been whether tyrosine nitration in specific proteins, can lead to substantial changes in their biological function and be part of pathophysiological processes. While it is relatively simple to demonstrate changes in protein function after nitration in vitro, the in vivo relevance of this process is a rather challenging task. First, it is arduous to unambiguously establish direct and quantitative relationships between extents of tyrosine nitration in specific proteins and biological responses in cells or tissues; secondly, the influence of protein tyrosine nitration in biological processes is usually obscured by a multiplicity of concurrent oxidative events. Possible biochemical consequences of protein tyrosine nitration involve changes in activity (either loss- or gain-of-function), eliciting of immunogenic responses, interference in tyrosine-kinase-dependent pathways, alteration of protein assembly and polymerization, facilitation of protein degradation (turnover) and participation in the creation of proteasome-resistant protein aggregates2. We have prudently compiled relevant examples of proteins reported to change their activities on tyrosine nitration in vitro and in vivo1–3; the list continues to rapidly grow4.
A large gap in the field exists in relation to the structural basis that determines how a relatively subtle post-translational modification (i.e. incorporation of -NO2 group) can sometimes trigger dramatic changes in protein activity. Recent structural biology studies have provided fresh insights into the factors that control the selectivity of protein tyrosine nitration and how the presence of 3-nitrotyrosine can lead to local or global structural changes that result in modification of protein function.
Thus, herein I will describe my current perspective and efforts to define 1) biologically-relevant mechanisms of protein tyrosine nitration and 2) how this modification can cause changes in protein structure and function at the molecular level.
BIOCHEMICAL PROPERTIES OF TYROSINE AND NITROTYROSINE
Tyrosine residues in proteins typically constitute in average 3–4 mol % and are located in different regions of the protein by virtue of the relatively large phenolic amphipathic side-chain capable to a) interact with water and participate in hydrogen bond formation and b) undergo cation-π and non-polar interactions5; typically, both solvent-exposed and buried residues coexist within a protein with, on average, only 15 % of tyrosine residues in proteins are at least 95 % buried6.
The versatile physico-chemical properties of tyrosine allows it to play a central role in conformation and molecular recognition5; moreover, tyrosine has special roles by virtue of the phenol functionality: 1) it can receive phosphate groups in target proteins by way of protein tyrosine kinases and 2) it participates in electron transfer processes with the intermediate formation of a tyrosyl radical (Tyr•)(Eo’ = +0.94 V)7.
The incorporation of a nitro group on the C-3 of the phenolic ring generates changes in key properties of the parent amino acid. Notably, a large decrease in the pKa of the −OH group is observed (ca. 10,0–10,3 to 7,2–7,5 for free tyrosine and 3-nitrotyrosine in water, respectively)2 (Fig. 1). Naturally, the pKa values of the phenolic group for tyrosine and 3-nitrotyrosine can vary within a protein compared to those of the free amino acids in solution, depending on the polarity of the medium and the influence of neighboring amino acids8. In addition, the nitro group represents a bulky and hydrophobic substituent that may create local steric restrictions, trigger conformational changes and impede tyrosine phosphorylation. The Eo’ values of the nitrotyrosine/nitrotyrosyl radical couple is 200–300 mV more positive than that of tyrosine/tyrosyl radical and therefore tyrosine nitration alters tyrosine-dependent intramolecular electron transfer processes in proteins8.
PROTEIN TYROSINE NITRATION AS A FREE RADICAL PROCESS
Current evidence indicates that the mechanism of protein tyrosine nitration in biological systems is mediated by free radical reactions, implying the intermediacy of Tyr• and subsequent reactions with either •NO or •NO21 (Fig. 2). There is no direct bimolecular reaction of tyrosine with peroxynitrite, but rather with peroxynitrite-derived radicals 1,9,10. Depending on the predominant nitrating species, mechanism and milieu, relevant one-electron oxidants are carbonate radicals (CO3•−), oxo-metal complexes (e.g. myeloperoxidase (MPO) compound I, hemin-Fe4+) and even lipid peroxyl radicals (LOO•), among others (Fig. 2)1,6,11. Protein Tyr· has been detected in vitro in a variety of nitration conditions by electron paramagnetic resonance (EPR)-spin trapping12,13. Interestingly, protein Tyr· formation in cells and tissues undergoing nitroxidative stress can be potentially detected in vivo immunochemically as the adduct of the spin trap 5,5 dimethyl-1-pyrroline-N-oxide (DMPO) with Tyr· evolves to a stable nitrone derivative that can be conveniently detected with antibodies (i.e. immunospintrapping)13.
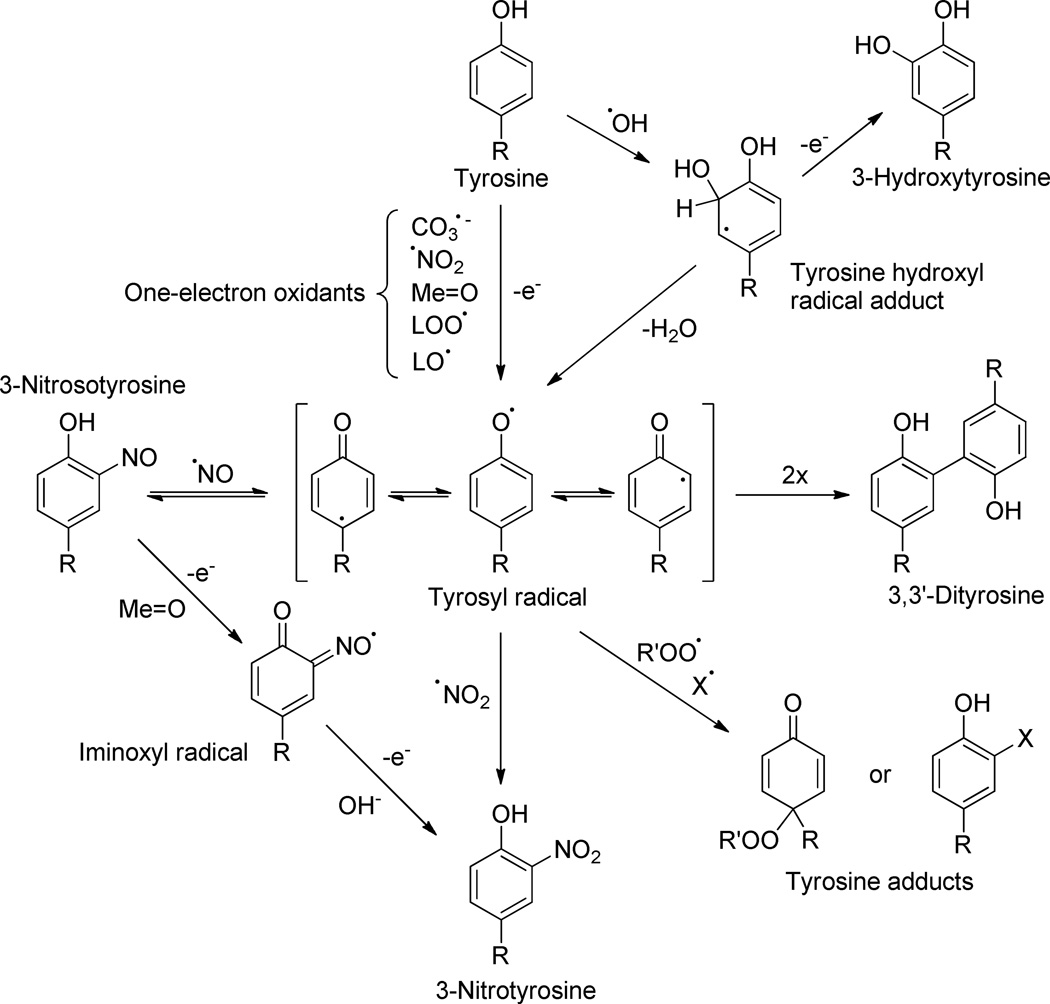
Figure 2. Free radical mechanisms of nitration and alternative fates of the tyrosyl radical.
One-electron oxidants promote the formation of the first intermediate, Tyr·. Nitration and other oxidative modifications, including hydroxylation and dimerization, are indicated. The scheme also shows the formation of novel Tyr· adducts in either positions 1- or 3-. R’OO• represents species such as lipid peroxyl radicals (LOO•) or O2•−, while X• exemplifies glutathionyl radical. Modified from 1.
The reaction of Tyr· with •NO2 (and •NO) is diffusion-controlled14,15 (Table I) and can lead to 3-nitrotyrosine (Fig. 2), but competing reactions can be also fast leading to other Tyr·-derived products, most notably, 3, 3’-dityrosine. While tyrosine nitration is kinetically favored over tyrosine dimerization, both processes can occur simultaneously, consistent with the usual finding of nitrated and cross-linked protein aggregates in tissues in degenerative disease conditions and the aging process2.
Table I.
Free radical mechanisms of tyrosine nitration. Rate constants and competing reactions.
| Reaction | Species (R) | k (M−1s−1) | Ref. |
|---|---|---|---|
| One-electron oxidation of tyrosine | |||
| TyrH + R• → Tyr• + RH | CO3•− | 4.5 × 107 | 38 |
| •NO2 | 3 × 105 | 38 | |
| aMe = O | 7.7 × 105 | 6,11 | |
| LOO• | 4.8 × 103 | 11 | |
| LO• | 3.5 × 105 | 21 | |
| •OH | 6.5 × 108 | 11 | |
| Formation of 3-nitrotyrosine | |||
| Tyr• + •NO2 → NO2Tyr | •NO2 | 3 × 109 | 14 |
| •NO | 1 × 109 | 15 | |
| Dimerization and adduct formation | |||
| Tyr• + •R → Tyr adduct | •OHb | 1.2 × 1010 | 11 |
| Tyr• | 2.3 × 108 | 38 | |
| O2•− | 1.5 × 109 | 39 | |
| LOO• | ND | - | |
| GS• | ND | - | |
| Tyrosyl radical reduction | |||
| Tyr• + RH → TyrH + R• | ascorbate | 4.4 × 108 | 18 |
| glutathione | 2 × 106 | 18 | |
For MPO compound I. MPO can readily oxidize free tyrosine that can serve as a “shuttle” for secondary oxidation of target protein tyrosines; alternatively, MPO can directly oxidize (expectedly with lower rate constants) protein tyrosines
The tyrosyl-hydroxyl radical adduct may evolve to Tyr· via a dehydration reaction (see Fig. 2)
ND, not determined
Tyrosine nitration reactions are strongly pH-dependent. This is partly due to the various possible mechanisms of peroxynitrite evolution to nitrating species (H+-, CO2- or transition metal-dependent)16, and the fact that one-electron oxidants typically react faster with the tyrosine phenolate. In addition, acid-catalyzed nitration by NO2− may be a relevant mechanism of tyrosine nitration in the gastric lumen, in a process that is likely to depend on free radical reactions involving the formation of •NO217.
Protein tyrosine nitration is observed in vivo in healthy tissues and cells, indicating that there is a “basal” flux of nitrating species; however, nitration yields are typically low due to factors such as competing reactions for the one-electron oxidants and nitrating species and the alternative fates of Tyr• and repair reactions (Figures 1 and 2). For instance, GSH (mM levels in cells) reacts fast with •NO2 and reduces Tyr· back to tyrosine (Fig. 2 and Table I), thus strongly inhibiting tyrosine nitration18,19. On the other hand, GSH does not react directly with •NO, suggesting that an alternative nitration route involving the intermediacy of 3-nitrosotyrosine and its further oxidation to 3-nitrotyrosine (Fig. 2) may be more relevant than currently appreciated up to date. Indeed, while the biological half-life of •NO2 is extremely short (i.e. 5–20 µs)19, that of •NO (i.e. > 100 ms) can be sufficiently long to favor its combination reaction with Tyr• vs other reactions.
CONNECTION BETWEEN LIPID PEROXIDATION AND PROTEIN TYROSINE NITRATION
We have recently assessed the mechanism of protein tyrosine nitration in lipid-rich biostructures6,11. Several tyrosine-nitrated proteins in vivo are associated to biomembranes or lipoproteins (e.g. apolipoprotein AI). However, the low polarity of the lipidic milieu, the abundance of unsaturated fatty acids (that will compete for the primary oxidizing species) and the restricted diffusion of protein Tyr• in these biostructures (that will hinder some of its potential fates) make the likelihood of the tyrosine nitration process differ to that in hydrophilic environments6. Additionally, molecules that critically modulate the degree of tyrosine nitration in aqueous phases such as CO3•− and GSH cannot penetrate lipid phases. Using hydrophobic-tyrosine analogs incorporated to liposomal and red cell membranes11 (and references therein), it was shown that tyrosine nitration could occur inside the lipid bilayer, and importantly, that the process is assisted during the lipid peroxidation reactions; indeed, lipid peroxyl radicals (LOO•) can oxidize tyrosine to Tyr· [eq. 1]. The reaction rate constant of ca. 5 × 103 M−1s−1 allows competition with the lipid peroxidation propagation reaction step (10–50 M−1s−1) [eq. 2].
| [1] |
| [2] |
This reported “connecting reaction” [eq. 1] is consistent with the contention that the reaction of peroxyl radicals with phenolic compounds is favored in nonpolar solvents11.
The effect of molecular oxygen in protein tyrosine nitration yields deserves a special analysis. In hydrophilic milieu, levels of molecular oxygen do not influence tyrosine nitration yields. Indeed, the reaction between Tyr• and molecular oxygen is rather slow (k < 103 M−1s−1)20. However, oxygen levels have a profound, yet indirect, effect on tyrosine nitration yields in hydrophobic structures. This influence is due to the essential role that molecular oxygen plays in the propagation phase of lipid peroxidation reactions (k = 3 × 108 M−1s−1) (eq. 4):
| [3] |
If LOO• are involved in the one-electron oxidation of tyrosine residues, then low oxygen levels will secondarily decrease tyrosine nitration yields11. Thus, a previously unexpected role of molecular oxygen in tyrosine oxidation has been unveiled, supporting that lipid peroxidation and molecular oxygen fuel protein tyrosine nitration in hydrophobic biostructures in vivo.
The lipid alkoxyl radical (LO•), an even stronger oxidant than LOO• formed during lipid peroxidation, may also participate in tyrosine oxidation. Indeed, a recent pulse and gamma-radiolysis study21 utilizing a model alkoxyl radical (t-butyl alkoxyl radical) has determined a significant rate of tyrosine oxidation (Table I):
| [4] |
Further studies should confirm whether reaction [4] is relevant during lipid and protein tyrosine oxidation/nitration processes in biomembranes.
SELECTIVITY IN PROTEIN TYROSINE NITRATION
With protein tyrosine nitration being a non-enzymatic mechanism based on free radical reactions, its selectivity for target residues in proteins is far from obvious. Due to the short biological half-life of nitrating species (ca. 5–20 ms for peroxynitrite16, nitrated proteins are normally in close vicinity of the subcellular or extracellular sites of reactive species generation22. A prime example of this contention are mitochondria, which are key loci for the formation of peroxynitrite23, usually contain a larger yield of nitrated proteins in comparison to other subcellular compartments3. Alternatively, MPO released by activated neutrophils during inflammatory processes associates to cells or lipoproteins and focuses nitration (e.g. nitration of vascular wall components, nitration of apoAI after MPO binding to HDL)1. Abundant proteins sometimes are more readily nitrated due to better competition for the nitrating species (e.g. Hsp90)24, although other kinetic and mechanistic aspects of the nitration process are relevant as well. Experimental strategies directed to define the contributing tyrosine nitration pathways under specific conditions have been presented in detail elsewhere1. For instance, the concomitant bioanalytical detection of other oxidative modifications in tyrosine such as 3-hydroxytyrosine or 3-chlorotyrosine (together with the immunochemical detection of MPO) points to the preferential participation of either peroxynitrite- or MPO-dependent nitration pathways, respectively. Similarly, nitration pathways in cells and animals can be successfully disclosed pharmacologically with, for example, the use of peroxynitrite decomposition catalysts and MPO inhibitors and genetically by studies with SOD and/or peroxiredoxin overexpressers (to inhibit peroxynitrite formation or promote its detoxification, respectively) or MPO knock-outs.
Peptide mapping studies have shown that one or two tyrosine residues in a protein become preferentially nitrated, the determinants of which depend on three main factors: a) protein structure, b) nitration mechanism, and c) environment where the protein is located. Some general tendencies on structural aspects in proteins that modulate tyrosine nitration have been reported elsewhere25,26 (e.g. favored nitration in tyrosines located near charged amino acids or on a loop structure, inhibited nitration by nearby cysteine residues), but none of them are conclusive. I would like to analyze additional mechanistic aspects which help to rationalize how nitrating species could be preferentially directed to specific solvent-exposed or buried tyrosines. A critical point is the site of formation of a relatively stable Tyr· within the protein. The given Tyr· could be formed by direct attack of a one-electron oxidant, or via intra- or intermolecular electron transfer. Regarding the direct attack, one-electron oxidants derived either from peroxynitrite decomposition in bulk solution or from MPO (Fig. 2) will preferentially react at variable rates with solvent-exposed tyrosines (Table I). On the other hand, in transition metal-containing proteins the reaction with peroxynitrite may generate oxidizing and nitrating species “in situ” (e.g. oxo-metal center plus •NO2) and favor the specific nitration of a nearby tyrosine residue (e.g. Tyr34 in MnSOD, Tyr430 in prostacyclin synthase)1. Similarly, the reaction of H2O2 with transition metal-containing proteins or compounds may lead to a Tyr·, that can undergo further reactions with either •NO or •NO2 and become nitrated (e.g. cytochrome c). As the Tyr· “traps” the proximal nitrating species, the process should be favored with longer-lived Tyr·.
Site-specificity in tyrosine nitration can be also obtained regardless of the initial attack by the one-electron oxidant (either a tyrosine or another amino acid), if short or long-range intramolecular electron transfer is possible. Indeed, some protein tyrosines act as “sinks” of oxidizing species, with intramolecular electron transfer rates estimated in the order of 103–104 s−17. In this context, the actual pKa value of the −OH group on specific tyrosines represents a relevant factor controlling the likelihood of nitration, as the tyrosine phenolate is a better electron donor. Thus, the influence of the protein microenvironment including amino acid composition, presence of transition metals and medium polarity on the selectivity of the tyrosine nitration sites is rapidly emerging.
PREPARATION OF NITRATED PROTEINS FOR STRUCTURAL AND FUNCTIONAL STUDIES
The formation of protein 3-nitrotyrosine was originally addressed in early protein chemistry studies with tetranitromethane aimed to establish the function of tyrosines in proteins27. Over three decades later, similar experiments were carried out using peroxynitrite as the nitrating agent. However, the precise contribution of tyrosine nitration and the structural basis involved in the changes in function have remained mostly elusive due to experimental limitations arising from most nitration protocols: first, nitration using excess amount of nitrating agent usually results in modification of several tyrosine residues and even other residues, making it impossible to draw direct conclusions about the effect of tyrosine nitration at a specific site only; alternatively, milder nitrating conditions‡ increase the selectivity of the process but leave a large amount of unmodified protein, making it difficult to establish the extent to which tyrosine nitration causes a change in function in the protein mixture. Thus, in the last years, we endeavored to prepare pure and well-characterized (i.e. with identified nitration site(s)) of mono-, di- or even poly-nitrated species to carry out structural and functional studies. This assessment is essential to unambiguously and precisely define the potential biological effects of tyrosine nitration in specific proteins. While the choice of the nitrating agent and experimental conditions to optimize nitration yields is variable, it is unavoidable to go through separation steps for both analytical characterization and preparative purposes. Indeed, chromatographed protein fractions derived from in vitro nitration should be characterized by mass spectrometry-peptide mapping analysis, and preferential nitration sites established. However, in vitro nitration does not always completely match the nitration sites identified in vivo, an aspect that warrants evaluation for each specific protein. In turn, identification of the nitration sites may aid in defining the actual nitrating agent in vivo.
In some reports, the heterogeneity of tyrosine nitrated protein species has been minimized by using single or multiple tyrosine to phenyalanine mutants, to eliminate some potential nitration sites28. While elegant, this approach may force the nitration reactions to remaining tyrosine residues and both other concurrent oxidative modifications and the mutations may obscure the potential effects of tyrosine nitration. Thus, the nitration data obtained with these mutated species should be ideally confirmed with purified nitrated wild type protein. An attractive alternative to generate site-specifically nitrated proteins has emerged with the discovery of a genetically evolved 3-nitrotyrosyl-tRNA syntethase/tRNA pair that allows the incorporation of the unnatural amino acid 3-nitrotyrosine to proteins co-translationally § 29. This novel approach has opened an important possibility for obtaining nitrated proteins from bacterial expression systems, if production yields can be significant. A caveat is that incorporation of 3-nitrotyrosine into the primary structure could lead to alternative folding pathways and/or variable incorporation of cofactors and metals, and therefore equivalence on structure and conformation between post- and co-translationally tyrosine nitrated proteins should be established before embarking on large structural or functional projects. Thus, chemical nitration followed by purification, remains at this time the most robust general method to provide the right protein samples to perform structural and functional studies in tyrosine nitrated proteins.
STRUCTURAL BIOLOGY OF NITRATED PROTEINS
A major gap on the field has been to understand how tyrosine nitration affects protein structure. This limitation has, in part, arisen due to the difficulty of working with generous amounts of pure and homogenously nitrated proteins. Herein, I will provide some new insights into factors that control the selectivity of protein tyrosine nitration and analyze structural data of individual nitrated proteins (i.e. cytochrome c and MnSOD, mitochondrial proteins) to illustrate how this post-translational modification can, by quite different mechanisms, lead to biologically-relevant gain- or loss-of function.
Mitochondrial protein tyrosine nitration is a well-documented phenomenon under both basal and disease conditions3,23. A seminal observation identified human MnSOD (homotetramer, 88 kD) as a key intramitochondrial target of nitrating species under inflammatory conditions: MnSOD tyrosine nitration leads to enzyme inactivation by peroxynitrite in vitro and in vivo30–32. In spite of containing nine tyrosine residues per monomer, peroxynitrite-dependent nitration in MnSOD occurs site-specifically at Tyr34 located 5 A from the active site (Fig. 3) via an Mn-catalyzed process (ca. 104–105 M−1s−1, Eq. 6). We propose that the “site-directed” nitration involves three redox steps as follows:
| [5] |
| [6] |
| [7] |
Nitration of tyrosines other than Tyr-34 via •NO2-dependent processes (e.g. MPO/H2O2/NO2−), which preferentially modify solvent-exposed residues, does not lead to inactivation. Thus, Tyr34 nitration fingerprints peroxynitrite as the proximal mitochondrial nitrating mechanism in vivo. Nitration and inactivation of MnSOD has been now reported under a variety of disease conditions and there is enough date to support the relevance of the phenomenon in vivo3. Peroxynitrite-dependent MnSOD nitration results in a gradual alteration of mitochondrial redox homeostasis, which in turn further amplifies the initial oxidative insult (and results in more enzyme inactivation), ultimately leading to mitochondrial dysfunction.
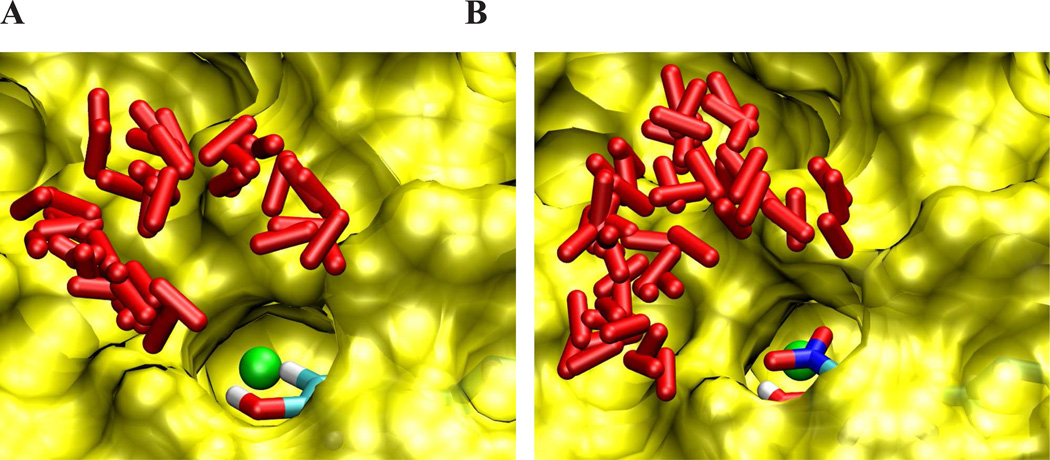
Figure 3. Schematic view of the access channel for superoxide radical in MnSOD and nitroTyr34-MnSOD.
The solvent accessible surface area of MnSOD is shown in yellow. Many different initial O2•− configurations (red rods) were used to build the free energy profile of O2•− entry as determined by multiple steered molecular dynamics simulations. A) MnSOD: the Mn atom and the Tyr34 −OH group are shown in the access channel; B) nitro-Tyr-74 MnSOD: a −NO2 group in Tyr34 ring blocks the access channel. Data obtained from 33.
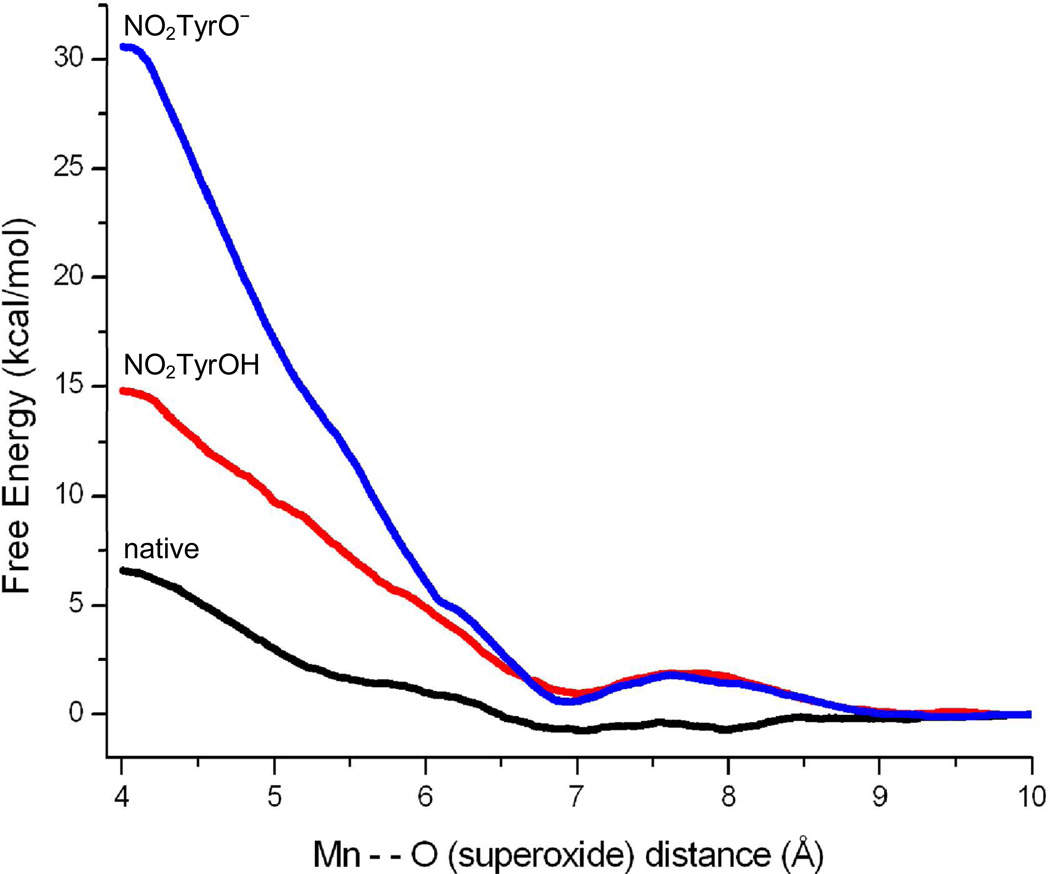
The structural basis by which Tyr34 nitration leads to enzyme inactivation have been recently solved by a combination of experimental and computational studies. Tyr34 helps to establish a hydrogen bond network around the Mn center that participates in proton transfer steps during catalysis33. Crystallographic data34 and molecular dynamics simulations33 indicate that Tyr34 nitration leads to enzyme inactivation via two processes that generate a large energy barrier for the entry of O2•− through the access channel of the enzyme (Fig. 4). On one hand, the −NO2 group imposes a steric restriction for the diffusion of the substrate towards the Mn center and, on the other hand, the drop in the pKa of the tyrosine −OH due to nitration, generates a negative charge in the active site, which causes electrostatic repulsion of O2•− (and possibly also alteration of the hydrogen bonding network and redox potential). While direct experimental measurements of the nitro-Tyr74 (MnSOD) phenol pKa are underway, we anticipate the value to be close to that of free nitrotyrosine due of its solvent accessibility33. Therefore, under the pH values of the mitochondrial matrix (ca. 7.8) over 50 % of nitro-Tyr74 would be dissociated as phenolate, further underscoring the relevance of the side chain negative charge on MnSOD inactivation.
Figure 4. Free energy profile for superoxide radical migration along the diffusion pathway in native and nitroTyr34-MnSOD.
The distance represents the driven coordinate from the Mn atom to one of the oxygen atoms in O2•−. Unmodified (native) and tyrosine-34 nitrated (in the phenol or phenolate forms) MnSOD are shown with black, red and blue lines, respectively. Modified from 33.
Mammalian cytochrome c, a 12 kD hemeprotein, contains four highly conserved tyrosines. The heme group in cytochrome c is hexacoordinated, in which the fifth and sixth ligands to the iron are His18 and Met80, respectively. The interaction of Fe with the S of Met80 is in part responsible for the redox potential value of the cyt c3+/cyt c2+ pair, ca. +250 mV. In addition, the hexacoordinated state of cytochrome c minimizes heme interactions with exogenous ligands, including H2O2, and therefore has a marginal peroxidatic activity (i.e. its capacity to react with peroxides and promote the oxidation of a second substrate).
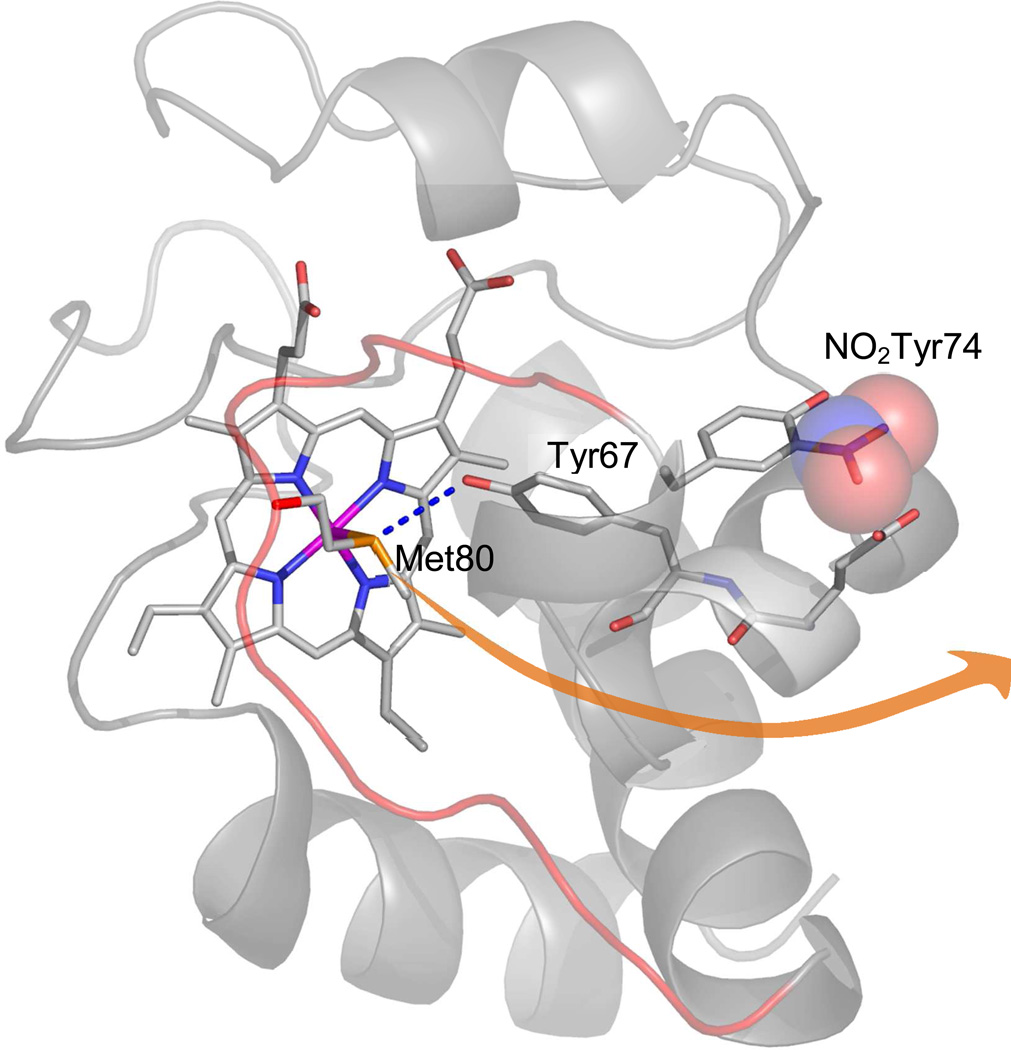
The tyrosine nitration of cytochrome c s has constituted a good case study to reveal the “site-specificity” of different nitrating mechanisms and to unravel how nitration of tyrosines distant from an active site can promote conformational changes leading to change in function. Exposure of cytochrome c to nitrating conditions, including peroxynitrite, NO2− or ·NO plus H2O22,35. results in cytochrome c tyrosine nitration at variable positions. Nitration by peroxynitrite leads to the preferential modification of Tyr74 or Tyr97, leading to mononitrated forms at tyrosines relatively distant from the active site (14.5 and 14 Å, respectively), compatible with preferential reactions of peroxynitrite-derived radicals with solvent-exposed tyrosines. On the other hand, nitration involving H2O2-dependent steps is likely to favor the mononitration of heme-adjacent Tyr67 (4.5 Å). In this case, H2O2 initially binds to the Fe after displacement of Met80. Then, a “compound I-like” of cytochrome c would be formed, which causes the one-electron oxidation of Tyr67, to yield a Tyr·. Once Tyr· is formed, it couples to ·NO to form 3-nitrosotyrosine that under excess H2O2, can be further oxidized to 3-nitrotyrosine (see also Fig. 2). In the presence of NO2−, its one electron oxidation leads to ·NO2, which also yields 3-nitrotyrosine.
The nitration of Tyr74 triggers a conformational change in cytochrome c, resulting in an alternative conformation lacking its normal electron transport capacities and with a gain-of-peroxidatic activity 35. The peroxidatic activity of cytochrome c is also elicited during its association with the mitochondrial phospholipid cardiolipin and triggers H2O2-dependent redox events relevant t cell death processes36. The conformational change induced in cytochrome c via Tyr74 nitration appears to be “wired” by the destabilization of a flexible omega loop encompassing amino acids 70 to 85, which then triggers the displacement of Met80 from its heme binding site, and promoting an alternative low spin conformation by replacement with Lys79 (Fig. 5). The conformational change induced by Tyr74 nitration is similar to that observed during the “alkaline transition” (pKa = 9.4) but, instead, it does so at a biologically-relevant pH (pKa = 7.4) 35. To note, the observed formation of dinitrated (Tyr74/Tyr67) cytochrome c species by peroxynitrite is compatible with an initial nitration at Tyr74 followed by the conformational change and a subsequent iron-catalyzed nitration at Tyr67. The functional consequences of this alternative cytochrome c conformation are related to an enhanced peroxidatic function, its facile translocation to the cytosol and nuclei and its inability to induce apoptosome activity36; the biological significance of these findings, including the possible consequences in redox signaling, await future studies.
Figure 5. Tyrosine nitration-triggered conformational change in cytochrome c.
The arrow indicates the movement of Met80 away from the heme upon Tyr74 nitration. A hydrogen bond between Met80 and Tyr67 present in the native conformation is also indicated. Modified from 35.
The studies on MnSOD and cytochrome c tyrosine nitration provide relevant examples on how this modification results in a loss-(MnSOD) or gain-of-function (cytochrome c) by rather contrasting mechanisms at the structural level.
FINAL CONSIDERATIONS AND PERSPECTIVES
The effect of protein tyrosine nitration on the biological actions of a number of proteins has been suggested, although much is yet to be known in terms of the specific effects on structure and activity, both in vitro and in vivo. Isolating and quantitating tyrosine nitration vs other oxidative modifications and relating it to the degree of functional change remain a difficult and challenging task. The data presented herein provided some examples and experimental possibilities to tackle these questions with pure tyrosine nitrated proteins of biological relevance.
A lesson from recent studies is that mapping of the nitrated protein tyrosine residues serves to establish the proximal nitrating agent in biological systems. This knowledge is applicable as one may want to interfere with a specific nitration pathway (e.g. peroxynitrite or hemeperoxidase-dependent) either pharmacologically or genetically. Identifying protein tyrosine nitration as a mediator in alterations of cell/tissue homeostasis can have relevant biomedical ramifications as therapeutic strategies directed to prevent protein tyrosine nitration may assist in inhibiting or delaying the evolution of a variety of disease conditions. Importantly, cell incorporation of tyrosine-containing peptides appears as a sound possibility to protect against the toxic effects of protein tyrosine nitration24, as the incorporated tyrosine peptides spare critical endogenous protein tyrosines. Some critically relevant issues in the field remain ill-defined. First, the role of tyrosine nitration as a physiological regulatory signal has not been demonstrated, although it may impact inflammatory signaling during pathologically-relevant conditions2,4. Second, while enzymatic processes for the reduction or removal of −NO2 from tyrosine have not been unambiguously established, some limited information indicates that some nitrated proteins are prone to “recovery” by “denitration”2,37. I find it difficult to envision a “general” denitrase or reductase activity, since nitrated tyrosines are present in rather heterogeneous regions of a large number of proteins, and such enzymatic machinery has not been isolated or characterized yet. A large experimental effort is needed to understand how tyrosine nitrated proteins are handled intra- and extra-cellularly by either proteolytic or repair systems (i.e. the fate of tyrosine nitrated proteins).
Overall, the accumulated biochemical data provide a sound foundation to further investigate the mechanisms by which protein tyrosine nitration in vivo may influence the progression of disease states associated with a disruption of •NO and redox metabolism.
ACKNOWLEDGEMENTS
I thank Drs. Silvina Bartesaghi, Valeria Valez, Gerardo Ferrer-Sueta, Matias Moller, Luciano Abriata, and Leonardo Boechi for helpful suggestions to this work.
This work was supported by grants from CSIC_Universidad de la Republica, ANII_Uruguay (FCE_2486), HHMI and NIH (R01AI095173).
Biography
Rafael Radi received his MD and PhD from Universidad de la República, Uruguay, in 1989 and 1991, respectively. He was a postdoctoral fellow at the University of Alabama at Birmingham. He has been faculty at the Department of Biochemistry, Facultad de Medicina, Universidad de la Republica for over two decades, now serving as Professor and Chairman. He has pioneered studies on the biological chemistry of peroxynitrite and free radical and redox mechanisms of oxidative signaling and injury.
Footnotes
The term peroxynitrite refers to both peroxynitrite anion and peroxynitrous acid. IUPAC-recommended names are oxoperoxonitrate (1-) and hydrogen oxoperoxonitrate, respectively.
Relevant hemeperoxidases include myeloperoxidase and eosinophil peroxidase.
e.g. Low peroxynitrite concentrations (i.e. 2–10 fold ratio respect to protein concentration); nitration yields are highly variable depending on experimental conditions, but at high protein concentration (e.g. 100 µM) can be in the order of 1–2 % respect to peroxynitrite (i.e. 10 µM nitrated protein using 1 mM peroxynitrite at pH 7.0)
Free 3-nitrotyrosine is not incorporated during protein synthesis
REFERENCES
- 1.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci U S A. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Souza JM, Peluffo G, Radi R. Protein tyrosine nitration-functional alteration or just a biomarker? Free Radic Biol Med. 2008;45:357–366. doi: 10.1016/j.freeradbiomed.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Castro L, Demicheli V, Tortora V, Radi R. Mitochondrial protein tyrosine nitration. Free Radic Res. 2011;45:37–52. doi: 10.3109/10715762.2010.516254. [DOI] [PubMed] [Google Scholar]
- 4.Molon B, Ugel S, Del Pozzo F, Soldani C, Zilio S, Avella D, De Palma A, Mauri P, Monegal A, Rescigno M, Savino B, Colombo P, Jonjic N, Pecanic S, Lazzarato L, Fruttero R, Gasco A, Bronte V, Viola A. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J Exp Med. 2011;208:1949–1962. doi: 10.1084/jem.20101956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koide S, Sidhu SS. The importance of being tyrosine: lessons in molecular recognition from minimalist synthetic binding proteins. ACS Chem Biol. 2009;4:325–334. doi: 10.1021/cb800314v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartesaghi S, Ferrer-Sueta G, Peluffo G, Valez V, Zhang H, Kalyanaraman B, Radi R. Protein tyrosine nitration in hydrophilic and hydrophobic environments. Amino Acids. 2007;32:501–515. doi: 10.1007/s00726-006-0425-8. [DOI] [PubMed] [Google Scholar]
- 7.Petruk AA, Bartesaghi S, Trujillo M, Estrin DA, Murgida D, Kalyanaraman B, Marti MA, Radi R. Molecular basis of intramolecular electron transfer in proteins during radical-mediated oxidations: Computer simulation studies in model tyrosine-cysteine peptides in solution. Arch Biochem Biophys. 2012;525:82–91. doi: 10.1016/j.abb.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokoyama K, Uhlin U, Stubbe J. Site-specific incorporation of 3-nitrotyrosine as a probe of pKa perturbation of redox-active tyrosines in ribonucleotide reductase. J Am Chem Soc. 2010;132:8385–8397. doi: 10.1021/ja101097p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alvarez B, Ferrer-Sueta G, Freeman BA, Radi R. Kinetics of peroxynitrite reaction with amino acids and human serum albumin. J Biol Chem. 1999;274:842–848. doi: 10.1074/jbc.274.2.842. [DOI] [PubMed] [Google Scholar]
- 10.Augusto O, Bonini MG, Amanso AM, Linares E, Santos CC, De Menezes SL. Nitrogen dioxide and carbonate radical anion: two emerging radicals in biology. Free Radic Biol Med. 2002;32:841–859. doi: 10.1016/s0891-5849(02)00786-4. [DOI] [PubMed] [Google Scholar]
- 11.Bartesaghi S, Wenzel J, Trujillo M, Lopez M, Joseph J, Kalyanaraman B, Radi R. Lipid peroxyl radicals mediate tyrosine dimerization and nitration in membranes. Chem Res Toxicol. 2010;23:821–835. doi: 10.1021/tx900446r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YR, Chen CL, Chen W, Zweier JL, Augusto O, Radi R, Mason RP. Formation of protein tyrosine ortho-semiquinone radical and nitrotyrosine from cytochrome c-derived tyrosyl radical. J Biol Chem. 2004;279:18054–18062. doi: 10.1074/jbc.M307706200. [DOI] [PubMed] [Google Scholar]
- 13.Romero N, Radi R, Linares E, Augusto O, Detweiler CD, Mason RP, Denicola A. Reaction of human hemoglobin with peroxynitrite. Isomerization to nitrate and secondary formation of protein radicals. J Biol Chem. 2003;278:44049–44057. doi: 10.1074/jbc.M305895200. [DOI] [PubMed] [Google Scholar]
- 14.Prutz WA, Monig H, Butler J, Land EJ. Reactions of nitrogen dioxide in aqueous model systems: oxidation of tyrosine units in peptides and proteins. Arch Biochem Biophys. 1985;243:125–134. doi: 10.1016/0003-9861(85)90780-5. [DOI] [PubMed] [Google Scholar]
- 15.Eiserich JP, Butler J, van der Vliet A, Cross CE, Halliwell B. Nitric oxide rapidly scavenges tyrosine and tryptophan radicals. Biochem J. 1995;310(Pt 3):745–749. doi: 10.1042/bj3100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrer-Sueta G, Radi R. Chemical biology of peroxynitrite: kinetics, diffusion, and radicals. ACS Chem Biol. 2009;4:161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 17.Rocha BS, Gago B, Barbosa RM, Lundberg JO, Radi R, Laranjinha J. Intragastric nitration by dietary nitrite: implications for modulation of protein and lipid signaling. Free Radic Biol Med. 2012;52:693–698. doi: 10.1016/j.freeradbiomed.2011.11.011. [DOI] [PubMed] [Google Scholar]
- 18.Folkes LK, Trujillo M, Bartesaghi S, Radi R, Wardman P. Kinetics of reduction of tyrosine phenoxyl radicals by glutathione. Arch Biochem Biophys. 2011;506:242–249. doi: 10.1016/j.abb.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Ford E, Hughes MN, Wardman P. Kinetics of the reactions of nitrogen dioxide with glutathione, cysteine, and uric acid at physiological pH. Free Radic Biol Med. 2002;32:1314–1323. doi: 10.1016/s0891-5849(02)00850-x. [DOI] [PubMed] [Google Scholar]
- 20.Hunter EP, Desrosiers MF, Simic MG. The effect of oxygen, antioxidants, and superoxide radical on tyrosine phenoxyl radical dimerization. Free Radic Biol Med. 1989;6:581–585. doi: 10.1016/0891-5849(89)90064-6. [DOI] [PubMed] [Google Scholar]
- 21.Folkes LK, Bartesaghi S, Trujillo M, Radi R, Wardman P. Kinetics of oxidation of tyrosine by a model alkoxyl radical. Free Radic Res. 2012 doi: 10.3109/10715762.2012.695868. [DOI] [PubMed] [Google Scholar]
- 22.Heijnen HF, van Donselaar E, Slot JW, Fries DM, Blachard-Fillion B, Hodara R, Lightfoot R, Polydoro M, Spielberg D, Thomson L, Regan EA, Crapo J, Ischiropoulos H. Subcellular localization of tyrosine-nitrated proteins is dictated by reactive oxygen species generating enzymes and by proximity to nitric oxide synthase. Free Radic Biol Med. 2006;40:1903–1913. doi: 10.1016/j.freeradbiomed.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Radi R, Cassina A, Hodara R, Quijano C, Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radic Biol Med. 2002;33:1451–1464. doi: 10.1016/s0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- 24.Ye Y, Quijano C, Robinson KM, Ricart KC, Strayer AL, Sahawneh MA, Shacka JJ, Kirk M, Barnes S, Accavitti-Loper MA, Radi R, Beckman JS, Estevez AG. Prevention of peroxynitrite-induced apoptosis of motor neurons and PC12 cells by tyrosine-containing peptides. J Biol Chem. 2007;282:6324–6337. doi: 10.1074/jbc.M610800200. [DOI] [PubMed] [Google Scholar]
- 25.Bayden AS, Yakovlev VA, Graves PR, Mikkelsen RB, Kellogg GE. Factors influencing protein tyrosine nitration--structure-based predictive models. Free Radic Biol Med. 2011;50:749–762. doi: 10.1016/j.freeradbiomed.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Souza JM, Daikhin E, Yudkoff M, Raman CS, Ischiropoulos H. Factors determining the selectivity of protein tyrosine nitration. Arch Biochem Biophys. 1999;371:169–178. doi: 10.1006/abbi.1999.1480. [DOI] [PubMed] [Google Scholar]
- 27.Sokolovsky M, Riordan JF, Vallee BL. Tetranitromethane. A reagent for the nitration of tyrosyl residues in proteins. Biochemistry. 1966;5:3582–3589. doi: 10.1021/bi00875a029. [DOI] [PubMed] [Google Scholar]
- 28.Ly HK, Utesch T, Diaz-Moreno I, Garcia-Heredia JM, De La Rosa MA, Hildebrandt P. Perturbation of the Redox Site Structure of Cytochrome c Variants upon Tyrosine Nitration. J Phys Chem B. 2012 doi: 10.1021/jp302301m. [DOI] [PubMed] [Google Scholar]
- 29.Neumann H, Hazen JL, Weinstein J, Mehl RA, Chin JW. Genetically encoding protein oxidative damage. J Am Chem Soc. 2008;130:4028–4033. doi: 10.1021/ja710100d. [DOI] [PubMed] [Google Scholar]
- 30.MacMillan-Crow, L. A. C. JP, Kerby JD, Beckman JS, Thompson JA. Nitration and inactivation of manganese superoxide dismutase in chronic rejection of human renal allgrafts. Proc Natl Acad Sci U S A. 1996;93:11853–11858. doi: 10.1073/pnas.93.21.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quijano C, Hernandez-Saavedra D, Castro L, McCord JM, Freeman BA, Radi R. Reaction of peroxynitrite with Mn-superoxide dismutase. Role of the metal center in decomposition kinetics and nitration. J Biol Chem. 2001;276:11631–11638. doi: 10.1074/jbc.M009429200. [DOI] [PubMed] [Google Scholar]
- 32.Yamakura F, Taka H, Fujimura T, Murayama K. Inactivation of Human Manganese-superoxide Dismutase by Peroxynitrite is caused by Exclusive Nitration of Tyrosine 34 to 3-Nitrotyrosine. J Biol Chem. 1998;273:14085–14089. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]
- 33.Moreno DM, Marti MA, De Biase PM, Estrin DA, Demicheli V, Radi R, Boechi L. Exploring the molecular basis of human manganese superoxide dismutase inactivation mediated by tyrosine 34 nitration. Arch Biochem Biophys. 2011;507:304–309. doi: 10.1016/j.abb.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Quint P, Reutzel R, Mikulski R, McKenna R, Silverman DN. Crystal structure of nitrated human manganese superoxide dismutase: mechanism of inactivation. Free Radic Biol Med. 2006;40:453–458. doi: 10.1016/j.freeradbiomed.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 35.Abriata LA, Cassina A, Tortora V, Marin M, Souza JM, Castro L, Vila AJ, Radi R. Nitration of solvent-exposed tyrosine 74 on cytochrome c triggers heme iron-methionine 80 bond disruption. Nuclear magnetic resonance and optical spectroscopy studies. J Biol Chem. 2009;284:17–26. doi: 10.1074/jbc.M807203200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Godoy LC, Munoz-Pinedo C, Castro L, Cardaci S, Schonhoff CM, King M, Tortora V, Marin M, Miao Q, Jiang JF, Kapralov A, Jemmerson R, Silkstone GG, Patel JN, Evans JE, Wilson MT, Green DR, Kagan VE, Radi R, Mannick JB. Disruption of the M80-Fe ligation stimulates the translocation of cytochrome c to the cytoplasm and nucleus in nonapoptotic cells. Proc Natl Acad Sci U S A. 2009;106:2653–2658. doi: 10.1073/pnas.0809279106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gorg B, Qvartskhava N, Voss P, Grune T, Haussinger D, Schliess F. Reversible inhibition of mammalian glutamine synthetase by tyrosine nitration. FEBS Lett. 2007;581:84–90. doi: 10.1016/j.febslet.2006.11.081. [DOI] [PubMed] [Google Scholar]
- 38.Bartesaghi S, Valez V, Trujillo M, Peluffo G, Romero N, Zhang H, Kalyanaraman B, Radi R. Mechanistic studies of peroxynitrite-mediated tyrosine nitration in membranes using the hydrophobic probe N-t-BOC-L-tyrosine tert-butyl ester. Biochemistry. 2006;45:6813–6825. doi: 10.1021/bi060363x. [DOI] [PubMed] [Google Scholar]
- 39.Winterbourn CC, Kettle AJ. Radical-radical reactions of superoxide: a potential route to toxicity. Biochem Biophys Res Commun. 2003;305:729–736. doi: 10.1016/s0006-291x(03)00810-6. [DOI] [PubMed] [Google Scholar]