Abstract
The promyelocytic zinc finger transcription factor (PLZF) is required for the development of activated phenotypes in NKT and other innate T lymphocytes. Although strong TCR stimulation has been implicated in the induction of PLZF, the factors regulating PLZF expression are incompletely understood. We show here that costimulation of pre-selection double positive thymocytes through the Signaling lymphocyte activation molecule (SLAM) family receptor Ly108 markedly enhanced PLZF expression compared to that induced by T cell receptor (TCR) stimulation alone. Costimulation with Ly108 increased expression of Egr-2 and binding of Egr-2 to the promoter of Zbtb16, which encodes PLZF, and resulted in PLZF levels similar to those seen in NKT cells. In contrast, costimulation with αCD28 failed to enhance Egr-2 binding and Zbtb16 expression. Moreover, mice lacking Ly108 showed decreased numbers of PLZF expressing CD4+ T cells. Together, these results support a potential role for Ly108 in the induction of PLZF.
INTRODUCTION
Innate-like T cells, including natural killer T (NKT) cells differ from their conventional counterparts in that they possess innate, memory-like, rather than naive phenotypes upon maturation, and rapidly secrete effector cytokines upon activation without need for prior differentiation. As such, innate-like T cells play an important role at the interface between the innate and adaptive immune systems (1).
Recent data demonstrate that the Promyelocytic Zinc Finger transcription factor (PLZF) is required for the acquisition of activated phenotypes by iNKT and other innate-like T lymphocytes. While ectopic expression of PLZF is sufficient to impart an activated phenotype to conventional T cells, its absence can impair both the expansion and effector differentiation of iNKT cells (2–6). Therefore, PLZF likely plays a critical role in setting the tone of immediate lymphocyte responses. However, the factors regulating PLZF expression are only minimally understood. While data suggest that strong TCR signals associated with high expression of the Ras- and Ca2+-dependent Early Growth Response transcription factors Egr-1 and-2 lead to PLZF induction (4, 7), it is not clear that PLZF expression is governed solely by signals from the TCR(8). Unlike conventional αβ TCR+ cells, iNKT and other innate-like T cells undergo a distinct pathway of selection that requires interactions with and selection on MHC and related molecules expressed on other DP thymocytes instead of thymic stroma, suggesting that distinct signaling pathways may be activated during their development (9). Indeed, PLZF can be induced in developing thymocytes by enforced T cell-T cell interactions (10, 11). These data suggest that PLZF expression may be regulated in part by receptor/ligands expressed specifically on DP cells rather than on thymic stroma.
Of interest in this regard are the Signaling Lymphocyte Activation Molecule (SLAM) family receptors, which are expressed at high levels on double-positive (DP) thymocytes, but are absent from the thymic stroma (12). Mutations affecting the SLAM associated protein (SAP), which is required for signaling downstream of SLAM family receptors (13), result in a drastic loss of iNKT and other innate T lymphocytes (14–19). Studies using mixed bone marrow chimeras have further implicated two SLAM members, SLAM and Ly108, in the development of iNKT cells (12). Nonetheless, how these receptors contribute to the development of iNKT and other innate T cell lineages is not well understood.
We have recently found that the homophilic SLAM family receptor Ly108, (CD352, encoded by Slamf6), is heavily phosphorylated in the thymus in a contact-dependent manner (20). Given the connection between Ly108 and innate T cells, we examined the effects of Ly108 on the expression of PLZF and other transcription factors required for innate T lymphocyte development. We find that Ly108 engagement amplifies TCR signaling, prolonging phosphorylation of LAT, PLC-γ and ERK and increasing expression of Egr-2. Strikingly, costimulation with Ly108, but not CD28, led to a dramatic induction of PLZF associated with increased Egr-2 binding to the promoter of Zbtb16, the gene encoding PLZF. Moreover, Ly108-deficient mice have decreased numbers of PLZF+CD4+ cells and conversely, a mouse model with enhanced Ly108 signaling (B6.Sle1b) showed increased numbers of PLZF+CD4+ cells. Our results suggest that Ly108 contributes a distinct signal that can potentiate TCR-mediated induction of PLZF.
MATERIALS AND METHODS
Mice
Mice were used in accordance with the Institutional Animal Care and Use Committee, NHGRI, NIH. Sh2d1a−/− (21) mice were backcrossed to C57Bl/6J for 10 generations and carry the C57Bl/6J-derived SLAM locus. C57BL/6J were from Jackson Laboratories. MHC Class I/Class II-deficient (B6.129-H2-Ab1tm1Gru B2mtm1Jae) (22) and β2m−/− (B2mtm1Jae) (23) mice were from Taconic. Egr1−/−Egr2fl/fl/LckCre mice were previously described (24–26). Slamf6−/− mice (Supplemental Fig. S2) were generated by introducing a stop codon into exon 2 and removing part of exons 2 and 3 of Slamf6 in HGTC-8 C57BL/6J-derived ES cells (27). B6.Sle1b mice have been previously described (28)
Pre-selection DP cell isolation
PS-DP thymocytes were isolated by negative selection (αFITC isolation kit, Miltenyi, CA) using FITC-αCD3, αCD25, and αCD44, which removed post-selection cells (CD3hi), CD25+ DN cells, as well as mature (CD44hi) innate T cells (eBiosciences, CA). Post-selection, cells were ≥98% CD4+CD8+, and ≥99% CD69lo and CD44lo. Additionally, over 98% of PLZF+ cells were removed by this treatment (Supplemental Fig. S1). In some experiments, α–γδTCR was also included in the negative selection step to deplete γδ T-cells. Both selection procedures gave comparable results.
Cell culture and staining
5×106 PS-DP thymocytes were stimulated in 0.5 mL complete RPMI plus 8% FBS, 1% pen/strep, 2mM L-glutamine and 0.05mM 2-ME in 24 well plates coated with plate-bound αCD3 (2C11, 2 µg/ml) ± αCD28 (PC61, 5µg/ml) Bio-X-Cell, NH) ± αLy108 (5 µg/ml 13G3-19D, eBiosciences, CA) or an isotype control for 18, 24 or 48h. B cell stimulation experiments were performed by using WT LPS-activated B cells (1µg/ml for 72 hrs) that were pre-incubated with αCD3 (2 µg/ml) for 10 minutes prior to addition of PS-DP thymocytes for 24–48 hrs. Ly108 interactions were blocked by pre-incubating thymocytes with Ly108-Fc fusion protein (Recombinant Mouse NTB-A/SLAMF6 Fc Chimera, R&D systems) for 10 minutes before adding B cells plus αCD3. Cells were harvested, stained for CD4 and CD8, then fixed with FOXP3 buffer (eBiosciences, CA) and stained with αPLZF (clone D-9, Santa Cruz Biotech, CA) followed by PE-αmouse Ig. Controls for staining specificity included staining with αPLZF isotype control (mouse IgG1) followed by PE-αmouse Ig or staining with PE-αmouse Ig alone. iNKT cells were identified by CD1d-PBS57 tetramer using unloaded tetramer as a control (NIAID tetramer facility, Atlanta, GA). For RNA extraction, cells were harvested and stored at −80°C in Trizol.
Real time PCR
RNA was isolated with Trizol and the RNeasy® mini kit (Qiagen). cDNA was generated by Taqman reverse transcription reagents (Applied Biosystems, CA). cDNA was amplified with Taqman probe primer mixes (Applied Biosystems, CA).
Chromatin Immunoprecipitations (ChIPs)
PS-DP thymocytes were harvested 18 h post-stimulation and ChIPs performed using Magna ChIP-A kit (Millipore, MA). DNA was sheared in 500 µl using a Misonix 3000 water bath sonicator (Output 5.5, 6 min 30 sec On, 30 sec Off) and 50 µl (1×106 cell equivalent) sheared DNA was immunoprecipitated and sequences amplified by SYBR green qPCR with primers: Egr-2 F: 5’ ATCCGGAGCAACAGTTCCCC 3’, Egr-2 R 5’ TGGGCGGTGCGAGTCCTTC 3’; CREB F 5’ AGCACTAAAGATGGAGAGGCG 3’, CREB R 5’ TCGCAGTACCCGCTCTCG 3’; NFAT F 5’ TTCTGACTCTGAGTTTGGGGATAAATGAC 3’, NFAT R 5’ TGGGTTTGCTAGTTATTAGTCAGAGATG 3’. ChIPs were normalized to input DNA and expressed as fold-enrichment relative to isotype control from non-stimulated (NS) thymocytes.
Stimulations and Protein analyses
Thymocytes were incubated with biotinylated αCD3ε, αCD4 (BD Biosciences, NJ) ± αLy108 (eBiociences, CA), washed, stimulated with streptavidin and stopped with 2X SDS loading buffer. Immunoblots were probed with 4G10, LAT and PLCγ-1 (Millipore, MA); pLAT Y191 and pPLCγ-1 Y783 (Biosource-Invitrogen, NY); pPKCθ Thr 538, pERK and ERK (Cell Signaling, MA), and PKCθ (Santa Cruz, CA) followed by TrueBlot® HRP-secondary Ab. For analysis of Ly108 phosphorylation in intact B6 and B6.Sle1b thymi, freshly harvested intact thymi were crushed in RIPA lysis buffer and Ly108 immunoprecipitated from cleared lysates for 2hrs at 4°C using αLy108 monoclonal (13G3-19D clone, eBiosciences) and Protein A agarose beads (Santa Cruz, CA). Immunoprecipitates were resolved on 10% Tris Glycine gels and were probed for phosphotyrosine using 4G10 and for total Ly108 protein using polyclonal αLy108 rabbit antibody (20). Western blots were developed using enhanced chemiluminescence detection reagents (GE Healthcare). Films were scanned and intensities quantitated using imageJ software (NIH, http://rsb.info.nih.gov/ij).
Ca2+ Flux
PS-DP thymocytes were incubated with Fluo-3 and Fura-red (Molecular Probes/Invitrogen) and incubated with biotinylated αCD3ε and αCD4±αLy108 for 30s, at 37°C, followed by crosslinking with streptavidin. Data were collected for 10 min (FACS Calibur, BD Biosciences, NJ), analyzed using FlowJo (Tree Star, Inc., CA), and plotted as the ratio of Fluo-3/Fura-red.
Statistical analysis
A two-tailed, paired Student’s t-test was used to calculate P values.
RESULTS
Ly108 costimulation enhances Zbtb16 induction in pre-selection DP thymocytes
To evaluate factors that regulate PLZF expression, we first isolated pre-selection DP (PS-DP) thymocytes by negative selection using antibodies to CD3, CD25, and CD44. (Supplemental Fig. S1A). This selection enriches for pre-selection thymocytes that have not upregulated their TCR and also depletes CD25+ DN cells as well as innate lymphocytes, which are CD44hi. After purification, the isolated cells were over 98% DP and greater than 99% CD69lo, CD44lo. Over 98% of pre-existing PLZF+ cells were removed by this treatment (Supplemental Fig. S1A–E). As seen for developing γδ cells (4), stimulation of PS-DP thymocytes with plate-bound αCD3, led to an induction of Zbtb16 (Fig. 1A). However, costimulation with both αCD3 and an αLy108 antibody capable of inducing Ly108 phosphorylation (Supplemental Fig. S1F, (20)), dramatically increased Zbtb16 expression above stimulation with αCD3 alone (Fig. 1B). This enhancement of Zbtb16 message required stimulation of both CD3 and Ly108, as αLy108 alone was unable to induce Zbtb16 expression above background.
Figure 1. Ly108 costimulation enhances PLZF expression in developing PS-DP thymocytes.

(A) Expression of Zbtb16 (encoding PLZF) evaluated by qRT-PCR of WT PS-DP thymocytes stimulated with media alone (NS; non-stimulated) or plate-bound αCD3 for 24 or 48h. (B) Zbtb16 expression of WT PS-DP thymocytes stimulated with media alone (NS), αCD3±αLy108 or αLy108 alone. RQ is relative quantitation. Values in A–B were normalized to NS WT samples. Similar results were obtained using freshly isolated PS-DP cells as normalization controls (Supplemental Fig. S2A). Data are the means ± SEM of 5 independent experiments, using cells from ≥3 mice each. (C) Zbtb16 expression in WT and β2m-deficient PS-DP thymocytes stimulated with plate-bound αCD3±αLy108. Data are from two independent experiments. Cells in these experiments were also negatively selected to remove TCRγδ+ cells. (D) Zbtb16 expression in WT and SAP-deficient (Sh2d1a−/−) PS-DP thymocytes in the presence or absence of Ly108Fc fusion protein were stimulated with LPS-activated B cells (which express high surface levels of Ly108) plus αCD3. Data are the means ± SEM of 3 independent experiments, using cells from ≥3 mice each.
To confirm that the induction in Zbtb16 did not result from a specific outgrowth or survival advantage of pre-existing Zbtb16-expressing population, we evaluated pre-selection thymocytes that were isolated from β2m-deficient mice, which lack most classical iNKT cells (29–31), and which were further negatively selected with αTCRγδ to minimize contribution from PLZF expressing γδ T cells. A similar induction of Zbtb16 was also seen in pre-selection thymocytes from these mice (Fig. 1C), consistent with the fact that we did not detect appreciable PLZF expression in freshly isolated WT PS-DP thymocytes prior to stimulation (Fig. S1D and S2A). Furthermore, Ly108 costimulation did not induce any appreciable proliferation of PS-DP thymocytes in plate-bound stimulations (Fig. S2B). Thus, the induction of Zbtb16 by Ly108 costimulation is unlikely to result from the expansion of Zbtb16 expressing cells that may have escaped the purification of pre-selection DP cells. Although αCD3 stimulation of DP thymocytes does induce cell death, particularly by 48h, Ly108 costimulation did not appear to further diminish cell viability (Fig. S2C), also suggesting that we were not enriching for a Zbtb16+ population upon Ly108 costimulation.
To determine whether the plate-bound Ly108 antibody stimulation assay could be recapitulated using the endogenous self-ligand, and to evaluate the effects of homotypic Ly108 interactions similar to those observed in vivo, we next stimulated thymocytes with LPS-activated B cells, which express high levels of Ly108 when activated (32), and induced TCR signaling with αCD3. Since Ly108 is a self-ligand, this stimulation allowed engagement of Ly108 on thymocytes. Costimulation by Ly108 presented by B cells induced Zbtb16 expression in WT PS-DP thymocytes, but not in those from mice deficient in SAP, which is required for Ly108 activation (Fig. 1D). Moreover, pre-incubation of thymocytes with a blocking Ly108-Fc fusion protein (33) prevented this induction of Zbtb16, supporting the idea that Ly108 engagement contributes to Zbtb16 expression.
Ly108 is required for full induction of PLF in vitro
To confirm that these effects were specific for Ly108, we evaluated thymocytes from mice deficient in Ly108, (Slamf6−/− mice), which we generated on a pure C57BL/6 background to eliminate potential effects of Slam gene family polymorphisms. These mice showed no expression of Ly108 as detected by immunoblotting or flow cytometry (Fig. S2D). As previously reported in a mixed genetic background, Slamf6−/− mice showed no gross defects in conventional CD4+ and CD8+ T cell development (Fig. 2A and Table 1). However, the effects of Ly108 on Zbtb16 expression were abrogated in Ly108-deficient thymocytes, as well as in thymocytes from mice deficient in SAP or Fyn, which are required for Ly108 phosphorylation (20, 34) (Fig. 2B and data not shown). Moreover, evaluation of PLZF protein, by intracellular staining, confirmed that Ly108 costimulation increased PLZF protein levels over that induced by CD3 stimulation alone in nearly the entire population of WT PS-DP cells (Fig. 2C–D and Supplemental Fig. S2E), reaching levels similar to those seen in iNKT cells (Fig. 2E). Thus, Ly108 costimulation strongly enhanced αCD3-induced PLZF expression, to levels observed in innate T cells.
Figure 2. Ly108 is required for full induction of PLZF in vitro.
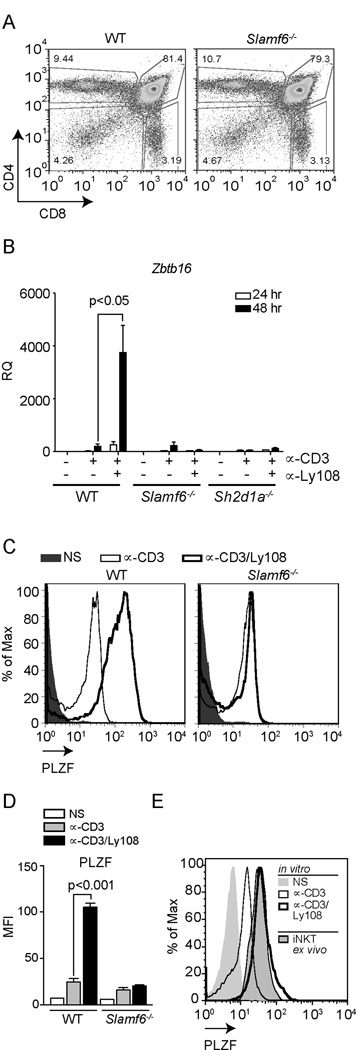
(A) Characterization of Ly108-deficient (Slamf6−/−) mice. Representative flow plots of thymocyte populations from WT and Slamf6−/− mice stained for CD4 and CD8. Frequencies and absolute numbers of thymocyte populations (N=6) stained for DP, DN CD4+ and CD8+ cells are depicted in Table 1. (B) Zbtb16 expression in WT, Slamf6−/− and SAP-deficient (Sh2d1a−/−) thymocytes stimulated as in Fig.1. Values were normalized to NS WT samples. Similar results were obtained using freshly isolated PS-DP cells as controls (Supplemental Fig. S2A). Data are the means ± SEM of 3 independent experiments, using cells from ≥3 mice each. (C) Representative histograms of intracellular PLZF staining in PS-DP thymocytes from WT and Slamf6−/− mice, NS (solid histogram) or stimulated with plate-bound αCD3±αLy108 for 48 h. Staining controls are shown in Supplemental Fig. S2E. (D) MFI ± SD of PLZF from 3 mice per genotype. (E) Intracellular PLZF expression in PS-DP thymocytes stimulated as in (C) compared to PLZF+CD1d-PBS57 tetramer+ iNKT cells. Histograms are representative of triplicates for each condition. Data in (E) were generated with a different antibody lot than in (C).
Table 1.
Conventional and PLZF+ T Cell populations in Slamf6−/− and B6.Sle1b mice
| DN % ± SEM (# × 106) |
DP % ± SEM (# × 106) |
CD8 SP % ± SEM (# × 106) |
CD4 SP % ± SEM (# × 106) |
PLZF+ % ± SEM (# × 105)* |
CD1d tet+ % ± SEM (# × 105)* |
|
|---|---|---|---|---|---|---|
| WT C57BL/6 N=6 (95.9×106 ±3.85) | 4.99±0.47 (4.79±0.57) |
80.9±0.29 (77.6±2.17) |
2.97±0.11 (2.84±0.16) |
9.47±0.23 (9.08±0.42) |
1.33±0.14 (1.21±0.18) |
1.85±0.05 (1.53±0.12) |
| Slamf6−/− N=6 (96.5×106 ±6.6) | 4.98±0.47 (4.78±0.48) |
81.8±1.14 (79.1±2.49) |
2.64±0.37 (2.55±0.33) |
8.91±0.84 (8.6±0.8) |
0.42±0.01 (0.36±0.02) |
1.06±0.07 (0.91±0.08) |
| WT C57BL/6# N=9 (71.7×106 ±6.31) | 3.08±0.26 (2.21±0.26) |
87.2±1.38 (62.5±3.2) |
1.46±0.18 (1.05±0.15) |
5.23±0.79 (3.75±0.63) |
0.76±0.18 (0.29±0.09) |
0.9±0.11 (0.34±0.06) |
| B6.Sle1b N=9 (99.5×106 ±5.44) | 3.63±0.66 (3.61±0.64) |
84.9±3.7 (84.5±6.61) |
2.43±0.59 (2.42±0.57) |
5.53±0.48 (5.5±0.58) |
1.91±0.24 (1.11±0.08) |
1.44±0.17 (0.79±0.15) |
Percentages are of gated CD4 single positive cells.
Controls for B6.Sle1b experiments (performed at a different time)
Ly108-deficient mice have fewer PLZF-expressing cells
To evaluate whether Ly108 contributes to PLZF expression in CD4+ T cells in vivo, we evaluated PLZF expressing thymocytes and thymic iNKT cell numbers from Ly108-deficient (Slamf6−/−) mice. As previously reported, we found that thymic iNKT cells were reduced by approximately 50% in Slamf6−/− mice (12). However, both the frequency and absolute numbers of CD4+PLZF+ thymocytes were even more compromised in Slamf6−/− mice (Fig. 3A–B and Table 1). Despite the reduction of PLZF positive thymocytes in Slamf6−/− mice, the intensity of PLZF in the remaining iNKT cells was similar between WT and Slamf6−/− thymocytes (Fig. 3C–D), suggesting compensating roles for other SLAM family members or other receptors in PLZF expression.
Figure 3. Ly108-deficient mice have fewer PLZF+CD4+ thymocytes.
Percentages (A) and absolute numbers (B) of CD1d-PBS57 Tetramer+ and PLZF+, CD4+ thymocytes in WT and Slamf6−/− mice. (C–D) PLZF levels in CD1d-PBS57 Tetramer+ iNKT cells from WT and Slamf6−/− mice. Data are the mean ± SD of 3 mice per group.
Increased PLZF expressing cells in a mouse model of enhanced Ly108 signaling
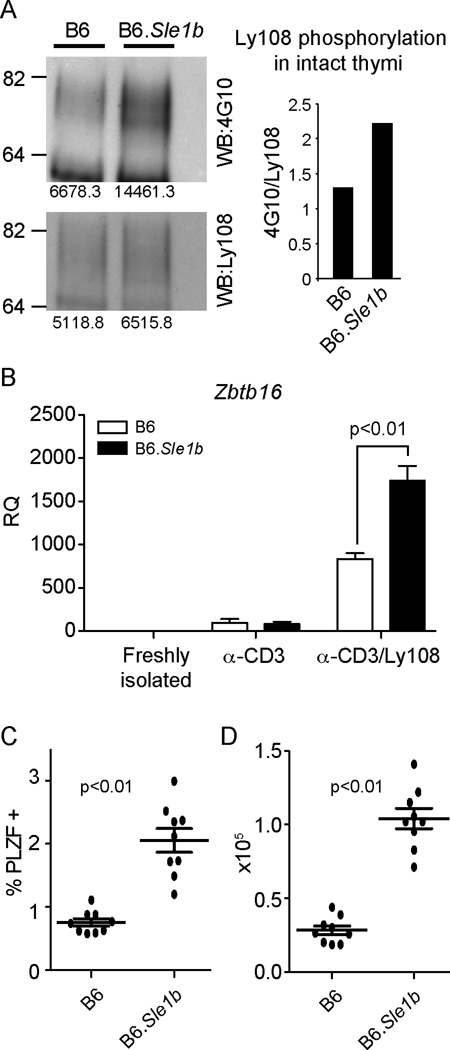
B6.Sle1b congenic mice carry the SLAM gene locus of lupus-prone NZM2410 mice introgressed into C57BL/6 background, and develop high levels of autoantibodies with age (28). This phenotype has been linked to higher expression of the Ly108-1 isoform, which is more heavily phosphorylated upon engagement (20, 28, 34). Consistent with these observations, we have found that lysates from intact thymi from B6.Sle1b mice show increased phosphorylation of Ly108 (Fig. 4A). Using plate-bound αCD3 ± αLy108 stimulation, we further found that B6.Sle1b PS-DP thymocytes showed higher levels of Zbtb16 induction upon Ly108 costimulation compared to C57BL/6 controls (Fig. 4B). Additionally, ex vivo staining of B6.Sle1b thymocytes also revealed increased percentages and numbers of PLZF positive cells compared to C57BL/6 controls (Fig. 4C–D and Table 1). Taken together, our results suggest that Ly108 signaling is a potent positive regulator of PLZF expressing cells.
Figure 4. Increased PLZF expressing cells observed in a mouse model with enhanced Ly108 signaling.
(A) Enhanced phosphorylation of Ly108 in intact thymi of B6.Sle1b mice. Ly108 was immunoprecipitated from lysates of intact thymi of C57Bl/6J (B6) and B6.Sle1b mice and probed for phosphotyrosine using 4G10 antibody (top) and for total Ly108 protein (bottom). Quantitation of relative tyrosine phosphorylation is shown on the right. (B) Zbtb16 expression in C57Bl/6J (B6) and B6.Sle1b PS-DP thymocytes stimulated for 48 hrs with plate-bound αCD3±αLy108 as in Figure 1. Data are representative of three independent experiments. Percentages (C) and absolute numbers (D) of PLZF+ CD4+ thymocytes in C57Bl/6J (B6) and B6.Sle1b mice.
Ly108 costimulation potentiates TCR signaling
The observation that the effect of Ly108 on PLZF expression requires stimulation with αCD3 suggests that Ly108 functions as a costimulatory molecule to potentiate TCR signaling. To evaluate whether Ly108 affects TCR signaling, we compared tyrosine-phosphorylated proteins from thymocytes stimulated with αCD3 (or αCD3+αCD4) ± αLy108. Ly108 stimulation alone elicited minor changes in tyrosine phosphorylation. However, αLy108 plus αCD3 (or αCD3/CD4), prolonged the tyrosine phosphorylation of multiple proteins, including an approximately 37 kDa protein corresponding to the molecular weight of LAT, in WT, but not Slamf6−/− thymocytes, compared to αCD3 (or αCD3/CD4) stimulation alone (Fig. 5A, Supplementary Fig. S3A). Although subtle, these effects were confirmed by probing for specific phospho-proteins, including LAT (Y191), PLCγ-1 (Y783), PKCθ (Thr538), and ERK (Fig. 5B and Supplementary Fig. S3B). To confirm that this increased signaling occurred in pre-selection thymocytes, we also evaluated the effects of Ly108 costimulation on thymocytes from MHC-class I/class II-deficient mice that lack mature conventional SP thymocytes. We observed a similar prolongation of phosphorylation of signaling proteins downstream of TCR upon Ly108 costimulation (Supplementary Fig. S3C). Evidence of prolonged signaling was also observed as a stronger resurgence of ERK phosphorylation after an hour of Ly108 costimulation (Fig. 5C).
Figure 5. Ly108 costimulation prolongs TCR signaling.
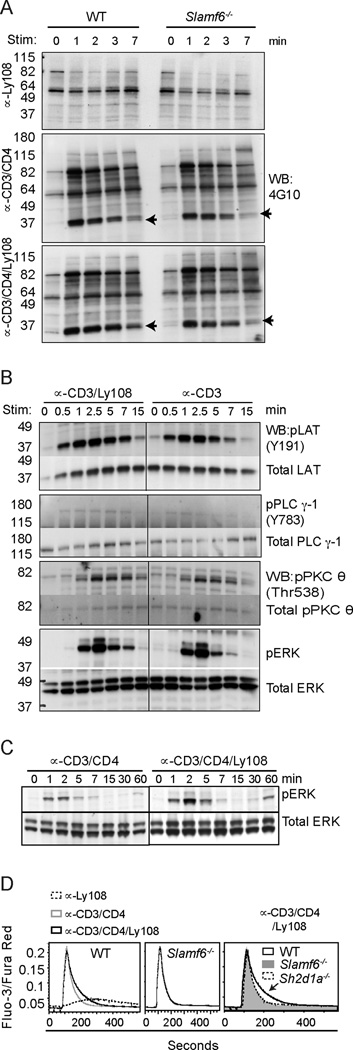
(A) Rested WT and Slamf6−/− thymocytes, pre-incubated with biotinylated αLy108 (top), αCD3+αCD4 (middle), and αCD3+αCD4+αLy108 (bottom), were stimulated with streptavidin for indicated times and lysates immunoblotted for α-phosphotyrosine (4G10). Arrows indicate predicted migration of LAT (left). Quantitation of 37 kDa band corresponding to LAT in (A) is shown in supplemental Fig. S3A (B) WT thymocytes were stimulated with αCD3±αLy108 as in (A) and lysates probed for pLAT (Y191), LAT; pPLCγ-1 (Y783), PLCγ-1; pPKCθ (Thr 538), PKCθ; pERK and ERK. Quantitation of band intensities are shown in supplemental Fig. S3B. (C) WT thymocytes were stimulated with αCD3+αCD4±αLy108 for extended times and probed for phospho and total Erk proteins. (D) Ca2+ flux of WT and Slamf6−/− PS-DP thymocytes stimulated with biotin-conjugated αCD3/CD4±αLy108 followed by streptavidin, as indicated by the ratio of Fluo-3/Fura-Red. Arrow indicates enhanced intracellular Ca2+ levels seen in WT, but not Ly108- or SAP-deficient thymocytes, upon αCD3/CD4/Ly108 stimulation. A–D are representative of 3 independent experiments.
In line with increased PLCγ–1 phosphorylation, Ly108 costimulation also slightly increased the prolonged phase of Ca2+ mobilization in WT, but not Slamf6−/− PS-DP thymocytes. In contrast, Ca2+ flux was minimal with αLy108 stimulation alone (Fig. 5D). Thus, although these effects are subtle, Ly108 costimulation enhanced signaling downstream of the TCR.
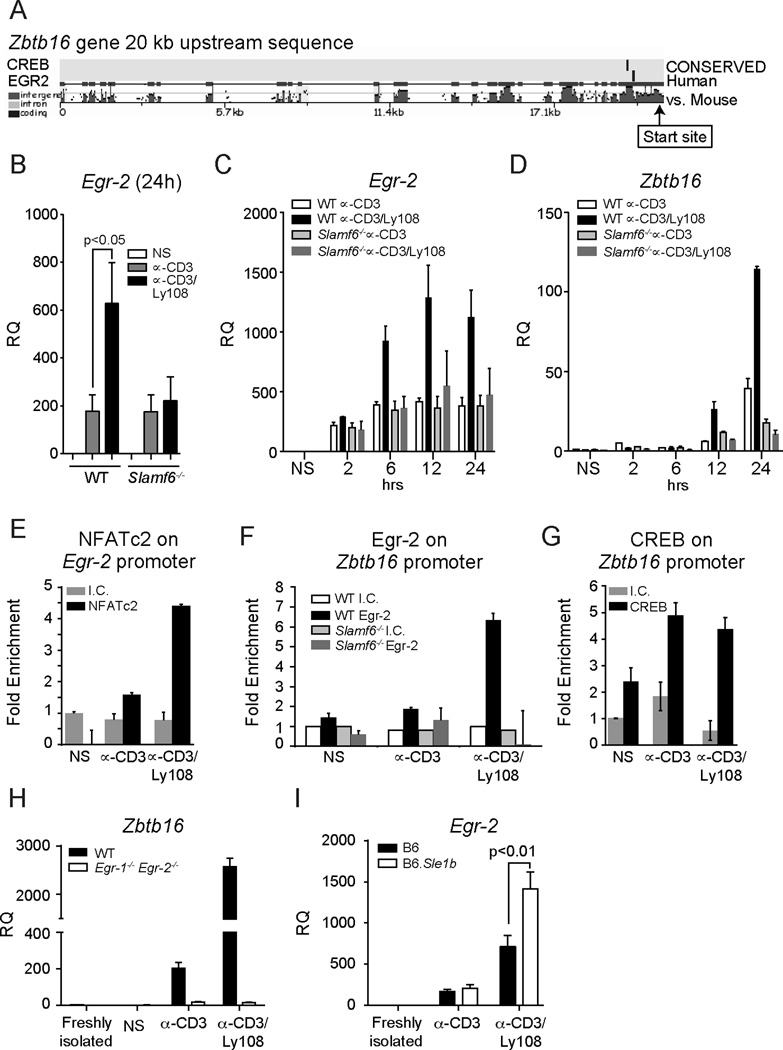
Egr-2 binds to PLZF promoter sequences and links Ly108 signaling to PLZF expression
The transcription factor Egr-2 is induced by TCR stimulation in a Ca2+ and NFAT-dependent manner and is important for both NKT cell development and expression of PLZF (7, 25, 35). Sequence analyses (NCBI; Mulan - http://mulan.dcode.org) revealed two conserved overlapping binding sites for Egr-2 in the Zbtb16 promoter (Fig. 6A). These sites were also found in a recent study using ChIP-Seq to evaluate potential Egr-2 binding sites, supporting a role for Egr-2 in regulating PLZF expression (7). Notably, we found that Ly108 costimulation enhanced CD3-induced Egr-2 expression in WT but not Ly108-deficient thymocytes (Fig. 6B). This effect was detectable as early as 6h and peaked by 12h (Fig. 6C), preceding the increase in Zbtb16 expression (Fig. 6D). Using Chromatin Immunoprecipitations (ChIPs), we further found that Ly108 costimulation enhanced NFATc2 binding at a conserved site upstream of Egr-2, suggesting that Ly108 costimulation increases Egr-2 expression via effects on NFATc2 (Fig. 6E).
Figure 6. Ly108 induces Egr-2 expression and binding to the Zbtb16 promoter.
(A) Mouse and human Zbtb16 sequences, 20 kb upstream of first exon, were aligned and analyzed for conserved binding sites for Egr-2 and CREB. (B) Egr-2 expression in WT or Slamf6−/− PS-DP thymocytes stimulated as in 1B for 24 h, normalized to NS controls. Data are the mean ± SEM from 6 independent experiments. Egr-2 (C) or Zbtb16 (D) expression by qRT-PCR from WT and Slamf6−/− PS-DP thymocytes stimulated for 2, 6, 12 or 24 h. Time courses are representative of 4 independent experiments. (E–G) Chromatin immunoprecipitations (ChIPs) from thymocytes stimulated as in 1B. (E) NFATc2 binding on the Egr-2 promoter. (F) Egr-2 binding on the Zbtb16 promoter. (G) CREB binding on the Zbtb16 promoter. ChIPs show average of 2 replicates ± SD from one of 2 or more independent experiments. (H) Zbtb16 expression in Egr-1−/−Egr-2fl/fl Lck Cre or littermate control PS-DP thymocytes stimulated as in Fig. 1B for 48 hours and normalized to NS controls. (I) Egr-2 expression in C57Bl/6J (B6) and B6.Sle1b PS-DP thymocytes stimulated with plate-bound αCD3±αLy108 for 48 hrs. Data are representative of three independent experiments.
Consistent with these observations, ChIP analyses revealed that Ly108 costimulation strongly enhanced Egr-2 binding to the conserved sites within the Zbtb16 promoter in WT but not Slamf6−/− thymocytes (Fig. 6F). In contrast, CREB, a TCR-activated transcription factor (36) that also has conserved binding sites on the Zbtb16 promoter (Fig 6A), failed to show increased Zbtb16 binding upon Ly108 costimulation (Fig. 6G). Thus, Ly108 costimulation enhanced Egr-2 expression and binding of Egr-2 to the PLZF promoter. Consistent with a critical role for Egr proteins in the regulation of PLZF, stimulation of PS-DP thymocytes from T cell-specific Egr1−/−Egr2−/− conditional mice showed only minimal CD3-mediated induction of PLZF, which did not increase upon Ly108 costimulation (Fig. 6H). Conversely, stimulation of PS-DP thymocytes from B6.Sle1b mice that have enhanced Ly108 phosphorylation also showed increased Ly108-induced prolonged Egr-2 expression when compared to C57Bl/6 controls (Fig.6I). Taken together our results support the hypothesis that Ly108 costimulation can increase and sustain Egr-2 expression and binding to the Zbtb16 promoter in developing thymocytes.
CD28 costimulation does not enhance PLZF expression
To test whether the enhancement of PLZF expression was specific for Ly108, we examined the effects of CD28, which also potentiates TCR signaling. To minimize CD28 costimulation-induced thymocyte death (37–39), analyses were performed after 18–24 h of stimulation (Supplementary Fig. S2C). While αCD28 costimulation slightly increased Egr-2 expression compared to αCD3 alone, it failed to increase Egr-2 expression to levels observed with Ly108 costimulation (Fig. 7A), nor did CD28 costimulation increase Egr-2 binding to the PLZF promoter (Fig. 7B). Notably, CD28 costimulation also failed to induce PLZF expression in PS-DP thymocytes (Fig. 7C). These data suggest that Ly108 provides a distinct signal that enhances PLZF expression in developing thymocytes.
Figure 7. CD28 costimulation does not enhance PLZF expression.
(A–C) PS-DP thymocytes were stimulated with αCD3±αCD28 or ±αLy108 and evaluated for Egr-2 expression by qRT-PCR at 18h (A), Egr-2 binding at the Zbtb16 promoter by ChIP at 18h (B), or Zbtb16 expression at 24h (C). Data show the mean of 2 replicates ± SD from one of two or more independent experiments.
DISCUSSION
Although SLAM family members have been implicated in iNKT and other innate T lymphocyte development, the functions of these receptors in this process have not been clear. In this paper, we demonstrate that Ly108 can potentiate CD3-induced expression of Egr-2 and PLZF, a master regulator of innate T lymphocyte phenotypes. Using either plate-bound antibody or the endogenous ligand, we found that costimulation with Ly108 in vitro potently enhanced TCR-induced PLZF expression in PS-DP thymocytes, reaching levels observed in iNKT cells. Although we cannot absolutely rule out outgrowth of a previously PLZF+ population, we have rigorously purified cells to minimize PLZF+ (and iNKT) cells in our starting cultures. Moreover, as far as we can detect, Ly108 stimulation did not induce proliferation, nor did it further enhance αCD3-induced cell death, suggesting that we were not enriching for a specific PLZF-expression population in our stimulations. Indeed, the PLZF+ cells in these cultures did not react with the CD-1d tetramer that recognizes iNKT cells.
Ly108 costimulation also increased expression of Egr-2, which is highly expressed in iNKT cells and has recently been shown to be critical for both iNKT development and PLZF expression (7, 25, 35). Supporting an important role for Ly108 in this pathway, we observed that Ly108 strongly increased Egr-2 binding to the Zbtb16 promoter and that Ly108 could not induce PLZF expression in thymocytes from mice deficient in Egr-1 and -2. Thus while high levels of Egr-2 in iNKT cells have been interpreted as indicative of these cells having received strong TCR signals (7), our data suggest that Ly108 is an important contributor to the induction of Egr-2, which is critical for PLZF expression. These findings were further supported by observations in B6.Sle1b mice, a model of enhanced Ly108 signaling, which show increased Egr-2 expression and increased numbers of PLZF expressing CD4+ cells.
While the observations that the iNKT cells that do develop in Ly108-deficient mice express normal levels of PLZF could be used to argue against a role for Ly108 in regulating PLZF expression, an alternative interpretation is that cells that successfully upregulate PLZF have a selective advantage and preferentially develop. This idea is consistent with data showing decreased iNKT cells in Zbtb16−/− animals (12). Alternatively, given that bone-marrow chimeras that generate a functional double-deficiency in both Ly108 and SLAM have a more complete block in the development of PLZF expressing iNKT cells (12), other SLAM family members (or other receptors) may compensate for the absence of Ly108. Consistent with this idea, we have seen that an α-SLAM (CD150) antibody can modestly increase expression of Egr-2 and PLZF in similar plate-bound stimulation assays, although not nearly to the level seen with α-Ly108 (data not shown). Whether this is the result of functional differences between SLAM and Ly108 or the relative efficiency of the particular stimulating antibodies requires further evaluation.
It has been argued that SLAM/SAP-mediated signaling does not play a major role in the regulation of PLZF in part because a) the defect in iNKT cell development occurs earlier in SAP-deficient than that in PLZF-deficient mice (5, 12); b) some PLZF induction is still seen in SAP-deficient stage 0 iNKT cells (3) and c) restoring PLZF expression by transgenic expression does not restore iNKT development in SAP-deficient mice (5). However, each of these arguments does not rule out a role for SLAM family members in the regulation of PLZF. Indeed, our results are consistent with the idea that SLAM family members affect multiple factors and stages in NKT cell differentiation, and with recent data showing that the absence of SAP leads to strong inhibitory signals that may differ from the lack of SLAM family signaling (40, 41)
Our results further suggest that the expression of PLZF can be induced in most (pre-selection) thymocytes that receive the appropriate signals. We have recently found that Ly108 is heavily phosphorylated in the thymus, but that phosphorylation of Ly108 is lost within minutes of dispersion of thymocytes into a single cell preparation. In contrast, α-Ly108-mediated induction of Ly108 tyrosine phosphorylation is relatively slow compared to the global patterns of tyrosine phosphorylation induced by αCD3 (20). These results suggest that there is dynamic contact-mediated regulation of Ly108 phosphorylation in the thymus. Given the dramatic effects of Ly108 on PLZF induction, the rapid dephosphorylation (with relatively slow induction of phosphorylation) suggests that Ly108 activity is tightly controlled, perhaps preventing the full induction of PLZF unless contact is maintained between developing DP cells, as may occur during the selection of iNKT and other hematopoietically-selected T cells. This rapid dephosphorylation could provide a safety mechanism that allows only certain cells to develop and maintain high PLZF expression, and thereby limits the number of CD4+ T cells that develop innate-like characteristics.
However, whether Ly108 acts solely by potentiating TCR signaling and Egr-2 expression is not clear. While the effects of Ly108 co-stimulation observed at the level of tyrosine phosphorylation and Ca2+ mobilization are subtle, these may affect duration of signals, which could lead to distinct outcomes in terms of gene expression and cell fate. However, the contrast with CD28 costimulation for the induction of PLZF expression suggests that the effects of Ly108 may go beyond that of boosting TCR signaling. The identification of distinct Ly108 signaling pathway(s) that contribute to PLZF expression therefore remains an important question.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank J. Reilley, R. Handon and J. Fekecs.
Abbreviations
- DP
double positive
- DN
double negative
- SP
single positive
- NKT
Nature Killer T
- SLAM
Signaling Lymphocyte Activation Molecule
- SAP
SLAM associated protein
- XLP
X linked lymphoproliferative disease
- PSDP
Pre-selection Double Positive
- PLZF
Promyelocytic Zinc Finger transcription factor
- Egr
Early growth response protein
- ChIP
Chromatin immunoprecipitation
- LAT
Linker of activated T cells
- PLC γ-1
Phospholipase C gamma -1
- PKCθ
Protein kinase C θ
Footnotes
This work was supported by intramural funding of NHGRI, NIH and R01AI081314.
The authors have no competing financial interests in this work.
REFERENCES
- 1.Subleski JJ, Jiang Q, Weiss JM, Wiltrout RH. The split personality of NKT cells in malignancy, autoimmune and allergic disorders. Immunotherapy. 2011;3:1167–1184. doi: 10.2217/imt.11.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovalovsky D, Uche OU, Eladad S, Hobbs RM, Yi W, Alonzo E, Chua K, Eidson M, Kim HJ, Im JS, Pandolfi PP, Sant'Angelo DB. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Savage AK, Constantinides MG, Han J, Picard D, Martin E, Li BF, Lantz O, Bendelac A. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreslavsky T, Savage AK, Hobbs R, Gounari F, Bronson R, Pereira P, Pandolfi PP, Bendelac A, von Boehmer H. TCR-inducible PLZF transcription factor required for innate phenotype of a subset of gammadelta T cells with restricted TCR diversity. Proc Natl Acad Sci U S A. 2009;106:12453–12458. doi: 10.1073/pnas.0903895106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kovalovsky D, Alonzo ES, Uche OU, Eidson M, Nichols KE, Sant'Angelo DB. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. Journal of immunology. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 6.Raberger J, Schebesta A, Sakaguchi S, Boucheron N, Blomberg KE, Berglof A, Kolbe T, Smith CI, Rulicke T, Ellmeier W. The transcriptional regulator PLZF induces the development of CD44 high memory phenotype T cells. P Natl Acad Sci USA. 2008;105:17919–17924. doi: 10.1073/pnas.0805733105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seiler M, Mathew R, Liszewski M, Spooner C, Barr K, Meng F, Singh H, Bendelac A. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat Immunol. 2012 doi: 10.1038/ni.2230. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma A, Chen Q, Nguyen T, Yu Q, Sen JM. T cell factor.1 and beta-catenin control the development of memory-like CD8 thymocytes. J Immunol. 2012;188:3859–3868. doi: 10.4049/jimmunol.1103729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 10.Lee YJ, Jeon YK, Kang BH, Chung DH, Park CG, Shin HY, Jung KC, Park SH. Generation of PLZF(+) CD4(+) T cells via MHC class II-dependent thymocyte-thymocyte interaction is a physiological process in humans. Journal of Experimental Medicine. 2010;207:237–246. doi: 10.1084/jem.20091519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Min HS, Lee YJ, Jeon YK, Kim EJ, Kang BH, Jung KC, Chang CH, Park SH. MHC class II-restricted interaction between thymocytes plays an essential role in the production of innate CD8+ T cells. Journal of immunology. 2011;186:5749–5757. doi: 10.4049/jimmunol.1002825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griewank K, Borowski C, Rietdijk S, Wang NH, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by slamf1 and slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cannons JL, Tangye SG, Schwartzberg PL. SLAM family receptors and SAP adaptors in immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- 14.Pasquier B, Yin L, Fondaneche MC, Relouzat F, Bloch-Queyrat C, Lambert N, Fischer A, de Saint-Basile G, Latour S. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichols KE, Hom J, Gong SY, Ganguly A, Ma CS, Cannons JL, Tangye SG, Schwartzberg PL, Koretzky GA, Stein PL. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 17.Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W, Sofi MH, Rietdijk S, Wang N, Terhorst C, Chang CH. The SLAM-associated protein signaling pathway is required for development of CD4+ T cells selected by homotypic thymocyte interaction. Immunity. 2007;27:763–774. doi: 10.1016/j.immuni.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP Protein-Dependent Natural Killer T-like Cells Regulate the Development of CD8(+) T Cells with Innate Lymphocyte Characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta M, Schwartzberg PL. Characterization of Ly108 in the thymus: evidence for distinct properties of a novel form of Ly108. J Immunol. 2012;188:3031–3041. doi: 10.4049/jimmunol.1103226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czar MJ, Kersh EN, Mijares LA, Lanier G, Lewis J, Yap G, Chen A, Sher A, Duckett CS, Ahmed R, Schwartzberg PL. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc Natl Acad Sci U S A. 2001;98:7449–7454. doi: 10.1073/pnas.131193098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grusby MJ, Auchincloss H, Jr, Lee R, Johnson RS, Spencer JP, Zijlstra M, Jaenisch R, Papaioannou VE, Glimcher LH. Mice lacking major histocompatibility complex class I and class II molecules. Proc Natl Acad Sci U S A. 1993;90:3913–3917. doi: 10.1073/pnas.90.9.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zijlstra M, Bix M, Simister NE, Loring JM, Raulet DH, Jaenisch R. Beta 2-microglobulin deficient mice lack CD4−8+ cytolytic T cells. Nature. 1990;344:742–746. doi: 10.1038/344742a0. [DOI] [PubMed] [Google Scholar]
- 24.Taillebourg E, Buart S, Charnay P. Conditional, floxed allele of the Krox20 gene. Genesis. 2002;32:112–113. doi: 10.1002/gene.10062. [DOI] [PubMed] [Google Scholar]
- 25.Hu T, Gimferrer I, Simmons A, Wiest D, Alberola-Ila J. The Ras/MAPK pathway is required for generation of iNKT cells. PLoS One. 2011;6:e19890. doi: 10.1371/journal.pone.0019890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. The Journal of biological chemistry. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Dutra A, Takesono A, Garrett-Beal L, Schwartzberg PL. Improved generation of C57BL/6J mouse embryonic stem cells in a defined serum-free media. Genesis. 2004;39:100–104. doi: 10.1002/gene.20031. [DOI] [PubMed] [Google Scholar]
- 28.Wandstrat AE, Nguyen C, Limaye N, Chan AY, Subramanian S, Tian XH, Yim YS, Pertsemlidis A, Garner HR, Jr, Morel L, Wakeland EK. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 29.Bendelac A, Killeen N, Littman DR, Schwartz RH. A subset of CD4+ thymocytes selected by MHC class I molecules. Science. 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- 30.Ohteki T, MacDonald HR. Major histocompatibility complex class I related molecules control the development of CD4+8− and CD4−8− subsets of natural killer 1.1+ T cell receptor-alpha/beta+ cells in the liver of mice. The Journal of experimental medicine. 1994;180:699–704. doi: 10.1084/jem.180.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coles MC, Raulet DH. Class I dependence of the development of CD4+ CD8− NK1.1+ thymocytes. The Journal of experimental medicine. 1994;180:395–399. doi: 10.1084/jem.180.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cannons JL, Qi H, Lu KT, Dutta M, Gomez-Rodriguez J, Cheng J, Wakeland EK, Germain RN, Schwartzberg PL. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. 2010;32:253–265. doi: 10.1016/j.immuni.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valdez PA, Wang H, Seshasayee D, van Lookeren Campagne M, Gurney A, Lee WP, Grewal IS. NTB-A, a new activating receptor in T cells that regulates autoimmune disease. J Biol Chem. 2004;279:18662–18669. doi: 10.1074/jbc.M312313200. [DOI] [PubMed] [Google Scholar]
- 34.Zhong MC, Veillette A. Control of T lymphocyte signaling by Ly108, a signaling lymphocytic activation molecule family receptor implicated in autoimmunity. J Biol Chem. 2008;283:19255–19264. doi: 10.1074/jbc.M800209200. [DOI] [PubMed] [Google Scholar]
- 35.Lazarevic V, Zullo AJ, Schweitzer MN, Staton TL, Gallo EM, Crabtree GR, Glimcher LH. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat Immunol. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haubert D, Weckbecker G. Vav1 couples the T cell receptor to cAMP response element activation via a PKC-dependent pathway. Cellular signalling. 2010;22:944–954. doi: 10.1016/j.cellsig.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Punt JA, Osborne BA, Takahama Y, Sharrow SO, Singer A. Negative selection of CD4+CD8+ thymocytes by T cell receptor-induced apoptosis requires a costimulatory signal that can be provided by CD28. The Journal of experimental medicine. 1994;179:709–713. doi: 10.1084/jem.179.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amsen D, Kruisbeek AM. CD28-B7 interactions function to co-stimulate clonal deletion of double-positive thymocytes. International immunology. 1996;8:1927–1936. doi: 10.1093/intimm/8.12.1927. [DOI] [PubMed] [Google Scholar]
- 39.Kishimoto H, Cai Z, Brunmark A, Jackson MR, Peterson PA, Sprent J. Differing roles for B7 and intercellular adhesion molecule-1 in negative selection of thymocytes. The Journal of experimental medicine. 1996;184:531–537. doi: 10.1084/jem.184.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao F, Cannons JL, Dutta M, Griffiths GM, Schwartzberg PL. Positive and negative signaling through SLAM receptors regulate synapse organization and thresholds of cytolysis. Immunity. 2012;36:1003–1016. doi: 10.1016/j.immuni.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36:986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.