Abstract
The nuclear-cytoskeleton connection influences many aspects of cellular architecture, including, nuclear positioning, the stiffness of the global cytoskeleton, and mechanotransduction. Central to all of these processes is the assembly and function of conserved SUN-KASH bridges, or LINC complexes, that span the nuclear envelope. Recent studies provide details of the higher order assembly and targeting of SUN proteins to the inner nuclear membrane. Structural studies characterize SUN-KASH interactions that form the central link of the nuclear-envelope bridge. KASH proteins at the outer nuclear membrane link the nuclear envelope to the cytoskeleton where forces are generated to move nuclei. Significantly, SUN proteins were recently shown to contribute to the progression of laminopathies.
Introduction
How the nucleus interacts with the cytoskeleton is central to positioning the nucleus, which functions in a wide variety of cellular processes including nuclear migration, nuclear anchorage, centrosome attachment to the nucleus, and mechanotransduction [1–4]. The machinery that positions nuclei also plays important functions in DNA repair and pairing of chromosomes in meiosis, which will not be discussed here [5–10]. A conserved bridge consisting of SUN and KASH proteins spans both membranes of the nuclear envelope [11–13] and is often referred to as the LINC complex because it is the linker of the nucleoskeleton to the cytoskeleton [14–16]. To form the bridge, SUN proteins in the inner nuclear membrane interact with lamins in the nucleoplasm and KASH proteins in the perinuclear space (Figure 1). KASH proteins are then recruited specifically to the outer nuclear membrane where they are positioned to interact with a wide variety of cytoskeletal components [11]. Mutations in mammalian SUN and KASH proteins lead to developmental defects in neurogenesis, gametogenesis, myogenesis, cilliogenesis, and retina formation and contribute to human diseases, including muscular dystrophy, ataxia, Progeria, lissencephaly, and cancer [11,17–20].
Figure 1.

SUN and KASH proteins span the nuclear envelope. A trimer of SUN proteins (light blue, dark blue, and grey) forms at the inner nuclear membrane (INM). SUN proteins interact with the KASH domain (orange) in the perinuclear space. Only a single KASH protein is shown for simplicity. KASH proteins cross the outer nuclear membrane (ONM) and extend into the cytoplasm to interact with the cytoskeleton. One class of KASH proteins (yellow) recruit microtubule motors dynein and kinesin to the surface of the nucleus, while a second class (light green) tethers nuclei to actin filaments.
The rapidly growing field of nuclear-cytoskeletal interactions has recently been reviewed [11–13]. Here, with apologies to the rest of the field, we focus on five major findings reported over the past two years. The first step of building the SUN-KASH bridge is recruiting SUN proteins to the inner nuclear membrane. Surprisingly, trafficking SUN proteins to the inner nuclear membrane involves multiple, partially redundant mechanisms [21–23]. The second step of bridge building, the formation of a physical interaction between SUN and KASH domains, was recently beautifully elucidated at a structural level [16]. Once KASH proteins are recruited to the surface of the nucleus, they interact with microtubule motors or flowing actin filaments to move nuclei [24–27]. A fourth group of studies demonstrated that SUN-KASH bridges transfer forces across the nuclear envelope [3,28]. Finally, our understanding of the role of SUN proteins in disease has been advanced with the surprising finding that the absence of Sun1 suppresses disease pathologies associated with defects in lamin A [20]. These exciting studies not only advance our understanding of how SUN-KASH bridges are assembled and function, but also open exciting avenues for continued research.
Building SUN-KASH bridges 1: Targeting SUN proteins to the inner nuclear membrane
To assemble the bridge, SUN proteins must first be targeted specifically to the inner nuclear membrane. Upwards of 100 proteins are specifically targeted to the inner nuclear membrane using a variety of different mechanisms [29]. Here we focus on recent reports elucidating mechanisms used to target SUN proteins, specifically mammalian Sun1 and Sun2, C. elegans UNC-84, and S. cerevisiae Mps3 to the inner nuclear membrane [21–23,30]. These reports show that SUN proteins are first actively trafficked from the ER to the nuclear envelope and then shuttled across nuclear pores by multiple mechanisms. Finally, SUN proteins are retained at the inner nuclear membrane through interactions with the nuclear lamina, chromatin, and/or KASH proteins (Figure 2).
Figure 2.
Three steps to targeting SUN proteins to the inner nuclear membrane (INM). First, signals including INM-SM and SUN-NELS recruit partners to move SUN proteins from the peripheral ER to the outer nucear membrane (ONM). ATP and the Golgi retrieval signal (4R) also participate in this first step. Second, importins (red), ATP, and/or histone H2A.Z (pink) help shuttle SUN proteins across the nuclear pore. Finally, SUN proteins are retained at the INM by interacting with lamins and forming bridges with KASH proteins.
The first step of trafficking to the inner nuclear membrane is to actively move SUN proteins from the peripheral ER toward the nuclear envelope, which employs a variable combination of signals (Figure 2). Both Sun2 and UNC-84 contain a predicted inner nuclear membrane-sorting motif (INM-SM). INM-SMs are found next to the cytoplasmic end of a transmembrane span and bind a truncated, membrane-associated importin α during translation to facilitate transport toward the nuclear envelope [31,32]. A novel SUN- Nuclear Envelope Localization Signal (SUN-NELS) is conserved between UNC-84 and Sun1; SUN-NELS binding partners have not been identified [22]. Mutating the INM-SM or the SUN-NELS in UNC-84 caused a significant delay in targeting to the nuclear envelope [22]. Likewise, a short region containing the SUN-NELS in Sun1 participates in localization [33]. An additional player in SUN trafficking is ATP, as depletion of ATP disrupted the mobility of Sun2 in the ER [30]. Finally, a Golgi retrieval signal further ensures that SUN proteins get to the correct compartment. Mutating the Golgi retrieval signal in Sun2 caused it to mislocalize [23]. Together, these data suggest that multiple mechanisms are required for presorting and trafficking SUN proteins toward the nuclear envelope. These signals likely work together, as multiple mutations worsened the trafficking defects [22,23].
Once enriched at the nuclear envelope, multiple mechanisms mediate the translocation of SUN proteins across the nuclear pore (Figure 2). The classical nuclear localization signal (cNLS) in Sun2 binds importins in a Ran-dependent manner and contributes to Sun2 localization [23]. ATP may also play a role in translocation across the nuclear pore [30]. The putative cNLSs in UNC-84 function in part redundantly with the SUN-NELS and INM-SM; only when all three signals are mutated does UNC-84 completely fail to localize to the inner nuclear membrane [22]. Independently of cNLSs, Mps3 uses its N-terminal acidic domain to interact with the histone variant H2A.Z to transverse the nuclear pore complex [21].
The final step of targeting SUN proteins is to retain them at the inner nuclear membrane (Figure 2). SUN proteins interact with multiple proteins in the nucleoskeleton, including lamins [34], which are strong candidates to retain SUN proteins at the inner nuclear membrane. An additional model postulates that the conserved SUN domain within the lumen aids in retention, presumably by forming nuclear envelope bridges [16,23].
Building SUN-KASH bridges 2: The interaction between SUN and KASH proteins in the perinuclear space
A direct interaction between SUN and KASH domains in the perinuclear space forms the central link of the nuclear envelope bridge [11]. Until recently, the oligomerization state of SUN proteins and the molecular interaction faces between SUN and KASH domains were not well understood [35,36]. Two new structural studies show that SUN domains assemble into clover-like trimers mediated by a triple-helix bundle of short, coiled regions at the amino end of the conserved SUN domain [16,37].
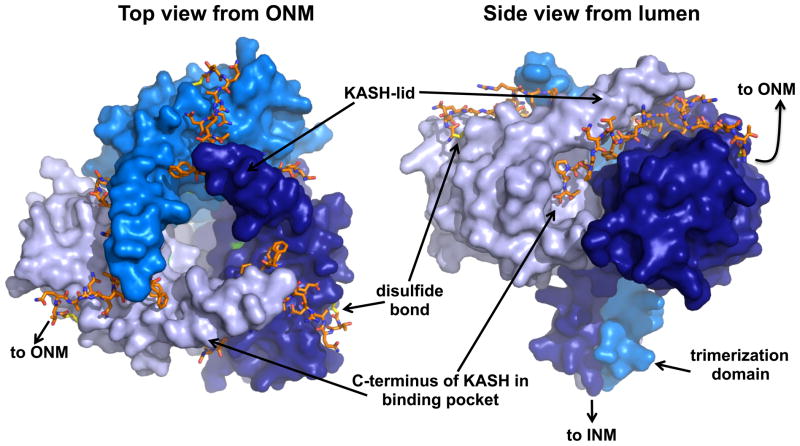
The most significant recent contribution to the understanding of the SUN-KASH bridge came when Sosa and Rothballer et al. [16] presented crystal structures of the interactions between human Sun2 and the KASH domains of Nesprin-1/-2 (Figure 3). They characterized extensive interaction faces among the three SUN protomers that create three independent KASH-binding sites. The C-terminal four residues of the KASH domain are buried in a pocket within the surface of one SUN protomer, which is supported by in vitro studies showing that the addition of a single alanine to the end of the KASH domain disrupts binding [16]. KASH domains then extend for 13 residues across a cleft formed by two SUN protomers and are clamped in place by a protruding β-sheet, or “KASH-lid”, from the first SUN protomer overlapping with its neighbor. Finally, the next six residues interact with the surface of the second SUN protomer. This interaction is further stabilized by the formation of an intermolecular disulfide bond between a cysteine 23 residues from the C-terminus of the KASH domain and a conserved cysteine on the surface of the SUN protein. Disulfide bond formation is dispensable for SUN-KASH binding, but is proposed to help withstand the forces transmitted across the bridge during nuclear migration or chromosome movement [16]. The remainder of the KASH domain extends away from the SUN trimer toward the outer nuclear membrane. This high-resolution view of SUN-KASH interactions gives us the first clear picture into how they tightly interact to perform their many functions.
Figure 3.
The crystal structure of the interaction between the SUN and KASH domains of human Sun2 and Nesprin-2. A surface representation of the three SUN protomers (shades of blue) and the backbone of the KASH peptide (orange) are shown from two angles. Adapted, with permission, from [16].
KASH proteins interact with microtubule motors or actin filaments to move nuclei
Once KASH proteins are recruited to the outer nuclear membrane, their cytoplasmic domains are free to mediate interactions between the nucleus and the cytoskeleton [11]. Two mechanisms for using KASH proteins to move nuclei were recently elucidated. In one mechanism, KASH proteins, including C. elegans UNC-83, Drosophila Klarsicht, and mammalian Nesprin-4 function as nuclear-specific adaptors to recruit motor proteins dynein and/or kinesin-1 to the surface of the nucleus [11,24,26]. In these three cases, kinesin-1 provides the major forces to move nuclei along polarized microtubules. Live imaging of C. elegans hypodermal nuclear migrations showed that kinesin-1 moves nuclei forward, while dynein is required to roll nuclei or to move them in the reverse direction to resolve cytoplasmic roadblocks [26]. In other systems, such as the C. elegans germline and early embryo, dynein is recruited to the nuclear envelope by the KASH protein ZYG-12 to position nuclei or to mediate meiotic chromosome movements and pairing [9,38]. Thus, the relative roles of the minus-end-directed microtubule-motor dynein vs. the plus-end-directed motor kinesin-1 vary.
The second mechanism for moving nuclei involves KASH proteins tethering nuclei to a moving actin network. In polarizing fibroblasts, actin filaments flow away from the wound edge. The KASH proteins Nesprin-1/-2 connect nuclei to the moving filaments [27]. Nesprin-1/-2, orthologs of C. elegans ANC-1 and Drosophila MSP-300, function to tether the outer nuclear membrane to actin [11]. In addition to Nesprins, SUN proteins, the inner nuclear membrane protein Samp1, and lamin also assemble into transmembrane actin-associated nuclear (TAN) lines to complete the connection between nuclei and moving actin filaments [27,39,40]. Together, these two examples demonstrate the variety of different mechanisms KASH proteins use to generate forces at the nuclear envelope.
The role of SUN-KASH bridges in mechanotransduction of forces across the nuclear envelope
Mechanotransduction is the translation of extracellular mechanical stimuli into chemical signals. Some mechanical stimuli are propagated through a pre-stressed cytoskeleton all the way to the nucleus [41]. SUN and KASH proteins have been hypothesized to propagate mechanical signals to the nucleus [42]. In support of this hypothesis, disruption of KASH proteins causes a loss of cellular mechanical stiffness, suggesting that nuclear envelope bridges organize the global cytoskeleton [43]. Two recent reports provide further evidence for the role of KASH proteins in mechanotransduction. First, a microneedle was used to physically pull on the cytoplasm and the displacement of the nucleus was used to approximate the strength of the mechanical coupling of the nucleus to the cytoskeleton [3]. The disruption of SUN or KASH proteins by dominant negative constructs reduced nuclear deformation, impaired intracellular force transduction, and affected cell migration and polarization, demonstrating that SUN-KASH bridges form a connection between the cytoskeleton and nucleus that is critical for intracellular force transmission [3]. The second report implicates the KASH protein Nesprin-3, which links intermediate filaments to the nucleus, in mechanotransduction [28]. In the presence of siRNA against Nesprin-3, cultured endothelial cells failed to polarize or migrate upon the induction of flow [28]. Together, these reports strongly support the hypothesis that SUN and KASH bridges function in mechanotransduction.
SUN-KASH bridges in human disease
Mutations in SUN and KASH proteins are thought to contribute to a wide variety of diseases, including cancer [11]. Here we focus on recent reports about the role of SUN proteins in laminopathies, a spectrum of diseases caused by mutations in lamins [44]. SUN and KASH proteins have long been postulated to contribute to the pathology of laminopathies [19,33,45]. A surprising report showed that knockout of mouse Sun1 reduced the severity of phenotypes associated with mutations in lamin A in mouse models for Emery-Dreifuss Muscular Dystrophy or Hutchinson-Gilford Progeria Syndrome [20]. Sun1, lamin A double mutant mice lived longer, grew larger, and had fewer defects in bone structure, muscle formation, senescence, heterochromatin marks, and the shape of nuclei than lamin A single mutant mice [20]. Similar results were observed in cells from Progeria patients treated with siRNA against Sun1 [20]. Thus, Sun1 enhances the defects associated with lamin A mutations in disease.
The mechanisms of how SUN proteins contribute to laminopathies are unknown, but multiple models have been proposed. Mutations in lamin A lead to less stiff nuclei [46,47], suggesting that the presence of Sun1 could lead to more pulling forces on the nuclear envelope and therefore more damage to a weakened nucleus. The other favored model is that mutations in lamins, and perhaps SUN proteins, lead to altered transcription patterns of important developmental factors [47,48]. An alternative model is that mutations in lamin A lead to overexpression of Sun1 and accumulation in the Golgi, which leads to toxicity [20]. The newest model is that overexpression of Sun1 in lamin A mutants causes toxicity by inducing hyperactivity in the DNA damage response [5,10].
Conclusions
Great progress has been made in the past two years in understanding how the LINC complex of SUN and KASH proteins is assembled, how it functions in nuclear migration, and how it participates in mechanotransduction. It has also become clear that LINC complexes play important roles in human disease. Despite this progress, many questions remain. Many players in SUN trafficking remain to be identified, including those utilizing ATP and proteins that bind the SUN-NELS. We do not understand when or where SUN multimerization and KASH binding occurs, how complexes are rearranged during important developmental switches, or how they participate in mechanotransduction. Finally, future experiments are required to determine the relative contributions of each of the proposed models for SUN and KASH proteins in disease progression. Continued research by basic and clinical scientists should translate these findings on nuclear-cytoskeletal interactions into the treatment of human disease.
Acknowledgments
We thank members of the Starr lab for insightful discussions. We apologize to those whose studies were not included due to space limitations. Studies in the Starr lab are supported by grant R01 GM073874 from the National Institutes of Health NIGMS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- 2.Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- *3.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. Journal of Biological Chemistry. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. This paper documents that SUN-KASH bridges are important components for nucleo-cytoskeletal force transmission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindeman RE, Pelegri F. Localized Products of futile cycle/lrmp Promote Centrosome-Nucleus Attachment in the Zebrafish Zygote. Curr Biol. 2012;22:843–851. doi: 10.1016/j.cub.2012.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes & development. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiraoka Y, Dernburg AF. The SUN rises on meiotic chromosome dynamics. Developmental Cell. 2009;17:598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 7.Harper NC, Rillo R, Jover-Gil S, Assaf ZJ, Bhalla N, Dernburg AF. Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Developmental Cell. 2011;21:934–947. doi: 10.1016/j.devcel.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Labella S, Woglar A, Jantsch V, Zetka M. Polo kinases establish links between meiotic chromosomes and cytoskeletal forces essential for homolog pairing. Developmental Cell. 2011;21:948–958. doi: 10.1016/j.devcel.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 9.Wynne DJ, Rog O, Carlton PM, Dernburg AF. Dynein-dependent processive chromosome motions promote homologous pairing in C. elegans meiosis. J Cell Biol. 2012;196:47–64. doi: 10.1083/jcb.201106022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei K, Zhu X, Xu R, Xu T, Zhuang Y, Han M. Inner nuclear envelope proteins SUN1 and SUN2 play a prominent role in the DNA damage response. Curr Biol. 2012 doi: 10.1016/j.cub.2012.06.043. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Developmental Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Razafsky D, Hodzic D. Bringing KASH under the SUN: the many faces of nucleo-cytoskeletal connections. J Cell Biol. 2009;186:461–472. doi: 10.1083/jcb.200906068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 Is a KASH Protein Required for Nuclear Migration and Is Recruited to the Outer Nuclear Membrane by a Physical Interaction with the SUN Protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **16.Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC Complexes Form by Binding of Three KASH Peptides to Domain Interfaces of Trimeric SUN Proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. This paper is the first to characterize the SUN-KASH interaction at a structural level, and shows that SUN protein trimerization mediates KASH protein binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gros-Louis F, Dupré N, Dion P, Fox MA, Laurent S, Verreault S, Sanes JR, Bouchard J-P, Rouleau GA. Mutations in SYNE1 lead to a newly discovered form of autosomal recessive cerebellar ataxia. Nature genetics. 2007;39:80–85. doi: 10.1038/ng1927. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Human molecular genetics. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **20.Chen C-Y, Chi Y-H, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang K-T. Accumulation of the inner nuclear envelope protein sun1 is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. This paper describes the surprising finding that the absence of Sun1 suppresses disease pathologies associated with mutations in lamin A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *21.Gardner JM, Smoyer CJ, Stensrud ES, Alexander R, Gogol M, Wiegraebe W, Jaspersen SL. Targeting of the SUN protein Mps3 to the inner nuclear membrane by the histone variant H2A.Z. J Cell Biol. 2011;193:489–507. doi: 10.1083/jcb.201011017. This paper shows that binding of a SUN protein to a chromatin factor, H2.AZ, facilitates active transport across the pore membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **22.Tapley EC, Ly N, Starr DA. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol Biol Cell. 2011;22:1739–1752. doi: 10.1091/mbc.E10-08-0733. This paper documents that SUN protein targeting requires an initial step that actively traffics the protein from the ER to the nuclear envelope. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **23.Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. The EMBO journal. 2010 doi: 10.1038/emboj.2010.119. This paper is the first to document that a Golgi retrieval pathway and the SUN domain contribute to inner nuclear membrane localization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux KJ, Crisp ML, Liu Q, Kim D, Kozlov S, Stewart CL, Burke B. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc Natl Acad Sci US A. 2009;106:2194–2199. doi: 10.1073/pnas.0808602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **26.Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. This paper employs live imaging to show that KASH proteins can interact with microtubule motors dynein and kinesin to mediate bidirectional movement of the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. This paper elucidates a novel mechanism to move nuclei by tethering nuclei to flowing actin filaments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Morgan JT, Pfeiffer ER, Thirkill TL, Peng G, Fridolfsson HN, Douglas GC, Starr DA, Barakat AI. Nesprin-3 Regulates Endothelial Cell Morphology, Perinuclear Cytoskeletal Architecture, and Flow-Induced Polarization. Mol Biol Cell. 2011 doi: 10.1091/mbc.E11-04–0287. This paper is the first to implicate the KASH protein Nesprin-3 in mechanotransduction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuleger N, Kerr ARW, Schirmer EC. Many mechanisms, one entrance: membrane protein translocation into the nucleus. Cell Mol Life Sci. 2012;69:2205–2216. doi: 10.1007/s00018-012-0929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *30.Zuleger N, Kelly DA, Richardson AC, Kerr ARW, Goldberg MW, Goryachev AB, Schirmer EC. System analysis shows distinct mechanisms and common principles of nuclear envelope protein dynamics. J Cell Biol. 2011;193:109–123. doi: 10.1083/jcb.201009068. This paper documents that SUN protein targeting to the inner nuclear membrane requires ATP at two distinct steps, and that multiple and distinct mechanisms are used to target inner membrane proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu D, Wu X, Summers MD, Lee A, Ryan KJ, Braunagel SC. Truncated isoforms of Kap60 facilitate trafficking of Heh2 to the nuclear envelope. Traffic. 2010;11:1506–1518. doi: 10.1111/j.1600-0854.2010.01119.x. [DOI] [PubMed] [Google Scholar]
- 32.Braunagel SC, Williamson ST, Ding Q, Wu X, Summers MD. Early sorting of inner nuclear membrane proteins is conserved. Proc Natl Acad Sci US A. 2007;104:9307–9312. doi: 10.1073/pnas.0703186104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **33.Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shannahan CM, Shackleton S. Mammalian SUN protein networks at the inner nuclear membrane and their role in laminopathy disease processes. Journal of Biological Chemistry. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. This paper identifies a link between Sun1 and laminopathies, as well as characterizes the physical interactions between Sun1/2 and their numerous binding partners. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Molecular and cellular biology. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu W, Gotzmann J, Sironi L, Jaeger VM, Schneider M, Luke Y, Uhlen M, Szigyarto CA, Brachner A, Ellenberg J, et al. Sun1 forms immobile macromolecular assemblies at the nuclear envelope. Biochim Biophys Acta. 2008;1783:2415–2426. doi: 10.1016/j.bbamcr.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Z, Du X, Cai Z, Song X, Zhang H, Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, et al. Structure of Sad1-UNC84 Homology (SUN) Domain Defines Features of Molecular Bridge in Nuclear Envelope. Journal of Biological Chemistry. 2012;287:5317–5326. doi: 10.1074/jbc.M111.304543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou K, Rolls MM, Hall DH, Malone CJ, Hanna-Rose W. A ZYG-12-dynein interaction at the nuclear envelope defines cytoskeletal architecture in the C. elegans gonad. J Cell Biol. 2009;186:229–241. doi: 10.1083/jcb.200902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Folker ES, Östlund C, Luxton GWG, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci US A. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- 41.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci US A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nature reviews. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- 43.Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Worman HJ. Nuclear lamins and laminopathies. [Internet] J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Human molecular genetics. 2007;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- 46.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. Journal of Clinical Investigation. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar D, Barkan R, Meshorer E, Gruenbaum Y. Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med. 2009;13:1059–1085. doi: 10.1111/j.1582-4934.2008.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson BR, Nitta RT, Frock RL, Mounkes L, Barbie DA, Stewart CL, Harlow E, Kennedy BK. A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc Natl Acad Sci US A. 2004;101:9677–9682. doi: 10.1073/pnas.0403250101. [DOI] [PMC free article] [PubMed] [Google Scholar]