Abstract
Microbial biodegradation and biotransformation reactions are essential to most bioremediation processes, yet the specific organisms, genes, and mechanisms involved are often not well understood. Stable isotope probing (SIP) enables researchers to directly link microbial metabolic capability to phylogenetic and metagenomic information within a community context by tracking isotopically labeled substances into phylogenetically and functionally informative biomarkers. SIP is thus applicable as a tool for the identification of active members of the microbial community and associated genes integral to the community functional potential, such as biodegradative processes. The rapid evolution of SIP over the last decade and integration with metagenomics provides researchers with a much deeper insight into potential biodegradative genes, processes, and applications, thereby enabling an improved mechanistic understanding that can facilitate advances in the field of bioremediation.
Keywords: bioremediation, biodegradation, stable isotope probing, metagenomics, sequence-based screening, function-based screening, gene-targeted metagenomics, high-throughput sequencing, high-throughput microarrays, carbon flow
Introduction
In addition to their critical role in global biogeochemical cycling, microbes play an essential role in the degradation, mineralization, or transformation of environmental pollutants and thus are potentially capable of restoring contaminated sites (Liu and Suflita, 1993, Diaz, 2004, Geoffrey, 2010). The natural biodegradative processes occurring in contaminated sites are known as intrinsic bioremediation or natural attenuation (Mulligan and Yong, 2004, Jørgensen, 2007). These microbial biodegradative processes can often be accelerated using various strategies, known as bioremediation. Together with phytoremediation, microbially mediated bioremediation is generally considered an environmentally friendly, inexpensive, publicly accepted and promising means to remove contaminants from the environment (Schnoor et al., 1995, Macek et al., 2000).
When designing a strategy for the bioremediation of any contaminated site, an understanding of the indigenous microbial community can be highly valuable. Information integral to the success of bioremediation strategies can include: (i) identification of the microorganisms present in the contaminated site, (ii) investigation of their metabolic capabilities, and (iii) understanding of potential microbial community shifts in response to changing environmental factors (Lovley, 2003). Much of this information, however, has been difficult to elucidate and is rarely achieved with traditional microbiological techniques. Until recently, knowledge of microbes involved in bioremediation processes had been based mostly on culture-dependent studies, which do not account for the fact that laboratory conditions differ substantially from the environment. Cultivation-based techniques have been shown to target only about 1% of microbes occurring in the environment (Torsvik and Øvreås, 2002, Lozupone and Knight, 2008, Zhang and Xu, 2008). More importantly, cultivation often fails to predict which microbes and which specific metabolic pathways will be active under realistic environmental conditions (Morales and Holben, 2011). The advent of molecular microbial ecology enabled culture-independent phylogenetic analyses of communities and functional genes, however, linking contaminant transformation to phylogenetic identity and specific genes/enzymes of metabolically active microbes remained a major challenge and still required cultivation. The development of stable isotope probing (SIP) was instrumental in circumventing the limitations of culture based investigations of biodegradation (Wellington et al., 2003, Friedrich, 2006). The coupling of SIP with the rapidly advancing field of metagenomics is further increasing its potential benefits to the field of bioremediation. This review aims to discuss the emerging trends in stable isotope probing techniques integrated with metagenomics, which can provide researchers with an unparalleled understanding of contaminant biodegradation and biotransformation processes in the environment.
Principles of stable isotope probing (SIP)
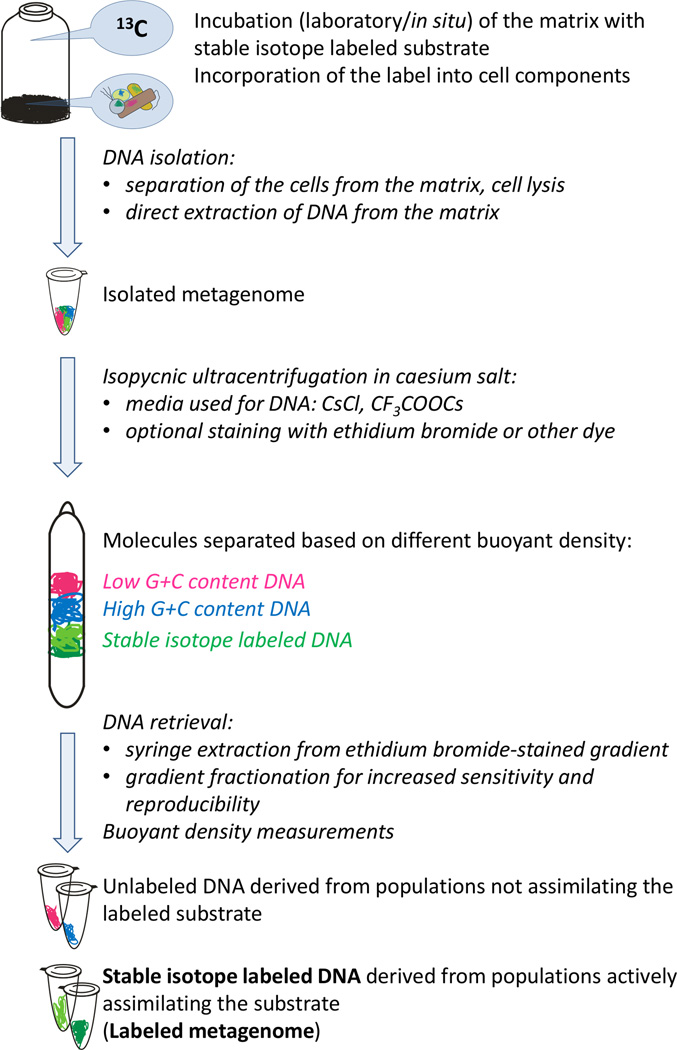
SIP tracks the incorporation of heavy stable isotopes, mainly 13C (Radajewski et al., 2003, Dumont and Murrell, 2005, Neufeld et al., 2007b, Uhlík et al., 2009a, Chen and Murrell, 2010, Madsen, 2010), 15N (Buckley et al., 2007b, Buckley et al., 2007a, Roh et al., 2009, Bell et al., 2011), or rarely 18O and 2H (Aanderud and Lennon, 2011, Woods et al., 2011), from specific substrates into phylogenetically informative biomarkers associated with microbes that assimilate the substrate. After stable isotopes have been pulsed into the environment and metabolically active cells have incorporated the label into their biomass, biomarkers are recovered and analyzed (Neufeld et al., 2007b, Dunford and Neufeld, 2010) (Figure 1). Therefore, SIP is an approach that can identify microbial populations with a defined function. The first biomarkers introduced in SIP studies were phospholipid-derived fatty acids (PLFA) (Boschker et al., 1998), followed by DNA (Radajewski et al., 2000), and rRNA (Manefield et al., 2002). Although it is the most sensitive of the three, PLFA-SIP is restricted to the classification of broad taxonomic groups (Table 1). Analyzing nucleic acids (RNA or DNA) can be far more informative taxonomically, with rRNA being a more responsive biomarker than DNA. This is due to the fact that the rates of rRNA synthesis are much higher than those of DNA, and rRNA labeling is independent of cellular replication (Manefield et al., 2002, Manefield et al., 2004, Whiteley et al., 2006). In addition, reverse transcription and sequence analysis of 16S rRNA provides equal resolution for phylogenetic identification as sequence analysis of 16S rRNA genes (DNA, Table 1). Ribosomal RNA-based analyses, however, cannot provide access to the functional genes responsible for the metabolic capabilities of the community, and generally 13C-labeled mRNA is challenging to isolate in sufficient quantities for SIP (Neufeld et al., 2007a, Simon and Daniel, 2009). However, success has been achieved with mRNA-SIP for detection of naphthalene dioxygenase transcripts following SIP incubations with naphthalene (Huang et al., 2009). Further research employing DNA-, mRNA-, and rRNA-SIP (Dumont et al., 2011) demonstrated higher rates of labeling of functional mRNAs than their genes, and hence higher sensitivity of mRNA-SIP compared to DNA-SIP (Table 1). In addition, protein-SIP has recently been developed, taking advantage of proteins as a combined indicator for a specific metabolic activity as well as for obtaining phylogenetic information (Table 1, Jehmlich et al., 2008a, Jehmlich et al., 2008b, Jehmlich et al., 2009).
Figure 1.
Scheme of DNA-based stable isotope probing (SIP) experiment with 13C-labeled substrate.
Table 1.
Comparison of methodological considerations for DNA-SIP, RNA-SIP, PLFA-SIP, and protein-SIP.
| Trait | Comparison of applicability of biomarkers |
Explanation |
|---|---|---|
| Sensitivity | protein > PLFA > RNA > DNA | DNA-SIP requires 15–20% isotopic enrichment, while protein-SIP only requires 1%. RNA labeling is 6.5 faster than that of DNA. |
| Incubation time | protein > PLFA > RNA > DNA | Incubation time is directly linked to sensitivity. DNA-SIP is the only technique that requires active cell division requiring the longest incubation periods potentially leading to biases. |
| Taxonomic resolution | DNA ≈ RNA > protein > PLFA | PLFA-SIP only distinguishes broader taxonomic groups, while DNA or RNA-SIP provides identification to the genus level or below. Databases for protein sequences are more limited than for 16S rRNA genes. |
| Indication of metabolic activity | protein > RNA > DNA | Proteins are the most explicit indicators of metabolic activity, while DNA only shows the metabolic potential. |
| Ease of isolation | DNA ≈ PLFA > RNA > protein | Isolation of PLFA and DNA are routinely performed in different matrices, but isolation of RNA and proteins from environmental samples can be very challenging. |
| Stability | DNA ≈ PLFA > protein > RNA | DNA or PLFA are fairly stable, but proteins may denature, and mRNA is very sensitive to degradation. |
| Application with ’omics‘ | DNA > RNA > protein | The application potential depends upon the developmental stage of the ’omics‘ methods. Currently, metagenomics is the most well-developed followed by metatranscriptomics and metaproteomics, respectively. |
To date, when functional gene analyses coupled to specific assimilatory processes are desired, DNA-SIP has been most often employed, due to the relative ease of detecting functional genes in 13C-DNA compared to mRNA. The use of either DNA-SIP or RNA-SIP can enable both the phylogenetic identification of those microbes performing the process as well as key metabolic genes possessed by the active populations likely to be involved in the process. When coupled with the rapidly expanding field of metagenomics, DNA-SIP has the potential for a focused, in-depth analysis of the collective genomes of the community active in a particular biodegradative process. This provides definitive information about the populations active in specific assimilatory processes and also requires considerably less sequencing effort than a full metagenomic analysis of the total community.
The metagenomics era
Metagenomics (also known as ecological genomics, community genomics, or environmental genomics) is a discipline that uses genomic methods to analyze natural ecological communities, namely the collective genomes in an environmental community (Handelsman et al., 1998, Riesenfeld et al., 2004). The major goal of metagenomics is to explicate the genomes of uncultured microbes, thereby permitting investigation of the broad diversity of taxonomically and phylogenetically relevant genes, individual catabolic genes, and whole operons (Schmeisser et al., 2007). Metagenomics itself was initially recognized for its potential to aid in discovery of novel biomolecules for biotechnological applications (Riesenfeld et al., 2004). Although the basic concept of metagenomics was first introduced at the end of last century (Handelsman et al., 1998), early forms of metagenomics had begun to emerge previously, with one example being the phylogenetic analyses of environmental microbial communities (Pace et al., 1985).
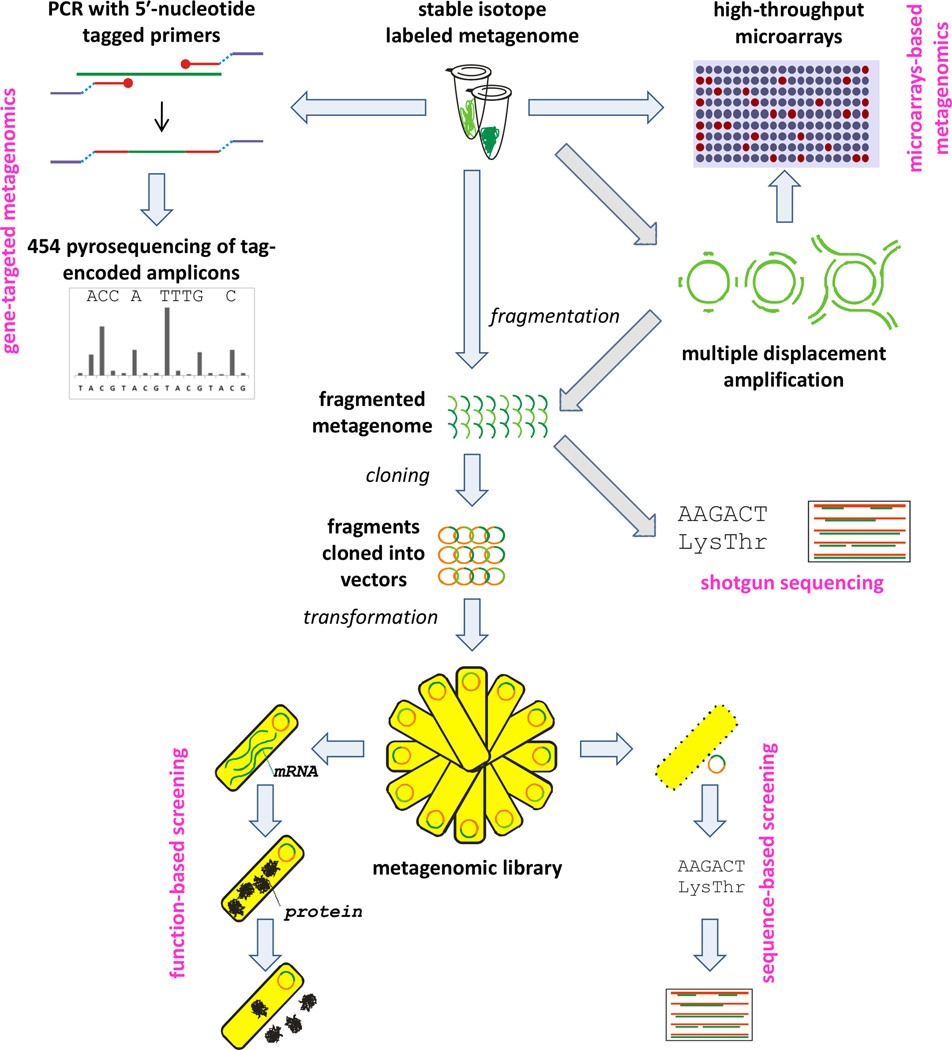
The approach introduced by Handelsman et al. (1998) involves extraction of the metagenome (genomic DNA from all organisms inhabiting the environment), its fragmentation, cloning, transformation, and subsequent screening of the constructed metagenomic library. The primary aim is to screen environmental communities for a specific biological activity and identify genes or gene clusters associated with it; also referred to as function-based screening (Yun and Ryu, 2005). The advent of high-throughput next generation DNA sequencing (e.g. 454 pyrosequencing, Illumina (Solexa) sequencing, SOLiD sequencing) gave rise to another approach to metagenomics, sequence-based screening. This method was first demonstrated by environmental genome shotgun sequencing of the Sargasso Sea (Venter et al., 2004), showing the potential of revealing the vast phylogenetic and metabolic diversity of microbial communities (Yun and Ryu, 2005). In addition to sequence-based screening of environmental metagenomic libraries, direct pyrosequencing of environmental communities is possible, bypassing cloning completely (Edwards et al., 2006). Because communities from habitats such as soils and sediments are often too diverse to permit screening in sufficient depth, even with the use of high-throughput sequencing, gene-targeted metagenomics techniques have emerged (Iwai et al., 2011), which are based on sequencing of PCR-generated amplicons (Figure 2).
Figure 2.
Overview of metagenomic approaches that can be used to analyze stable isotope labeled metagenomes.
In addition to high-throughput sequencing, metagenomic analyses have recently been performed with the use of high-throughput microarrays (Figure 2). These have been used to analyze microbial communities and monitor environmental biogeochemical processes. GeoChip microarrays (He et al., 2007, He et al., 2010) currently contain 83,992 50-meric sequences covering approximately 152,414 genes encoding for enzymes responsible for biogeochemical (C, N, P, S) cycling, metabolic processes, heavy metal resistance, antibiotic resistance, degradation of pollutants, and gyrB genes (Hazen et al., 2010, Lu et al., 2012). Marker gyrB encoding for the gyrase β-subunit is used instead of the more common 16S rRNA genes as probes for 16S rRNA usually do not provide resolution below genus level. gyrB can be used to differentiate even closely related species (He et al., 2010). GeoChip microarrays can be therefore used to study structure, dynamics, and potential metabolic activity of microbial communities and their variations depending on different stimuli. Another type of microarray valuable to microbial ecology and contaminant biodegradation is the PhyloChip, which is used for high throughput phylogenetic analyses of microbial communities (Brodie et al., 2006), and has been used for a variety of applications including assessing microbial community responses to petroleum contamination (Hazen et al., 2010, DeAngelis et al., 2011).
With the first applications of metagenomic techniques, it became apparent that they enable the discovery of genomic and metabolic diversity that had not been previously imagined (Schloss and Handelsman, 2005). As research progressed, however, the main drawbacks of metagenomics were realized: the inability to link specific functions to individual populations and to achieve full sequence coverage in more complex communities (Vieites et al., 2009). By combining metagenomic techniques with SIP, these limitations can be significantly reduced. SIP experiments are designed to provide a targeted analysis of the active populations, omitting the inactive majority that is not the focus of the study (Figure 2).
Integration of SIP with metagenomics
One of the main drawbacks of total community metagenomics is the unlikelihood of detecting particular genes of interest due to the extremely vast diversity and abundance of microbial genes occurring in most ecosystems, even when function-based metagenomic screening is used. Targeting metagenomics to specific subpopulations that are likely to contain the genes of interest, as with SIP, may overcome this obstacle (Schloss and Handelsman, 2003). This was demonstrated by Schwarz and colleagues (2006) who isolated genes encoding for coenzyme B12-dependent glycerol dehydratases. The source for this key enzyme for the anaerobic dehydration of glycerol was the enrichment cultures of 13C-glycerol-fermenting microorganisms from a sediment sample. When metagenomic library construction was preceded by SIP, the frequency of clones bearing target genes was increased by almost four fold. However, expressing target genes successfully in vitro, such as in metagenomic libraries, can be very challenging in some instances. Using high-throughput sequencing technologies for direct shotgun sequencing of SIP-derived metagenomes can aid in overcoming this obstacle. Selective enrichment of targeted populations, whose diversity is much less complex than that of total communities, with subsequent isolation of the particular functional metagenome of interest increases the feasibility of achieving coverage and assembly of individual genomes with significantly reduced efforts and sequencing cost. Shotgun sequencing of SIP-derived metagenomes can also help ensure that portions of the microbial community that have low abundance but are integral to the metabolic processes of interest will not be overlooked (Wellington et al., 2003). The major drawback of this approach is usually the recovery of DNA in quantities too small to be sufficient for shotgun sequencing. Advances in multiple displacement amplification over the last years (Binga et al., 2008) have helped to overcome this limitation. Although no bioremediation studies have yet been published performing direct shotgun sequencing of SIP-derived metagenomes, they are very likely to arise in the near future.
SIP and metagenomics to study biodegradation of ecologically significant C1 compounds
One of the pilot studies integrating DNA-SIP with metagenomics revealed a complete methane monooxygenase operon in forest soils (Dumont et al., 2006). Methane and other one-carbon (C1) compounds are of global ecological significance because they can affect climate change, influence atmospheric and marine chemistry, and impact cloud formation. In the context of bioremediation, methanotrophs and/or methylotrophs have been implicated in the degradation of trichloroethylene and cis-1,2-dichloroethylene (Little et al., 1988, Arai et al., 1999, Shigematsu et al., 1999, Takeuchi et al., 2005), insecticides (Topp et al., 1993), nitro-substituted explosives (Van Aken et al., 2004), methyl halides (Warner et al., 2005), methyl tert-butyl ether (Nakatsu et al., 2006, Kane et al., 2007), and other xenobiotic compounds. Therefore, investigating the metabolism of C1 compounds is also potentially valuable for bioremediation.
Dumont and colleagues (2006) were the first to apply SIP in combination with function- and sequence-based metagenomic library screening in this field. After the incubation of a soil sample with 13CH4, 13C-DNA was used for the construction of a metagenomic library using a bacterial artificial chromosome (BAC). Subsequent screening of the library for key methylotrophy genes resulted in the discovery of a clone carrying a pmoCAB operon, encoding for the particulate methane monooxygenase. A complete sequence of the operon was determined by shotgun sequencing. The sequence of the identified pmoA gene was almost identical to a Methylocystis sp. sequence which had been previously detected in this soil (Radajewski et al., 2002). Additionally, 12 other putative genes were detected on the same clone (Dumont et al., 2006), 3 of which take part in C1 metabolism. Labeled methylotroph populations were also analyzed phylogenetically by 16S rRNA gene DGGE fingerprints and subsequent sequencing of dominant DGGE bands. In addition to Methylocystis, the methanotrophic genera Methylobacter and Methylocella were identified together with sequences similar to Bacteroidetes and γ-Proteobacteria.
One of the main criticisms associated with SIP is considered to be inappropriately high concentrations of labeled substrates introduced during incubations compared to concentrations that occur in situ. However, these high concentrations were necessary to achieve sufficient yields of labeled DNA for metagenomic analysis. Murrell and colleagues were the first to resolve this issue (Neufeld et al., 2008). Their strategy was the application of multiple displacement amplification to “bridge the gap” between the picogram quantities of labeled DNA and required microgram quantities for subsequent metagenomic analyses. Their study found Methylophaga spp. to be involved in oceanic methanol cycling and detected a 9-kb DNA fragment that encoded for the enzymes involved in methanol dehydrogenase synthesis, regulation, and assembly. Similarly, the techniques described were used for the analysis of as yet uncultured Methylocystis-related populations in acidic peatlands (Chen et al., 2008). These populations, which had been found to be dominant in the majority of acidic peatlands sampled, were further confirmed to be actively involved in oxidizing methane by SIP-based investigations. After 13CH4 had been assimilated, 13C-labeled DNA containing the genome of Methylocystis spp. was used for a construction of a metagenomic library and screened for the presence of key methylotrophic genes. Shotgun sequencing of a clone containing methanol dehydrogenase gene permitted the researchers to assemble a gene cluster encoding polypeptides involved in methanol utilization (mxaFJGIRSAC). These reports (Dumont et al., 2006, Chen et al., 2008, Neufeld et al., 2008) were the first ones to show that retrieval of targeted genetic information can be achieved with minimal sequencing effort. At the same time, the authors proposed early sequencing of complete genomes of microbial populations directly from the environment (Dumont et al., 2006). Not long after, a nearly complete genome was obtained of a novel uncultured methylotrophic bacterium Methylotenera mobilis from the water and sediments of Lake Washington, WA, USA (Kalyuzhnaya et al., 2008). This proof-of-principle study shows that genomes of ecologically relevant subpopulations can be reassembled after whole genome shotgun sequencing of stable isotopically labeled DNA. Additional results of this study revealed several clades of bacteria involved in C1 substrate metabolism; some were traditional methylotrophs such as Methylobacter, Methylomonas, Methyloversatilis, or Ralstonia, and others clustered with Verrucomicrobia, Nitrospirae, and Planctomycetes, clades not commonly associated with methylotrophs.
SIP and metagenomics to study biodegradation of anthropogenic compounds
Some of the most widespread and environmentally significant xenobiotics are polychlorinated biphenyls (PCBs) (Breivik et al., 2002). Correspondingly, studies combining SIP and metagenomics to study PCB-degrading bacteria are common. The first such study performed DNA-SIP integrated with GeoChip-mediated metagenomic analysis of bacteria in the root zone of an Austrian pine (Pinus nigra L.) growing naturally in PCB-contaminated soil using 13C-biphenyl, a PCB analogue, as a substrate (Leigh et al., 2007). The findings of this study pointed to novel populations of biphenyl-utilizing bacteria, including Pseudonocardia, Nocardioides, Kribella, Variovorax, and Polaromonas in addition to previously known PCB-degrading Sphingomonas spp. GeoChip analyses of 13C-DNA detected 30 genes associated with organic contaminant degradation in the 13C-DNA, majority of which were associated with the degradation of aromatics, including biphenyl, benzoate, catechol, protocatechuate, naphthalene, phenol, dibenzofuran, and phenylpropionate. The presence of these genes in biphenyl-labeled populations suggests that they have the potential to degrade several aromatic substrates. In addition, genes of the β-ketoadipate pathway were detected indicating potential abilities of the populations to mineralize monoaromatics once biphenyl has been transformed into monoaromatic intermediates. Only four genes, however, were detected from the biphenyl upper pathway (bph operon) associated with rhodococci and bacilli, suggesting that only a tiny fraction of the actual diversity in the upper biphenyl pathway genes had been revealed. This hypothesis was supported by PCR amplification and sequence analyses of genes encoding for aromatic ring hydroxylating dioxygenases (ARHD), all of which shared homology but were not identical to those previously deposited in GenBank. Thus, these novel genes were undetected using the microarray. Moreover, some of the sequences did not cluster with any known ARHDs and represented a novel clade.
Sul and colleagues (2009) applied metagenomics directly to isolate a novel biphenyl dioxygenase (bphA) gene from PCB-contaminated river sediment bacteria enriched by the incubation with 13C-biphenyl. Biphenyl dioxygenase (BphAE), a multicomponent enzyme catalyzing the activation of biphenyl ring by insertion of two oxygen atoms, is crucial for biodegradation of biphenyl. Degradation of PCBs is permitted by relaxed substrate specificity of the enzyme, which has been determined to be closely connected with its primary structure (Vézina et al., 2007, 2008). The dioxygenase sequence detected by Sul et al. (2009) was highly similar to that in Pseudomonas sp. Cam-1 and Pseudomonas pseudoalcaligenes KF707. Although in most laboratory PCB-degrading strains, genes bphAE are organized in operons with other enzymes for subsequent transformation of dihydroxylated biphenyl, this clone only contained bphAE genes. The authors ascribe this phenomenon to an acquisition of the genes from another microorganism during exposure of the sediment to PCBs, possibly by horizontal gene transfer. This hypothesis was supported by a different G+C content of bphAE than the average for the cloned fragment. The activity of BphAE was tested after expression of the genes bphAE from the cosmid clone along with bphFGBC from Burkholderia xenovorans LB400. The spectrum of metabolized PCB congeners was similar to that of P. pseudoalcaligenes KF707, transforming only the congeners without chloro substitutions at the 2,3 positions.
The identification of previously characterized organisms and genes by Sul et al. (2009) may be due to the long SIP incubation and enrichment of fast-growing organisms that would be amenable to detection using cultivation-based approaches. In the future, combining direct shotgun metagenomic sequencing or function-based screening with DNA-SIP could enable the discovery of highly novel degradative genes, rather than those with similarity to known sequences as were detected with Leigh et al. (2007) and Sul et al. (2009) using PCR-based or microarray-based detection. Although Sul et al. (2009) did not discover a dioxygenase with a broader substrate specificity than had been observed previously, this study contributes to our understanding of genomic features of degradative populations. This particular case suggests that the gene organization of bph genes in nature might be scattered rather than clustered in operons. The idea of catabolic genes being dispersed on chromosomes and plasmids was supported by another paper which discusses the organization of aromatic degradation pathway genes (Suenaga et al., 2009). Thirty-eight fosmid clones were analyzed carrying genes for extradiol dioxygenases, and only two of the clones contained complete degradation pathways that are commonly found in aromatic compound-utilizing isolates. The other clones contained only subsets of the pathway genes with novel gene arrangements.
Recent results (Uhlík et al., 2012) demonstrated the ability of biphenyl-metabolizing bacteria to utilize other aromatic compounds in contaminated soil; populations of Rhodanobacter, Burkholderia, Pandoraea, Dyella and other Proteobacteria were observed to derive carbon from benzoate and naphthalene in addition to biphenyl. This study combined SIP with sequence analysis of 16S rRNA gene pyrotags amplified from 13C-DNA to identify taxa associated with the biodegradation of pollutants. Results of a few recently published bioremediation-related SIP studies reveal bacteria that had not been associated with utilization of the substrates before. Examples include newly associated populations of Pusillimonas or Rhodanobacter with the degradation of biphenyl (Lee et al., 2011, Uhlík et al., 2012) and Thermincola with the degradation of toluene (Pilloni et al., 2011). In addition, many unclassified 16S rRNA gene sequences were retrieved from 13C-DNA labeled by different substrates pointing to novel yet-to-be described bacterial taxa involved in biodegradation of biphenyl, benzoate, naphthalene, or toluene (Lee et al., 2011, Pilloni et al., 2011, Uhlík et al., 2012).
Potential contributions of SIP and metagenomics to bioremediation
Fundamental research on microbial aspects of bioremediation improves understandings of processes and could potentially improve bioremediation technologies. Thorough analysis of the labeled metagenomes of bioremediative populations can provide valuable information for assessing bioremediation potential of autochthonous microorganisms as well as designing and monitoring engineered bioremediation strategies.
Assessing bioremediation potential
Before bioremediation strategies are applied to a contaminated site, the bioremediation potential of the indigenous microflora should be assessed. In this case, SIP can be valuable for determining whether organisms capable of metabolizing the contaminant are already present at the site. If so, then biostimulation would likely be a viable bioremediation strategy. For these purposes, SIP incubations can be performed either in vitro using microcosms constructed from field-collected samples (Leigh et al., 2007, Uhlík et al., 2009b, Winderl et al., 2010) or directly in situ (Padmanabhan et al., 2003, DeRito et al., 2005, Mahmood et al., 2005, Liou et al., 2008, Pumphrey and Madsen, 2008, Bombach et al., 2010). Metagenomic functional gene analyses of SIP studies are particularly valuable in the case of biodegradation of xenobiotics that occur as mixtures, such as PCBs, since biphenyl dioxygenase enzymes vary widely in their capability to degrade different lower chlorinated PCB congeners (Erickson and Mondello, 1993, Mondello et al., 1997). Analyzing the SIP-labeled dioxygenase gene sequence in relation to known enzymes with known substrate specificities can help predict which of the congeners are likely to be degraded by the microbial community (Barriault et al., 2002, Vézina et al., 2008). The absence or inactivity of dioxygenases with appropriate congener specificity for the PCBs on-site might indicate that an anaerobic treatment to promote dehalogenation would be appropriate (Wiegel and Wu, 2000, Smidt and de Vos, 2004). In addition, metagenomic exploration of active populations can clarify the metabolic capabilities as well as regulatory mechanisms within microbes, such as through sequence-based detection of regulatory elements, and these properties can be subjected to genetic manipulations with the aim of improving the efficacy of bioremediation through bioaugmentation.
Bioaugmentation
SIP is ideal for identifying microbes optimal for bioaugmentation for several reasons. First, SIP-metagenomics has the potential to reveal the identity of microbial species that are metabolically active under a variety of environmental conditions (type of matrix, temperature, moisture, oxygen levels, etc.) or that derive carbon from more than one contaminant. Second, once bioaugmentation with exogenous organisms has been applied on a contaminated site, SIP can help determine whether the added microorganisms have adapted to new conditions and survived and, more importantly, if they are actively biodegrading the contaminant(s).
Understanding the mechanisms underlying biostimulation and phytoremediation technologies
Biostimulation methods seek to address limitations in environmental conditions and/or bioavailability in order to enable indigenous microbial communities to more rapidly biodegrade the contaminant. One approach to biostimulation is the use of plants to promote biodegradative activity in the root zone, which is a form of phytoremediation known as rhizoremediation (Kuiper et al., 2004, Macková et al., 2006, Gerhardt et al., 2009, Macek et al., 2009). Plants can rhizostimulate contaminant biodegradation through a variety of mechanisms. Root exudation and root decay provides rhizosphere microorganisms with growth substrates and secondary compounds that may function as cometabolites or inducers of biodegradative pathways, or surfactants and phytochemicals that increase the bioavailability of poorly soluble pollutants (Leigh et al., 2002, Bertin et al., 2003, Singer et al., 2003, Singer et al., 2004, Leigh et al., 2006, Yi and Crowley, 2007, Toussaint et al., 2012). In addition, the rhizosphere is also richer in oxygen essential for the activity of aerobic organisms and their dioxygenases and monooxygenases that are often involved in biodegradation processes of organic contaminants (Leigh et al., 2002). Understanding how the microbial community responds to biostimulation is important to improving bioremediation technologies. SIP and metagenomics approaches could provide novel and important insight into the mechanisms underlying rhizoremediation (Prosser et al., 2006) by identifying contaminant degraders active under different environmental and biostimulatory conditions.
Plants also host many bacteria in their endosphere. Emerging research indicates that they contribute to biodegradation of toxic organic compounds in contaminated soil and could have potential for improving phytoremediation (Newman and Reynolds, 2005, McGuinness and Dowling, 2009, Weyens et al., 2009, Weyens et al., 2011). Although no studies have as of yet employed SIP-metagenomics to study microbial metabolism inside plants, this topic is likely to be investigated with respect to bioremediation in the future.
Some plants contribute to remediation more directly, by taking up and transforming contaminants or storing them in above-ground parts (Macek et al., 2000). Studies have been published showing that plants, however, have limited abilities to mineralize pollutants. In case of PCBs, for instance, monohydroxylated and/or dihydroxylated derivatives are formed without the aromatic ring being cleaved (Rezek et al., 2008). In connection with improving phytoremediation efficiency, novel genes revealed by SIP-metagenomics can be used in order to prepare genetically modified plants with abilities to cleave aromatic structures (Macek et al., 2008, Nováková et al., 2009, Sylvestre et al., 2009, Van Aken et al., 2010).
SIP appears to be an ideal tool for use during laboratory tests evaluating how different biostimulation strategies alter biodegradative microbial populations, and the resultant information could also potentially be useful for creating and monitoring engineered bioremediation treatments.
Carbon flow through contaminated systems
One of the major drawbacks of DNA-SIP is the detection of cross-feeding populations in addition to primary utilizers of a substrate (Neufeld et al., 2007c). Experimental data have shown a very rapid sequestration of carbon in some microbial communities (Lueders et al., 2006). In addition, mineralization of 13C-labeled substrates generates 13CO2, which can further be taken up by autotrophic bacteria and incorporated into their biomass biasing thereby the conclusions of a study. Therefore, different strategies have been suggested to minimize the effect of cross-feeding detection by DNA-SIP or to clearly distinguish it from primary utilization. These include time-course experiments (Leigh et al., 2007), shortening the incubation times and addition of 13C-carrier DNA in the density gradient separation (Gallagher et al., 2005), or combination of DNA-SIP with RNA-SIP (Dumont et al., 2011). Yet in the context of bioremediation, cross-feeding populations may also be important contributors. For instance, intermediate metabolites or incompletely-degraded substrates released by the biodegradative population may be removed from the environment by cross-feeding microbes. Monitoring cross-feeding also has the potential to identify all microorganisms essential to complete degradation of contaminants and to reveal the flow of carbon through the microbial food web in contaminated environments.
Recently, a new SIP methodology has been developed which is ideal for determining carbon flow through food webs. Known as stable isotope switching (SIS) (Maxfield et al., 2012), the approach is based on the incubation of the matrix with 13C-labeled substrate that is switched for an unlabeled substrate when full labeling is achieved. Such a setting allows for monitoring carbon uptake, turnover, release, and sequestration.
Limitations
Despite the great potential contribution of SIP and metagenomics to microbial ecology and biotechnology, one has to realistically evaluate the limitations associated with each of the approaches. A major challenge associated with SIP is a very limited availability and high cost of labeled substrates. SIP is also very labor-intensive and low-throughput using current techniques. These factors may prohibit performing a sufficient number of replicates to enable statistically valid comparisons among treatments. In the future, the use of robotics and automation could assist in increasing the throughput of SIP gradient fractionation and assessments. Another challenge of SIP is the need to combine many techniques and technologies spanning many disciplines, which may be an obstacle for researchers without experience in these techniques (reviewed by Chen and Murrell, 2010).
When designing SIP-based experiments, it is also important to use realistic substrate concentrations and incubation times that are of appropriate duration to enable labeling yet minimize potential cross-feeding and over-enrichment (Neufeld et al., 2007a). Inappropriately high concentrations of a labeled substrate can lead to population shifts, such as opportunistic growth of some degraders, suppression of populations adapted to only low concentrations of the substrate, or shifts caused by accumulation of inhibitory or toxic intermediates.
It has been proposed earlier (Madsen, 2006) that, especially for bioremediation purposes, SIP assay conditions should ideally match the conditions in the field. Field conditions are constantly changing (temperature, humidity, etc.) and may be affected by larger scale processes such as living plant roots, and these factors are challenging to adequately represent under laboratory microcosms. Microcosms often create other unrealistic conditions associated with depletion of oxygen, increased concentrations of CO2, unrealistic substrate concentrations, or unrealistic availability of the substrate (Friedrich, 2006). Therefore, laboratory microcosms need to be recognized as model systems, with the caveat that they will not fully represent field conditions. Thorough bioremediation surveys require either performing SIP directly in situ (Padmanabhan et al., 2003, DeRito et al., 2005, Mahmood et al., 2005, Liou et al., 2008, Pumphrey and Madsen, 2008, Bombach et al., 2010) or a very careful design and monitoring of microcosm experiments.
One of the major limitations of SIP-metagenomics for bioremediation purposes is the restriction of its utility solely to assimilatory processes (Andreoni and Gianfreda, 2007). Yet many bioremediation processes are dissimilatory, for example, with the contaminant serving as the terminal electron acceptor. Many anthropogenic compounds are halogenated (polychlorinated dioxins, dibenzofurans, biphenyls, or chlorinated solvents), and their bioremediation relies on dehalorespiration (i.e., utilization of a halogenated compound as a terminal electron acceptor to yield energy) (Futagami et al., 2008). These processes can be monitored through different stable isotope analyses (for review, see Ruess and Chamberlain, 2010, Boecklen et al., 2011, Braeckevelt et al., 2012).
Metagenomics techniques are also not exempt from limitations. Rapidly advancing high-throughput sequencing technologies have substantially reduced the cost of sequencing efforts over traditional Sanger sequencing. Sequence-based screening of metagenomic data, however, relies on databases that are far from being completely or accurately annotated. The incompleteness of genetic databases means that about 30% of the genes sequenced can remain unassigned (Harrington et al., 2007), which prevents metagenomics from achieving its full potential. Other potential problems are associated with the function-based screening of metagenomic libraries. These include the necessity of analyzing a large number of clones to recover positive ones (Sul et al., 2009), possible inabilities of the host cells (usually E. coli) to express foreign genes and to form active proteins, or problems associated with cloning of large gene clusters which requires high-molecular weight DNA of high purity (Daniel, 2005, Simon and Daniel, 2009). At the same time, problems associated with gene-targeted metagenomics have been identified. These include errors introduced during PCR by DNA polymerase, generation of chimeric sequences when heterologous templates are used, or errors introduced during pyrosequencing. As a result of these phenomena, diversity is overestimated (Kunin et al., 2010) unless appropriate tools are used for sequence processing. Introduction of several denoising algorithms (Quince et al., 2009, Reeder and Knight, 2010, Quince et al., 2011), improved operational taxonomic unit (OTU) clustering (Huse et al., 2010), or their combination (Schloss et al., 2011, Uhlík et al., 2012) have resulted in much more accurate diversity estimations. Additionally, the most commonly used phylogenetic marker gene is 16S rRNA, which is present in different copy numbers in different species, ranging from 1 to 15 (Pei et al., 2010). As a result, the relative abundance of 16S rRNA genes in a sequence library does not necessarily correspond to the number of cells, and may distort the apparent community structure.
New frontiers of SIP
Realizing the limitations of DNA-SIP and metagenomics gives rise to alternative approaches being considered. One of these is SIP-metatranscriptomics. mRNA-SIP benefits from high sensitivity and the fact that mRNA bears functional information. Studies carried out so far by Huang et al. (2009) and Dumont et al. (2011) focused on one transcript each – naphthalene dioxygenase and methane monooxygenase, respectively. The rapidly advancing field of metatranscriptomics (Filiatrault, 2011, Simon and Daniel, 2011, Su et al., 2012) seems to have overcome some of the major obstacles linked with mRNA, such as low recovery of high-quality environmental transcripts, instability of mRNA, or difficulties in separation of mRNA from other RNA species (Mou et al., 2011, Ottesen et al., 2011, Feike et al., 2012, Marchetti et al., 2012, Rinta-Kanto et al., 2012, Stewart et al., 2012). Therefore, it is only a matter of time before mRNA-SIP is combined with metatranscriptomic analyses to provide comprehensive information on the actively transcribed genes associated with the utilization of a specific substrate.
Another group of biomarkers bearing both phylogenetic and functional information are proteins. Protein-SIP was initially applied to track carbon flow in pure cultures (Jehmlich et al., 2008a, Jehmlich et al., 2008b), however the authors proposed its use for the analysis of phylogenetically diverse microbial communities as well, which was reported shortly thereafter (Bastida et al., 2010, Bastida et al., 2011, Pan et al., 2011). More recently, protein-SIP was used for quantitative analysis of induced proteins in substrate shift experiments (Taubert et al., 2011). Protein-SIP is currently very challenging due to the inability to assign even a putative function to many proteins. Yet protein-SIP is very promising as it may provide a more substantial access to real microbial activity, since proteins are the most explicit indicators of metabolic activity. Therefore, in the future, with more complete databases, better modelling systems, and further development of metaproteomics techniques, protein-SIP is expected to widely expand and advance the field of microbial ecology.
Alternatives to SIP
Although SIP is the main focus of this review, other technologies expanding the scope of isotope labeling experiments have been developed that can potentially link microbial phylogeny with metabolic activity (Gutierrez-Zamora and Manefield, 2010). Most of these methods involve fluorescence in situ hybridization (FISH), which is a technique commonly applied to taxonomically identify microbial cells using rRNA-targeted oligonucleotide probes (Wagner et al., 2003). FISH developed into a function-identity method after it was combined with (i) microautoradiography (FISH-MAR), which uses radioactive isotopes to monitor microbial uptake of labeled substrates (Lee et al., 1999); (ii) secondary ion mass spectrometry (FISH-SIMS) and nanometer-scale secondary ion mass spectrometry (FISH-nanoSIMS), which determines the isotopic composition of the FISH-identified targeted cells (Orphan et al., 2001, Kuypers and Jørgensen, 2007, Li et al., 2008); (iii) Raman microspectroscopy (Raman-FISH), which detects vibrational shifts of covalent bonds in molecules of FISH-labeled cells (Huang et al., 2007); or (iv) immunomagnetic cell capture (magneto-FISH), which permits targeted magnetic capture of FISH-labeled microorganisms and cell aggregates (Pernthaler et al., 2008). Behrens et al. (2008) reported a new combination of FISH and nanoSIMS for linking microbial phylogeny to metabolic activity at the single cell level by applying enhanced element labeling of microbial cells by FISH (EL-FISH). Such a modification allowed them to increase the sensitivity of cell detection and broadened the applicability of the methodology for environmental studies. Some of these techniques, mainly FISH-MAR, have been employed in bioremediation studies (Yang et al., 2003, Hesselsoe et al., 2008).
In addition to FISH, other isotope-based techniques have been used such as isotope arrays (Adamczyk et al., 2003) or shotgun isotope arrays (Tobino et al., 2011), small subunit-isotope ratio mass spectrometry (SSU-IRMS) (MacGregor et al., 2002, MacGregor et al., 2006), or radioactive isotope probing (RIP) (Nikolausz et al., 2007). The advantages and limitations of these methods as well as their comparison to SIP are discussed elsewhere (Wagner et al., 2006, Neufeld et al., 2007c, Gutierrez-Zamora and Manefield, 2010).
Recently some FISH-based methods have been combined with SIP in order to link microbial phylogeny to metabolic activity at the single-cell level. For example, Huang and colleagues combined rRNA and mRNA-SIP with single-cell Raman-FISH (Huang et al., 2009). The main advantage of such a combined approach is that it allows for determination and quantification of in situ functions of a microbial community. RNA-SIP has also been combined with magnetic-bead capture hybridization (MacGregor et al., 2006, Miyatake et al., 2009). This approach is applicable for linking phylogeny with metabolic activity at the level of class or family while requiring approximately 10,000 times less stable isotope enrichment of RNA, allowing thus the use of environmentally relevant concentrations of the isotopically labeled target substrate. The principle of RNA-SIP combined with magnetic-bead capture hybridization lies in extracting RNA from the matrix after it has been stable isotope labeled, hybridizing the RNA with probes, capturing those hybrids with beads, and collecting beads with a magnet. Captured rRNA is then released and analyzed by isotope ratio mass spectrometry (IRMS). This approach thus eliminates possible biases associated with the use of artificially high substrate concentrations during incubations relative to those in situ, which is one of the major criticisms of SIP (Dumont and Murrell, 2005, Neufeld et al., 2008).
Conclusions
Since the first development of SIP more than a decade ago (Boschker et al., 1998, Radajewski et al., 2000), this technique has progressed rapidly and has significantly broadened the field of microbial ecology, first by linking phylogenetic identity with function, including that of novel clades with no cultured representatives, and later by linking function with novel functional gene sequences. The recent integration of SIP with metagenomics has enabled a more comprehensive understanding of the functional community dynamics of entire microbial systems. An improved mechanistic understanding of microbial ecological function has the potential to enable new breakthroughs in bioremediation technologies such as biostimulation, bioaugmentation and phytoremediation. The great advantage of SIP is its ability to enable a focused detection and analysis of only the organisms active in the utilization of a specific substrate, either directly or indirectly through the food web. In the context of bioremediation, SIP-metagenomics is invaluable for revealing the identity of contaminant-degraders and their functional genes, and this information may be used to assess their response to biostimulation methods or to identify organisms and genes useful for bioaugmentation. As with any method, SIP has limitations, such as false positives, in which cross-feeding organisms may be mistaken for primary utilizers, and the potential for microbial community change during incubations. However if these factors are taken into account, SIP combined with metagenomics has the potential to provide novel insights to the intricate interactions within microbial ecosystems. Limitations like cross-feeding can also be advantageous, providing valuable new insight into the flow of carbon derived from contaminants through the microbial food web. As cultivation-independent methods like SIP and metagenomic studies advance, it is important to note that cultivation-based techniques are still a crucial means to verify and investigate the physiology and genetics of individual contaminant-degrading microorganisms, to facilitate bioaugmentation, and to enable improved annotation of metagenomic databases. The integration of SIP with the rapidly advancing field of metagenomics is opening new windows into microbial processes and interactions, which may enable major breakthroughs in the field of bioremediation.
Acknowledgements
The authors acknowledge the support of Czech Ministry of Education, Youth and Sports (grant ME09024), and the 7th Framework Programme (grant MINOTAURUS, no: 265946). This publication was made possible by grants from the national center for research resources (5P20RR016466-12) and the national institute of general medical sciences (8P20GM103395-12) from the national institutes of health. MCL acknowledges the David L. Boren National Security Education Program (NSEP) Fellowship for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Ondrej Uhlik, Email: ondrej.uhlik@vscht.cz.
Mary-Cathrine Leewis, Email: mcleewis@alaska.edu.
Michal Strejcek, Email: michal.strejcek@vscht.cz.
Lucie Musilova, Email: lucie.musilova@vscht.cz.
Martina Mackova, Email: martina.mackova@vscht.cz.
Mary Beth Leigh, Email: mbleigh@alaska.edu.
Tomas Macek, Email: tomas.macek@vscht.cz.
References
- Aanderud ZT, Lennon JT. Validation of heavy-water stable isotope probing for the characterization of rapidly responding soil bacteria. Appl Environ Microbiol. 2011;77:4589–4596. doi: 10.1128/AEM.02735-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamczyk J, Hesselsoe M, Iversen N, Horn M, Lehner A, Nielsen PH, Schloter M, Roslev P, Wagner M. The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl Environ Microbiol. 2003;69:6875–6887. doi: 10.1128/AEM.69.11.6875-6887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoni V, Gianfreda L. Bioremediation and monitoring of aromatic-polluted habitats. Appl Microbiol Biotechnol. 2007;76:287–308. doi: 10.1007/s00253-007-1018-5. [DOI] [PubMed] [Google Scholar]
- Arai K, Tsubone T, Takechi T, Inoue T. Bioremediation of trichloroethylene and cis-1,2-dichloroethylene-contaminated groundwater by methane-utilizing bacteria. J Vet Med Sci. 1999;61:861–863. doi: 10.1292/jvms.61.861. [DOI] [PubMed] [Google Scholar]
- Barriault D, Plante MM, Sylvestre M. Family shuffling of a targeted bphA region to engineer biphenyl dioxygenase. J Bacteriol. 2002;184:3794–3800. doi: 10.1128/JB.184.14.3794-3800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastida F, Jechalke S, Bombach P, Franchini AG, Seifert J, von Bergen M, Vogt C, Richnow HH. Assimilation of benzene carbon through multiple trophic levels traced by different stable isotope probing methodologies. FEMS Microbiol Ecol. 2011;77:357–369. doi: 10.1111/j.1574-6941.2011.01118.x. [DOI] [PubMed] [Google Scholar]
- Bastida F, Rosell M, Franchini AG, Seifert J, Finsterbusch S, Jehmlich N, Jechalke S, Von Bergen M, Richnow HH. Elucidating MTBE degradation in a mixed consortium using a multidisciplinary approach. FEMS Microbiol Ecol. 2010;73:370–384. doi: 10.1111/j.1574-6941.2010.00889.x. [DOI] [PubMed] [Google Scholar]
- Behrens S, Losekann T, Pett-Ridge J, Weber PK, Ng WO, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol. 2008;74:3143–3150. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TH, Yergeau E, Martineau C, Juck D, Whyte LG, Greer CW. Identification of nitrogen-incorporating bacteria in petroleum-contaminated Arctic soils by using 15N DNA-based stable isotope probing and pyrosequencing. Appl Environ Microbiol. 2011;77:4163–4171. doi: 10.1128/AEM.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin C, Yang XH, Weston LA. The role of root exudates and allelochemicals in the rhizosphere. Plant Soil. 2003;256:67–83. [Google Scholar]
- Binga EK, Lasken RS, Neufeld JD. Something from (almost) nothing: the impact of multiple displacement amplification on microbial ecology. ISME J. 2008;2:233–241. doi: 10.1038/ismej.2008.10. [DOI] [PubMed] [Google Scholar]
- Boecklen WJ, Yarnes CT, Cook BA, James AC. On the use of stable isotopes in trophic ecology. Annu Rev Ecol Evol Syst. 2011;42:411–440. [Google Scholar]
- Bombach P, Chatzinotas A, Neu TR, Kästner M, Lueders T, Vogt C. Enrichment and characterization of a sulfate-reducing toluene-degrading microbial consortium by combining in situ microcosms and stable isotope probing techniques. FEMS Microbiol Ecol. 2010;71:237–246. doi: 10.1111/j.1574-6941.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- Boschker HTS, Nold SC, Wellsbury P, Bos D, de Graaf W, Pel R, Parkes RJ, Cappenberg TE. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature. 1998;392:801–805. [Google Scholar]
- Braeckevelt M, Fischer A, Kaestner M. Field applicability of Compound-Specific Isotope Analysis (CSIA) for characterization and quantification of in situ contaminant degradation in aquifers. Appl Microbiol Biotechnol. 2012;94:1401–1421. doi: 10.1007/s00253-012-4077-1. [DOI] [PubMed] [Google Scholar]
- Breivik K, Sweetman A, Pacyna JM, Jones KC. Towards a global historical emission inventory for selected PCB congeners - a mass balance approach. 1. Global production and consumption. Sci Total Environ. 2002;290:181–198. doi: 10.1016/s0048-9697(01)01075-0. [DOI] [PubMed] [Google Scholar]
- Brodie EL, Desantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, Hazen TC, Richardson PM, Herman DJ, Tokunaga TK, Wan JM, Firestone MK. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DH, Huangyutitham V, Hsu SF, Nelson TA. Stable isotope probing with 15N2reveals novel noncultivated diazotrophs in soil. Appl Environ Microbiol. 2007a;73:3196–3204. doi: 10.1128/AEM.02610-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DH, Huangyutitham V, Hsu SF, Nelson TA. Stable isotope probing with 15N achieved by disentangling the effects of genome G+C content and isotope enrichment on DNA density. Appl Environ Microbiol. 2007b;73:3189–3195. doi: 10.1128/AEM.02609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dumont MG, Neufeld JD, Bodrossy L, Stralis-Pavese N, McNamara NP, Ostle N, Briones MJ, Murrell JC. Revealing the uncultivated majority: combining DNA stable-isotope probing, multiple displacement amplification and metagenomic analyses of uncultivated Methylocystis in acidic peatlands. Environ Microbiol. 2008;10:2609–2622. doi: 10.1111/j.1462-2920.2008.01683.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Murrell JC. When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 2010;18:157–163. doi: 10.1016/j.tim.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Daniel R. The metagenomics of soil. Nat Rev Microbiol. 2005;3:470–478. doi: 10.1038/nrmicro1160. [DOI] [PubMed] [Google Scholar]
- DeAngelis KM, Wu CH, Beller HR, Brodie EL, Chakraborty R, DeSantis TZ, Fortney JL, Hazen TC, Osman SR, Singer ME, Tom LM, Andersen GL. PCR Amplification-Independent Methods for Detection of Microbial Communities by the High-Density Microarray PhyloChip. Appl Environ Microbiol. 2011;77:6313–6322. doi: 10.1128/AEM.05262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRito CM, Pumphrey GM, Madsen EL. Use of field-based stable isotope probing to identify adapted populations and track carbon flow through a phenol-degrading soil microbial community. Appl Environ Microbiol. 2005;71:7858–7865. doi: 10.1128/AEM.71.12.7858-7865.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz E. Bacterial degradation of aromatic pollutants: a paradigm of metabolic versatility. Int Microbiol. 2004;7:173–180. [PubMed] [Google Scholar]
- Dumont MG, Murrell JC. Stable isotope probing - linking microbial identity to function. Nat Rev Microbiol. 2005;3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- Dumont MG, Pommerenke B, Casper P, Conrad R. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ Microbiol. 2011;13:1153–1167. doi: 10.1111/j.1462-2920.2010.02415.x. [DOI] [PubMed] [Google Scholar]
- Dumont MG, Radajewski SM, Miguez CB, McDonald IR, Murrell JC. Identification of a complete methane monooxygenase operon from soil by combining stable isotope probing and metagenomic analysis. Environ Microbiol. 2006;8:1240–1250. doi: 10.1111/j.1462-2920.2006.01018.x. [DOI] [PubMed] [Google Scholar]
- Dunford EA, Neufeld JD. DNA stable-isotope probing (DNA-SIP) J Vis Exp. 2010;42:2027. doi: 10.3791/2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM, Saar MO, Alexander S, Alexander EC, Rohwer F. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics. 2006;7 doi: 10.1186/1471-2164-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson BD, Mondello FJ. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl Environ Microbiol. 1993;59:3858–3862. doi: 10.1128/aem.59.11.3858-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feike J, Jurgens K, Hollibaugh JT, Kruger S, Jost G, Labrenz M. Measuring unbiased metatranscriptomics in suboxic waters of the central Baltic Sea using a new in situ fixation system. ISME J. 2012;6:461–470. doi: 10.1038/ismej.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filiatrault MJ. Progress in prokaryotic transcriptomics. Curr Opin Microbiol. 2011;14:579–586. doi: 10.1016/j.mib.2011.07.023. [DOI] [PubMed] [Google Scholar]
- Friedrich MW. Stable-isotope probing of DNA: insights into the function of uncultivated microorganisms from isotopically labeled metagenomes. Curr Opin Biotechnol. 2006;17:59–66. doi: 10.1016/j.copbio.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Futagami T, Goto M, Furukawa K. Biochemical and genetic bases of dehalorespiration. Chem Rec. 2008;8:1–12. doi: 10.1002/tcr.20134. [DOI] [PubMed] [Google Scholar]
- Gallagher E, McGuinness L, Phelps C, Young LY, Kerkhof LJ. 13C-carrier DNA shortens the incubation time needed to detect benzoate-utilizing denitrifying bacteria by stable-isotope probing. Appl Environ Microbiol. 2005;71:5192–5196. doi: 10.1128/AEM.71.9.5192-5196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffrey MG. Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology. 2010;156:609–643. doi: 10.1099/mic.0.037143-0. [DOI] [PubMed] [Google Scholar]
- Gerhardt KE, Huang XD, Glick BR, Greenberg BM. Phytoremediation and rhizoremediation of organic soil contaminants: Potential and challenges. Plant Sci. 2009;176:20–30. [Google Scholar]
- Gutierrez-Zamora ML, Manefield M. An appraisal of methods for linking environmental processes to specific microbial taxa. Rev Environ Sci Biotechnol. 2010;9:153–185. [Google Scholar]
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol. 1998;5:R245–R249. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- Harrington ED, Singh AH, Doerks T, Letunic I, von Mering C, Jensen LJ, Raes J, Bork P. Quantitative assessment of protein function prediction from metagenomics shotgun sequences. Proc Natl Acad Sci USA. 2007;104:13913–13918. doi: 10.1073/pnas.0702636104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D'Haeseleer P, Holman HYN, Osman S, Lu ZM, Van Nostrand JD, Deng Y, Zhou JZ, Mason OU. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, Li X, Wu L, Gentry TJ, Yin Y, Liebich J, Hazen TC, Zhou J. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010;4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, Huang Z, Wu W, Gu B, Jardine P, Criddle C, Zhou J. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 2007;1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- Hesselsoe M, Bjerring ML, Henriksen K, Loll P, Nielsen JL. Method for measuring substrate preferences by individual members of microbial consortia proposed for bioaugmentation. Biodegradation. 2008;19:621–633. doi: 10.1007/s10532-007-9167-x. [DOI] [PubMed] [Google Scholar]
- Huang WE, Ferguson A, Singer AC, Lawson K, Thompson IP, Kalin RM, Larkin MJ, Bailey MJ, Whiteley AS. Resolving genetic functions within microbial populations: in situ analyses using rRNA and mRNA stable isotope probing coupled with single-cell raman-fluorescence in situ hybridization. Appl Environ Microbiol. 2009;75:234–241. doi: 10.1128/AEM.01861-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WE, Stoecker K, Griffiths R, Newbold L, Daims H, Whiteley AS, Wagner M. Raman-FISH: combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol. 2007;9:1878–1889. doi: 10.1111/j.1462-2920.2007.01352.x. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S, Chai B, Jesus EdC, Penton CR, Lee TK, Cole JR, Tiedje JM. Gene-Targeted Metagenomics (GT Metagenomics) to Explore the Extensive Diversity of Genes of Interest in Microbial Communities. Handbook of Molecular Microbial Ecology I: John Wiley & Sons, Inc.; 2011. pp. 235–243. [Google Scholar]
- Jehmlich N, Schmidt F, Hartwich M, von Bergen M, Richnow HH, Vogt C. Incorporation of carbon and nitrogen atoms into proteins measured by protein-based stable isotope probing (Protein-SIP) Rapid Commun Mass Spectrom. 2008a;22:2889–2897. doi: 10.1002/rcm.3684. [DOI] [PubMed] [Google Scholar]
- Jehmlich N, Schmidt F, Taubert M, Seifert J, von Bergen M, Richnow HH, Vogt C. Comparison of methods for simultaneous identification of bacterial species and determination of metabolic activity by protein-based stable isotope probing (Protein-SIP) experiments. Rapid Commun Mass Spectrom. 2009;23:1871–1878. doi: 10.1002/rcm.4084. [DOI] [PubMed] [Google Scholar]
- Jehmlich N, Schmidt F, von Bergen M, Richnow HH, Vogt C. Protein-based stable isotope probing (Protein-SIP) reveals active species within anoxic mixed cultures. ISME J. 2008b;2:1122–1133. doi: 10.1038/ismej.2008.64. [DOI] [PubMed] [Google Scholar]
- Jørgensen KS. In Situ Bioremediation. Adv Appl Microbiol. 2007;61:285–305. doi: 10.1016/S0065-2164(06)61008-3. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Lapidus A, Ivanova N, Copeland AC, McHardy AC, Szeto E, Salamov A, Grigoriev IV, Suciu D, Levine SR, Markowitz VM, Rigoutsos I, Tringe SG, Bruce DC, Richardson PM, Lidstrom ME, Chistoserdova L. High-resolution metagenomics targets specific functional types in complex microbial communities. Nat Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- Kane SR, Chakicherla AY, Chain PS, Schmidt R, Shin MW, Legler TC, Scow KM, Larimer FW, Lucas SM, Richardson PM, Hristova KR. Whole-genome analysis of the methyl tert-butyl ether-degrading beta-proteobacterium Methylibium petroleiphilum PM1. J Bacteriol. 2007;189:1931–1945. doi: 10.1128/JB.01259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper I, Lagendijk EL, Bloemberg GV, Lugtenberg BJ. Rhizoremediation: a beneficial plant-microbe interaction. Mol Plant Microbe Interact. 2004;17:6–15. doi: 10.1094/MPMI.2004.17.1.6. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Kuypers MMM, Jørgensen BB. The future of single-cell environmental microbiology. Environ Microbiol. 2007;9:6–7. doi: 10.1111/j.1462-2920.2006.01222_5.x. [DOI] [PubMed] [Google Scholar]
- Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer KH, Wagner M. Combination of fluorescent in situ hybridization and microautoradiography - a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol. 1999;65:1289–1297. doi: 10.1128/aem.65.3.1289-1297.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TK, Lee J, Sul WJ, Iwai S, Chai BC, Tiedje JM, Park J. Novel biphenyl-oxidizing bacteria and dioxygenase genes from a Korean tidal mudflat. Appl Environ Microbiol. 2011;77:3888–3891. doi: 10.1128/AEM.00023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh MB, Fletcher JS, Fu X, Schmitz FJ. Root turnover: an important source of microbial substrates in rhizosphere remediation of recalcitrant contaminants. Environ Sci Technol. 2002;36:1579–1583. doi: 10.1021/es015702i. [DOI] [PubMed] [Google Scholar]
- Leigh MB, Pellizari VH, Uhlík O, Sutka R, Rodrigues J, Ostrom NE, Zhou J, Tiedje JM. Biphenyl-utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs) ISME J. 2007;1:134–148. doi: 10.1038/ismej.2007.26. [DOI] [PubMed] [Google Scholar]
- Leigh MB, Prouzová P, Macková M, Macek T, Nagle DP, Fletcher JS. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol. 2006;72:2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Wu TD, Mazeas L, Toffin L, Guerquin-Kern JL, Leblon G, Bouchez T. Simultaneous analysis of microbial identity and function using NanoSIMS. Environ Microbiol. 2008;10:580–588. doi: 10.1111/j.1462-2920.2007.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou JS, DeRito CM, Madsen EL. Field-based and laboratory stable isotope probing surveys of the identities of both aerobic and anaerobic benzene-metabolizing microorganisms in freshwater sediment. Environ Microbiol. 2008;10:1964–1977. doi: 10.1111/j.1462-2920.2008.01612.x. [DOI] [PubMed] [Google Scholar]
- Little CD, Palumbo AV, Herbes SE, Lidstrom ME, Tyndall RL, Gilmer PJ. Trichloroethylene biodegradation by a methane-oxidizing bacterium. Appl Environ Microbiol. 1988;54:951–956. doi: 10.1128/aem.54.4.951-956.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Suflita JM. Ecology and evolution of microbial populations for bioremediation. Trends Biotechnol. 1993;11:344–352. doi: 10.1016/0167-7799(93)90157-5. [DOI] [PubMed] [Google Scholar]
- Lovley DR. Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol. 2003;1:35–44. doi: 10.1038/nrmicro731. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. Species divergence and the measurement of microbial diversity. FEMS Microbiol Rev. 2008;32:557–578. doi: 10.1111/j.1574-6976.2008.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZM, Deng Y, Van Nostrand JD, He ZL, Voordeckers J, Zhou AF, Lee YJ, Mason OU, Dubinsky EA, Chavarria KL, Tom LM, Fortney JL, Lamendella R, Jansson JK, D'Haeseleer P, Hazen TC, Zhou JZ. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J. 2012;6:451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders T, Kindler R, Miltner A, Friedrich MW, Kaestner M. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl Environ Microbiol. 2006;72:5342–5348. doi: 10.1128/AEM.00400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macek T, Kotrba P, Svatoš A, Nováková M, Demnerová K, Macková M. Novel roles for genetically modified plants in environmental protection. Trends Biotechnol. 2008;26:146–152. doi: 10.1016/j.tibtech.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Macek T, Macková M, Káš J. Exploitation of plants for the removal of organics in environmental remediation. Biotechnol Adv. 2000;18:23–34. doi: 10.1016/s0734-9750(99)00034-8. [DOI] [PubMed] [Google Scholar]
- Macek T, Uhlík O, Ječná K, Nováková M, Lovecká P, Rezek J, Dudková V, Štursa P, Vrchotová B, Pavlíková D, Demnerová K, Macková M. Advances in phytoremediation and rhizoremediation. In: Varma A, editor. Soil Biology. 17 ed. Berlin, Germany: Springer; 2009. pp. 257–277. [Google Scholar]
- MacGregor BJ, Boschker HT, Amann R. Comparison of rRNA and polar-lipid-derived fatty acid biomarkers for assessment of 13C-substrate incorporation by microorganisms in marine sediments. Appl Environ Microbiol. 2006;72:5246–5253. doi: 10.1128/AEM.00423-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor BJ, Bruchert V, Fleischer S, Amann R. Isolation of small-subunit rRNA for stable isotopic characterization. Environ Microbiol. 2002;4:451–464. doi: 10.1046/j.1462-2920.2002.00324.x. [DOI] [PubMed] [Google Scholar]
- Macková M, Dowling D, Macek T. Focus on Biotechnology. Dordrecht, Netherlands: Springer; 2006. Phytoremediation and Rhizoremediation. Theoretical Background; p. 300. [Google Scholar]
- Madsen EL. The use of stable isotope probing techniques in bioreactor and field studies on bioremediation. Curr Opin Biotechnol. 2006;17:92–97. doi: 10.1016/j.copbio.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Madsen EL. Stable Isotope Probing Techniques and Bioremediation. In: Murrell JC, Whiteley A, editors. Stable Isotope Probing and Related Technologies. Washington, DC: ASM Press; 2010. pp. 165–202. [Google Scholar]
- Mahmood S, Paton GI, Prosser JI. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ Microbiol. 2005;7:1349–1360. doi: 10.1111/j.1462-2920.2005.00822.x. [DOI] [PubMed] [Google Scholar]
- Manefield M, Whiteley AS, Bailey MJ. What can stable isotope probing do for bioremediation? Int Biodeterior Biodegrad. 2004;54:163–166. [Google Scholar]
- Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol. 2002;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, Morales R, Allen AE, Armbrust EV. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci USA. 2012;109:E317–E325. doi: 10.1073/pnas.1118408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield PJ, Dildar N, Hornibrook ERC, Stott AW, Evershed RP. Stable isotope switching (SIS): a new stable isotope probing (SIP) approach to determine carbon flow in the soil food web and dynamics in organic matter pools. Rapid Commun Mass Spectrom. 2012;26:997–1004. doi: 10.1002/rcm.6172. [DOI] [PubMed] [Google Scholar]
- McGuinness M, Dowling D. Plant-associated bacterial degradation of toxic organic compounds in soil. Int J Environ Res Public Health. 2009;6:2226–2247. doi: 10.3390/ijerph6082226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyatake T, MacGregor BJ, Boschker HT. Linking microbial community function to phylogeny of sulfate-reducing Deltaproteobacteria in marine sediments by combining stable isotope probing with magnetic-bead capture hybridization of 16S rRNA. Appl Environ Microbiol. 2009;75:4927–4935. doi: 10.1128/AEM.00652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondello FJ, Turcich MP, Lobos JH, Erickson BD. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63:3096–3103. doi: 10.1128/aem.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales SE, Holben WE. Linking bacterial identities and ecosystem processes: can ‘omic’ analyses be more than the sum of their parts? FEMS Microbiol Ecol. 2011;75:2–16. doi: 10.1111/j.1574-6941.2010.00938.x. [DOI] [PubMed] [Google Scholar]
- Mou XZ, Vila-Costa M, Sun SL, Zhao WD, Sharma S, Moran MA. Metatranscriptomic signature of exogenous polyamine utilization by coastal bacterioplankton. Environ Microbiol Rep. 2011;3:798–806. doi: 10.1111/j.1758-2229.2011.00289.x. [DOI] [PubMed] [Google Scholar]
- Mulligan CN, Yong RN. Natural attenuation of contaminated soils. Environ Int. 2004;30:587–601. doi: 10.1016/j.envint.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Nakatsu CH, Hristova K, Hanada S, Meng XY, Hanson JR, Scow KM, Kamagata Y. Methylibium petroleiphilum gen. nov., sp. nov., a novel methyl tert-butyl ether-degrading methylotroph of the Betaproteobacteria. Int J Syst Evol Microbiol. 2006;56:983–989. doi: 10.1099/ijs.0.63524-0. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Chen Y, Dumont MG, Murrell JC. Marine methylotrophs revealed by stable-isotope probing, multiple displacement amplification and metagenomics. Environ Microbiol. 2008;10:1526–1535. doi: 10.1111/j.1462-2920.2008.01568.x. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Dumont MG, Vohra J, Murrell JC. Methodological considerations for the use of stable isotope probing in microbial ecology. Microb Ecol. 2007a;53:435–442. doi: 10.1007/s00248-006-9125-x. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Vohra J, Dumont MG, Lueders T, Manefield M, Friedrich MW, Murrell JC. DNA stable-isotope probing. Nat Protoc. 2007b;2:860–866. doi: 10.1038/nprot.2007.109. [DOI] [PubMed] [Google Scholar]
- Neufeld JD, Wagner M, Murrell JC. Who eats what, where and when? Isotope-labelling experiments are coming of age. ISME J. 2007c;1:103–110. doi: 10.1038/ismej.2007.30. [DOI] [PubMed] [Google Scholar]
- Newman LA, Reynolds CM. Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol. 2005;23:6–8. doi: 10.1016/j.tibtech.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Nikolausz M, Palatinszky MA, Rusznyak A, Richnow HH, Kappelmeyer U, Kastner M. Novel approach using substrate-mediated radiolabelling of RNA to link metabolic function with the structure of microbial communities. FEMS Microbiol Lett. 2007;274:154–161. doi: 10.1111/j.1574-6968.2007.00821.x. [DOI] [PubMed] [Google Scholar]
- Nováková M, Macková M, Chrastilová Z, Viktorová J, Szekeres M, Demnerová K, Macek T. Cloning the bacterial bphC gene into Nicotiana tabacum to improve the efficiency of PCB phytoremediation. Biotechnol Bioeng. 2009;102:29–37. doi: 10.1002/bit.22038. [DOI] [PubMed] [Google Scholar]
- Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science. 2001;293:484–487. doi: 10.1126/science.1061338. [DOI] [PubMed] [Google Scholar]
- Ottesen EA, Marin R, Preston CM, Young CR, Ryan JP, Scholin CA, DeLong EF. Metatranscriptomic analysis of autonomously collected and preserved marine bacterioplankton. ISME J. 2011;5:1881–1895. doi: 10.1038/ismej.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR, Stahl DA, Lane DJ, Olsen GJ. Analyzing natural microbial populations by rRNA sequences. ASM News. 1985;51:4–12. [Google Scholar]
- Padmanabhan P, Padmanabhan S, DeRito C, Gray A, Gannon D, Snape JR, Tsai CS, Park W, Jeon C, Madsen EL. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl Environ Microbiol. 2003;69:1614–1622. doi: 10.1128/AEM.69.3.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan CL, Fischer CR, Hyatt D, Bowen BP, Hettich RL, Banfield JF. Quantitative tracking of isotope flows in proteomes of microbial communities. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei AY, Oberdorf WE, Nossa CW, Agarwal A, Chokshi P, Gerz EA, Jin Z, Lee P, Yang L, Poles M, Brown SM, Sotero S, Desantis T, Brodie E, Nelson K, Pei Z. Diversity of 16S rRNA genes within individual prokaryotic genomes. Appl Environ Microbiol. 2010;76:3886–3897. doi: 10.1128/AEM.02953-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from-deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilloni G, von Netzer F, Engel M, Lueders T. Electron acceptor-dependent identification of key anaerobic toluene degraders at a tar-oil-contaminated aquifer by Pyro-SIP. FEMS Microbiol Ecol. 2011;78:165–175. doi: 10.1111/j.1574-6941.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- Prosser JI, Rangel-Castro JI, Killham K. Studying plant-microbe interactions using stable isotope technologies. Curr Opin Biotechnol. 2006;17:98–102. doi: 10.1016/j.copbio.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Pumphrey GM, Madsen EL. Field-based stable isotope probing reveals the identities of benzoic acid-metabolizing microorganisms and their in situ growth in agricultural soil. Appl Environ Microbiol. 2008;74:4111–4118. doi: 10.1128/AEM.00464-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. Removing noise from pyrosequenced amplicons. BMC Bioinformatics. 2011;12 doi: 10.1186/1471-2105-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radajewski S, Ineson P, Parekh NR, Murrell JC. Stable-isotope probing as a tool in microbial ecology. Nature. 2000;403:646–649. doi: 10.1038/35001054. [DOI] [PubMed] [Google Scholar]
- Radajewski S, McDonald IR, Murrell JC. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr Opin Biotechnol. 2003;14:296–302. doi: 10.1016/s0958-1669(03)00064-8. [DOI] [PubMed] [Google Scholar]
- Radajewski S, Webster G, Reay DS, Morris SA, Ineson P, Nedwell DB, Prosser JI, Murrell JC. Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology. 2002;148:2331–2342. doi: 10.1099/00221287-148-8-2331. [DOI] [PubMed] [Google Scholar]
- Reeder J, Knight R. Rapidly denoising pyrosequencing amplicon reads by exploiting rank-abundance distributions. Nat Methods. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezek J, Macek T, Macková M, Tríska J, Růžičková K. Hydroxy-PCBs, methoxy-PCBs and hydroxy-methoxy-PCBs: metabolites of polychlorinated biphenyls formed in vitro by tobacco cells. Environ Sci Technol. 2008;42:5746–5751. doi: 10.1021/es800445h. [DOI] [PubMed] [Google Scholar]
- Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- Rinta-Kanto JM, Sun SL, Sharma S, Kiene RP, Moran MA. Bacterial community transcription patterns during a marine phytoplankton bloom. Environ Microbiol. 2012;14:228–239. doi: 10.1111/j.1462-2920.2011.02602.x. [DOI] [PubMed] [Google Scholar]
- Roh H, Yu CP, Fuller ME, Chu KH. Identification of hexahydro-1,3,5-trinitro-1,3,5-triazinedegrading microorganisms via 15N-stable isotope probing. Environ Sci Technol. 2009;43:2505–2511. doi: 10.1021/es802336c. [DOI] [PubMed] [Google Scholar]
- Ruess L, Chamberlain PM. The fat that matters: Soil food web analysis using fatty acids and their carbon stable isotope signature. Soil Biol Biochem. 2010;42:1898–1910. [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE. 2011;6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Biotechnological prospects from metagenomics. Curr Opin Biotechnol. 2003;14:303–310. doi: 10.1016/s0958-1669(03)00067-3. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-8-229. 229-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser C, Steele H, Streit WR. Metagenomics, biotechnology with non-culturable microbes. Appl Microbiol Biotechnol. 2007;75:955–962. doi: 10.1007/s00253-007-0945-5. [DOI] [PubMed] [Google Scholar]
- Schnoor JL, Licht LA, Mccutcheon SC, Wolfe NL, Carreira LH. Phytoremediation of organic and nutrient contaminants. Environ Sci Technol. 1995;29:A318–A323. doi: 10.1021/es00007a747. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Waschkowitz T, Daniel R. Enhancement of gene detection frequencies by combining DNA-based stable-isotope probing with the construction of metagenomic DNA libraries. World J Microbiol Biotechnol. 2006;22:363–367. [Google Scholar]
- Shigematsu T, Hanada S, Eguchi M, Kamagata Y, Kanagawa T, Kurane R. Soluble methane monooxygenase gene clusters from trichloroethylene-degrading Methylomonas sp. strains and detection of methanotrophs during in situ bioremediation. Appl Environ Microbiol. 1999;65:5198–5206. doi: 10.1128/aem.65.12.5198-5206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Daniel R. Achievements and new knowledge unraveled by metagenomic approaches. Appl Microbiol Biotechnol. 2009;85:265–276. doi: 10.1007/s00253-009-2233-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C, Daniel R. Metagenomic analyses: Past and future trends. Appl Environ Microbiol. 2011;77:1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer AC, Crowley DE, Thompson IP. Secondary plant metabolites in phytoremediation and biotransformation. Trends Biotechnol. 2003;21:123–130. doi: 10.1016/S0167-7799(02)00041-0. [DOI] [PubMed] [Google Scholar]
- Singer AC, Thompson IP, Bailey MJ. The tritrophic trinity: a source of pollutant-degrading enzymes and its implications for phytoremediation. Curr Opin Microbiol. 2004;7:239–244. doi: 10.1016/j.mib.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Smidt H, de Vos WM. Anaerobic microbial dehalogenation. Annu Rev Microbiol. 2004;58:43–73. doi: 10.1146/annurev.micro.58.030603.123600. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- Su C, Lei LP, Duan YQ, Zhang KQ, Yang JK. Culture-independent methods for studying environmental microorganisms: methods, application, and perspective. Appl Microbiol Biotechnol. 2012;93:993–1003. doi: 10.1007/s00253-011-3800-7. [DOI] [PubMed] [Google Scholar]
- Suenaga H, Koyama Y, Miyakoshi M, Miyazaki R, Yano H, Sota M, Ohtsubo Y, Tsuda M, Miyazaki K. Novel organization of aromatic degradation pathway genes in a microbial community as revealed by metagenomic analysis. ISME J. 2009;3:1335–1348. doi: 10.1038/ismej.2009.76. [DOI] [PubMed] [Google Scholar]
- Sul WJ, Park J, Quensen JF, III, Rodrigues JL, Seliger L, Tsoi TV, Zylstra GJ, Tiedje JM. DNA-stable isotope probing integrated with metagenomics for retrieval of biphenyl dioxygenase genes from polychlorinated biphenyl-contaminated river sediment. Appl Environ Microbiol. 2009;75:5501–5506. doi: 10.1128/AEM.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sylvestre M, Macek T, Macková M. Transgenic plants to improve rhizoremediation of polychlorinated biphenyls (PCBs) Curr Opin Biotechnol. 2009;20:242–247. doi: 10.1016/j.copbio.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Nanba K, Iwamoto H, Nirei H, Kusuda T, Kazaoka O, Owaki M, Furuya K. In situ bioremediation of a cis-dichloroethylene-contaminated aquifer utilizing methane-rich groundwater from an uncontaminated aquifer. Water Res. 2005;39:2438–2444. doi: 10.1016/j.watres.2005.04.041. [DOI] [PubMed] [Google Scholar]
- Taubert M, Jehmlich N, Vogt C, Richnow HH, Schmidt F, von Bergen M, Seifert J. Time resolved protein-based stable isotope probing (Protein-SIP) analysis allows quantification of induced proteins in substrate shift experiments. Proteomics. 2011;11:2265–2274. doi: 10.1002/pmic.201000788. [DOI] [PubMed] [Google Scholar]
- Tobino T, Kurisu F, Kasuga I, Furumai H. Shotgun isotope array for rapid, substrate-specific detection of microorganisms in a microbial community. Appl Environ Microbiol. 2011;77:7430–7432. doi: 10.1128/AEM.00121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp E, Hanson RS, Ringelberg DB, White DC, Wheatcroft R. Isolation and characterization of an N-methylcarbamate insecticide-degrading methylotrophic bacterium. Appl Environ Microbiol. 1993;59:3339–3349. doi: 10.1128/aem.59.10.3339-3349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsvik V, Øvreås L. Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol. 2002;5:240–245. doi: 10.1016/s1369-5274(02)00324-7. [DOI] [PubMed] [Google Scholar]
- Toussaint J-P, Pham T, Barriault D, Sylvestre M. Plant exudates promote PCB degradation by a rhodococcal rhizobacteria. Appl Microbiol Biotechnol. 2012;95:1589–1603. doi: 10.1007/s00253-011-3824-z. [DOI] [PubMed] [Google Scholar]
- Uhlík O, Ječná K, Leigh MB, Macková M, Macek T. DNA-based stable isotope probing: a link between community structure and function. Sci Total Environ. 2009a;407:3611–3619. doi: 10.1016/j.scitotenv.2008.05.012. [DOI] [PubMed] [Google Scholar]
- Uhlík O, Ječná K, Macková M, Vlček C, Hroudová M, Demnerová K, Pačes V, Macek T. Biphenyl-metabolizing bacteria in the rhizosphere of horseradish and bulk soil contaminated by polychlorinated biphenyls as revealed by stable isotope probing. Appl Environ Microbiol. 2009b;75:6471–6477. doi: 10.1128/AEM.00466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlík O, Wald J, Strejček M, Musilová L, Rídl J, Hroudová M, Vlček Č, Cardenas E, Macková M, Macek T. Identification of bacteria utilizing biphenyl, benzoate, and naphthalene in long-term contaminated soil. PLoS ONE. 2012;7:e40653. doi: 10.1371/journal.pone.0040653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken B, Correa PA, Schnoor JL. Phytoremediation of polychlorinated biphenyls: new trends and promises. Environ Sci Technol. 2010;44:2767–2776. doi: 10.1021/es902514d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken B, Yoon JM, Schnoor JL. Biodegradation of nitro-substituted explosives 2,4,6-trinitrotoluene, hexahydro-1,3,5-trinitro-1,3,5-triazine, and octahydro-1,3,5,7-tetranitro-1,3,5-tetrazocine by a phytosymbiotic Methylobacterium sp. associated with poplar tissues (Populus deltoides x nigra DN34) Appl Environ Microbiol. 2004;70:508–517. doi: 10.1128/AEM.70.1.508-517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers YH, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Vézina J, Barriault D, Sylvestre M. Family shuffling of soil DNA to change the regiospecificity of Burkholderia xenovorans LB400 biphenyl dioxygenase. J Bacteriol. 2007;189:779–788. doi: 10.1128/JB.01267-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vézina J, Barriault D, Sylvestre M. Diversity of the C-terminal portion of the biphenyl dioxygenase large subunit. J Mol Microbiol Biotechnol. 2008;15:139–151. doi: 10.1159/000121326. [DOI] [PubMed] [Google Scholar]
- Vieites JM, Guazzaroni ME, Beloqui A, Golyshin PN, Ferrer M. Metagenomics approaches in systems microbiology. FEMS Microbiol Rev. 2009;33:236–255. doi: 10.1111/j.1574-6976.2008.00152.x. [DOI] [PubMed] [Google Scholar]
- Wagner M, Horn M, Daims H. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr Opin Microbiol. 2003;6:302–309. doi: 10.1016/s1369-5274(03)00054-7. [DOI] [PubMed] [Google Scholar]
- Wagner M, Nielsen PH, Loy A, Nielsen JL, Daims H. Linking microbial community structure with function: fluorescence in situ hybridization-microautoradiography and isotope arrays. Curr Opin Biotechnol. 2006;17:83–91. doi: 10.1016/j.copbio.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Warner KL, Larkin MJ, Harper DB, Murrell JC, McDonald IR. Analysis of genes involved in methyl halide degradation in Aminobacter lissarensis CC495. FEMS Microbiol Lett. 2005;251:45–51. doi: 10.1016/j.femsle.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Wellington EMH, Berry A, Krsek M. Resolving functional diversity in relation to microbial community structure in soil: exploiting genomics and stable isotope probing. Curr Opin Microbiol. 2003;6:295–301. doi: 10.1016/s1369-5274(03)00066-3. [DOI] [PubMed] [Google Scholar]
- Weyens N, Truyens S, Saenen E, Boulet J, Dupae J, Taghavi S, van der Lelie D, Carleer R, Vangronsveld J. Endophytes and their potential to deal with co-contamination of organic contaminants (toluene) and toxic metals (nickel) during phytoremediation. Int J Phytoremediat. 2011;13:244–255. doi: 10.1080/15226511003753920. [DOI] [PubMed] [Google Scholar]
- Weyens N, van der Lelie D, Taghavi S, Vangronsveld J. Phytoremediation: plant-endophyte partnerships take the challenge. Curr Opin Biotechnol. 2009;20:248–254. doi: 10.1016/j.copbio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Whiteley AS, Manefield M, Lueders T. Unlocking the 'microbial black box' using RNA-based stable isotope probing technologies. Curr Opin Biotechnol. 2006;17:67–71. doi: 10.1016/j.copbio.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Wiegel J, Wu Q. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol Ecol. 2000;32:1–15. doi: 10.1111/j.1574-6941.2000.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Winderl C, Penning H, von Netzer F, Meckenstock RU, Lueders T. DNA-SIP identifies sulfate-reducing Clostridia as important toluene degraders in tar-oil-contaminated aquifer sediment. ISME J. 2010;4:1314–1325. doi: 10.1038/ismej.2010.54. [DOI] [PubMed] [Google Scholar]
- Woods A, Watwood M, Schwartz E. Identification of a toluene-degrading bacterium from a soil sample through H2 18O DNA stable isotope probing. Appl Environ Microbiol. 2011;77:5995–5999. doi: 10.1128/AEM.05689-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YR, Zarda A, Zeyer J. Combination of microautoradiography and fluorescence in situ hybridization for identification of microorganisms degrading xenobiotic contaminants. Environ Toxicol Chem. 2003;22:2840–2844. doi: 10.1897/02-423. [DOI] [PubMed] [Google Scholar]
- Yi H, Crowley DE. Biostimulation of PAH degradation with plants containing high concentrations of linoleic acid. Environ Sci Technol. 2007;41:4382–4388. doi: 10.1021/es062397y. [DOI] [PubMed] [Google Scholar]
- Yun J, Ryu S. Screening for novel enzymes from metagenome and SIGEX, as a way to improve it. Microb Cell Fact. 2005;4:8. doi: 10.1186/1475-2859-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu Z. Assessing bacterial diversity in soil. J Soil Sediment. 2008;8:379–388. [Google Scholar]