Abstract
The human chromosome 15q11-q13 region hosts a wide variety of coding and non-coding RNAs, and is also the site of nearly every imaginable type of RNA processing. To deepen the intrigue, the transcripts in the human chromosome 15q11-q13 region are subject to regulation by genomic imprinting, and some of these transcripts are imprinted in a tissue-specific manner. Since the region is critically important for three human neurogenetic disorders, Angelman syndrome, Prader-Willi syndrome, and 15q duplication syndrome, there is intense interest in understanding the types of RNA and RNA processing that occurs among the imprinted genes. This review summarizes what is known about the various RNAs within the imprinted domain, including a novel type of RNA that was only very recently identified.
THE CHROMOSOME 15Q11-Q13 REGION AND ASSOCIATED HUMAN DISORDERS
The human chromosome 15q11-q13 region is a 6 Mb segment of the genome that is centrally involved in three human neurogenetic disorders: Prader-Willi syndrome (PWS), Angelman syndrome (AS) and 15q duplication syndrome (reviewed in1). In addition to its relevance for human disease, this region hosts a variety of unique gene regulation paradigms, the most notable of which is genomic imprinting, an epigenetic phenomenon by which genes are expressed preferentially from only one parental allele The genomic organization and regulation of imprinted expression is conserved on the syntenic region of mouse chromosome 7C2,3. Through a variety of studies in both mouse and human systems, much has been learned about this region.
Imprinted genes are usually clustered and utilize a variety of mechanisms to establish and maintain their imprinted status, including differential DNA methylation, differential histone modification, and antisense transcription. Within the 15q11-q13 region there are at least 14 imprinted transcripts: MKRN3, MAGEL2, NDN, PWRN1, C15ORF2/NPAP1, SNURF-SNRPN, SNORD107/HBII-436, SNORD64/HBII-13, SNORD108/HBII437, SNORD109A/HBII438A, SNORD116/HBII-85, IPW, SNORD115/HBII-52, SNORD109B/HBII-438B, UBE3A, and ATP10A. Most of these genes are expressed exclusively from the paternally inherited allele in somatic tissues. UBE3A and ATP10A show brain-specific imprinted expression and are the only imprinted genes in the region that are expressed from the maternally inherited allele. Moreover, ATP10A does not show imprinted expression in all individuals4.
The imprinted domain is flanked by non-imprinted genes: TUBCGP5, CYFIP1, NIPA2, and NIPA1 that are proximal to the imprinted domain5 and GABRB3, GABRA5, GABRG3, OCA2, and HERC2 that are distal to the imprinted domain6. Most individuals with PWS and AS have a large deletion of the 15q11-q13 region7. Depending on their specific deletion breakpoints, individuals with PWS or AS also may be lacking one copy of some or all of the non-imprinted genes in the region8–10. In this review, we focus only on the imprinted domain of the chromosome 15q11-q13 region.
Genomic imprinting at chromosome 15q11-q13
Genomic imprinting in the chromosome 15q11-q13 region is controlled by a bipartite imprinting center (IC)11,12. One element in this imprinting center, termed the Prader-Willi syndrome imprinting center, or PWS-IC, includes the major promoter and exon 1 of the SNURF-SNRPN gene. Within the PWS-IC lies a differentially-methylated CpG island that is methylated on the maternally-inherited allele and unmethylated on the paternally-inherited allele. PWS individuals with small deletions that include the PWS-IC and mouse models have functionally defined the PWS-IC and determined it to lie within an approximately 5 kb region (4.3 kb in humans and 6 kb in mouse)12,13. In addition to the SNURF-SNRPN major promoter and exon 1, this region is thought to include an additional element that is necessary for full PWS-IC function14.
The second element in the bipartite imprinting center is the Angelman syndrome imprinting center, or the AS-IC. While the mechanism by which this element functions is not known, the AS-IC is thought to repress the activity of the PWS-IC in the maternal germline, thereby facilitating the methylation of the maternal allele of the PWS-IC in this lineage and during subsequent development. Recent data using the mouse model system postulates that the AS-IC establishes the maternal imprint by driving expression from the upstream exons of SNURF-SNRPN in the maternal germline15.
The bipartite imprinting center executes its function amidst several allele-specific covalent histone modifications and other chromatin features. The maternally inherited PWS-IC is marked with tri-methylated histone H3, lysine 9 (H3K9me3), while the paternally inherited PWS-IC is marked with di- and tri-methylated histone H3 lysine 4(H3K4me2 and me3) as well as acetylated histones H3 and H416,17. Mouse studies have determined that H3K36me3 unexpectedly marks the repressed maternal allele, along with the expected repressive histone modifications, H4K20me3, H3K9me3, and H3K79me318.
Human-Mouse shared synteny
A region on mouse chromosome 7C is orthologous to the human chromosome 15q11-q13 region3 (Figure 1). The gene structure and regulation of genomic imprinting is exquisitely maintained between human and mouse, with a few exceptions. One protein-coding gene, Frat3, is imprinted in mouse, but not present in human19. Conversely, C15ORF2/NPAP1 is imprinted in humans, but is not present in mouse20. Another gene ATP10A, is imprinted in some human individuals, but not imprinted in mouse4,21. There are several annotated non-coding RNAs, however, that are not conserved between the two species. Whether these RNAs exist in both species, but are poorly annotated or are structurally different and therefore not recognized as being the same ncRNA is unclear. These ncRNAs will be further discussed below.
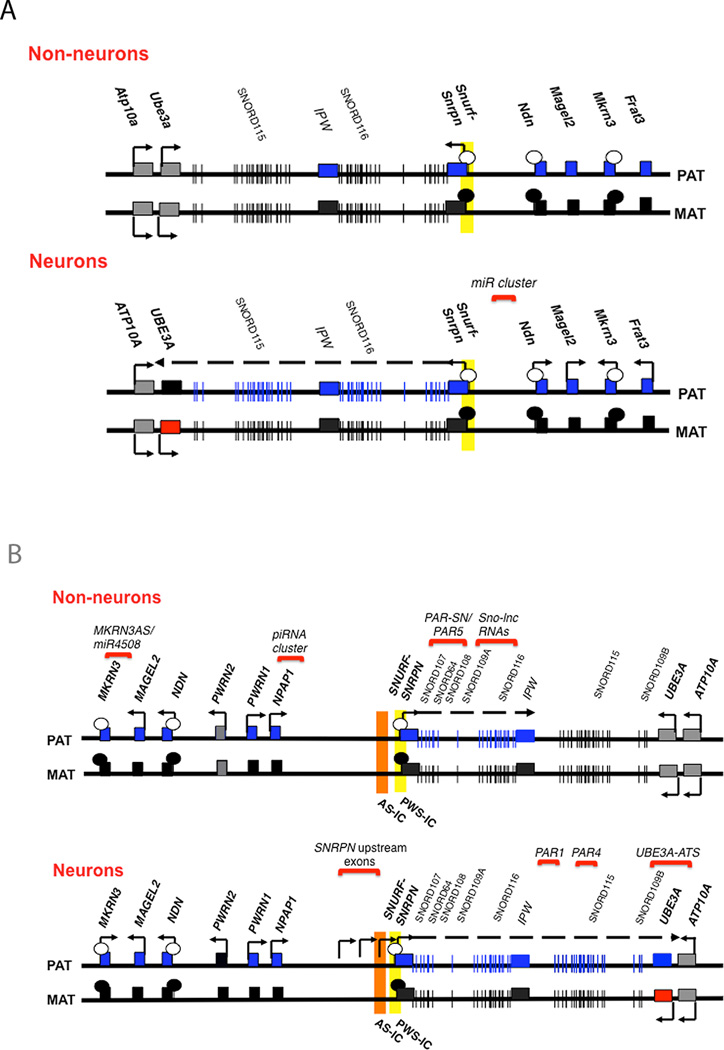
Figure 1. Maps of the human chromosome 15q11-q13 region and the mouse syntenic region on chromosome 7C.
A.) Map of the mouse chromosome 7C region indicating gene expression patterns in non-neurons (top) and neurons (bottom). B.) Map of the human chromosome 15q11-q13 imprinted region indicating gene expression patterns in non-neurons (top) and neurons (bottom). Blue rectangles represent imprinted transcripts that are paternally expressed, red rectangles represent imprinted transcripts that are maternally expressed, black rectangles represent the repressed alleles of imprinted genes, and grey rectangles represent transcripts that are not imprinted. Maps are not to scale and not intended to be all-inclusive.
The expression pattern of the genes also differs between mouse and human (compare Figure 1A and B). In mouse, only the protein-coding portion of Snurf-Snrpn is expressed in multiple tissues; all other known paternally expressed genes in the region, are expressed exclusively in the brain. Therefore, most of the murine genes in the mouse 7C imprinted region exhibit brain-specific expression (Figure 1A). In human, the protein coding SNURF-SNRPN transcript, as well as the ncRNAs between the end of the protein coding gene and IPW are expressed in a variety of cell types. Furthermore, the proximal genes, NDN and MKRN3 are also expressed in a variety of cell types. Only the portion of the SNURF-SNRPN transcript distal to IPW, some upstream exons of SNURF-SNRPN, and MAGEL2 are brain-specific (Figure 1B).
GENES AND GENE EXPRESSION
Nearly every type of RNA and/or gene can be found in the chromosome 15q11-q13 imprinted region. These RNAs employ a variety of processing mechanisms, including alternative splicing, alternative polyadenylation, and exonucleolytic processing from introns, as well as undetermined mechanisms involving antisense transcription and resulting gene silencing. The details of the RNAs and what is known about their processing is described below and summarized in Tables 1–3.
Table 1. Protein-coding genes.
| Gene name | Position | Function | Ref. |
|---|---|---|---|
| Peg12/Frat3 | Mm. chr7:69,606,757-69,609,396 | GSK3-binding protein family members | 19 |
| MKRN3 | Hs. chr15:23,810,454-23,813,166 Mm. chr7:69,562,479-69,565,025 |
Makorin-ring protein, probable ubiquitin ligase | 21,24 |
| MAGEL2 | Hs. chr15:23,888,696-23,892,993 Mm. chr7:69,521,865-69,526,526 |
Melanoma antigen gene-encoding (MAGE)-like protein | 25–28 |
| NDN | Hs. chr15:23,930,554-23,932,450 Mm. chr7:69,493,163-69,494,813 |
May suppress growth in post-mitotic neurons | 29–40 |
| NPAP1/C15ORF2 | Hs. chr15:24,920,541-24,928,593 | Nuclear pore-associated protein | 41–44 |
| SNURF | Hs. chr15:25,200,070-25,213,976 Mm. chr7:67,140,305-67,150,042 |
Function unknown | 46 |
| SNRPN* | Hs. chr15:25,200,070-25,223,729 Mm. chr7:67,127,388-67,150,042 |
Member of the small nuclear ribonucleoprotein complex | 46,47 |
| UBE3A | Hs. chr15:25,582,396-25,684,175 Mm. chr7:66,484,120-66,562,097 |
Ubiquitin E3 ligase | 51–61 |
| ATP10A | Hs. chr15:25,923,860-26,108,349 Mm. chr7:65,913,572-66,084,796 |
phospholipid-transporting ATPase | 4,20 |
Table 3. Small non-coding RNAs.
Sno-lnc RNAs range in size from 1,178 nucleotides to 2,859 nucleotides, and are therefore not technically small non-coding RNAs, however, due to their relationship to snoRNAs, they are included in this table.
| Gene name | Position | copies | Ref. |
|---|---|---|---|
| snoRNAs | 50,67, 73–75 | ||
| SNORD107/HBII-436 | Hs. chr15:25,227,141-25,227,215 | 1 | |
| SNORD64/HBII-13 | chr15:25,230,247-25,230,313 | 1 | |
| SNORD108/HBII-437 | chr15:25,232,072-25,232,140 | 1 | |
| SNORD109A/HBII-438A | chr15:25,287,121-25,287,187 | 1 | |
| SNORD116/HBII-85* | chr15:25,296,701-25,352,633 | 29 | |
| SNORD115/HBII-52* | chr15:25,417,020-25,516,600 | 48 | |
| SNORD109B/HBII-438B | chr15:25,523,490-25,523,556 | 1 | |
| sno-lncRNAs | 78 | ||
| SNOLNCRNA1 | Hs. chr15:25,310,172-25,313,029 | 1 | |
| SNOLNCRNA2 | chr15:25,324,204-25,325,380 | 1 | |
| SNOLNCRNA3 | chr15:25,330,531-25,331,761 | 1 | |
| SNOLNCRNA4 | chr15:25,332,808-25,334,043 | 1 | |
| SNOLNCRNA5 | chr15:25,344,645-25,346,822 | 1 | |
| miRNAs | 79,81 | ||
| miR4508 | Hs. chr15:23,807,209-23,807,278 | 1 | |
| miR344 | Mm. chr7:68,830,112-69,194,754 | 10 | |
| piRNAs | Hs. chr15:24,920,541-24,928,593 | 17 | 42,80 |
Protein coding transcripts
Intronless genes located in proximal 15q11-q13
MKRN3, MAGEL2, NDN, and C15ORF2/NPAP1 are located in a cluster at the proximal end of human chromosome 15q11-q13 and at the distal end of the syntenic region on mouse chromosome 7C. MKRN3 and NDN each have differentially methylated CpG islands at their respective promoters that are methylated on the maternally inherited allele and unmethylated on the paternally inherited allele22,23. Data from individuals with atypical deletions or translocations suggest that these three genes are not responsible for the major features associated with PWS24. One PWS individual was reported to have a deletion that does not include the MKRN3, MAGEL2, or NDN genes, and showed NDN expression in a patient-derived lymphoblastoid cell line. DNA methylation at the NDN promoter was also consistent with expression from the paternally inherited allele. Another individual was reported with a translocation that resulted in the deletion of the paternal copies of MKRN3, MAGEL2, and NDN, however, this individual did not have PWS24.
MKRN3 (makorin ring finger protein 3) encodes a probable ubiquitin ligase and is one of four members of the makorin family of zinc finger proteins25. MKRN3 has a typical C3HC4 zinc finger, which comprises the RING domain and confers ubiquitin ligase activity. Notably, MKRN3 is specific to therian mammals, where it is imprinted22,25. There is also a ncRNA that is antisense to MKRN3.
MAGEL2 encodes a melanoma antigen gene-encoding (MAGE)-like protein that is also known as Necdin-like 1 (NDNL1) due to its similarity to NDN26. Magel2 has been disrupted by targeted mutation in mouse27. Mice deficient for Magel2 exhibit neonatal growth retardation, excessive weight gain, increased adiposity, hypophagia, altered metabolism, disrupted circadian rhythms, and impaired reproductive function27–29.
NDN (Necdin) encodes another MAGE-like protein that is thought to function as a growth suppressor in neuronal and non-neuronal cells30. Ndn binds to the transactivation domain of E2F1 and is thought to facilitate exit of the cell from the cell cycle through this mechanism31. However, NDN also interacts with SIRT1 and mediates the deacetylation of p53, which may also mechanistically explain the regulation of the cell cycle32. Disruption of the murine Ndn gene has led to conflicting results. One group reported that Ndn-deficient mice exhibited postnatal lethality, skin scraping, and increased spatial learning33. Another group reported postnatal lethality due to respiratory defects34. A third group reported no obvious phenotype associated with targeted mutation of Ndn35. Using these mouse models, roles for Ndn in myoblast differentiation, axonal outgrowth and migration, adipogenesis, tumor suppression, and modulation of the thyroid axis have all been reported36–41.
C15ORF2/NPAP1 is a protein-coding gene that is primate-specific and has undergone positive selection during evolution42. C15ORF2/NPAP1 consists of a single exon of 8 kb that is predicted to encode a 1156 amino acid protein. The presence of a long terminal repeat sequence in the 3’ end of the gene suggests that it may have arisen in the primate lineage due to retroviral integration43. The mRNA is expressed in brain and testes20. Preliminary work using an antibody raised against C15ORF2/NPAP1 suggests that it is expressed in various regions of the brain, including the hypothalamus, and is slightly larger than 130 kDa42. The function of the protein is not known, although it was recently shown to be associated with the nuclear pore complex44.
While MKRN3, MAGEL2, and NDN are conserved between human and mouse, Frat3 is not. Frat3 arrived in the imprinted domain on mouse chromosome 7C by a recent retrotransposition event and is most likely imprinted due to its fortuitous location in an imprinted region19. Very little is known about Frat3, however it is most similar to Frat1 and Frat2, GSK3-binding protein family members that are known to positively regulate Wnt signaling45. Frat3 does not exist in the human genome19.
SNURF-SNRPN
The SNURF-SNRPN gene encodes a bicistronic transcript that produces two proteins: SNURF and SNRPN46. SNRPN is a small nuclear ribonucleoprotein that functions in pre-mRNA processing, and perhaps alternative splicing47. SNURF is a nuclear localized protein of unknown function that is produced from the first three exons of the SNURF-SNRPN transcript46. The major promoter and first exon of SNURF-SNRPN comprise the PWS-IC and is necessary for the expression of all of the paternally expressed genes in the 15q11-q13 imprinted region13. Targeted mutations of the Snurf and Snrpn genes have no obvious phenotype, as long as they do not disrupt the imprinting center48,49. However, a 6 kb deletion that includes the promoter and first exon of the Snurf-Snrpn transcript is sufficient to lead to an imprinting defect in mice and loss of expression from all paternally expressed genes in the region13. The SNURF-SNRPN gene also produces a long, ncRNA and is subject to extensive alternative splicing at the 5’ and 3’ ends50. The SNURF-SNRPN transcript will be further discussed below.
UBE3A
The UBE3A gene encodes an E3 ubiquitin ligase, known as E6-associated protein (E6AP)51. The UBE3A mRNA has at least 7 reported isoforms that arise from alternative splicing52. Most of the transcript variation resides in the 5’ end of the transcript. There is an antisense transcript to UBE3A (UBE3A-ATS, discussed further below) that most likely regulates its imprinted expression by leading to repression of paternal UBE3A53–55. The loss function from the maternal allele of UBE3A causes Angelman syndrome56.
The E6AP protein is the prototypical HECT (homologous to E6AP carboxyl terminus) domain ubiquitin ligase. It functions to recognize target substrates and transfer ubiquitin moieties from the E2 ubiquitin-conjugating enzymes to them. E6AP has been found in the cytoplasm and nucleus and is expressed in a broad range of tissues. Three different protein isoforms, differing in the extreme N-termini have been reported and are hypothesized to mediate different target substrate specificities, although this has not been demonstrated57. Mice deficient for Ube3a have seizures (strain dependent), deficits in LTP and LTD, poor performance on the rotarod, and loss of plasticity in the visual cortex58–61.
Non-coding RNAs
The most intriguing aspect of the chromosome 15q11-q13 region is the variety of non-coding RNAs (ncRNAs) that can be found. Long non-coding RNAs (lncRNAs), small nucleolar RNAs (snoRNAs), small nucleolar long non-coding RNAs (snolncRNAs), microRNAs (miRNAs), and PIWI-interacting RNAs (piRNAs) have all been identified in the imprinted domain. There are also at least two known cases of antisense transcription, and function has been ascribed to one of them. Furthermore, both alternative splicing and alternative promoter usage appear to play roles in regulation of expression within the region.
lncRNAs
The annotated human lncRNAs include PWRN2, PWRN1, PAR-SN, PAR5, HBT8, IPW, PAR1, and PAR4. Additionally, three other lncRNAs, MKRN3AS, PWCR1, and UBE3A-ATS, have been reported in the literature, but are currently not annotated in RefSeq or the UCSC Genome Browser. All but two of these lncRNAs (MKRN3AS and PWRN2) are part of the non-coding portion of the SNURF-SNRPN transcript. PWRN1, PAR-SN, PAR5, HBT8, IPW, PAR4, and UBE3A-ATS have all been reported to be part of the SNURF-SNRPN transcript, and thus may be nothing more than spliced or unspliced variants of that transcript20,62,63. However, since the processing of the SNURF-SNRPN transcript is not fully understood, it is not clear whether any of these transcripts have independent functions. The lncRNAs are not as conserved in mouse: only four of the human lncRNAs MKRN3AS, PWCR1, IPW, and UBE3A-ATS are present in the mouse chromosome 7C region, and they have very little sequence conservation. Conversely, putative murine lncRNAs include twelve annotated, but un-named transcripts in addition to the aforementioned transcripts. Seven of 16 murine lncRNAs appear to be derived from the Snurf-Snrpn transcript.
MKRN3AS is the most proximal lncRNA and its function is unknown. The mRNA for MKRN3AS has been reported to be approximately 7.1 kb, containing two exons22. MKRN3 and MKRN3AS overlap completely, with 1 kb of the 3’ end of MKRN3 extending into the intron of MKRN3AS. Murine Mkrn3as has also been identified. The splice acceptor site is conserved between mouse and human, but a full-length transcript has not been identified64.
PWRN2 and PWRN1 are proximal to the PWS-IC in humans, but do not exist in mice. PWRN2 and PWRN1 are alternatively spliced and polyadenylated non-coding RNAs that lie between C15ORF2 and NDN20. PWRN2 and PWRN1 were originally identified using in silico searches of two databases. PWRN2 spans 7.2 kb of genomic DNA consisting of three exons. PWRN1 spans 160 kb of genomic DNA and 26 exons. Both genes have been partially duplicated: PWRN1 and PWRN2 each have 5 partially duplicated copies. The PWRN1 copies lie proximal to the PWRN1 gene, and the PWRN2 copies lie both proximal and distal to PWRN2. Both genes are most highly expressed in testes, but PWRN1 can also be detected at low levels in prostate, heart, liver, lung, trachea, spinal cord, and fetal brain20. The 3’ exons of PWRN1 have also been identified as part of the SNURF-SNRPN transcript in brain, and thus PWRN1 may represent more upstream exons of this very long transcript42. PWRN2 is expressed from both parental alleles in testes, and therefore does not seem to be imprinted. However, this tissue has undergone resetting of the imprint, so it is not known whether PWRN2 bears the epigenetic marks of imprinted genes or whether it is imprinted in other tissues that have not been analyzed to date20.
The SNURF-SNRPN transcript is complicated and unique. It not only produces a bicistronic message that encodes the SNURF and SNRPN proteins (discussed above)46, but it also comprises many other lncRNAs. Altogether, the human SNURF-SNRPN transcript spans approximately 600 kb of genomic DNA and has in excess of 148 exons50. SNURF-SNRPN undergoes extensive alternative splicing, the extent of which is not fully understood.
At the 5’ end, alternative upstream exons or alternative promoters are preferentially used in brain and perhaps germ cells15,65–68. As many as 9 upstream exons have been identified that span approximately 150kb of genomic distance upstream of SNURF-SNRPN exon 167. These upstream exons are similar to one another and also harbor CpG islands, two of which show the same methylation pattern as the PWS-IC (maternally methylated and paternally unmethylated), while the third shows the opposite pattern of methylation, being maternally unmethylated and paternally methylated67. One putative function of these upstream exons may be to regulate expression of the SNURF-SNRPN non-coding RNAs50,68. Interestingly, one of the alternative upstream exons is included within the smallest region of deletion overlap for AS imprinting center mutations, and may play a role in the function of the AS-IC67. A recent publication suggests that the AS-IC may function by promoting transcription from the upstream alternative exons in the female germline15.
How the SNRPN transcript extends distally of the polyadenylation site(s) at the 3’ end of the coding portion of the gene is not known. Transcription through the polyadenylation site(s) of SNRPN occurs in most tissues in humans, but does not occur in mouse, except for in brain. In most human tissues, the non-coding RNAs downstream of SNRPN, but ending with IPW are expressed50,69–71. While it is known that much of this non-coding transcript serves as a host for snoRNAs70 and a newly identified class of RNAs termed snolncRNAs(further discussed below), there are also transcripts of unknown function, including PAR-SN, PAR5, and HBT8. These RNAs are annotated to be approximately 1.7, 3.4, and 4.1 kb in size, respectively, although by northern blot, a PAR5 probe recognizes an approximately 12kb band in skeletal muscle and a diffuse smear in brain62, and a PAR-SN probe recognizes a 3kb band in brain, heart, and skeletal muscle63. HBT8 was identified as an mRNA that encodes a short peptide expressed in brain, however the protein-coding potential of this transcript has not been verified. While these transcripts do seem to be part of distinct non-coding RNAs, it is also possible that they are simply non-functional exons within the SNURF-SNRPN non-coding transcript.
IPW is a non-coding transcript that is present in both mouse and human. The non-coding SNURF-SNRPN transcript is terminated at or upstream of IPW in most human cell types. In brain only, the transcript extends distal to IPW50,69–71. The IPW RNA itself is spliced and consists of three exons in humans and six exons in mouse69,72. The human IPW gene produces two transcripts of 2.4 kb and 1.3 kb in most tissues. In brain, however, only the 2.4 kb band is detectable69. Analogous RNAs in mouse have not been identified—by northern blot, murine Ipw seems to exist as a high-molecular weight smear. IPW is poorly conserved in mammals, except for an approximately 2 kb portion, which shows considerable conservation72. The function of IPW is also not known. It may only be an exon within the SNURF-SNRPN transcript50.
The 3’ end of the SNURF-SNRPN non-coding transcript, distal to IPW, is only expressed in brain. While this portion of the transcript largely serves as a host transcript for the SNORD115/HBII-52 snoRNAs70 (further discussed below), other non-coding RNAs are generated from this transcript, including PAR1, PAR4, and UBE3A-ATS. PAR1 and PAR4 are intronless non-coding RNAs of unknown function that are annotated to be 2.4 kb and 342 bp, respectively. Northern blot analysis using a probe for PAR1 detects a high molecular weight smear in brain only62, and therefore may simply be one exon in the brain-specific portion of the SNURF-SNRPN non-coding transcript, although it does not seem to be part of any known splice forms in the region. PAR4 is located in the midst of the SNORD115/HBII-52 cluster and in fact contains SNORD115-23. Portions of PAR4 are repeated throughout much of the SNORD115/HBII-52 cluster, suggesting that PAR4, too, may be little more than one exon within this transcript.
The generation of UBE3A-ATS may be the evolutionary purpose for the brain-specific portion of the SNURF-SNRPN transcript. UBE3A-ATS refers to the part of the SNURF-SNRPN transcript that overlaps the UBE3A gene. UBE3A-ATS was first observed as an unspliced portion of the UBE3A transcript that was reduced in PWS brain, but strongly present in AS brain53. Subsequently, murine Ube3a-ats was found to be controlled by the PWS-IC, and mice with a paternal deletion of PWS-IC showed reactivation of paternal Ube3a in the brain54. These data suggest that expression of the UBE3A-ATS leads to the repression of paternal UBE3A in the brain. A recent paper by Meng et al. confirmed this finding by showing that termination of murine Ube3a-ats results in reactivation of paternal Ube3a55. This paper also characterized the murine transcript. They reported that Ube3a-ats is a non-polyadenylated RNA polymerase II transcript with a relatively short half-life. This and other reports showed that Ube3a-ats is transcribed across the entire Ube3a gene, even overlapping the promoter55,65,73. There are several mechanism by which UBE3A-ATS could repress paternal UBE3A, including the following possibilities: 1.) UBE3A-ATS RNA could recruit repressive histone modifications to the paternal UBE3A promoter (or other regulatory element), 2.) the UBE3A-ATS transcript could establish a repressive three-dimensional chromatin structure across the paternal allele, or 3.) the act of transcription through the paternal UBE3A locus in an antisense direction could cause transcriptional interference. These possibilities are not necessarily mutually exclusive and are an intense area of investigation.
snoRNAs
One major function of the SNURF-SNRPN non-coding transcript is to generate 72 C/D box small nucleolar RNAs (snoRNAs)50,70. These snoRNAs are processed from the introns of the host transcript and are conserved between mouse and human. The snoRNAs fall into 6 groups: SNORD107/HBII-436, SNORD64/HBII-13, SNORD108/HBII-437, SNORD109/HBII-438 (2 members), SNORD116/HBII-85 (29 members), and SNORD115/HBII-52 (48 members). SnoRNAs feature a C box denoted by the sequence UGAUGA and a D box denoted by the sequence CUGA, and often they contain C’ and D’ boxes, which are replicas of the C and D boxes, respectively. Immediately adjacent to the D and D’ boxes lie antisense boxes. SnoRNAs typically add 2’-O-methyl groups or pseudouridine to ribosomal RNAs and/or spliceosomal RNAs in a sequence-specific manner that is encoded by the sequence of the antisense box. The antisense boxes for the snoRNAs within the human chromosome 15q imprinted region and syntenic mouse region on chromosome 7C are not obviously complimentary to ribosomal RNAs or to spliceosomal RNAS, and thus they have been deemed orphan snoRNAs. Instead, it has been suggested that they may modify mRNAs70.
Little is known about most of the snoRNAs in this region. The most highly studied snoRNA, SNORD115/HBII-52, has an antisense box that is identical among all 48 members of the cluster70. Furthermore, this antisense box sequence is also completely conserved in the mouse. The reason for the extensive conservation may be that this snoRNA has been shown to bind and alter post-transcriptional modification of the RNA encoding the serotonin receptor, HTR2C70,74. It has been proposed that SNORD115/HBII-52 may work by preventing ADAR2-mediated RNA editing at specific sites in the HTR2C RNA75. However, it is not clear whether SNORD115/HBII-52 is performing this function in humans76.
It has also been proposed that the chromosome 15q11-q13 snoRNAs may be further processed into sno-derived RNAs, or sdRNAs77. These sdRNAs show complementarity to sequences near splice junctions, and thus have been hypothesized to influence alternative splicing of specific genes outside of the chromosome 15 region. Recently, it was shown that in addition to the non-canonical snoRNAs that likely modify mRNAs in trans, that the chromosome15q snoRNAs also make canonical snoRNAs that modify ribosomal RNAs78.
Sno-lncRNAs
A novel form of small RNA, termed sno-lncRNA (small nucleolar long non-coding RNA), has been very recently discovered in the human chromosome 15q11-q13 region79. Sno-lncRNAs are non-coding RNAs that are flanked by snoRNAs. These unique RNAs form where two snoRNAs lie within a single intron. SnoRNAs are processed from introns by exonucleolytic cleavage that is limited by the snoRNPs (small nucleolar ribonucleoproteins) that are recruited to and assembled onto the snoRNA. Sno-lncRNAs are processed from introns similarly, however, the intervening sequence between the two snoRNAs is also protected from exonucleolytic cleavage. The result is a highly stable, non-coding RNA of variable length that has snoRNA sequences and bound snoRNP proteins at either end. The function of these sno-lncRNAs is unknown, however, the sno-lncRNAS in the chromosome 15 region appear to bind the RNA binding protein, RBFOX2, and may influence alternative splicing by titrating the available levels of this known alternative splice factor79.
miRNAs and piRNAs
There is one known miRNA80 and a cluster of 17 piRNAs located81 within the chromosome 15q11-q13 imprinted region in humans. The human miRNA, miR4508 lies proximal to MKRN3 and may be transcribed as part of the MKRN3AS transcript. This miRNA is not conserved in mouse, however. The human piRNA cluster is located within the C15ORF2/NPAP1 gene. Three piRNAs in this cluster match the long terminal repeat sequence in the 3’ end of the gene, suggesting that C15ORF2/NPAP1 may be recognized as an invasive gene, and may be silenced by RNA mediated mechanisms43. In mouse, a cluster of 10 miRNAs reside in approximately 400 kilobases of genomic sequence between Ndn and Snurf-Snrpn82. The expression patterns and functions of these miRNAs are not known.
CONCLUSIONS
The imprinted region of chromosome 15q11-q13 harbors a wide variety of RNAs from canonical protein-coding transcripts to novel non-coding RNAs of unknown function. The SNURF-SNRPN gene alone accounts for a bi-cistronic transcript, an antisense transcript, lncRNAs of unknown function, snoRNAs, and a novel RNA species called sno-lncRNAs, and is important for three human neurogenetic disorders, Prader-Willi syndrome, Angelman syndrome, and 15q duplication associated autism. How the processing of these multitude RNAs are involved in the human disorders is still not fully known. Studies comparing the neuronal versus non-neuronal RNAs across the region in both human and mouse will undoubtedly shed some light on this complex and interesting genetic region.
Table 2. Large non-coding RNAs.
SNURF-SNRPN, IPW, and UBE3A-ATS sizes are approximate.
| Gene name | Position | Size | Ref. |
|---|---|---|---|
| MKRN3-AS | Hs. unknown Mm. unknown |
Unknown Unknown |
21,62 |
| PWRN2 | Hs. chr15:24,409,926-24,415,053 | 5,128 bp | 43 |
| PWRN1 | Hs. chr15:24,803,304-24,832,926 | 29,623 bp | 43 |
| SNURF-SNRPN | Hs. chr15:25,068,063-25,687,473 Mm.chr7:66,484,149-67,293,246 |
~616,500 bp ~809,000 bp |
15, 46–50, 63–66 |
| PAR-SN** | Hs. chr15:25,227,141-25,228,937 | 1,797 bp | 69 |
| PAR5** | Hs. chr15:25,230,007-25,233,379 | 3,373 bp | 68 |
| HBT8** | Hs. chr15:25,277,534-25,281,635 | 4,102 bp | http://genome.ucsc.edu |
| IPW** | Hs. chr15:25,361,711-25,367,655 Mm. chr7:66,875,161-66,937,474 |
5945 bp 62,314 bp |
70–71 |
| PAR1** | Hs. chr15:25,380,789-25,383,200 | 2,412 bp | 68 |
| PAR4** | Hs. chr15:25,456,839-25,457,180 | 342 bp | 68 |
| UBE3A-ATS** | Hs. chr15:25,582,436-25,684,806 Mm. chr7:66,484,028-66,562,559 |
~102,500 bp ~78,500 bp |
53–55 63,72 |
denotes lncRNAs that are part of the SNURF-SNRPN transcriptional unit, therefore, the size of the SNURF-SNRPN RNA includes the lncRNAs marked with two asterisks.
Acknowledgements
The author would like to thank Marc Lalande, Kristen Martins-Taylor, Ivy Chen, and Jack Hsiao for critical reading of this manuscript. This work is supported by the Raymond and Beverly Sackler Foundation and NIH/NICHD grant R01HD068730.
REFERENCES
- 1.Chamberlain SJ, Lalande M. Neurodevelopmental disorders involving genomic imprinting at human chromosome 15q11-q13. Neurobiology of disease. 2010;39:13–20. doi: 10.1016/j.nbd.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Cattanach BM, et al. A candidate model for Angelman syndrome in the mouse. Mamm Genome. 1997;8:472–478. doi: 10.1007/s003359900479. [DOI] [PubMed] [Google Scholar]
- 3.Cattanach BM, et al. A candidate mouse model for Prader-Willi syndrome which shows an absence of Snrpn expression. Nat Genet. 1992;2:270–274. doi: 10.1038/ng1292-270. [DOI] [PubMed] [Google Scholar]
- 4.Hogart A, Patzel KA, LaSalle JM. Gender influences monoallelic expression of ATP10A in human brain. Hum Genet. 2008;124:235–242. doi: 10.1007/s00439-008-0546-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chai JH, et al. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader-Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am J Hum Genet. 2003;73:898–925. doi: 10.1086/378816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji Y, Eichler EE, Schwartz S, Nicholls RD. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 2000;10:597–610. doi: 10.1101/gr.10.5.597. [DOI] [PubMed] [Google Scholar]
- 7.Cassidy SB, Dykens E, Williams CA. Prader-Willi and Angelman syndromes: sister imprinted disorders. Am J Med Genet. 2000;97:136–146. doi: 10.1002/1096-8628(200022)97:2<136::aid-ajmg5>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 8.Amos-Landgraf JM, et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am J Hum Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christian SL, et al. Integrated YAC contig map of the Prader-Willi/Angelman region on chromosome 15q11-q13 with average STS spacing of 35 kb. Genome Res. 1998;8:146–157. doi: 10.1101/gr.8.2.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13) Hum Mol Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- 11.Reis A, et al. Imprinting mutations suggested by abnormal DNA methylation patterns in familial Angelman and Prader-Willi syndromes. Am J Hum Genet. 1994;54:741–747. [PMC free article] [PubMed] [Google Scholar]
- 12.Saitoh S, et al. Minimal definition of the imprinting center and fixation of chromosome 15q11-q13 epigenotype by imprinting mutations. Proc Natl Acad Sci U S A. 1996;93:7811–7815. doi: 10.1073/pnas.93.15.7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubose AJ, Smith EY, Yang TP, Johnstone KA, Resnick JL. A new deletion refines the boundaries of the murine Prader-Willi syndrome imprinting center. Human Molecular Genetics. 2011;20:3461–3466. doi: 10.1093/hmg/ddr262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Jato S, Nicholls RD, Driscoll DJ, Yang TP. Characterization of cis- and trans-acting elements in the imprinted human SNURF-SNRPN locus. Nucleic Acids Res. 2005;33:4740–4753. doi: 10.1093/nar/gki786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith EY, Futtner CR, Chamberlain SJ, Johnstone KA, Resnick JL. Transcription is required to establish maternal imprinting at the Prader-Willi syndrome and Angelman syndrome locus. PLoS genetics. 2011;7:e1002422. doi: 10.1371/journal.pgen.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin Z, Allis CD, Wagstaff J. Parent-specific complementary patterns of histone H3 lysine 9 and H3 lysine 4 methylation at the Prader-Willi syndrome imprinting center. Am J Hum Genet. 2001;69:1389–1394. doi: 10.1086/324469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fulmer-Smentek SB, Francke U. Association of acetylated histones with paternally expressed genes in the Prader--Willi deletion region. Human Molecular Genetics. 2001;10:645–652. doi: 10.1093/hmg/10.6.645. [DOI] [PubMed] [Google Scholar]
- 18.Chantalat S, et al. Histone H3 trimethylation at lysine 36 is associated with constitutive and facultative heterochromatin. Genome research. 2011;21:1426–1437. doi: 10.1101/gr.118091.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chai JH, Locke DP, Ohta T, Greally JM, Nicholls RD. Retrotransposed genes such as Frat3 in the mouse Chromosome 7C Prader- Willi syndrome region acquire the imprinted status of their insertion site. Mamm Genome. 2001;12:813–821. doi: 10.1007/s00335-001-2083-1. [DOI] [PubMed] [Google Scholar]
- 20.Buiting K, et al. C15orf2 and a novel noncoding transcript from the Prader-Willi/Angelman syndrome region show monoallelic expression in fetal brain. Genomics. 2007;89:588–595. doi: 10.1016/j.ygeno.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Dubose AJ, Johnstone KA, Smith EY, Hallett RA, Resnick JL. Atp10a, a gene adjacent to the PWS/AS gene cluster, is not imprinted in mouse and is insensitive to the PWS-IC. Neurogenetics. 2009 doi: 10.1007/s10048-009-0226-9. [DOI] [PubMed] [Google Scholar]
- 22.Jong MT, et al. A novel imprinted gene, encoding a RING zinc-finger protein, and overlapping antisense transcript in the Prader-Willi syndrome critical region. Hum Mol Genet. 1999;8:783–793. doi: 10.1093/hmg/8.5.783. [DOI] [PubMed] [Google Scholar]
- 23.Jay P, et al. The human necdin gene NDN, is maternally imprinted and located in the Prader-Willi syndrome chromosomal region. Nat Genet. 1997;17:357–361. doi: 10.1038/ng1197-357. [DOI] [PubMed] [Google Scholar]
- 24.Kanber D, et al. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader-Willi syndrome. Eur J Hum Genet. 2009;17:582–590. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bohne A, et al. The vertebrate makorin ubiquitin ligase gene family has been shaped by large-scale duplication and retroposition from an ancestral gonad-specific, maternal-effect gene. BMC genomics. 2010;11:721. doi: 10.1186/1471-2164-11-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boccaccio I, et al. The human MAGEL2 gene and its mouse homologue are paternally expressed and mapped to the Prader-Willi region. Human Molecular Genetics. 1999;8:2497–2505. doi: 10.1093/hmg/8.13.2497. [DOI] [PubMed] [Google Scholar]
- 27.Bischof JM, Stewart CL, Wevrick R. Inactivation of the mouse Magel2 gene results in growth abnormalities similar to Prader-Willi syndrome. Hum Mol Genet. 2007;16:2713–2719. doi: 10.1093/hmg/ddm225. [DOI] [PubMed] [Google Scholar]
- 28.Kozlov SV, et al. The imprinted gene Magel2 regulates normal circadian output. Nature Genetics. 2007;39:1266–1272. doi: 10.1038/ng2114. [DOI] [PubMed] [Google Scholar]
- 29.Mercer RE, Wevrick R. Loss of magel2, a candidate gene for features of Prader-Willi syndrome, impairs reproductive function in mice. PLoS One. 2009;4:e4291. doi: 10.1371/journal.pone.0004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi Y, Matsuyama K, Takagi K, Sugiura H, Yoshikawa K. Arrest of cell growth by necdin, a nuclear protein expressed in postmitotic neurons. Biochemical and biophysical research communications. 1995;213:317–324. doi: 10.1006/bbrc.1995.2132. [DOI] [PubMed] [Google Scholar]
- 31.Taniura H, Taniguchi N, Hara M, Yoshikawa K. Necdin, a postmitotic neuron-specific growth suppressor, interacts with viral transforming proteins and cellular transcription factor E2F1. The Journal of biological chemistry. 1998;273:720–728. doi: 10.1074/jbc.273.2.720. [DOI] [PubMed] [Google Scholar]
- 32.Hasegawa K, Yoshikawa K. Necdin regulates p53 acetylation via Sirtuin1 to modulate DNA damage response in cortical neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2008;28:8772–8784. doi: 10.1523/JNEUROSCI.3052-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muscatelli F, et al. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader-Willi syndrome. Hum Mol Genet. 2000;9:3101–3110. doi: 10.1093/hmg/9.20.3101. [DOI] [PubMed] [Google Scholar]
- 34.Gerard M, Hernandez L, Wevrick R, Stewart CL. Disruption of the mouse necdin gene results in early post-natal lethality. Nature Genetics. 1999;23:199–202. doi: 10.1038/13828. [DOI] [PubMed] [Google Scholar]
- 35.Tsai TF, Armstrong D, Beaudet AL. Necdin-deficient mice do not show lethality or the obesity and infertility of Prader-Willi syndrome. Nat Genet. 1999;22:15–16. doi: 10.1038/8722. [DOI] [PubMed] [Google Scholar]
- 36.Deponti D, et al. Necdin mediates skeletal muscle regeneration by promoting myoblast survival and differentiation. The Journal of cell biology. 2007;179:305–319. doi: 10.1083/jcb.200701027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bush JR, Wevrick R. The Prader-Willi syndrome protein necdin interacts with the E1A-like inhibitor of differentiation EID-1 and promotes myoblast differentiation. Differentiation; research in biological diversity. 2008;76:994–1005. doi: 10.1111/j.1432-0436.2008.00281.x. [DOI] [PubMed] [Google Scholar]
- 38.Bush JR, Wevrick R. Loss of the Prader-Willi obesity syndrome protein necdin promotes adipogenesis. Gene. 2012;497:45–51. doi: 10.1016/j.gene.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 39.Hasegawa K, et al. Necdin controls Foxo1 acetylation in hypothalamic arcuate neurons to modulate the thyroid axis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:5562–5572. doi: 10.1523/JNEUROSCI.0142-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniura H, Matsumoto K, Yoshikawa K. Physical and functional interactions of neuronal growth suppressor necdin with p53. The Journal of biological chemistry. 1999;274:16242–16248. doi: 10.1074/jbc.274.23.16242. [DOI] [PubMed] [Google Scholar]
- 41.Lee S, et al. Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Human Molecular Genetics. 2005;14:627–637. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- 42.Wawrzik M, et al. The C15orf2 gene in the Prader-Willi syndrome region is subject to genomic imprinting and positive selection. Neurogenetics. 2010;11:153–161. doi: 10.1007/s10048-009-0231-z. [DOI] [PubMed] [Google Scholar]
- 43.Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biology of the cell / under the auspices of the European Cell Biology Organization. 2008;100:149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- 44.Neumann LC, et al. The imprinted NPAP1/C15orf2 gene in the Prader-Willi syndrome region encodes a nuclear pore complex associated protein. Human Molecular Genetics. 2012 doi: 10.1093/hmg/dds228. [DOI] [PubMed] [Google Scholar]
- 45.van Amerongen R, van der Gulden H, Bleeker F, Jonkers J, Berns A. Characterization and functional analysis of the murine Frat2 gene. The Journal of biological chemistry. 2004;279:26967–26974. doi: 10.1074/jbc.M400439200. [DOI] [PubMed] [Google Scholar]
- 46.Gray TA, Saitoh S, Nicholls RD. An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5616–5621. doi: 10.1073/pnas.96.10.5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huntriss JD, Latchman DS, Williams DG. The snRNP core protein SmB and tissue-specific SmN protein are differentially distributed between snRNP particles. Nucleic Acids Research. 1993;21:4047–4053. doi: 10.1093/nar/21.17.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang T, et al. A mouse model for Prader-Willi syndrome imprinting-centre mutations. Nature Genetics. 1998;19:25–31. doi: 10.1038/ng0598-25. [DOI] [PubMed] [Google Scholar]
- 49.Bressler J, et al. The SNRPN promoter is not required for genomic imprinting of the Prader-Willi/Angelman domain in mice. Nat Genet. 2001;28:232–240. doi: 10.1038/90067. [DOI] [PubMed] [Google Scholar]
- 50.Runte M, et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 51.Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 53.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 54.Chamberlain SJ, Brannan CI. The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics. 2001;73:316–322. doi: 10.1006/geno.2001.6543. [DOI] [PubMed] [Google Scholar]
- 55.Meng L, Person RE, Beaudet AL. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Human Molecular Genetics. 2012 doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 57.Yamamoto Y, Huibregtse JM, Howley PM. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 1997;41:263–266. doi: 10.1006/geno.1997.4617. [DOI] [PubMed] [Google Scholar]
- 58.Jiang YH, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 59.Miura K, et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9:149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- 60.Yashiro K, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12:777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sato M, Stryker MP. Genomic imprinting of experience-dependent cortical plasticity by the ubiquitin ligase gene Ube3a. Proc Natl Acad Sci U S A. 2010;107:5611–5616. doi: 10.1073/pnas.1001281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sutcliffe JS, et al. Deletions of a differentially methylated CpG island at the SNRPN gene define a putative imprinting control region. Nature Genetics. 1994;8:52–58. doi: 10.1038/ng0994-52. [DOI] [PubMed] [Google Scholar]
- 63.Ning Y, et al. Identification of a novel paternally expressed transcript adjacent to snRPN in the Prader-Willi syndrome critical region. Genome research. 1996;6:742–746. doi: 10.1101/gr.6.8.742. [DOI] [PubMed] [Google Scholar]
- 64.Jong MT, et al. Imprinting of a RING zinc-finger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Human Molecular Genetics. 1999;8:795–803. doi: 10.1093/hmg/8.5.795. [DOI] [PubMed] [Google Scholar]
- 65.Castle JC, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buiting K, et al. Disruption of the bipartite imprinting center in a family with Angelman syndrome. Am J Hum Genet. 2001;68:1290–1294. doi: 10.1086/320120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farber C, Dittrich B, Buiting K, Horsthemke B. The chromosome 15 imprinting centre (IC) region has undergone multiple duplication events and contains an upstream exon of SNRPN that is deleted in all Angelman syndrome patients with an IC microdeletion. Hum Mol Genet. 1999;8:337–343. doi: 10.1093/hmg/8.2.337. [DOI] [PubMed] [Google Scholar]
- 68.Landers M, et al. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wevrick R, Kerns JA, Francke U. Identification of a Novel Paternally Expressed Gene in the Prader- Willi-Syndrome Region. Human Molecular Genetics. 1994;3:1877–1882. doi: 10.1093/hmg/3.10.1877. [DOI] [PubMed] [Google Scholar]
- 70.Cavaille J, et al. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci U S A. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chamberlain SJ, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci U S A. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wevrick R, Francke U. An imprinted mouse transcript homologous to the human imprinted in Prader-Willi syndrome (IPW) gene. Human Molecular Genetics. 1997;6:325–332. doi: 10.1093/hmg/6.2.325. [DOI] [PubMed] [Google Scholar]
- 73.Numata K, Kohama C, Abe K, Kiyosawa H. Highly parallel SNP genotyping reveals high-resolution landscape of mono-allelic Ube3a expression associated with locus-wide antisense transcription. Nucleic Acids Research. 2011;39:2649–2657. doi: 10.1093/nar/gkq1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 75.Vitali P, et al. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. The Journal of cell biology. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glatt-Deeley H, Bancescu DL, Lalande M. Prader-Willi syndrome, Snord115, and Htr2c editing. Neurogenetics. 2009 doi: 10.1007/s10048-009-0209-x. [DOI] [PubMed] [Google Scholar]
- 77.Kishore S, et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Human Molecular Genetics. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bortolin-Cavaille ML, Cavaille J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Research. 2012 doi: 10.1093/nar/gks321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yin Q-F, Yang L, Zhang Y, Xiang J-F, Wu Y-W, Carmichael GG, Chen L-L. Long noncoding RNAs with snoRNA ends. Molecular Cell. 2012 doi: 10.1016/j.molcel.2012.07.033. In press. [DOI] [PubMed] [Google Scholar]
- 80.Jima DD, et al. Deep sequencing of the small RNA transcriptome of normal and malignant human B cells identifies hundreds of novel microRNAs. Blood. 2010;116:e118–e127. doi: 10.1182/blood-2010-05-285403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 82.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes & Development. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]