Abstract
Introduction
Mucin 1 (MUC1) is a high molecular weight transmembrane glycoprotein with an aberrant expression profile in various malignancies including breast cancer. Its increased overexpression and underglycosylation in breast cancer is associated with tumor invasiveness and metastatic potential. In this report we made the next step towards establishing MUC1 as a potential diagnostic, prognostic and therapeutic target by investigating its expression and post-translational modification (glycosylation/sialation).
Materials and Methods
In these studies we used a breast cancer tissue microarray and freshly frozen multistage breast cancer tissues. We analyzed in detail the expression of normal and underglycosylated/sialated MUC1 by immunohistochemistry, real time qRT-PCR and various analytical techniques.
Results
We found that changes in cellular localization as well as in upregulation and/or underglycosylation of MUC1 were associated with higher tumor grade. A key finding in this study was that underglycosylated MUC1 overexpression and sialation were observed in tissues adjacent to tumor but identified as “normal” on pathology reports.
Conclusion
These findings suggest that uMUC1 can indeed be used as an early diagnostic marker and provide additional insights into breast cancer management.
Keywords: glucosylation, sialation, malignant transformation, cancer biomarker, tumor grade
Introduction
Mucin 1 (MUC1 heterodimer) is a highly o-glycosylated transmembrane protein predominantly expressed on the apical surface of glandular epithelial cells (1, 2). It is involved in normal lubrication and protects the apical border against microorganisms and damage induced by cellular events (3, 4). MUC1 has a large extracellular domain consisting predominantly of variable numbers of O-glycosylated 20 amino acid tandem repeats (VNTR), a transmembrane domain and a 72 amino acid cytoplasmic tail (5–7). Within each repeat sequence, there are five potential glycosylation sites. Cellular distribution of MUC1 is allocated to the cytoplasm, as well as to the cell surface (8). In normal tissues MUC1 is heavily glycosylated (carbohydrates contribute 50–90% of its molecular mass) (9). During tumorogenesis, expression of uMUC1 is no longer restricted to the apical surface of plasma membranes and covers the entire cell surface leading to interaction of its cytoplasmic tail with signaling molecules (10, 11). Furthermore, in neoplastic tissues, MUC1 is underglycosylated (uMUC1, (9)) because of the premature termination of the carbohydrate chains by the addition of sialic acids limiting their branching potential (12, 13).
Such abnormal expression has been associated with cellular growth, transformation, adhesion, invasion and immune cells responsiveness and tolerance (14, 15). It has been shown that MUC1 is overexpressed on almost all human epithelial cell adenocarcinomas including ovarian (16–18), pancreatic (19), colorectal (20), lung (21, 22), prostate (23), colon (19) and gastric carcinomas (24, 25). Furthermore, MUC1 expression has been demonstrated in non-epithelial cancer cell lines (astrocytoma, melanoma and neuroblastoma (26)), and in hematological malignancies such as multiple myeloma and some B-cell non-Hodgkin lymphomas (27–30), bringing the number to over 50% of all cancers in humans (31).
In breast adenocarcinomas uMUC1 plays a crucial role, with over 90% of these cancers overexpressing its underglycosylated form (32–34). It has been directly linked to tumor invasiveness and metastatic potential (35–37). It has been recently demonstrated that uMUC1 is one of the seven highly expressed marker genes identified in ductal carcinoma in situ (DCIS) and in invasive ductal carcinoma (IDC) in human and murine samples (38). Involvement of uMUC1 in tumorogenesis also leads to induced expression of genes predictive of the outcome of chemotherapy and overall clinical outcome in breast cancer patients (39) which makes it an attractive target for both therapy (40) and diagnostic imaging (41, 42). In fact, it has been shown that mammary tumor progression is delayed in MUC1 null mice (43).
Increased sialic acid addition is particularly important in breast cancer transformation and progression to metastasis. Sialyltransferases (ST) are responsible for early transfer of sialic acid from CMP-sialic acid to carbohydrate groups on uMUC1, which reduces branching of sugar moieties (13). It has been shown that ST expression is altered during cancer progression and metastasis (44, 45). In breast cancer high level of sialyltransferses such as ST3Gal-I (46) and ST6Gal-I (47) has been correlated with shorter survival and poor prognosis (48). Among sialated antigens, sialyl-Tn (STn) is a mucin-type O-glycan, which is associated with 30% of all breast carcinoma and is correlated with a decrease of overall survival of patients (49). STn has recently been used as a target for cancer immunotherapy (50).
Since uMUC1 seems to play one of the key roles in breast cancer progression, in this report we made the next step towards establishing it as a potential diagnostic, prognostic and therapeutic target by investigating its expression and post-translational modification (glycosylation/sialation) in tissue microarrays and in the freshly frozen tissue samples obtained after surgical rejection.
Materials and Methods
Tissue specimens
Human tissue microarrays (TMAs) were obtained from the Cooperative Human Tissue Network (CHTN) of the National Cancer Institute (NCI), the National Institutes of Health, Bethesda, MD (http://faculty.virginia.edu/chtn-tma/home.html). The TMAs contained 56 formalin fixed paraffin embedded samples of human breast epithelium representing various stages of tumor progression in breast adenocarcinoma including: 1) non-neoplastic breast epithelium from healthy subjects (NB-NC); 2) non-neoplastic breast epithelium from patients with breast cancer (NB-C); 3) ductal carcinoma in situ - low grade (DCIS-L); 4) ductal carcinoma in situ - high grade (DCIS-H); 5) invasive ductal adenocarcinoma - low grade (IDC-L); 6) invasive ductal adenocarcinoma- high grade (IDC-H); 7) metastatic carcinoma to regional lymph nodes (LNM). In addition, freshly frozen individual human breast tissue samples of all stages of cancer were also collected from CHTN (n=48). All studies were conducted in accordance with IRB #2009-P-001766 approved by the review board at Massachusetts General Hospital.
The sample cohort consisted of adjacent normal (16), DCIS (6), Stage I/IDC (4), Stage II/IDC (12), Stage III/IDC (5), Stage IV/metastatic (6) cancer and five healthy control samples. For histological classification such as TNM stage, receptor status etc., tissue samples were examined by the pathologist at the CHTN participating hospital.
Immunohistochemistry (IHC)
The TMAs of thin paraffin sections (7µm) of breast tissues were heated to 60°C for 15 min to improve tissue adhesion to the charged glass slides (Fisher Plus). Before immunostaining, TMA slides were deparaffinized in xylene (2×5min) and then gradually rehydrated in a series of alcohol baths (100, 95, and 75%) and in distilled water. To improve antigen retrieval, the samples were treated with 10mM sodium citrate buffer (pH 6.0) at 100°C for10 min. Endogenous peroxidase activity was blocked by treating with 3% hydrogen peroxide. The samples were washed with phosphate buffered saline (PBS; pH 7.2) and incubated in 3% blocking serum (goat or horse) for 1h. The tissue samples were then incubated either with an anti-MUC1 mouse monoclonal antibody (clone VU4H5; Zymed Lab, Invitrogen, 1:100 dilution), which stains tandem repeat portion of the antigen or hamster anti-human MUC1AB-5 to the cytoplasmic tail of MUC1 (clone MH1 or CT2; Thermo Fisher, 1:100 dilution) overnight at 4°C. After washing in PBS slides were incubated with biotinylated secondary antibody (horse anti-mouse or goat anti-hamster immunoglobulins respectively (DAKO) for 45 min at RT. Next, the sections were incubated with an avidin–biotin–peroxidase complex (ABC) reagent (Vectastain ABC Elite kit; Vector Laboratories, Burlingame, CA) and developed using DAB (ImmPACT DAB; DAKO). Slides were counterstained with hematoxylin (Sigma), dehydrated and mounted using Permount (Sigma). Control experiments included incubating slides with nonimmune serum and by omitting the primary antibody.
Scoring of immunoreactivity
The stained slides were visually scored in a double-blind fashion by two independent investigators. Expression of MUC1 in the normal tissue and ductal lesions was scored as negative or (heterogeneously) positive. Immunohistochemical localisation of the antibody was divided into apical (luminal surface) and entire cell membrane staining. Aberrant expression was defined as expression of MUC1 on the entire cell membrane and/or expression of underglycosylated forms of MUC1. The number of low power (×10) optical fields in a specimen that were positively stained was expressed as a percentage of the total number of optical fields contained in the tissue. Evaluation of MUC1 semiquantitative immunostaining was carried out by assessing the intensity of apical and cytoplasmic staining giving a score of 0 for negative, + for weak, ++ for moderate, +++ for strong and ++++ for very strong staining. Scoring was performed by three independent operators.
Real-time qRT-PCR
Frozen breast tissues were sectioned into pieces and homogenized on ice using a mechanical glass-Teflon homogenizer set at 3,000 rpm. Total RNA was extracted from breast tissue lysates using the Qiagen RNeasy Mini Kit (Qiagen, Inc., Valencia, CA). Quantitative qRT-PCR was performed using total RNA as described (42). TaqMan real time PCR analysis was done using an ABI Prism 7700 sequence detection system (PE Applied Biosystems). The PCR primers and TaqMan probe specific for MUC1 mRNA were designed using Primer express software 1.5. Primer and probe sequences were as follows: forward primer, 5’-ACAGGTTCTGGTCATGCAAGC-3’ (nucleotides 64–84 in the 5’-nonrepetitive region); reverse primer, 5’-CTCACAGCATTCTTCTCAGTAGAGCT- 3’ (nucleotides 139–164 in the 5’-nonrepetitive region); and TaqMan probe, 5’-FAM-TGGAGAAAAGGAGACTTCGGCTACCCAGA-TAMRA-3’ (nucleotides 96–124 in the 5’-nonrepetitive region). Eukaryotic 18S rRNA TaqMan PDAR Endogenous Control reagent mix (PE Applied Biosystems) was used to amplify 18S rRNA as an internal control according to the manufacturer’s protocol. At the RT step, incubation was done at 48°C for 30 min followed by 95°C for 10min for enzyme inactivation. The PCR reaction was run for 40 cycles (two steps) at 95°C for 15 sec and at 60°C for 60 sec. Relative expression was calculated according to the delta delta CT methods (51).
Western Blotting
Frozen tumor specimens were thawed and homogenized in tissue protein extraction lysis buffer (Tissue-PE LB from G-Biosciences, St Louis, MO) containing 1mM PMSF and proteinase inhibitor cocktails (Sigma). Protein content was determined with the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). Twenty micrograms of total protein from each sample was applied onto SDS-polyacrylamide gel (4–20%) under reducing conditions. Rainbow marker (Amersham Life Science, Arlington Heights, IL) was used as a high molecular weight standard. Resolved proteins were transferred electrophoretically to nitrocellulose membranes and pre-blocked with 5% nonfat dry milk in Tris-buffered saline (TBS) for 1h at room temperature. After blocking, the membrane was incubated overnight at 4°C in 1% milk/TBS containing either monoclonal anti-human MUC1 antibody (1µg/ml; C595 (NCRC48), Abcam, Cambridge, MA) or monoclonal β-actin (1µg/ml; Applied Biosystem). The membrane was then washed 3 times with 0.05% Tween TBS (TBST) and incubated with horseradish peroxidase-conjugated goat anti-mouse IgG (Invitrogen, Camarillo, CA) for 60 minutes at room temperature, followed by washing 3 times with TBST and one time with TBS. Membranes were then developed using an ECL Plus Western Blotting detection reagent kit (GE Healthcare, Piscataway, NJ) according to manufacturer’s specifications.
Neuraminidase treatments
In order to remove sialic acid from MUC1 antigen, total lysates from extracted breast tissue (20µg) were digested overnight at 37°C with neuraminidase from Clostridium perfringens (New England Biolabs, Ipswitch, MA) at a final concentration (1U/µl) in 50mM sodium citrate (pH 6.0). Preparations without the enzyme served as controls. The samples were then boiled with 2X reducing sample buffer (BIO-RAD), subjected to SDS-PAGE (4–20%) followed by Western blotting using anti-MUC1 antibody as described above.
Sialyltransferase assay
To evaluate sialyltransferase activity we utilized a fluorescence assay based on the method described by Gross et al. with minor modification (52). The standard reaction mixture (30µl) contained a 62.5mM sodium cacodylate buffer, pH 6.5, 1.66mg/ml asialofetuin (exogenous acceptor) and 166µM CMP-fluoresceinyl-AcNeu. The latter was obtained by labeling CMP-ac-Neu (EMD Biosciences) with FITC using FITC labeling kit (Calbiochem) followed by HPLC purification. The reaction was initiated by adding 25µg of proteins from breast tumor lysates. After incubation at 37°C for 1 h in the dark, the reaction was terminated by adding 10µl of a sample buffer (4×; non-reducing; Bio-RAD) followed by incubation for 2 min at 100°C. The reaction products were separated using 10% SDS-PAGE. After migration fluorescently labeled glycoproteins were detected using an IVIS imaging system (Caliper Life Science/Perkin Elmer, Hopkinton, MA) equipped with 500nm excitation and 540nm emission filters. Background fluorescence level was obtained using controls without protein lysates. A region of interest (ROI) was manually selected over relevant regions of fluorescence intensity. The area of the ROI was kept constant, and the intensity (Total radiant efficiency) was recorded as maximum photon counts within an ROI. The higher radiant efficiency represented the higher enzyme activity in the samples as indicated in the figures.
Immunohistochemical detection of MUC1 STn antigen
Tumor tissue sections selected for STn expression, were deparaffinized in xylene and rehydrated in a series of ethanols. Sections were incubated with mouse monoclonal antibody to STn (CA 72-4 Ab-1; clone B72.3; Thermo scientific, Hudson, NH) at 4°C overnight. After washing in PBS, sections were incubated with biotinylated horse anti-mouse IgG (DAKO) diluted 1:200 and Strept ABC complex/HRP (DAKO). The remaining steps were carried out as described above for IHC.
Statistical analysis
All data were represented as mean +/ SD. Statistical analysis was done using a two-tailed Student’s t test and linear regression where indicated. P<0.05 was considered statistically significant.
Results
MUC1 detection in multi-stage human breast cancer
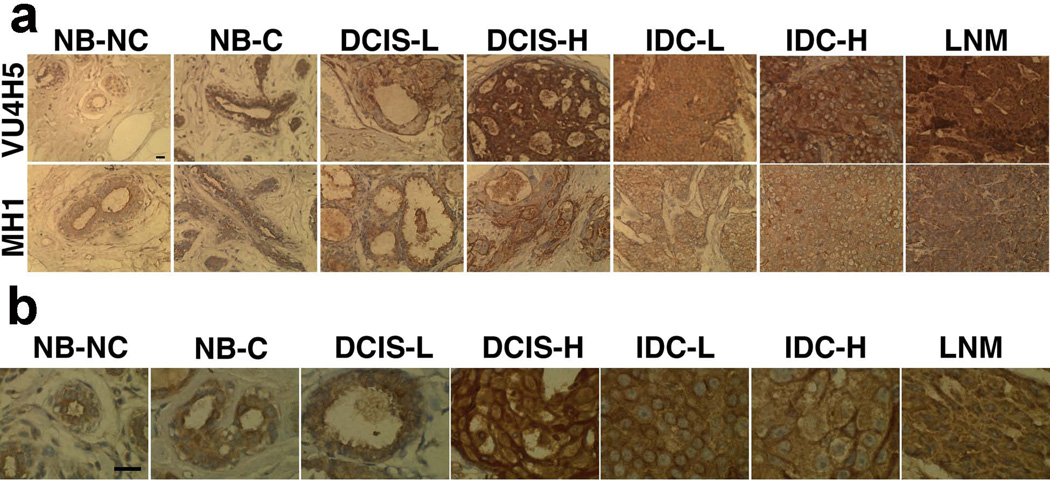
Tissue distribution of MUC1 was examined by light microscopy of a TMA containing 56 human breast tissue sections. Tissue sections were incubated separately with two primary antibodies to the variable underglycosylated extracellular portion of uMUC1 (VU4H5 clone) and to the non-variable cytoplasmic tail of MUC1 (MH1 clone). Samples from patients with no history of breast cancer (NB-NC) showed normal glandular architecture with very weak staining with VU4H5 antibody as well as with MH1 antibody (Fig. 1a; enlarged view of staining with VU4H5 is shown in Fig. 1b; overview is shown in Supplemental Fig. 1). Off note, the VU4H5 antibody reflects the posttranslational modification of the antigen, because it binds to the tandemly repeated protein backbone (this backbone is differentially exposed for antibody binding by the differential glycosylation of the tandem repeat). By contrast, the MH1 antibody binds the non-variable, non-glycosylated cytoplasmic tail and reflects the absolute expression of the antigen. Non-neoplastic breast epithelium from patients with breast cancer (NB-C) showed clear glandular architecture with intermediate staining with both VU4H5 and MH1. Staining appears to be both cytosolic and membranous with some elements restricted to the apical surface of the glandular epithelium (Fig. 1b). Tissue samples from those patients with low-grade ductal carcinoma in situ (DCIS-L) showed heterogeneous glandular architecture that appears normal in some cases but disordered in others. MUC1 staining with both VU4H5 and MH1 antibodies showed intermediate intensity with the distribution restricted to the apical surface of the glandular epithelium. Staining appears to be both cytosolic and membranous (Fig. 1b), with obvious enlargement of the glands. Staining of samples from high-grade ductal carcinoma in situ (DCIS-H) revealed strong staining with both VU4H5 and MH1 antibodies localized to both apical and basolateral surface of the glandular epithelium. Clearly disordered glandular architecture was observed in these samples though the glands were still distinguishable. Staining appears to be both cytosolic and membranous (Fig. 1a, b). Specimens from low-grade invasive ductal adenocarcinoma (IDC-L) exhibited strong staining with both VU4H5 and MH1 antibody and disordered, barely distinguishable glandular architecture with ubiquitous MUC1 distribution. In some of the tissues there is a clearer appearance of glandular tissue with apical and basolateral staining. Staining appears both cytosolic and on the cell-surface.
Fig 1.
Immunohistochemical staining of a human breast tissue microarray (TMA) representative of tumor progression. The breast TMAs were incubated with anti-MUC1 (clone VU4H5) antibody that binds underglycosylated MUC1 and MH1 (CT-2) antibody to cytoplasmic tail of MUC1. a) uMUC1 expression was evident in all tumor stages including adjacent normal tussue. Magnification bar = 10 µm; b) Transition of uMUC1 distribution from apical localization (NB-NC, normal, non-cancerous tissue, arrow) to randomized cytoplasmic/ membranous staining with increasing tumor grade. Note that NB-C tissue showed mixed staining (cytoplasmic/ membranous with some apical localization). Magnification bar = 50 µm.
Finally, specimen from invasive ductal adenocarcinoma- high grade (IDC-H) showed strong ubiquitous staining that revealed the complete loss of glandular architecture. Staining appears both cytosolic and cell-surface.
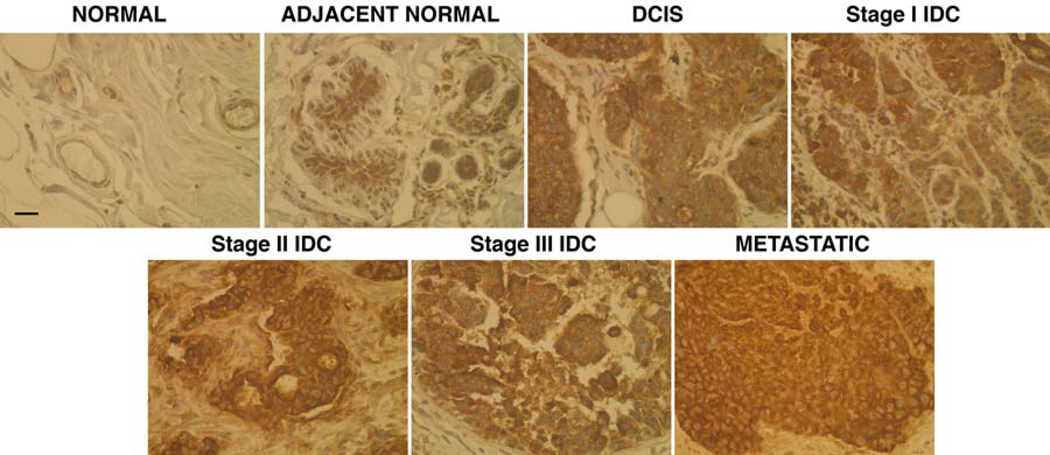
Metastatic carcinoma from regional lymph nodes (LNM) shows strong ubiquitous staining with both antibodies indicating a high level of MUC1 expression distributed throughout the entire tissue and a complete absence of glandular architecture. The majority of the staining was detected within cytosol and the entire cell membrane. Next we validated our results from TMA tissue analysis by performing IHC on freshly frozen biopsy samples (n=48) representing multi-stage breast cancer. Sections stained with VU4H5 clone (Fig. 2) showed that the degree of expression and cellular localization of uMUC1 varied significantly between tumor and normal tissues. In accordance with TMA data, in higher tumor grades major MUC1 expression was localized predominantly to cytosol and cell membrane. There was a clear association between subcellular uMUC1 expression and tumor staging obtained by pathological assessment. In case of non-neoplastic tissues in adjacent normal regions, uMUC1 stained strongly in apical and moderately in cytosolic compartments indicating that molecular changes in this tissue begin at the very early stages. The results from IHC studies are summarized in Table 1. The overall total staining indicated the presence of the highest levels of uMUC1 in both DCIS and Stage I, which were steadily maintained until the metastatic stage. In addition, similar to results from TMA staining, adjacent normal tissues also contained significant levels of uMUC1 (p<0.0002).
Fig 2.
IHC for uMUC1 in freshly frozen breast tumor tissues from patients with various tumor stages determined by pathological assessment. There was increasing staining for uMUC1 antigen with the higher tumor stage. Magnification bar = 50 µm.
Table 1.
Muc-1 expression in breast cancer tissues (IHC).
| Stages | Total staining (p<0.0002) |
Degree of basolateral staining |
Degree of cytosolic vs membrane staining |
|---|---|---|---|
| Normal (no cancer history) | 1.00±0.00 | 1.83±1.61 | 1.33±1.15 |
| Adjacent normal (cancer history) | 2.04±0.75 | 2.58±0.63 | 2.68±0.46 |
| DCIS | 4.17±0.76 | 3.67±0.58 | 3.00±1.00 |
| Stage I | 3.88±0.25 | 3.88±0.25 | 3.38±0.48 |
| Stage II | 3.08±1.28 | 3.25±0.61 | 2.42±0.38 |
| Stage III | 3.00±0.00 | 3.50±0.71 | 3.00±0.00 |
| Metastatic | 3.75±1.77 | 2.50±0.71 | 2.00±0.00 |
Data represented as average ± stddiv
Molecular analysis of uMUC1 in malignant breast tissues
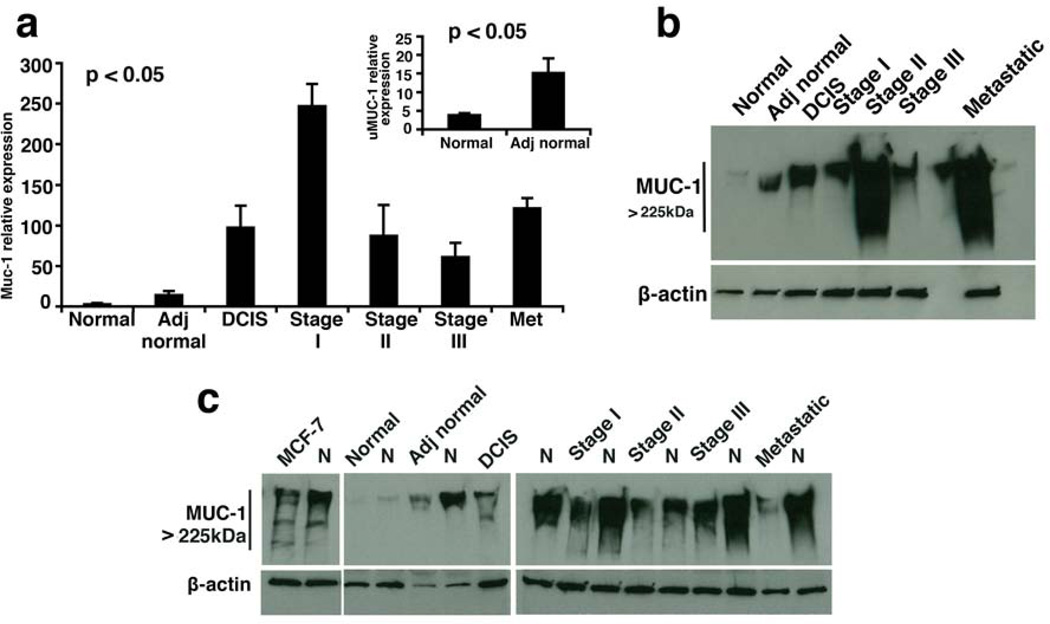
Our microscopy studies confirmed upregulation of uMUC1 with disease progression. To verify these findings at the transcription level, we performed real-time qRT-PCR to detect relative expression of muc1 gene. As shown in Fig. 3a there was already a significant increase in MUC1 mRNA expression starting in adjacent normal tissue (p<0.05) followed by DCIS. The level of expression peaked at stage I IDC and then dropped to a maintenance level throughout the disease progression. Two conclusions can be drawn from these results. First, it is evident that an upregulation of MUC1 coincides with the tissue acquiring a more malignant phenotype. Second, this upregulation was already apparent in adjacent normal tissue of patients with breast cancer.
Fig 3.
uMUC1 expression in tissues from multi-stage breast cancer patients. a) MUC1 mRNA levels were measured by real-time qRT-PCR. Tissues of all stages showed increased MUC1 expression. Note that expression in adjacent normal tissue was >3 times higher than in normal tissue (p<0.05, insert). b) Western blot analysis of MUC1 protein levels. There was a significantly higher protein expression in tissues of all stages including adjacent normal compared to normal tissue. c) Analysis of uMUC1sialation levels in tissues of multistage breast cancer patients. Increased availability of MUC1 epitopes after neuraminidase treatment (N) was demonstrated in all samples including adjacent normal tissue compared to normal tissue. MCF-7 cell lysates served as a MUC1 positive control.
To correlate the results from PCR to protein expression, extracts prepared from the same tissues were analyzed by Western blot. We first performed systematic investigation of uMUC1 availability in tissue extracts from patients representing normal breast epithelium and the different stages of adenocarcinoma. As expected, we observed a clear increase in the amount of uMUC1 in adjacent normal tissue from patients with breast cancer relative to normal tissue from patients without a history of breast cancer (Fig. 3b). In addition, we observed an increase in the abundance of uMUC1 in all stages of disease relative to normal tissue (Fig. 3b). This pattern is reflective of both the absolute expression of the muc1 gene, as shown by qRT-PCR and the level of underglycosylated MUC1 protein obtained by Western blot.
Next, we investigated the sialation status of uMUC1 in these tissues by treating them with pan-neuraminidase, an enzyme that specifically removes sialic acids from the core peptide of MUC1. We hypothesized that desialylation of MUC1 would allow for the antibody to get a better access to MUC1 epitopes. A marked increase in band density has been observed in all neuraminidase-treated tissue lysates including adjacent normal tissue. This trend was especially pronounced in the metastatic samples, consistent with multi-fold increased underglycosylation (Fig. 3c). Treating of normal tissue (no cancer history) with neuraminidase did not produce any staining. These observations were also confirmed with an additional uMUC1 antibody (VU4H5) to validate our findings (Data not shown).
Early molecular changes in uMUC1 expression in normal tissues adjacent to cancer lesions
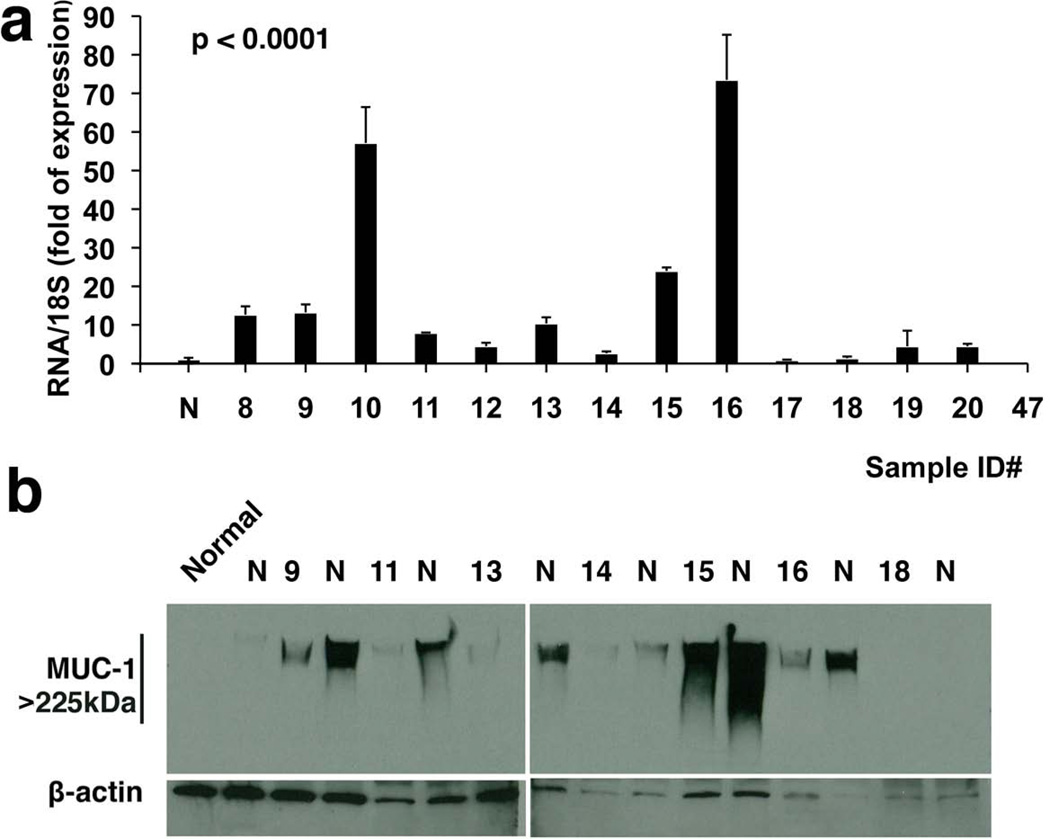
A detailed RNA expression analysis of uMUC1 in adjacent normal tissues from patients with cancer history is shown in Fig. 4a. More than 85% (12 out of 14 tested) of mRNA samples demonstrated the higher level of uMUC1 mRNA expression compared to healthy controls. These data clearly correlate with protein expression in samples as detected by IHC. Neuraminidase treatment of these samples analyzed by Western blotting (Fig. 4b) indicated that in all samples (except for one) uMUC1 was present in a sialated form similar to what was shown with tumors of different stages (Fig. 3c). These results are significant because they confirm our previous observations by light microscopy of tissue microarrays indicating the emergence of an abnormal uMUC1 phenotype in apparently normal tissue from breast cancer patients. This phenomenon may be a representation of the so called “field effect” of carcinogenesis and suggests that MUC1 is a very early marker of transformation.
Fig 4.
Analysis of altered uMUC1 expression in human breast tissue from adjacent normal with cancer history. a) The levels of MUC1 transcript were measured by real-time qRT-PCR. All samples (except for samples ##17 and 47) exhibited significantly higher levels of MUC1 mRNA compared to normal tissue (p< 0.05). b) Western blot analysis of sialated uMUC1 proteins from adjacent normal tissues demonstrated increased epitope availability upon treatment with neuroaminidase (N). Sample IDs in Western blot correspond to those shown in panel a).
Sialyltransferase activity (ST) is elevated in breast tumor tissues
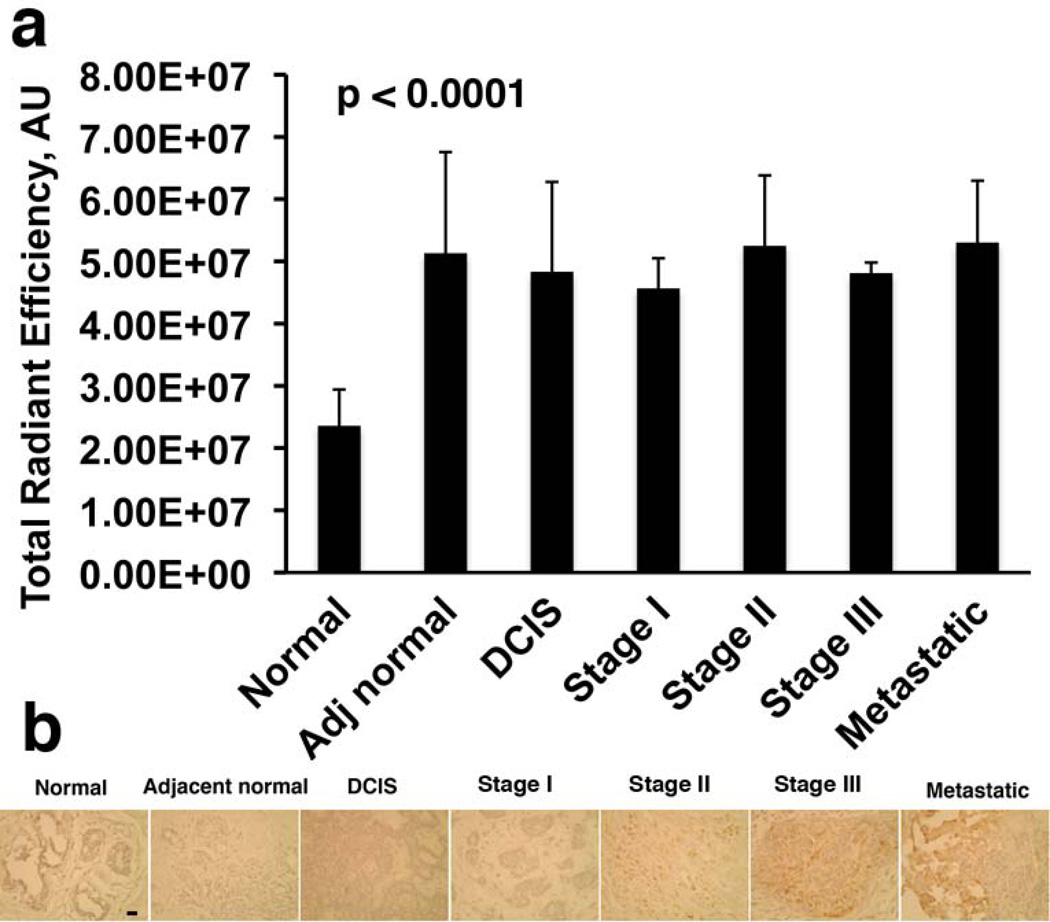
Next we confirmed that the presence of sialated uMUC1 in breast cancer tissue samples concomitant with the disease initiation was due to ST enzyme activity. The experiment was performed by analyzing the transfer of FITC-sialic acid (CMP-FITC-Neu as a donor) to asialofetuin (serving as an acceptor). As shown in Fig. 5a, a strong ST activity was present in tissue extracts from various stages of breast cancer including adjacent normal samples from patients with a breast cancer history compared to healthy normal tissue (p<0.0001). Measured ST activity by this method represents a total pan-ST enzyme activity including three breast tumor associated ST enzymes, ST3Gal-1, ST6Gal-1 and ST6GalNAc-I. The latter is responsible for the synthesis of mucin-carried sialated STn, the most common glycoprotein and independent predictor of poor prognosis in ~30% of breast cancer patients (53). We hypothesized that upregulated ST6GalNAc-I activity might elevate the synthesis of STn. To analyze STn formation, tumor sections were incubated with mucin-carried-STn epitope-specific mouse monoclonal antibody. Fig. 5b demonstrated the presence of typical STn staining in tumors with a more invasive phenotype at higher stages of disease. Metastatic tissue here showed the strongest antibody staining whereas staining of normal breast tissue was negative.
Fig 5.
Detection of sialyltransferase activity in breast tissue lysates. a) Enzyme activity was assayed using asialofetuin as acceptor and CMP-FITC NeuAc as a sugar donor as described in Materials and Methods. Increased ST activity was detected in all tissues. b) Distribution of mucin-carried-sialylated-Tn (STn) epitopes caused by ST6GalNAc-I activation in breast tissue sections detected by IHC. There was a clear increase in expression of STn in invasive (metastatic) tissues. Magnification bar = 10 µm.
Discussion and Conclusion
Post-translational modification of proteins is essential for cellular functions particularly in relation to interactions with other cells and the extracellular matrix. Altered glycosylation/sialation among cell surface proteins is common and important in cancer- related transformation and metastasis. Aberrant glycosylation frequently observed in breast cancer results in the synthesis of multiple cancer-associated glycan including STn, an O-linked antigen carried by mucins and other glycoproteins.
There is a lack of consensus in the literature on the relevance of the uMUC1 antigen to breast cancer emergence and progression. In this study, we analyzed expression of underglycosylated MUC1 and its transformation in various stages of breast cancer using both tissues in TMA and freshly frozen tissue from patients. We utilized various analytical techniques (qRT-PCR, IHC, Western blotting and underglycosylation and sialation assays) to identify changes in MUC1 expression and distribution with disease progression. In agreement with previous studies (54–57) we found that apical expression of uMUC1 was associated predominantly with lower tumor grade whereas in the higher grade tumors its distribution acquired a cytoplasmic pattern.
Analysis of qRT-PCR results indicates that MUC1 expression peaks during the early stages of invasive ductal carcinoma but then levels off or even decreases during the later more invasive stages of the disease. This observation brings up the possibility that in more invasive forms of breast cancer the expression of uMUC1 antigen may be lower while the levels of underglycosylation may be more dramatic. This would reflect a two-stage process of carcinogenesis in which there is an initial upregulation of the antigen, followed by replacement of more heavily glycosylated forms of the molecule with substantially more underglycosylated forms. Considering that sialation and the associated underglycosylation of MUC1 enhances tumor cell binding to selectins and dissemination of these cells into the bloodstream, our hypothesis would be consistent with the transition to post-translational modification of MUC1 in more invasive stages.
A reduced level of glycosylation followed by sialation terminates oligosaccharide chains of glycoproteins. In breast cancer aberrant glycosylation of MUC1 begins with alpha2-3-sialation due to the elevation of ST3Gal-1 (46) followed by ST6Gal-1 mediated alpha2-6-sialation (47), whereas alpha2-6-linkage to GalNAc-1 moiety (Sialyl-Tn) is mediated by the elevated ST6GalNAc-1 enzyme, which is critical for metastasis (58). To clarify the actual incidence of STn expression associated with tumor-specific ST activity we analyzed total ST activity and the downstream formation of STn in breast tumor tissues. The results presented here showed increasing sialyltransferase activity in breast tumor tissues and in adjacent normal regions whereas STn expression with distinct morphology was restricted to tissues with higher tumor grades and a highly invasive phenotype as in metastatic samples.
Two key findings emerged from our results. First, upregulation of uMUC1 is associated with tumors acquiring a more malignant phenotype. This was accompanied by redistribution of uMUC1 cellular expression from the apical surface to cytoplasm and cell membrane. Importantly, uMUC1 upregulation was significant in normal tissues adjacent to tumors. Second, it is apparent that MUC1 becomes underglycosylated very early in pathology. This was clearly seen not only in low grade DCIS but in normal tissues of patients with breast cancer history. There are several important implications of these findings. Adjacent tissues that were identified as “normal” on pathology reports showed significant aberrations in uMUC1 expression, distribution and sialation. There is a possibility that these abnormalities can be associated with more “systemic” molecular transformation of the entire breast epithelium suggesting that uMUC1 can indeed be used as an early diagnostic marker. However, we should not rule out the possibility that while adjacent tissues were identified as “normal” by certified pathologists, there is a chance that a few breast cancer cells might have been missed during the exam. In terms of treatment, more radical surgical intervention might be necessary to clear the tissue with aberrant uMUC1 expression, which could cause breast cancer recurrence in the future. This consideration actually points to the wider implications of our study for breast cancer management. Specifically, MUC1 might be considered as a diagnostic biomarker for non-invasive cancer detection. With the development of molecular imaging techniques for cancer diagnosis and image-guided therapy, the utility of uMUC1 antigen as a targeted cancer biomarker has been already demonstrated by us in animal studies (41, 42, 59, 60). In the future, targeting of uMUC1 antigen with specific imaging probes could assist in identifying areas of breast tissue that are subject to resection or monitoring. The former is feasible since surgery-guided imaging systems have already been introduced in clinic for breast cancer treatment (61). Furthermore, identification of uMUC1 antigen in pre-malignant tissue could lead to its targeting for therapeutic purposes as have been recently shown by devising inhibitors to its transmembrane MUC1-C-terminal subunit (62).
Supplementary Material
Immunohistochemical staining of human breast tissue microarray (TMA) representative of tumor progression. The breast TMAs were incubated with anti-MUC1 (clone VU4H5) antibody that binds underglycosylated MUC1 and MH1 (CT-2) antibody to cytoplasmic tail of MUC1. uMUC1 expression was evident in all tumor stages including adjacent normal tissue. Magnification bar = 50 µm.
Clinical Practice Points.
It is known that underglycosylated mucin 1 antigen is aberrantly expressed in 90% of breast cancers. Such abnormal expression has been associated with cellular growth, transformation, adhesion, invasion and immune cells responsiveness and tolerance. It has been directly linked to tumor invasiveness and metastatic potential. It has been recently demonstrated that uMUC1 is one of the seven highly expressed marker genes identified in ductal carcinoma in situ (DCIS) and in invasive ductal carcinoma (IDC) in human and rodent tissues. uMUC1 is an attractive target for therapy since it is involved in induced expression of genes predictive of the outcome of chemotherapy and overall clinical outcome in breast cancer patients. Our studies clearly demonstrated that its expression and glycosylation vary with the more aggressive forms of breast cancer. We also state that these changes appear early in the pathology. Further, our findings that tissues defined as “normal” by pathology report had elevated levels of uMUC1 point to a possibility that more aggressive intervention is needed in certain clinical cases. The availability of clinical cancer imaging modalities would be instrumental in detecting patients with aberrant uMUC1 expression in normal tissues and deciding on the individualized course of therapy.
Acknowledgements
This study was supported in part by the NIH grant award 1RO1CA135650 to A.M and Z.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All authors have no conflicts of interest.
References
- 1.Zotter S, Hageman P, Lossnitzer A, et al. Tissue and tumour distribution of human polymorphic epithelial mucin. Cancer Rev. 1988;11–12:55–101. [Google Scholar]
- 2.Patton S, Gendler SJ, Spicer AP. The epithelial mucin, MUC1, of milk, mammary gland and other tissues. Biochim Biophys Acta. 1995;1241:407–423. doi: 10.1016/0304-4157(95)00014-3. [DOI] [PubMed] [Google Scholar]
- 3.Gendler SJ. MUC1, the renaissance molecule. J Mammary Gland Biol Neoplasia. 2001;6:339–353. doi: 10.1023/a:1011379725811. [DOI] [PubMed] [Google Scholar]
- 4.Brayman M, Thathiah A, Carson DD. MUC1: a multifunctional cell surface component of reproductive tissue epithelia. Reprod Biol Endocrinol. 2004;2:4. doi: 10.1186/1477-7827-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wesseling J, van der Valk SW, Vos HL, et al. Episialin (MUC1) overexpression inhibits integrin-mediated cell adhesion to extracellular matrix components. J Cell Biol. 1995;129:255–265. doi: 10.1083/jcb.129.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wesseling J, van der Valk SW, Hilkens J. A mechanism for inhibition of E-cadherin-mediated cell-cell adhesion by the membrane-associated mucin episialin/MUC1. Mol Biol Cell. 1996;7:565–577. doi: 10.1091/mbc.7.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohlgraf KG, Gawron AJ, Higashi M, et al. Contribution of the MUC1 tandem repeat and cytoplasmic tail to invasive and metastatic properties of a pancreatic cancer cell line. Cancer Res. 2003;63:5011–5020. [PubMed] [Google Scholar]
- 8.Perey L, Hayes DF, Kufe D. Effects of differentiating agents on cell surface expression of the breast carcinoma-associated DF3-P epitope. Cancer Res. 1992;52:6365–6370. [PubMed] [Google Scholar]
- 9.Lloyd KO, Burchell J, Kudryashov V, et al. Comparison of O-linked carbohydrate chains in MUC-1 mucin from normal breast epithelial cell lines and breast carcinoma cell lines. Demonstration of simpler and fewer glycan chains in tumor cells. J Biol Chem. 1996;271:33325–33334. doi: 10.1074/jbc.271.52.33325. [DOI] [PubMed] [Google Scholar]
- 10.Hattrup CL, Gendler SJ. MUC1 alters oncogenic events and transcription in human breast cancer cells. Breast Cancer Res. 2006;8:R37. doi: 10.1186/bcr1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carson D. The cytoplasmic tail of MUC1: a very busy place. Sci Signal. 2008;1 doi: 10.1126/scisignal.127pe35. pe35. [DOI] [PubMed] [Google Scholar]
- 12.Barratt-Boyes S. Making the most of mucin: a novel target for tumor immunotherapy. Cancer Immunol Immunother. 1996;43:142–151. doi: 10.1007/s002620050315. [DOI] [PubMed] [Google Scholar]
- 13.Storr SJ, Royle L, Chapman CJ, et al. The O-linked glycosylation of secretory/shed MUC1 from an advanced breast cancer patient's serum. Glycobiology. 2008;18:456–462. doi: 10.1093/glycob/cwn022. [DOI] [PubMed] [Google Scholar]
- 14.Singh R, Bandyopadhyay D. MUC1: a target molecule for cancer therapy. Cancer Biol Ther. 2007;6:481–486. doi: 10.4161/cbt.6.4.4201. [DOI] [PubMed] [Google Scholar]
- 15.Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9:874–885. doi: 10.1038/nrc2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Hof A, Molthoff C, Davies Q, et al. Biodistribution of (111)indium-labeled engineered human antibody CTMO1 in ovarian cancer patients: influence of protein dose. Cancer Res. 1996;56:5179–5185. [PubMed] [Google Scholar]
- 17.Avichezer D, Taylor-Papadimitriou J, Arnon R. A short synthetic peptide (DTRPAP) induces anti-mucin (MUC-1) antibody, which is reactive with human ovarian and breast cancer cells. Cancer Biochem Biophys. 1998;16:113–128. [PubMed] [Google Scholar]
- 18.Hough C, Sherman-Baust C, Pizer E, et al. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- 19.Budrick M, Harris A, Reid C, et al. Oligosaccarides expressed on MUC1 produced by pancreatic and colon tumour cell lines. J Biol Chem. 1997;272:24198–24202. doi: 10.1074/jbc.272.39.24198. [DOI] [PubMed] [Google Scholar]
- 20.Aoki R, Tanaka S, Haruma K, et al. MUC-1 expression as a predictor of the curative endoscopic treatment of submucosally invasive colorectal carcinoma. Dis Colon Rectum. 1998;41:1262–1272. doi: 10.1007/BF02258227. [DOI] [PubMed] [Google Scholar]
- 21.Willsher P, Xing P, Clarke C, et al. Mucin 1 antigens in the serum and bronchial lavage fluid of patients with lung cancer. Cancer. 1993;72:2936–2942. doi: 10.1002/1097-0142(19931115)72:10<2936::aid-cncr2820721013>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 22.Maeshima A, Miyagi A, Hirai T, et al. Mucin-producing adenocarcinoma of the lung, with special reference to goblet cell type adenocarcinoma: immunohistochemical observation and Ki-ras gene mutation. Pathol Int. 1997;47:454–460. doi: 10.1111/j.1440-1827.1997.tb04524.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Zhang S, Reuter V, et al. Expression of potential target antigensfor immunotherapy on primary and metastatic prostate cancers. Clin Cancer Res. 1998;4:295–302. [PubMed] [Google Scholar]
- 24.Strous G, Dekker J. Mucin-like glycoproteins. Crit Rev Biochem Mol Biol. 1992;27:57–92. doi: 10.3109/10409239209082559. [DOI] [PubMed] [Google Scholar]
- 25.Medina M, Velez D, Asenjo J, et al. Human colon adenocarcinomas express a MUC1-associated novel carbohydrate epitope on core mucin glycans defined by a monoclonal antibody (A10) raised against murine Ehrlich tumor cells. Cancer Res. 1999;59:1061–1070. [PubMed] [Google Scholar]
- 26.Oosterkamp H, Scheiner L, Stefanova M, et al. Comparison of MUC-1 mucin expression in epithelial and non-epithelial cancer cell lines and demonstration of a new short variant form (MUC-1/Z) Int J Cancer. 1997;72:87–94. doi: 10.1002/(sici)1097-0215(19970703)72:1<87::aid-ijc13>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 27.Burton J, Mishina D, Cardillo T, et al. Epithelial mucin-1 (MUC1) expression and MA5 anti-MUC1 monoclonal antibody targeting in multiple myeloma. Clin Cancer Res. 1999;5:3065s–3072s. [PubMed] [Google Scholar]
- 28.Treon S, Mollick J, Urashima M, et al. Muc-1 core protein is expressed on multiple myeloma cells and is induced by dexamethasone. Blood. 1999;93:1287–1298. [PubMed] [Google Scholar]
- 29.Dyomin V, Palanisamy N, Lloyd K, et al. MUC1 is activated in a B-cell lymphoma by the t(1:14)(q21;q32) translocation and is rearranged and amplified in B-cell lymphoma subsets. Blood. 2000;95:2666–2671. [PubMed] [Google Scholar]
- 30.Brossart P, Schneider A, Dill P, et al. The epithelial tumor antigen MUC1 is expressed in hematological malignancies and is recognized by MUC1-specific cytotoxic T-lymphocytes. Cancer Res. 2001;61:6846–6850. [PubMed] [Google Scholar]
- 31.Greenlee R, Murray T, Bolden S, et al. Cancer statistics. CA Cancer J Clin. 2000;50:7. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- 32.Hayes D, Mesa-Tejada R, Papsidero L, et al. Prediction of prognosis in primary breast cancer by detection of a high molecular weight mucin-like antigen using monoclonal antibodies DF3, F36/22, and CU18: a Cancer and Leukemia Group B study. J Clin Oncol. 1991;9:1113–1123. doi: 10.1200/JCO.1991.9.7.1113. [DOI] [PubMed] [Google Scholar]
- 33.Perey L, Hayes D, Kufe D. Effects of differentiating agents of cell surface expression of the breast carcinoma-associated DF3-P epitope. Cancer Res. 1992;52:6365–6370. [PubMed] [Google Scholar]
- 34.Nacht M, Ferguson A, Zhang W, et al. Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res. 1999;59:5464–5470. [PubMed] [Google Scholar]
- 35.van der Vegt B, de Roos MA, Peterse JL, et al. The expression pattern of MUC1 (EMA) is related to tumour characteristics and clinical outcome of invasive ductal breast carcinoma. Histopathology. 2007;51:322–335. doi: 10.1111/j.1365-2559.2007.02757.x. [DOI] [PubMed] [Google Scholar]
- 36.de Roos MA, van der Vegt B, Peterse JL, et al. The expression pattern of MUC1 (EMA) is related to tumour characteristics and clinical outcome in 'pure' ductal carcinoma in situ of the breast. Histopathology. 2007;51:227–238. doi: 10.1111/j.1365-2559.2007.02754.x. [DOI] [PubMed] [Google Scholar]
- 37.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- 38.Kretschmer C, Sterner-Kock A, Siedentopf F, et al. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15–26. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khodarev N, Pitroda S, Beckett M, et al. MUC1-induced transcriptional programs associated with tumorigenesis predict outcome in breast and lung cancer. Cancer Res. 2009;69:2833–2837. doi: 10.1158/0008-5472.CAN-08-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raina D, Ahmad R, Joshi MD, et al. Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69:5133–5141. doi: 10.1158/0008-5472.CAN-09-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore A, Medarova Z, Potthast A, et al. In vivo targeting of underglycosylated MUC-1 tumor antigen using a multimodal imaging probe. Cancer Res. 2004;64:1821–1827. doi: 10.1158/0008-5472.can-03-3230. [DOI] [PubMed] [Google Scholar]
- 42.Medarova Z, Rashkovetsky L, Pantazopoulos P, et al. Multiparametric monitoring of tumor response to chemotherapy by noninvasive imaging. Cancer Res. 2009;69:1182–1189. doi: 10.1158/0008-5472.CAN-08-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spicer AP, Rowse GJ, Lidner TK, et al. Delayed mammary tumor progression in Muc-1 null mice. J Biol Chem. 1995;270:30093–30101. doi: 10.1074/jbc.270.50.30093. [DOI] [PubMed] [Google Scholar]
- 44.Harduin-Lepers A, Recchi MA, Delannoy P. 1994, the year of sialyltransferases. Glycobiology. 1995;5:741–758. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- 45.Harduin-Lepers A, Mollicone R, Delannoy P, et al. The animal sialyltransferases and sialyltransferase-related genes: a phylogenetic approach. Glycobiology. 2005;15:805–817. doi: 10.1093/glycob/cwi063. [DOI] [PubMed] [Google Scholar]
- 46.Burchell J, Poulsom R, Hanby A, et al. An alpha2,3 sialyltransferase (ST3Gal I) is elevated in primary breast carcinomas. Glycobiology. 1999;9:1307–1311. doi: 10.1093/glycob/9.12.1307. [DOI] [PubMed] [Google Scholar]
- 47.Lin S, Kemmner W, Grigull S, et al. Cell surface alpha 2,6 sialylation affects adhesion of breast carcinoma cells. Exp Cell Res. 2002;276:101–110. doi: 10.1006/excr.2002.5521. [DOI] [PubMed] [Google Scholar]
- 48.Recchi MA, Hebbar M, Hornez L, et al. Multiplex reverse transcription polymerase chain reaction assessment of sialyltransferase expression in human breast cancer. Cancer Res. 1998;58:4066–4070. [PubMed] [Google Scholar]
- 49.Sewell R, Backstrom M, Dalziel M, et al. The ST6GalNAc-I sialyltransferase localizes throughout the Golgi and is responsible for the synthesis of the tumor-associated sialyl-Tn O-glycan in human breast cancer. J Biol Chem. 2006;281:3586–3594. doi: 10.1074/jbc.M511826200. [DOI] [PubMed] [Google Scholar]
- 50.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nature reviews. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Gross HJ, Sticher U, Brossmer R. A highly sensitive fluorometric assay for sialyltransferase activity using CMP-9-fluoresceinyl-NeuAc as donor. Anal Biochem. 1990;186:127–134. doi: 10.1016/0003-2697(90)90585-w. [DOI] [PubMed] [Google Scholar]
- 53.Cazet A, Julien S, Bobowski M, et al. Consequences of the expression of sialylated antigens in breast cancer. Carbohydr Res. 2010;345:1377–1383. doi: 10.1016/j.carres.2010.01.024. [DOI] [PubMed] [Google Scholar]
- 54.Rakha EA, Boyce RW, Abd El-Rehim D, et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol. 2005;18:1295–1304. doi: 10.1038/modpathol.3800445. [DOI] [PubMed] [Google Scholar]
- 55.Rahn JJ, Dabbagh L, Pasdar M, et al. The importance of MUC1 cellular localization in patients with breast carcinoma: an immunohistologic study of 71 patients and review of the literature. Cancer. 2001;91:1973–1982. doi: 10.1002/1097-0142(20010601)91:11<1973::aid-cncr1222>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 56.Chu JS, Chang KJ. Mucin expression in mucinous carcinoma and other invasive carcinomas of the breast. Cancer Lett. 1999;142:121–127. doi: 10.1016/s0304-3835(99)00161-5. [DOI] [PubMed] [Google Scholar]
- 57.Nishidate T, Katagiri T, Lin ML, et al. Genome-wide gene-expression profiles of breast-cancer cells purified with laser microbeam microdissection: identification of genes associated with progression and metastasis. Int J Oncol. 2004;25:797–819. [PubMed] [Google Scholar]
- 58.Yoon WH, Park HD, Lim K, et al. Effect of O-glycosylated mucin on invasion and metastasis of HM7 human colon cancer cells. Biochem Biophys Res Commun. 1996;222:694–699. doi: 10.1006/bbrc.1996.0806. [DOI] [PubMed] [Google Scholar]
- 59.Medarova Z, Pham W, Kim Y, et al. In vivo imaging of tumor response to therapy using a dual-modality imaging strategy. Int J Cancer. 2006;118:2796–2802. doi: 10.1002/ijc.21672. [DOI] [PubMed] [Google Scholar]
- 60.Kumar M, Yigit M, Dai G, et al. Image-guided breast tumor therapy using a small interfering RNA nanodrug. Cancer Res. 2010;70:7553–7561. doi: 10.1158/0008-5472.CAN-10-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hutteman M, Mieog J, van der Vorst J, et al. Randomized, double-blind comparison of indocyanine green with or without albumin premixing for near-infrared fluorescence imaging of sentinel lymph nodes in breast cancer patients. Breast Cancer Res Treat. 2011;127:163–170. doi: 10.1007/s10549-011-1419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kufe DW. MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene. 2012 May 14; doi: 10.1038/onc.2012.158. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunohistochemical staining of human breast tissue microarray (TMA) representative of tumor progression. The breast TMAs were incubated with anti-MUC1 (clone VU4H5) antibody that binds underglycosylated MUC1 and MH1 (CT-2) antibody to cytoplasmic tail of MUC1. uMUC1 expression was evident in all tumor stages including adjacent normal tissue. Magnification bar = 50 µm.