Abstract
Centrioles are the key foundation of centrosomes and cilia, yet a molecular understanding of how they form has only recently begun to emerge. Building a fully functional centriole that can form a centrosome and cilium requires two cell cycles. Centriole building starts with procentriole nucleation, a process that is coordinated by the conserved proteins Plk4/Zyg-1, and Asterless/Cep152. Subsequently, Sas-6, a conserved procentriole protein, self-assembles to provide nine-fold symmetry to the centriole scaffold. The procentriole then continues to elongate into a centriole, a process controlled by Sas-4/CPAP and CP110. Then, centrioles recruit Sas-4-mediated pre-assembled centrosomal complexes from the cytoplasm to form the pericentriolar material (PCM). Finally, CP110, together with its interacting proteins, are also involved in controlling the timing of centriole templating of the cilium.
Introduction
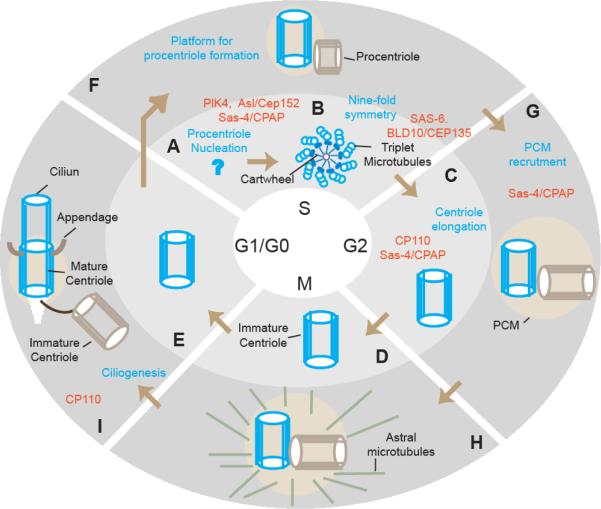
Centrioles are microtubule-based, conserved eukaryotic cell structures measuring ~200 by 500 nm in size that have a distinctive radial nine-fold symmetry. Typically, an animal cell requires two cell cycles to build a fully functional centriole, which is then required to form an independent centrosome and a cilium (Figure 1). In the first cell cycle, the building of a centriole starts at S phase with the formation of a procentriole (Figure 1A–B). At its core, the procentriole harbors a central tubule that radiates nine spokes, which together resemble a cartwheel (Figure 1B). The cartwheel spokes are connected to nine triplet microtubules that form the centriole wall. In G2, M and G1 phases, the centriole elongates to ~500 nm and is referred to as an immature centriole. During this time, the immature centriole appears to be non-functional.
Fig 1. Building of a centriole.
Depiction of the structural and molecular events taking place during the formation of one of the centrioles in a cell (depicted in blue) through two consecutive cell cycles. During the first cell cycle (light gray background, A–E), the basic structure of the centriole is formed. During second cell cycle (darker gray background, F–I), the immature centriole acquires functions in a step-by-step manner until it become fully mature and functional (H). A second centriole formed near the original centriole is depicted in light brown. Major events in the formation of the centriole are noted in blue. Key proteins are indicated in orange. Centrioles are depicted as they would appear from a cross section (B) and a side view (C–I).
As the immature centriole proceeds through the second cell cycle, it starts to acquire characteristics of a mature centriole in a step-by-step manner. In S phase, the immature centriole serves as a platform for the formation of a procentriole (Figure 1F). In G2, the immature centriole (together with the procentriole attached to it) acquires large PCM, starts to nucleate astral microtubules, and in general, begins to function as an independent centrosome (Figure 1G). However, this centrosome is functionally distinct from the second preexisting mature centriole [1,2]. In G1, the now mature centriole has acquired distal appendages and docks to the plasma membrane, where it templates the formation of the ciliary axoneme to initiate ciliogenesis (Figure 1I). Thus, a centriole requires two cell cycles from its initial formation to become fully mature.
Over the last decade, many of the centriolar proteins involved in building the centriole have been identified and have began to be characterized. Here, we describe insights into centriole biogenesis gained in the last few years, limiting our discussion to the proteins that function in nucleating of the procentriole, establishing the centriole's nine-fold symmetry, centriole elongation, and the recruitment of PCM.
Procentriole nucleation
A single procentriole forms at a right angle to the proximal segment of the preexisting centriole (Figure 1F), Despite being one of the most conserved aspects of centriole duplication, the mechanisms that govern this restricted pattern remain enigmatic. A major difficulty in addressing this question is that it is hard to monitor the early events that take place in procentriole formation (i.e. procentriole nucleation) in sufficient detail (Figure 1A). Previous studies in various model organisms have suggested that the PCM is essential for procentriole formation [3] and that Caenorhabditis elegans Zyg-1 (and its Human analog Plk4) [4,5], Drosophila asterless (and its mammalian ortholog Cep152) [6], and Sas-6 [4] are early players essential to this process.
While many cell cycle kinases regulate centriole duplication, Plk4/Zyg-1 is thought to be the key kinase responsible for the initiation of centriole duplication. Consistent with this idea, Plk4 is found in the PCM where the procentriole nucleates [7]; Plk4 binds to or phosphorylates several centriolar proteins including the procentriole nucleation protein Asl/Cep152 [7–9] and GCP6, a component of the γ-TuRC microtubule nucleating complex [10]. Plk4 also phosphorylates regulators of centriolar proteins including the E3-ubiquitin ligase FBXW5, which targets the cartwheel protein Sas-6 for degradation [11]. These events likely orchestrate the initiation of procentriole nucleation.
The PCM protein Asl and its ortholog Cep152 is also found where the procentriole nucleates and is critical early in centriole formation, leading to the idea that Asl/Cep152 nucleates procentriole formation [6]. Recently, a model was proposed where the interaction of Asl/Cep152 with Plk4 and Sas-4/CPAP provides a scaffold for procentriole formation [7,8]. However, a more recent study in Drosophila suggests that the interaction of Asl with Sas-4 is important for Asl localization in the PCM, and that this localization is not essential for initiating centriole formation [12]. How exactly the PCM proteins Plk4 and Asl/Cep152 coordinate the nucleation of a single procentriole near to the preexisting centriole remains unclear. One possibility is that PLk4 activates a yet unidentified complex of Asl/Cep152, which then triggers procentriole formation.
Centriole nine-fold symmetry
Centrioles display a conserved structural signature of nine-fold radial symmetry (Figure 1B). Since centrioles template ciliogenesis, the symmetry of the centriole is also translated to cilium. Based on earlier electron microscopy studies, it has long been evident that the first structure in the developing centriole that displays nine-fold symmetry is the cartwheel [13] (Figure 1B). Based on this structural observation, it is apparent that the cartwheel is a structural scaffold in establishing the centriole and, as a result, the nine-fold symmetry of the cilium. Several candidate proteins including Sas-6, Bld10/Cep135, Poc1, and Sas-5/Ana-2/STIL were previously proposed to be cartwheel proteins, but only Sas-6 [14] and Bld10 [15] have so far been shown to be essential to organize the nine-fold symmetry of the centriole.
Recently, Sas-6 was shown to self-assemble into a central tubule-like structure, and analysis of the Sas-6 crystal structure suggests that Sas-6 polymers display the nine-fold symmetry found in the cartwheel central tubule [16–18]. Sas-6 interacts with Sas-5, a protein that is thought to be the ortholog of Drosophila Ana2 and mammalian STIL. Indeed Sas-5 [19], Ana-2 [20] and STIL [21,22] were reported to be essential for centriole formation. Strikingly, over expression of Drosophila Sas-6 and Ana-2 resulted in the formation of large aggregates composed of cartwheel-like structures [23], arguing that Sas-5/Ana-2/ STIL and Sas-6 are limiting factors in cartwheel formation. Still, it is not clear if Sas-6 and Sas-5/Ana-2/STIL are the only players in this process, as it is likely that other proteins participate in nucleating and capping the cartwheel.
Centriole elongation and size control
Centriole elongation takes place in at least 3 stages along centriole formation. First, the procentriole exists only for a brief period of time during S phase (Figure 1A–B), forming at approximately 200 nm in length. Second, the procentriole elongates through G2 to M phase until it reaches its final size (Figure 1C). Finally, the mature centriole further elongates to form the axoneme of the cilium (Figure 1I). The molecular players and mechanisms that determine the size control of a centriole have just recently started to emerge. These include CPAP/Sas-4, Bld10/Cep135, Poc1, CP110, Cep97, and Kinesin-13 homologues Kif24 and Klp10A. These proteins appear to play a role in multiple elongation stages, but this may be because it is difficult to dissect the stages of the elongation process independently.
Sas-4/CPAP has been implicated in centriole elongation by the observation that Drosophila sas-4 mutants have short centrioles [24], as well as by over expression studies of Sas-4 in cell culture that produced elongated centrioles [25–28]. Interestingly, over expression of a CPAP mutant that cannot bind tubulin does not induce centriole elongation, and it was argued that this suggests that CPAP promotes centriole elongation by stabilizing centriolar microtubules [27]. However, there is evidence that CPAP binds tubulin dimers in a way that interferes with microtubule elongation [29,30], and that its tubulin binding capacity functions to regulate Sas-4/CPAP recruitment to the centrosome [31]. Regardless, it is likely that CPAP is only involved in determining the size of the procentriole or proximal end of mature centrioles, as Sas-4/CPAP normally localizes only to these locations [5,24]. Indeed, it was recently found that PLK2 phosphorylation is critical for the function of CPAP in procentriole formation and centriole elongation [32].
Like Drosophila sas-4 mutants, Drosophila bld10/cep135 and poc1 mutants have short centrioles [24,33], and Poc1 over expression in cell culture was reported to induce centriole elongation [34]. However, unlike Sas-4, which is restricted to the proximal end of the centriole, Bld10/Cep135 and Poc1 can be found all along the centriole's length [24,33]. Therefore, it is possible that these proteins are involved in determining centriole size by stabilizing the centriole along its length.
CP110 localizes to distal centrioles and its depletion results in centriole elongation and the formation of a cilium, suggesting that CP110 blocks centriole elongation and prevents ciliogenesis [35]. CP110 interacts with multiple proteins such as Cep97, Cep290, as well as the Kinesin-13 homologs Kif24 and Klp10A. All four proteins are localized to the distal centriole, but they have differential roles in centriole formation and ciliogenesis. Cep97 recruits CP110 to centrosomes and, like CP110 depletion, its depletion result in centriole elongation, and the formation of an aberrant cilium [35]. In mammalian cells, loss of Kif24 leads to the disappearance of CP110 from mother centrioles, resulting in defective cilium assembly. However, unlike CP110 and Cep97 loss, the absence of Kif24 does not promote the growth of abnormally long centrioles [36]. Similarly, in Drosophila, CP110 appears to cooperate with Klp10A in restricting centriole elongation [37]. CP110 also interacts with Cep290 and Rab8a, which are also essential for ciliogenesis, but their depletion does not result in the aberrant formation of centrioles or cilia [38]. Interestingly, CP110 forms at least two discrete complexes that appear to participate in distinct functions [38]. It is possible that CP110 and its interacting proteins function as a capping complex at the distal end of the centriole to block or allow centriole elongation based on cell cycle cues.
PCM formation
Traditionally, the PCM is thought to be a distinct structure surrounding the centriole that is involved in nucleating and organizing cytoplasmic microtubules (Figure 1G). However, recent studies also suggest an intimate involvement of the PCM in building the centriole [3,39] and common mechanisms for centriole and PCM formation [31]. In the past, several mechanisms were proposed to mediate PCM formation [40,41]. Recently, it was suggested that the centriolar and PCM protein Sas-4 plays a role in PCM recruitment [12,31]. Originally, Sas-4 was reported to be essential for both centriole and PCM recruitment in Caenorhabditis elegans [42,43]. However, upon reduction of Sas-4 levels to varying degrees, a correlation has been observed between centriole defects and the amount of PCM recruitment, leading to the idea that PCM recruitment defects are a consequence of structurally-compromised centrioles [42]. This idea was reinforced by the observation that in Caenorhabditis elegans, Sas-4 is a stable component of the centriole that is incorporated only during initial centriole formation [43].
However, studies in human cells and Drosophila show that Sas-4 and CPAP (one of the two human orthologs of Sas-4) have distinct characteristics, suggesting that they may have specific roles in PCM function. For example, Sas-4 and CPAP are not only found in the centriole's proximal region but can also be found in the PCM [12]. CPAP is dynamically exchanged throughout the cell cycle between the centrosome and cytosol [44], and centrosomal levels of Sas-4 in vivo change along the cell cycle [12]. CPAP levels are cell cycle-dependent and are at their highest in G2 when PCM recruitment takes place [25,27]. Finally, Sas-4 forms complexes with PCM proteins such as ! -tubulin [45], Asl/Cep152 [7,8], CNN/Cep215, and Drosophila Pericentrin-like-protein [12,46].
Direct evidence for the role of Sas-4 in PCM formation has come from studies of sas-4 point mutations in Drosophila. In these studies, Sas-4 mutants are expressed in the background of a sas-4 null mutant to avoid complications raised by the presence of wild type protein. This approach also allows the expression of Sas-4 via its own promoter, avoiding artifacts that result from Sas-4 overexpression. It was found that eliminating the conserved PN2–3 segment of Sas-4 leads to a reduction in PCM recruitment without blocking centriole duplication [12]. On the other hand, mutating the Sas-4 tubulin binding site resulted in an increase in PCM recruitment without effecting centriole number [31].
Additional evidence for the role of Sas-4 in PCM formation has come from biochemical studies of the interaction between Sas-4, tubulin dimers, PCM proteins, and whole centrosomes. It was found that Sas-4 scaffolds complexes of PCM proteins and can tether them to purified centrosomes that have been stripped of PCM [12]. This function is regulated by a guanine nucleotide that binds to the tubulin dimer attached to Sas-4 [31]. Tubulin-GDP favors binding to Sas-4 and promotes PCM complex formation while binding of tubulin to a GTP analog blocks this process. Similarly, the binding of tubulin-GDP to Sas-4 promotes Sas-4-PCM complex tethering to the centrosome while the tubulin-GTP analog blocks this tethering activity. These studies have led to a model in which tubulin dimers regulate Sas-4-mediated PCM recruitment to the centrosome [31].
Summary
Classically, the centrioles and their surrounding PCM have been thought to be structurally and functionally distinct entities. However, recent studies aiming to understand how the centriole is built point to a more holistic point of view where centriole and PCM formation are interdependent. In addition, our initial simplistic understandings of centriole and centrosome building as linear processes, where centriole formation precedes PCM formation, may not necessarily be the case. Finally, a common way of thinking is that centriole proteins mediate centriole formation once they are recruited to the centriole where they play structural and regulatory functions. However, studies that are even more recent point to an alternative and equally important function for centriolar proteins in the PCM and the cytosol. A good example of this holistic model can be found in considering the important function of Sas-4, a protein that was initially thought to only be important for centriole formation [42,43] and now found to also have function in the cytosol and PCM [12,31].
The complexity in how centriole proteins function, together with the small size of the centriole, the low abundance of centriolar proteins, and the speed in which procentriole formation takes place, are ongoing challenges in understanding how the centriole and centrosome are built. However, recent developments that employ a combination of technologies such as sub-diffraction light microscopy, less invasive genetic approaches, and the development of biochemical approaches towards reconstituting centriole formation provide an exciting avenue by which the long lasting question of how the centriole is built can be addressed.
Acknowledgements
We apologize that due to space constraint many important contributions could not be acknowledged. We would like to thank Marcus Basiri for editing the manuscript. This work was supported by a grant (R01GM098394) from NIH and National Institute of General Medical Sciences as well as grant 1121176 from National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest The author(s) have no conflict of interest to declare.
References
- 1.Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat Commun. 2011;2(243) doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]; * Studying Drosophila neuroblasts using photo converted centrioles and a daughter-centriole-specific markers,the authors show that upon asymmetric mitosis, the old centrosome is inherited by the differentiating daughter cell while the stem cells inherit the new centriole. This demonstrates that old and new centrioles are functionally distinct, but this distinction is used differently from one cell type to the other
- 2.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315(5811):518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7(6):815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 4.Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in caenorhabditis elegans. Nature. 2006;444(7119):619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 5.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate cep152 are orthologs essential for centriole duplication. Genetics. 2008;180(4):2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I. Cep152 acts as a scaffold for recruitment of plk4 and cpap to the centrosome. J Cell Biol. 2010;191(4):731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467(7316):714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 9.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with plk4 and is required for centriole duplication. J Cell Biol. 2010;191(4):721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bahtz R, Seidler J, Arnold M, Haselmann-Weiss U, Antony C, Lehmann WD, Hoffmann I. Gcp6 is a substrate of plk4 and required for centriole duplication. J Cell Sci. 2012;125(Pt 2):486–496. doi: 10.1242/jcs.093930. [DOI] [PubMed] [Google Scholar]
- 11.Puklowski A, Homsi Y, Keller D, May M, Chauhan S, Kossatz U, Grunwald V, Kubicka S, Pich A, Manns MP, Hoffmann I, et al. The scf-fbxw5 e3-ubiquitin ligase is regulated by plk4 and targets hssas-6 to control centrosome duplication. Nat Cell Biol. 2011;13(8):1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 12.Gopalakrishnan J, Mennella V, Blachon S, Zhai B, Smith AH, Megraw TL, Nicastro D, Gygi SP, Agard DA, Avidor-Reiss T. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat Commun. 2011;2(359) doi: 10.1038/ncomms1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson RG, Brenner RM. The formation of basal bodies (centrioles) in the rhesus monkey oviduct. J Cell Biol. 1971;50(1):10–34. doi: 10.1083/jcb.50.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. Sas-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17(24):2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Bld10p, a novel protein essential for basal body assembly in chlamydomonas: Localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol. 2004;165(5):663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Breugel M, Hirono M, Andreeva A, Yanagisawa HA, Yamaguchi S, Nakazawa Y, Morgner N, Petrovich M, Ebong IO, Robinson CV, Johnson CM, et al. Structures of sas-6 suggest its organization in centrioles. Science. 2011;331(6021):1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]; ** Using structural biology, the authors of this paper solved the structure of Sas-6 domains and predicted that it organize to a central tubule with 9-fold symmetry.
- 17.Kitagawa D, Vakonakis I, Olieric N, Hilbert M, Keller D, Olieric V, Bortfeld M, Erat MC, Fluckiger I, Gonczy P, Steinmetz MO. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144(3):364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Using structural biology, the authors of this paper solved the structure of Sas-6 domains and predicted that it organize to a central tubule with 9-fold symmetry.
- 18.Gopalakrishnan J, Guichard P, Smith AH, Schwarz H, Agard DA, Marco S, Avidor-Reiss T. Self-assembling sas-6 multimer is a core centriole building block. J Biol Chem. 2010;285(12):8759–8770. doi: 10.1074/jbc.M109.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P. Centriolar sas-5 is required for centrosome duplication in c. Elegans. Nat Cell Biol. 2004;6(7):656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Li S, Januschke J, Rossi F, Izumi Y, Garcia-Alvarez G, Gwee SS, Soon SB, Sidhu HK, Yu F, Matsuzaki F, et al. An ana2/ctp/mud complex regulates spindle orientation in drosophila neuroblasts. Dev Cell. 2011;21(3):520–533. doi: 10.1016/j.devcel.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK. The human microcephaly protein stil interacts with cpap and is required for procentriole formation. EMBO J. 2011;30(23):4790–4804. doi: 10.1038/emboj.2011.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castiel A, Danieli MM, David A, Moshkovitz S, Aplan PD, Kirsch IR, Brandeis M, Kramer A, Izraeli S. The stil protein regulates centrosome integrity and mitosis through suppression of chfr. J Cell Sci. 2011;124(Pt 4):532–539. doi: 10.1242/jcs.079731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stevens NR, Roque H, Raff JW. Dsas-6 and ana2 coassemble into tubules to promote centriole duplication and engagement. Dev Cell. 2010;19(6):913–919. doi: 10.1016/j.devcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blachon S, Cai X, Roberts KA, Yang K, Polyanovsky A, Church A, Avidor-Reiss T. A proximal centriole-like structure is present in drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 2009;182(1):133–144. doi: 10.1534/genetics.109.101709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gonczy P. Overly long centrioles and defective cell division upon excess of the sas-4-related protein cpap. Curr Biol. 2009;19(12):1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by cpap and cp110. Curr Biol. 2009;19(12):1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. Cpap is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11(7):825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 28.Kim MK, Dudognon C, Smith S. Tankyrase 1 regulates centrosome function by controlling cpap stability. EMBO Rep. 2012;13(8):724–732. doi: 10.1038/embor.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu WB, Hung LY, Tang CJ, Su CL, Chang Y, Tang TK. Functional characterization of the microtubule-binding and -destabilizing domains of cpap and d-sas-4. Exp Cell Res. 2008;314(14):2591–2602. doi: 10.1016/j.yexcr.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 30.Cormier A, Clement MJ, Knossow M, Lachkar S, Savarin P, Toma F, Sobel A, Gigant B, Curmi PA. The pn2-3 domain of centrosomal p4.1-associated protein implements a novel mechanism for tubulin sequestration. J Biol Chem. 2009;284(11):6909–6917. doi: 10.1074/jbc.M808249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gopalakrishnan J, Chim YC, Ha A, Basiri ML, Lerit DA, Rusan NM, Avidor-Reiss T. Tubulin nucleotide status controls sas-4-dependent pericentriolar material recruitment. Nat Cell Biol. 2012;14(8):865–873. doi: 10.1038/ncb2527. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Using a combination of genetics and biochemistry, the authors demonstrate that tubulin regulates the formation of Sas-4-PCM complexes and thereby PCM recruitment to the centrosome. This is the first study that demonstrates that tubulin nucleotide status can serve as a switch in a process other than microtubule formation.
- 32.Chang J, Cizmecioglu O, Hoffmann I, Rhee K. Plk2 phosphorylation is critical for cpap function in procentriole formation during the centrosome cycle. EMBO J. 2010;29(14):2395–2406. doi: 10.1038/emboj.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mottier-Pavie V, Megraw TL. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell. 2009;20(10):2605–2614. doi: 10.1091/mbc.E08-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keller LC, Geimer S, Romijn E, Yates J, 3rd, Zamora I, Marshall WF. Molecular architecture of the centriole proteome: The conserved wd40 domain protein poc1 is required for centriole duplication and length control. Mol Biol Cell. 2009;20(4):1150–1166. doi: 10.1091/mbc.E08-06-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and cp110 suppress a cilia assembly program. Cell. 2007;130(4):678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T, Tsang WY, Li J, Lane W, Dynlacht BD. Centriolar kinesin kif24 interacts with cp110 to remodel microtubules and regulate ciliogenesis. Cell. 2012;145(6):914–925. doi: 10.1016/j.cell.2011.04.028. [DOI] [PubMed] [Google Scholar]; ** This study shows that the kinesin-13 homolog, Kif24, is a centriolar kinesin that interacts with CP110 and Cep97, and regulates centriole elongation and ciliogenesis. This study suggests a mechanistic difference between different stages of centriole elongation and cilia.
- 37.Delgehyr N, Rangone H, Fu J, Mao G, Tom B, Riparbelli MG, Callaini G, Glover DM. Klp10a, a microtubule-depolymerizing kinesin-13, cooperates with cp110 to control drosophila centriole length. Curr Biol. 2012;22(6):502–509. doi: 10.1016/j.cub.2012.01.046. [DOI] [PubMed] [Google Scholar]
- 38.Tsang WY, Bossard C, Khanna H, Peranen J, Swaroop A, Malhotra V, Dynlacht BD. Cp110 suppresses primary cilia formation through its interaction with cep290, a protein deficient in human ciliary disease. Dev Cell. 2008;15(2):187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10(3):322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blagden SP, Glover DM. Polar expeditions--provisioning the centrosome for mitosis. Nat Cell Biol. 2003;5(6):505–511. doi: 10.1038/ncb0603-505. [DOI] [PubMed] [Google Scholar]
- 41.Doxsey S. Re-evaluating centrosome function. Nat Rev Mol Cell Biol. 2001;2(9):688–698. doi: 10.1038/35089575. [DOI] [PubMed] [Google Scholar]
- 42.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. Sas-4 is a c. Elegans centriolar protein that controls centrosome size. Cell. 2003;112(4):575–587. doi: 10.1016/s0092-8674(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 43.Leidel S, Gonczy P. Sas-4 is essential for centrosome duplication in c elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4(3):431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 44.Kitagawa D, Kohlmaier G, Keller D, Strnad P, Balestra FR, Fluckiger I, Gonczy P. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins cpap and stil. J Cell Sci. 2012;124(Pt 22):3884–3893. doi: 10.1242/jcs.089888. [DOI] [PubMed] [Google Scholar]
- 45.Hung LY, Tang CJ, Tang TK. Protein 4.1 r-135 interacts with a novel centrosomal protein (cpap) which is associated with the gamma-tubulin complex. Mol Cell Biol. 2000;20(20):7813–7825. doi: 10.1128/mcb.20.20.7813-7825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conduit PT, Brunk K, Dobbelaere J, Dix CI, Lucas EP, Raff JW. Centrioles regulate centrosome size by controlling the rate of cnn incorporation into the pcm. Curr Biol. 2010;20(24):2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]