Abstract
The innate immune system is important for control of infections, including herpesvirus infections. Intracellular DNA potently stimulates antiviral IFN responses. It is known that plasmacytoid dendritic cells sense herpesvirus DNA in endosomes via TLR9, and that non-immune tissue cells can sense herpesvirus DNA in the nucleus. However, it remains unknown how and where myeloid cells, like macrophages and conventional dendritic cells, detect infections with herpesviruses. Here we demonstrate that the HSV-1 capsid was ubiquitinated in the cytosol and degraded by the proteasome, hence releasing genomic DNA into the cytoplasm for detection by DNA sensors. In this context, the DNA sensor IFI16 is important for induction of IFN-β in human macrophages after infection with HSV-1 and CMV. Viral DNA localized to the same cytoplasmic regions as IFI16, with DNA sensing being independent of viral nuclear entry. Thus, proteasomal degradation of herpesvirus capsids releases DNA to the cytoplasm for recognition by DNA sensors.
Introduction
The innate immune system represents a first line of defense against infections, including viral infections (1,2), and utilizes a limited set of pattern recognition receptors (PRR)s3 to sense pathogen-associated molecular patterns (PAMP)s, which are either microbe-specific molecules or molecules with abnormal location of chemical modifications (3-6). A subset of PRRs stimulate expression of the antiviral type I IFNs in response to PAMP recognition (1,2), and absence of proper IFN responses have been shown to lead to severely impaired defense against viral infections in humans and mice (7-9).
Herpesviruses are a large family of double-stranded (ds) DNA viruses, which are the causative agents of disease, including encephalitis, genital herpes (HSV), congenital disorders and various conditions in immunocompromised individuals (CMV). Following entry of the virus, either by direct fusion with the plasma membrane or via endocytosis, productive infection is initiated by transport of the DNA-containing capsid along microtubules to the nucleus where the viral DNA is delivered (10). However, many virus particles do not lead to productive infection even in highly permissive cells (11), and cell types differ with respect to permissiveness for herpesvirus infections. Therefore, knowledge of innate immune response to viral infection, requires understanding of both productive and non-productive infections as well as the issue of cell type specificity.
DNA represents a potent PAMP stimulating IFN responses in many cell types (12-14). About ten intracellular DNA sensors have been proposed to date, yet the specific role for most of these sensors remains unclear. TLR9 is predominantly expressed by plasmacytoid dendritic cells (pDC)s and localizes to endosomes where it senses DNA including herpesvirus DNA (15-17). In non-pDCs, DNA is sensed in other subcellular locations. One proposed DNA sensor is IFN gamma-inducible (IFI)16, which is mainly localized to the nucleus, where this protein has long been known to play a role in DNA damage response, p53 signaling, and apoptosis (18). It has recently been reported that HSV-1 DNA interacts with IFI16 in the nucleus of the human osteosarcoma cell line U2OS and that induction of IFN-β by HSV-1 in HEK293 cells is dependent on the nuclear localization of IFI16 (19). This has subsequently been reported to be counteracted by the HSV-1 protein infected cell protein 0 (20). A previous report has demonstrated nuclear sensing of Kaposi's sarcoma-associated herpesvirus (KSHV) in human microvascular endothelial cells (21). In some cell types, including myeloid cells, a small portion of the cellular pool of IFI16 is localized in the cytoplasm (22), and most of the other proposed DNA sensors, including the helicase DDX41, which is involved in DNA sensing in conventional DCs (cDCs), localize to the cytoplasm (23). However, there is no knowledge on the subcellular site of herpesvirus DNA sensing in myeloid cells, and how the viral genomic material is made accessible for DNA sensors in these cells. Myeloid cells like macrophages and cDCs play important roles in innate control of virus infections, and are important producers of type I IFN during infection (24,25). In addition, it has been reported that cDCs activated by cytosolic DNA sensing potently activate the adaptive immune response (26).
Common for IFI16, DDX41 and other proposed DNA sensors is the requirement for stimulator of interferon genes (STING) for downstream signalling stimulating IFN expression (22,23,27,28). Upon DNA sensing, STING re-localizes to an, as yet, uncharacterized cytoplasmic foci, believed to serve as assembly platforms for signaling (28). It was recently reported that the C-terminal region of STING is responsible for assembly of the signaling complex activating the transcription factor IFN regulatory factor (IRF)-3, which drives transcription of IFN-β and IFN-stimulated genes (ISG) (29).
Here we demonstrate that HSV-1 and CMV infection induce IFI16-dependent IFN-β expression in human macrophages and that the infections mobilize IFI16 and STING to re-localize to the same subcellular regions. Moreover, IFI16 also associated with the same regions as the viral DNA genomes. In the macrophages, the ability of HSV-1 to induce IFN responses was independent of the ability of the virus to deliver its DNA into the nucleus, and the IFI16-DNA association was strictly cytoplasmic. Finally, we report that the HSV-1 capsid becomes ubiquitinated in macrophages and describe a vital role for the ubiquitin-proteasome pathway in degradation of HSV-1 capsids allowing for exposure of viral DNA for immediate sensing by DNA sensors. This study provides a mechanism for how viral DNA is made accessible to cytosolic DNA sensors to stimulate antiviral and inflammatory responses.
Materials and Methods
Cells
THP1 cells, cultured as non-adherent monocyte like cells, Vero, and U2OS cells were grown in RPMI (Invitrogen), with 10% FCS, 600 μg/ml Glutamine, 200 IU/ml of penicillin and 100μg/ml streptomycin (Gibco). Human forskin fibroblasts (HFF)s were grown in DMEM supplemented with 15% FCS and antibiotics as described above. THP1 cells were differentiated into macrophage like cells by addition of 100nM PMA (Sigma-Aldrich). All presented data with THP1 cells were based on PMA-differentiated cells. Buffy coats from Aarhus University Hospital blood bank were used to collect PBMCs by Ficoll Paque (GE Healthcare) gradient centrifugation. Monocytes were purified by plastic adherence using 1×108 PBMC seeded in 6cm UpCell plates (Nunc, Thermo-Scientific) pre-coated with Poly-L-Lysine (0.01% w/v, Cultrex) and allowed to stabilize over-night in Iscove's Modified Dulbecco's Medium supplemented with 10% FCS, 600 μg/ml Glutamine, 200 IU/ml of penicillin and 100μg/ml streptomycin. The following day, unattached cells were washed away and adherent cells were differentiated into monocyte-derived macrophages (MDM)s by culturing for additional 5 days in culture media: Iscove's Modified Dulbecco's Medium supplemented with 10% (vol/vol) pooled AB+ human sera (Invitrogen), 600 μg/ml Glutamine, 200 IU/ml of penicillin and 100μg/ml streptomycin (Gibco), and 20 ng/ml of macrophage colony-stimulating factor (Sigma-Aldrich). 0.5×106 MDMs were reseeded on glass coverslips for immunofluorescence studies. MDMs for IFI16 knock-down were generated as previously described (30). Bone marrow-derived macrophages were generated as previously described (31).
Antibodies and reagents
The antibodies used were: anti-IFI16 N-terminal (sc-8023), anti-ubiquitin (sc-34870), anti-RCC1 (sc-1161) (all Santa Cruz Biotechnology), anti-K48 polyubiquitin (Cell Signaling Technology, # 4289), anti-STING (Imgenex, IMG-6485A), anti-Vp5 (Ab6508), anti-proteasome p20S subunit (ab3325) anti-beta actin (Ab49900) (AbCam), anti-CMV minor capsid protein p28 (GenWay, 20-251-400023), anti-DDX41 (Sigma, SAB-2100554), Peroxidase-conjugated F(ab′)2 Donkey anti-Mouse IgG (H+L) and Peroxidase-conjugated affinipure F(ab′)2 Donkey anti-Rabbit IgG (H+L) (both Jacksons ImmunoResearch) and Polyclonal Rabbit anti-Goat immunoglobulin (DakoCytomation). Anti-IFI16 C-terminal was produced as described (32). The proteasome inhibitors used were MG132 (Merck Millipore), PI-083 (Merck Millipore), and PS-341 (Lifesensors). The proteasome inhibitors were shown to block lipopolysaccharide-induced IκBα degradation at the concentrations used in this study (data not shown). Leptomycin B (LMB) was obtained from Tocris Bioscience and shown to block nucleocytoplasmic shuttling of IκBα (33) (data not shown). The synthetic dsDNA 60mer derived from the HSV-1 genome nt. 144107-144166 was obtained from DNA Technology (22).
Viruses
HSV-1 strains 17+, HFEM, and the HFEM-derived mutant tsB7 were grown as previously described (34). Viruses were used at; HSV-1 MOI 3-10, CMV MOI 5. Human CMV strain AD169 was cultivated as previously described (30). HSV-1 and CMV were titrated on Vero cells and HFFs (30). The virus preparations were tested for endotoxin content, and found all to contain similar low levels (~ 0.75 endotoxin units/ml). Virus was inactivated with UVC light irradiation for 1 min (wavelength = 253.7 nm) with an intensity of 450 μW/cm2, which we have previously shown to reduce HSV replication by more 106 fold (35).
siRNA-mediated silencing
For transient knock-down of IFI16 in MDMs 100 nM of IFI16 siRNAs (Stealth RNAi mix; HSS105205, HSS105206, and HSS105207, Invitrogen) and recommended controls (Stealth RNAi control, Invitrogen) were transfected using HiPerfect (Qiagen) according to the manufacturer's instructions at day 5. The cells were incubated with the HiPerfect-siRNA mix for 4 h at 37°C before changing the media to fresh media including GM-CSF. Forty-eight hour after siRNA transfections, the macrophages were used for infection experiments. Two hours before infection macrophage were supplied with fresh media excluding GM-CSF.
shRNA mediated silencing
The lentiviral shRNA expression plasmid pLKO.1 was utilized for generating stable gene expression knockdown in THP1 cells (OpenBiosystems, Huntsville, USA). The targeting shRNA sequence was: IFI16 clone ID # TCRN0000019079; ddx41-1 clone ID # TRCN0000104010; ddx41-4 clone ID# TRCN0000104013. The control shRNA vector was an empty vector pLKO.1 with a 18nt shuttle sequence instead of the hairpin sequences. shRNA plasmids were amplified in TOP10 E.coli (Invitrogen, Denmark) and purified using Qiagen plasmid Plus kits (Qiagen, Denmark). Virus-like particles were produced using Fugene 6 (Roche) transfection of HEK293T cells with the packaging system pMDIg/p-RRE, pRSV-rev, pMG.2 and shRNA vector plasmid. Virus supernatants were harvest after 48 hours and filtrated through a 0.45um membrane. Undifferentiated THP1 cells were infected with titrating amounts of virus supernatant and after two days post-infection placed under selection with puromycin at 1ug/ml (Sigma). Level of knockdown was determined on differentiated THP1 cells by Western blot analysis after seven to ten days.
RT-PCR and primers
IFNβ and IFI16 gene expression were determined by real time PCR, using TaqMan and SYBR green detection systems, respectively (Applied Biosystems, Qiagen). Expression levels were normalized to β-Actin or GAPDH, expression and data presented as the fold induction over un-treated controls for each phenotype. Data represent the mean +/- SD from either biological replicates or technical replicates. PCR primers: IFI16 - Forward, TAG GCC CAG CTG AGA GCC ATC C; Reverse, TGA GGT CAC TCT GGG CAC TGT CTT, ICP27 – Forward, AGA CCA GAC GGA TCC CCT GGG AAA CCT; Reverse, AAA CAC GAA GGA TCC AAT GTC CTT AAT, GAPDH - Forward CGACCACTTTGTCAAGCTCA; Reverse, GGTGGTCCAGGGGTCTTACT (all DNA technology), IFNβ Applied Biosystems TaqMan Assay Hs01077958_s1, DDX41 Applied Biosystems TaqMan Assay Hs00169602_m1, AIM2 Applied Biosystems TaqMan Assay Hs00915710_m1, b-Actin Applied Biosystems TaqMan Assay Hs99999903_m1.
Confocal microscopy
For visualization of IFI16, following infection with viruses at indicated times, cells were fixed and permeabilized with methanol at -20°C and labeled with antibodies against IFI16 or STING. For visualization of ubiquitination and proteasome, BMM were fixed with 4% formaldehyde and permeabilized with 0.2% Triton x100 and stained with antibodies specific for ubiquitin and the proteasome p20S subunit. Images were acquired on Zeiss LSM 710 confocal microscope, using 63x 1.4 oil objective or Deltavision RT Core fluorescent microscope using a 100x 1.4 oil objective, fitted with Cascade 650 CCD camera. Image processing was performed using Zen 2010 (Zeiss) and ImageJ. Deconvolution of images acquired with Deltavision RT Core was performed using Softworx software. All images are representative of at least 2-3 independent experiments.
Fluorescence in situ hybridization
Visualization of HSV-1 genomic DNA by FISH was performed as described (22). CMV genomic DNA was visualized using the same protocol. The fluorescein-labeled CMV-specific probes used were Probe 1 – TAG CGG GGG GGT GAA ACT TGG AGT TGC GTG TGT GGA CGG CGA CTA GTT GCG TGT GGT G and Probe 2 – TTG GCA GGG TGT GTC AGG GTG TGT CGC GGG CGT GTG CCG GGT GTG TCG TGC CGG GTG TGT (DNA technology Aarhus Denmark). Briefly, cells were left untreated or, pre-treated with LMB (10 ng/ml), or MG132 (10 μg/ml) Genomic viral DNA was labeled with genome-specific probes and the capsid was labeled with antibodies specific for Vp5 (HSV-1) or p28 CMV. In addition, cells were also stained with antibodies specific for IFI16 (32). Images were acquired as described above. All images are representative of at least 3 independent experiments.
ELISA and Luminex
Cells were infected as described above, and supernatants were harvested and CXCL10 levels were measured by ELISA (RnD Systems). Phospho-IκBα were determined by Luminex technology using kits from BioRad.
Isolation of nuclear extracts
To isolate nuclear proteins, cells were washed twice in ice-cold PBS, scraped into 5 ml PBS and centrifuged for 1 min at 2000xg. The cells were resuspended in hypotonic lysis buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, and protease inhibitors) and left for 15 min on ice, after which NP40 was added to 0.6%, and the suspension was vortexed vigorously for 15 s. The nuclei were recovered by centrifugation (10000xg for 1 min) and resuspended in 40 μl extraction buffer (20 mM HEPES pH 7.9, 20% glycerol, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, 0.2% NP40, and protease inhibitors). Supernatants containing nuclear proteins were isolated after 30 min of rocking at 4 °C.
Immunoprecipitation and Western Blotting
Cells were washed twice in PBS and lysed in 850 μl of lysis buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM EDTA, 10% (v/v) glycerol, 0.5% (v/v) Nonidet P-40 containing Complete protease intibitor cocktail (Roche), and 1 mM sodium orthovanadate). For immunoprecipitation, anti-Vp5 was precoupled to protein A-Sepharose beads (Sigma-Aldrich) overnight at 4 °C. The beads were then washed twice in lysis buffer and incubated with 1.5 mg of cell lysate/sample overnight at 4 °C. The immune complexes were washed three times in lysis buffer, boiled, and analyzed by standard SDS-PAGE and Western blotting. Blots were visualized using an ImageQuant LAS 4000 mini Luminescent Image Analyzer (GE Healthcare).
Statistical analysis
Students T-test was performed for all statistical analysis.
Reproducibility of data
All data presented are representative of at least 3 independent experiments.
Results
Essential role for IFI16 in herpesvirus-induced IFN-β expression in human macrophages
Type I IFNs are important for control of herpesvirus infections (4,7-9). In the human monocyte-like cell line THP1 differentiated into a macrophage-like phenotype with PMA we found that both HSV-1 and CMV induced IFN-β expression and also activation of signal transduction to the NF-κB pathway and induction of the ISG, CXCL10 (Fig. 1A-E). To explore the role of DNA sensors in induction of this response, we generated a THP1-derived cell line stably transfected with control shRNA or shRNA targeting IFI16 (Supplemental Fig. 1A, B). Infection of these cells with HSV-1 or CMV revealed a requirement for IFI16 for IFN-β induction by these viruses (Fig. 1F, G). Interestingly, knockdown of DDX41 also affected the IFN-β response to HSV-1 infection in the PMA-differentiated THP1 cells (Supplemental Fig. 1C, D, E), which could indicate that these two proposed DNA sensors act either sequentially, in distinct pathways, or cooperatively in the same pathway. Finally, we wanted to validate the data above by testing the role of IFI16 in primary human MDMs. Importantly, similar to what we found in the cell line, we observed virus-induced IFN-β expression, which was decreased following siRNA knockdown of IFI16 (Fig. 1H, I and Supplemental Fig. 1F).
FIGURE 1.
Herpesviruses induce IFN responses in macrophages - dependent on IFI16. PMA-differentiated THP1 macrophages were infected with (A,C) HSV-1; or (B,D) CMV. (A, B) Total RNA was harvested 6 h post infection, and IFNβ mRNA was determined by RT-PCR. (C, D) Supernatants from cells treated for 16 h as indicated were analyzed for CXCL10 protein levels by ELISA. (E) Whole cell extracts from PMA-differentiated THP1 macrophages, treated as indicated were isolated and phospho-IκBα was determined by Luminex. (F, G) PMA-differentiated THP1 macrophages stably transfected with control vector (EV) or IFI16 shRNA (sh-IFI16) or (H, I) MDMs transfected with si-CTRL or si-IFI16 RNA were infected with (F, H) HSV-1 or (G, I) CMV. Total RNA was harvested 6 h post infection, and IFNβ mRNA was determined by RT-PCR. Data represent mean +/- SD of duplicates. * p<0.05.
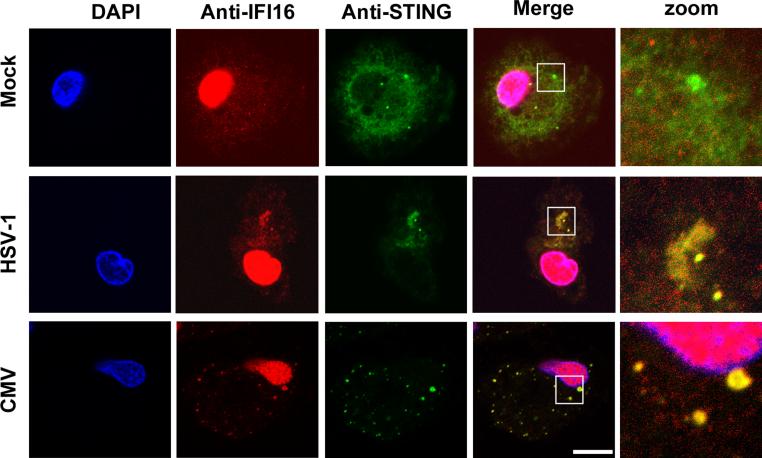
IFI16-induced IFN responses are dependent on STING (22), which relocates to discrete punctuate structures after DNA recognition (28). Using MDMs and confocal microscopy we were able to demonstrate that both IFI16 and STING relocated to such structures after infection with HSV-1 or CMV with a clear colocalization between IFI16-positive and STING-positive structures (Fig. 2). Over two donors we found that 20% and 51% of STING foci were also positive for IFI16 after infection with HSV-1 and HCMV, respectively. The constitutive expression of IFI16 in MDMs and PMA-differentiated THP1 cells (Fig. 2 and Supplemental Fig. 2A, B) suggests a role for IFI16 as an early sensor of DNA. This was further underscored by co-localization of synthetic dsDNA and IFI16 as early as 30 min after DNA transfection, and prior to the co-localization of dsDNA and STING, which was observed 60 min after transfection (Supplemental Fig. 2C). Thus, HSV-1 and CMV mobilize STING in human macrophages and induce IFN-β expression in an IFI16-dependent manner.
FIGURE 2.
IFI16 and STING co-localize following infection with herpesviruses. MDMs were mock infected or infected with HSV-1 or CMV for 4 h. Subcellular distribution of IFI16 and STING was determined by confocal microscopy. White box indicates area displayed in zoom column, scale bar 10 μm.
IFI16 senses herpesvirus DNA in the cytoplasm to drive IFN expression
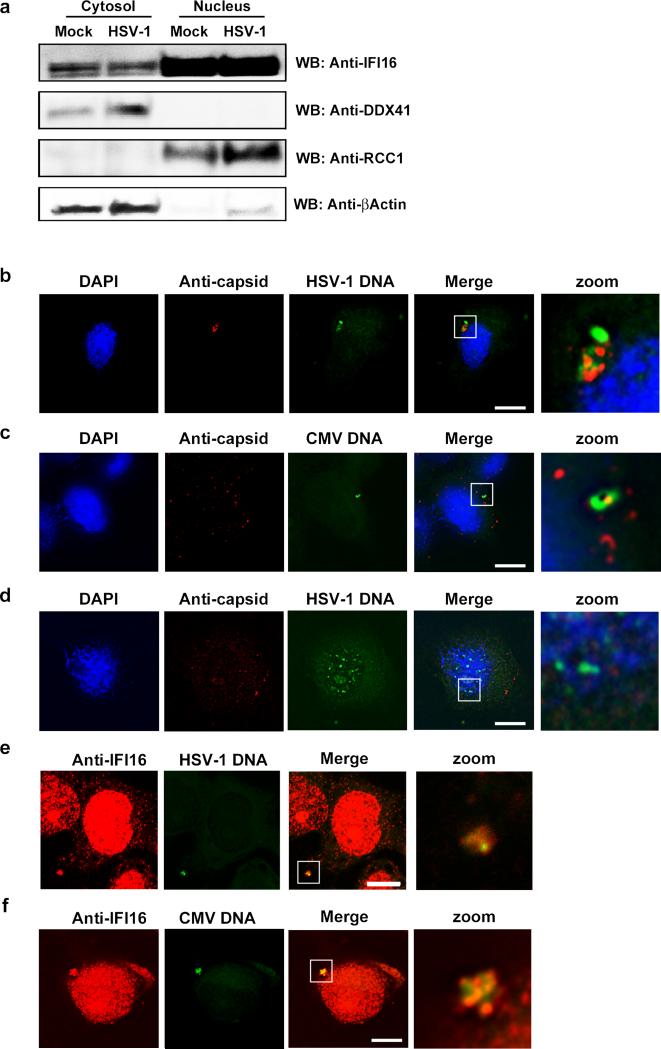
In order to examine for the subcellular localization of cellular DNA sensors in macrophages, we isolated nuclear and cytosolic fractions and performed Western blotting. We blotted for IFI16 and DDX41 as well as RRC1 (regulator of chromosome condensation 1, a nuclear protein) and β-actin (a cytoplasmic protein). IFI16 was mainly found in the nuclear fraction, but a small pool of the cellular IFI16 localized to the cytoplasm in macrophages (Fig. 3A). By contrast DDX41 was exclusively found in the cytoplasm (Fig. 3A). To look for the subcellular localization of viral DNA in the cells, HSV-1- and CMV-infected macrophages were probed with genome-specific probes, and visualized by fluorescence. In the infected cells, the HSV-1 and CMV genomes were detected in the cytoplasm (Fig. 3B, C). In the case of HSV-1, most of the viral DNA (29 +/- 5 genome-positive spots per 100 cells) co-localized with the capsid protein Vp5 2 h after infection (data not shown), but not at 4 h post infection (Fig. 3B, C). At 6 h post infection, very little staining for HSV-1 DNA was observed (data not shown). The cytoplasmic localization of herpesvirus DNA in the macrophages, was in contrast to the predominantly nuclear localization of the viral genome in Vero cells (Fig. 3D), consistent with potent viral transcription and replication in the Vero cells (Supplemental Fig. 3A, B). Importantly, in macrophages IFI16 co-localized with the HSV-1 and CMV genomic DNA in the cytoplasm (Fig. 3E, F), with over 40% colocalization between DNA and IFI16-positive cytoplasmic foci (42 % +/- 7%). The co-localization of IFI16 and HSV-1 DNA in the cytoplasm was not inhibited by LMB (Fig. 4A), which inhibits CRM1-mediated nuclear-export-signal-mediated export of proteins from the nucleus. This treatment also did not inhibit the detection of capsid-free HSV-1 DNA spots in the cytoplasm of infected macrophages (Fig. 4B, C), as determined by the application of a Hi-Lo look-up table (LUT) to each image, to allow for the observation of individual foci. These data indicate sensing of herpesvirus DNA to occur in the cytoplasm in macrophages and to be independent of nucleus-to-cytoplasm translocation of proteins.
FIGURE 3.
Herpesvirus DNA is present in the cytosol of infected macrophages and co-localizes with IFI16. (A) PMA-differentiated THP1 cells were mock-infected or infected with HSV-1 at MOI 3 for 6 h. Cytosolic and nuclear extracts were isolated and analyzed for IFI16, DDX41, RCC1, and β-actin by Western blotting. (B, C) PMA-differentiated THP1 cells were infected with (B) HSV-1 (C) CMV, and (D) Vero cells were infected with HSV-1 for 4 h and viral DNA was visualized by virus specific FISH probes (green) and co-stained with anti-capsid specific antibodies. White box indicates area displayed in zoom column, scale bar 10 μm. (E, F) MDMs were infected with HSV-1 or CMV for 4 hr and viral DNA was visualized by virus specific FISH probes (green) and co-stained with anti-IFI16 specific antibodies. White box indicates area displayed in zoom column, scale bar 10 μm.
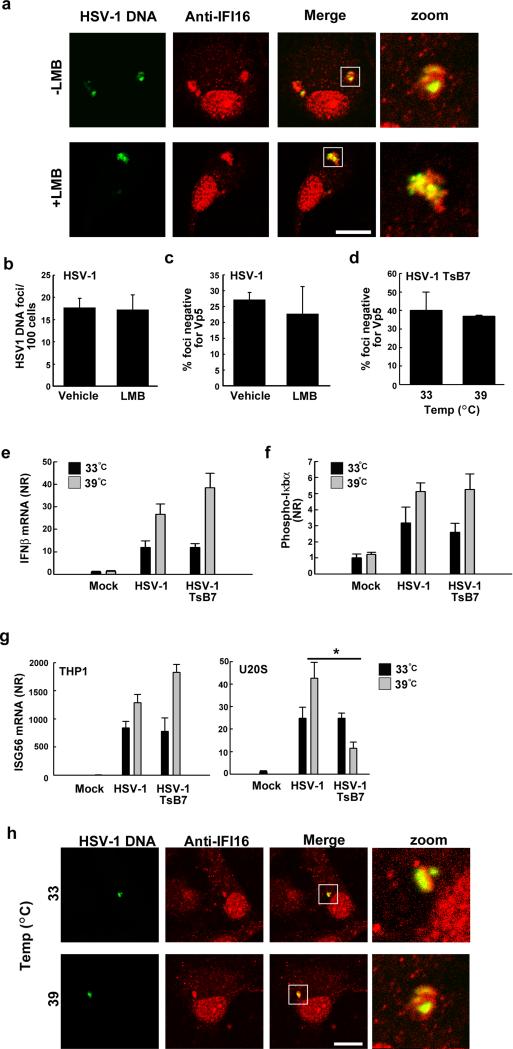
FIGURE 4.
Induction of type I IFN response and IFI16 co-localisation is independent of nuclear delivery of the viral genome. (A). MDMs were treated with vehicle or the nuclear export inhibitor LMB and infected with HSV-1 for 4 h. Viral DNA was visualized by HSV-1 specific FISH probes (green) and co-stained with anti-IFI16 specific antibodies, scale bar 10 μm. PMA-differentiated THP1 macrophages were (B, C) pre-treated with LMB and infected with HSV-1 for 4 h. (B) HSV-1 DNA foci per 100 cells and (C) percentage DNA foci negative for Vp5 were determined, data represent mean +/- SD. (D) PMA-differentiated THP1 macrophages were infected with the HSV-1 mutant TsB7 at 33°C (permissive) or 39°C (non-permissive) for 4 h and the percentage of HSV-1 DNA foci negative for Vp5 determined, data represent mean +/- SD. (E-F) PMA-differentiated THP1 macrophages were infected with TsB7 and HSV-1 for 6 hr and total RNA and whole-cell extracts were isolated for measurement of IFNβ mRNA (RT-qPCR) and phospho-IκBα (Luminex), respectively, data represent mean +/- SD of duplicates. (G) PMA-differentiated THP1 macrophages and U20S cells were infected with TsB7 and HSV-1 for 6 hr at 33°C or 39°C. For THP1 cells, live virus was used, and for U2OS cells UV-inactivated virus was used. Total RNA was isolated for measurement of ISG56 mRNA (RT-qPCR) data represent mean +/- SD of triplicates. * p<0.05 (H) MDMs were infected with TsB7 at 33°C or 39°C for 4 hr and viral DNA visualized by HSV-1 specific FISH probes (green) and co-stained with anti-IFI16 specific antibodies, scale bar 10 μm.
To further investigate the role of the nucleus in sensing HSV-1 DNA for stimulation of IFNβ expression, we utilized a temperature sensitive HSV-1 mutant TsB7. At the permissive temperature of 33°C TsB7 behaves as a wild type virus. However, at the non-permissive temperature of 39°C, the TsB7 HSV-1 is unable to deliver its genome to the nucleus (36). We confirmed that infection with wildtype HSV-1 was able to stimulate expression of ICP27 at both 33°C and 39°C, whereas infection with TsB7 led to ICP27 expression only at 33 °C (Supplemental Fig. 3C). Detection of capsid-free TsB7 genomes in the cytosol was not affected following incubation at the non-permissive temperature, compared to the permissive temperature (Fig. 4D), further supporting the contention that HSV-1 DNA does not need to enter the nucleus prior to cytosolic delivery. In addition, nuclear exclusion of HSV-1 DNA at the non-permissive temperature did not diminish, but in fact increased the phosphorylation of IκBα and also the expression of IFNβ mRNA (Fig. 4E, F). At both the permissive and non-permissive temperature, no statistically significant differences were observed between the responses to wildtype and TsB7 HSV-1 (Fig. 4E, F). The IFNβ responses to the viruses at either temperature were not inhibited by pretreatment with inhibitory oligonucleotides, hence excluding a role for TLR9 in the innate response at the abnormal temperatures (data not shown).
In cells highly permissive for HSV-1 infection, like Vero cells and U2OS cells, where we observed viral DNA in the nucleus (Fig. 3D), we were unable to observe significant IFN-β induction by the live virus (data not shown). However, using UV-inactivated HSV-1 we did observe an induction of the IFN-inducible gene ISG56 in U2OS cells (Fig. 4G). Importantly, the ISG56 response was elevated in these cells at 39°C after infection with HSV-1 but significantly inhibited after infection with TsB7 (p= 0.003) (Fig. 4G).
Finally, HSV-1 DNA was detectable in the cytoplasm of the differentiated THP1 cells co-localizing with IFI16 after infection with either HSV-1 or TsB7 at either 33°C or 39°C (Fig. 4H). These results demonstrate that recognition of HSV-1 DNA in the cytoplasm is responsible for IFN-β induction in macrophages.
Release of viral DNA into the cytoplasm is dependent on proteasomal capsid degradation
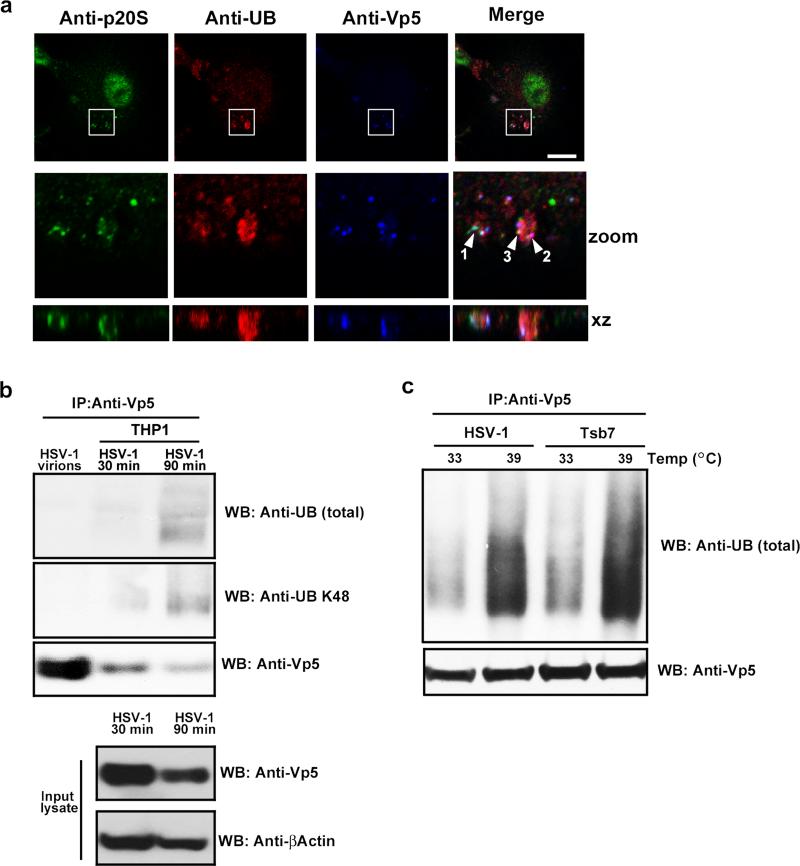
The mechanism through which herpesvirus DNA is exposed for sensing by PRRs in the cytosol of myeloid cells is unclear. We analyzed the potential role for the ubiquitin-proteasome pathway in this process (37,38). Prior to infection, ubiquitin was distributed throughout the cytosol and nucleus, the proteasome subunit p20S exhibited a punctuate distribution in the cytosol, with more pronounced staining in the nucleus, and no staining for Vp5 was observed (data not shown). Following infection with HSV-1, ubiquitin and p20S localized to discrete areas of the cytosol, with some HSV-1 capsid staining clearly co-localizing with ubiquitin and the proteasome (Fig. 5A). By counting over 100 Vp5 positive foci from 3 different experiments, we found that 18% +/- 2.5% of Vp5 foci co-localized with p20S at 2 h post-infection.
FIGURE 5.
Ubiquitination of the HSV-1 capsid. (A) BMMs were infected with HSV-1 (MOI 3) for 2 hr and co-stained with Anti-Vp5, anti-ubiquitin (Ub), and anti-proteasome (p20S). White box indicates area displayed in zoom row Arrows indicates the following co-localizations: 1, capsid and p20S; 2, caspid and Vp5; and 3, capsid p20S and Vp5 scalebar: 20 μm. (B) Lysates from PMA-differentiated THP1 macrophages infected for 30 or 90 min with 300 MOI of HSV-1 were subjected to IP using anti-Vp5-coupled beads. Total Ubiquitin and K48-coupled ubiquitin in the precipitate was detected using Western blotting. Levels of Vp5 and βactin in the input lysate used for IP was determined by Western blotting. (C) Lysates from PMA-differentiated THP1 macrophages infected for 90 min with 300 MOI of HSV-1 or the HSV-1 mutant TsB7 at either 33 °C or 39 °C were subjected to IP using anti-Vp5-couples beads. Total Ubiquitin in the precipitate was detected using Western blotting.
In order to examine whether the viral capsid was ubiquitinated, we precipitated Vp5 from HSV-1-infected THP1 macrophages and analyzed ubiquitination by Western blotting. Interestingly, whereas purified HSV-1 virions did not contain detectable levels of ubiquitin, Vp5 precipitated from infected macrophages did give rise to a clear signal for both total ubiquitin and K48-linkage polyubiquitin chains (Fig. 5B). Blotting with anti-K63 ubiquitin did not reveal detectable levels of this form of ubiquitin on either the virion capsid or capsids isolated from infected macrophages (data not shown). To determine whether the capsid ubiquitination was indeed targeting the incoming capsids in the cytoplasm and not empty capsids that had already delivered viral DNA to the nucleus, we utilized TsB7. Importantly, we found that Vp5 ubiquitination, which was higher at 39°C as compared to 33°C, was indistinguishable between HSV-1 and TsB7 (Fig. 5C).
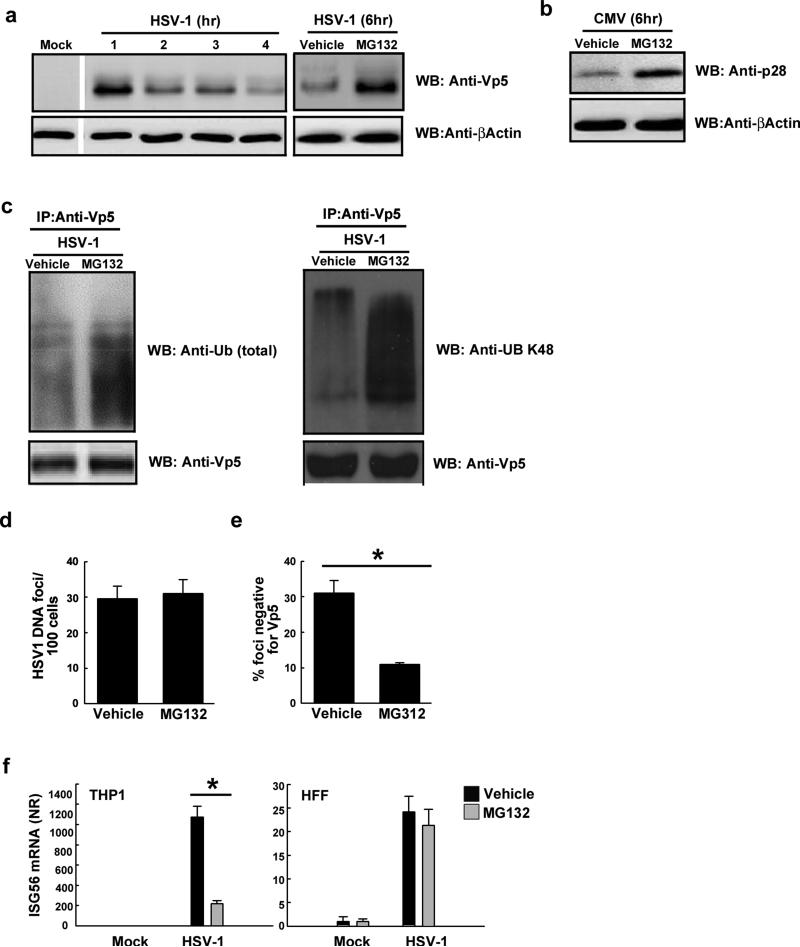
In order to examine the importance of Vp5 ubiquitin and proteasomal co-localisation for capsid degradation, the proteasome was inhibited with MG132. Readouts used were Vp5 levels in cytosolic extracts and quantification of Vp5-negative HSV-1 DNA spots in macrophages (as a measure of capsid-free DNA). Following infection of PMA-differentiated THP1 macrophages with HSV-1, intact Vp5 expression steadily decreased over 6h (Fig. 6A). This decrease in expression could be halted by pre-treatment of the cells with MG132 (Fig. 6A), and was also seen using two different proteasome inhibitors (Supplemental Fig. 4A). MG132 also extended the half-life of the minor CMV capsid protein p28 (Fig. 6B). Importantly, the ubiquitination of Vp5 was amplified if proteasome activity was inhibited by MG132 (Fig. 6C). The consequence of proteasomal degradation would likely be the release of viral DNA into the cytosol, and this was visualized using HSV-1-specific FISH probes (22,39). To allow for the quantification of HSV-1 DNA release, the number of HSV-1 DNA foci was determined and expressed as the HSV-1 DNA foci per 100 cells as above (Fig. 4B, C). Chemical inhibition of the proteasome did not affect the efficiency of viral entry, as determined by total DNA foci/100 cells (Fig. 6D), but led to a significant decrease in the amount of capsid-free viral DNA in primary macrophages (Fig. 6E) and THP1 cells (data not shown). Importantly, MG132 treatment of macrophages prevented HSV-1-induced expression of ISG56 (Fig. 6F), which is driven by IRF-3 (40). This was observed after infection at MOIs of either 3 or 300 (Fig. 6F and data not shown). Inhibition of the proteasome did not lead to accumulation of viral DNA in the nucleus and did not affect expression of HSV-1 ICP27 gene expression (data not shown). In contrast to the observations in macrophages, inhibition of the proteasome had only a minor effect on HSV-1-induced ISG56 expression in HFFs and U2OS cells (Fig. 6F and Supplemental Fig. 4B) where ISG induction was dependent on delivery of the viral genome to the nucleus (Fig. 4G)(20).
FIGURE 6.
The HSV-1 capsid is degraded by the proteasome. (A, B) PMA-differentiated THP1 macrophages were un-treated or pre-treated with MG132 (10 μg/ml) and infected with HSV-1 or CMV (MOI 3). Total cell lysates were isolated at the indicated time points post infection and the capsid protein Vp5 (A) or CMV p28 (B) were determined by Western blot. (C) Lysates from PMA-differentiated THP1 macrophages infected for 90 min with HSV-1 (MOI 300) in the presence or absence of MG132 were subjected to IP using anti-Vp5-coupled beads. Total and K48 Ubiquitin in the precipitate was detected using Western blotting. (D, E) BMMs were pretreated with MG132 and infected with HSV-1 (MOI 3) for 4 h. The cells were fixed and probed with anti-Vp5 and a HSV-1-specific FISH probe. The data are presented as (D) number of HSV-1 DNA foci per100 cells and (E) percentage HSV-1 DNA foci negative for Vp5. Data represents mean +/- SD, * p<0.05. (F) PMA-differentiated THP1 macrophages or HFFs were treated with vehicle or pre-treated with MG132 and infected with live (THP1 cells) or UV-inactivated (HFFs) HSV-1 (MOI 3) for 6 h and total RNA was isolated for measurement of ISG56 mRNA (RT-qPCR) data represent mean +/- SD of triplicates. * p<0.05.
Cumulatively, these results indicate that HSV-1 capsids are subjected to ubiquitination in macrophages and degraded by the proteasome, leading to the release of viral DNA to the cytosol for recognition by PRRs.
Discussion
DNA is a potent PAMP demonstrated to be involved in stimulation of both protective immunity to infections and excess inflammation (41,42). Herpesviruses are DNA viruses that stimulate innate immune responses mainly through nucleic acids, and these events are important for prevention of disease (17,22,23,41,43,44). While pDCs sense herpesvirus DNA in endosomes via TLR9 (15-17), recent reports demonstrate a role IFI16-mediated sensing of herpesvirus DNA in the nucleus of non-immune tissue cells (19,21). However, it remains unresolved how herpesvirus DNA is exposed to PRRs for sensing in myeloid cells. Our present study demonstrates that the HSV-1 capsid becomes ubiquitinated in the cytoplasm of macrophages and degraded via the ubiquitin-proteasome pathway, hence exposing viral genomic DNA for recognition by DNA sensors. Thus HSV-1 and CMV infection of macrophages leads to viral DNA recognition by DDX41 and IFI16, STING mobilization, and IFN-β expression.
Previously it has been reported that the proposed murine orthologue of IFI16, p204, is important for IFN induction during herpesvirus infection (22,45), and that HEK293 cells overexpressing IFI16 induce IFN-β in an IFI16-dependent manner (19). Using siRNA, we provide the first data for a role for IFI16 in HSV-induced IFN-β induction in a primary human cell type. We also observed that IFI16 was constitutively expressed to high levels and localized mainly in the nucleus with a significant sub-fraction localizing to the cytosol. IFI16 co-localised with viral DNA and relocated to the STING-postive foci, proposed to be key signaling platforms after DNA sensing (28). Using synthetic DNA we observed IFI16 to associate with DNA very early after transfection and prior to co-localization with STING. These data strongly support the conclusion that IFI16 is a bona fide IFN-inducing DNA sensor involved in early detection of foreign DNA in myeloid cells. The data also argue against the previously proposed role for IFI16 as a secondary amplifier of the IFN response (23). It is of note that HSV-1-induced IFN-β expression was potently reduced by knock-down of either DDX41 or IFI16. This raises the question as to how these two DNA sensors interact, and urges for more knowledge on the mechanism of action of the DNA sensors with respect to both actual DNA recognition and signal transduction. In particular it will be interesting to learn whether IFI16 and DDX41 work in the same pathway or in separate pathways merging on STING.
Two papers have recently addressed the role of IFI16 in recognition of HSV-1 and KSHV infections and both reported that the nucleus was the site of herpesvirus DNA recognition (19,21). These studies were both conducted in cells permissive for the viruses used, with accompanying nuclear localization of viral DNA and activation of viral gene expression. In this work we were able to confirm the nuclear location of HSV-1 DNA in the permissive cell line Vero, which was in contrast to the cytoplasmic localization of viral DNA in the macrophages. Moreover, we found that nuclear delivery of viral DNA was essential for induction of ISGs in these cells. By contrast, in the macrophages, IFI16 associated with DNA in the cytoplasm in a manner not inhibited by the nuclear export inhibitor leptomycin B and induced IFN-β expression in a manner independent of delivery of viral DNA to the nucleus. Thus, it seems that IFI16 can recognize DNA both in the nucleus and the cytoplasm and that the subcellular site of DNA recognition depends on where the DNA is delivered to. In the case of macrophages this is mainly the cytoplasm due to an active cytoplasmic system to detect and degrade herpesvirus capsids. It remains to be determined why permissive cells such as Vero cells, U2OS cells, and HFFs do not sense viral DNA in the cytosol after capsid degradation. One possibility is lack of a capsid sensing system or efficient viral evasion in these cell types. It will also be interesting to learn if there are qualitative differences in the cellular response to DNA sensing in the nucleus versus the cytoplasm.
A key finding of the present study is the ubiquitination of the HSV-1 capsid and degradation of the major capsid protein Vp5 in a proteasome-dependent manner, leading to exposure of viral DNA and recognition by DNA sensors and induction of innate responses such as IFNβ. It has previously been reported that the capsids of adeno-associated virus type 2 and 5 are ubiquitinated and that proteasome inhibitors increased transduction efficiency with these viruses (37). This indicates that the ubiquitin-proteasome pathway may represent an important antiviral pathway, which both degrades viral capsids and exposes viral nucleic PAMPs to cytosolic PRRs for induction of IFNs. With the data from the present work, and previous reports from others, there is now knowledge on how DNA derived from viruses, bacteria and the host genome may be delivered to the cytoplasm for detection by DNA sensors and induction of protective and pathological responses (42,46).
Pertel et al. have recently demonstrated that the cytosolic protein Trim5α, which has long been known to detect retroviral capsids and target them for degradation (38), also acts as a cytosolic PRR, stimulating ubiquitination of host proteins and establishment of an anti-viral state (47). Our present data demonstrating that herpesvirus capsids become ubiquitinated and subject to proteasomal degradation strongly suggest that the herpesvirus capsid is also recognized within the cytosol. Interestingly, Trim5α has recently been reported to inhibit HSV-1 and -2 replication at an early stage of the infection cycle (48), suggesting a role for this or a related protein in cytosolic sensing of herpesvirus capsids. Early work by Sodeik et al demonstrated that empty HSV-1 capsids (indicating delivery of DNA to the nucleus) start to accumulate inside Vero cells about 1 h post infection, and reach a level of 60% of all intracellular capsids at 4 h post infection (49). Such data suggest that the cytosolic innate immune surveillance pathway has a time window of between 1-4 hours to detect and degrade viral capsids in order to avoid nuclear entry and productive replication.
In conclusion, we propose that during herpesvirus infection in myeloid cells, the viral DNA is released into the cytosol after degradation of the viral capsid via the ubiquitin-proteasome pathway. This leads to cytosolic herpesvirus DNA sensing by IFI16 and IFN-β expression through the STING pathway. Together with the knowledge on herpesvirus DNA sensing in endosomes of pDCs and recent work on innate sensing of herpesviruses in the nucleus of non-immune tissue cells (19), this work demonstrates that the subcellular localization of innate DNA sensing is highly cell type specific, and most likely determined by the cellular sensing and trafficking systems, and the ability of microbes to interfere with these systems.
Supplementary Material
Acknowledgments
The technical assistance of Kirsten Stadel Petersen is greatly appreciated.
Footnotes
This work was funded by The Danish Medical Research Council (grant no 09-072636), The Novo Nordisk Foundation, The Velux Foundation, The Lundbeck Foundation (grant no R34-3855), Elvira og Rasmus Riisforts almenvelgørende Fond, and Fonden til Lægevidenskabens Fremme (all awarded to S.R.P.). K.A.H. was funded by a Marie Curie Incoming International Fellowship. This work is supported by grants from the NIH (AI083713 and AI083215 awarded to K.A.F and NIH grants DE018281 and CA019014 awarded to B.D.).
cDC, conventional dendritic cells; IFI, IFN-gamma inducible gene; IRF, IFN regulatory factor; ISG, IFN-stimulated gene; KSHV, Kaposi's sarcoma-associated herpesvirus; MDM, Monocyte-derived macrophage; PAMP, pathogen associated molecular patterns, pDC, plasmacytoid dendritic cell, PRR, pattern recognition receptor; STING, stimulator of IFN Genes.
References
- 1.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nature Immunology. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 2.Paludan SR, Bowie AG, Horan KA, Fitzgerald KA. Recognition of herpesviruses by the innate immune system. Nat.Rev.Immunol. 2011;11:143–154. doi: 10.1038/nri2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb.Symp.Quant.Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Poltorak A, He X, Smirnova I, Liu M, van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1999;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 5.Imai Y, Kuba K, Neely GG, Yaghubian-Malhami R, Perkmann T, van Loo G, Ermolaeva M, Veldhuizen R, Leung YH, Wang H, Liu H, Sun Y, Pasparakis M, Kopf M, Mech C, Bavari S, Peiris JS, Slutsky AS, Akira S, Hultqvist M, Holmdahl R, Nicholls J, Jiang C, Binder CJ, Penninger JM. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thurston TLM, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–U1515. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dupuis S, Jouanguy E, Al Hajjar S, Fieschi C, Al Mohsen IZ, Al Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P, Al Ghonaium A, Tufenkeji H, Frayha H, Al Gazlan S, Al Rayes H, Schreiber RD, Gresser I, Casanova JL. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat.Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 8.Leib DA, Harrison TE, Laslo KM, Machalek MA, Moorman NJ, Virgin HW. Interferons regulate the phenotype of wild-type and mutant herpes simplex viruses in vivo. J.Exp.Med. 1999;189:663–672. doi: 10.1084/jem.189.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ank N, Iversen MB, Bartholdy C, Staeheli P, Hartmann R, Jensen UB, Dagnaes-Hansen F, Thomsen AR, Chen Z, Haugen H, Klucher K, Paludan SR. An important role for Type III interferon (IFN-lambda/IL-28) in Toll-like receptor-induced antiviral activity. J.Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- 10.Pellet PE, Roizman B. The Family Herpesviridae: A brief introduction. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Field's Virology. Lippincott-Williams and Wilkins; New York: 2007. pp. 2479–2499. [Google Scholar]
- 11.Harland J, Brown SM. HSV Growth, Preparation, and Assay. Methods Mol Med. 1998;10:1–8. doi: 10.1385/0-89603-347-3:1. [DOI] [PubMed] [Google Scholar]
- 12.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 13.Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, Sato S, Yamamoto M, Uematsu S, Kawai T, Takeuchi O, Akira S. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat.Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 14.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J.Exp.Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krug A, Luker GD, Barchet W, Leib DA, Akira S, Colonna M. Herpes simplex virus type 1 activates murine natural interferon-producing cells through toll-like receptor 9. Blood. 2004;103:1433–1437. doi: 10.1182/blood-2003-08-2674. [DOI] [PubMed] [Google Scholar]
- 17.Krug A, French AR, Barchet W, Fischer JA, Dzionek A, Pingel JT, Orihuela MM, Akira S, Yokoyama WM, Colonna M. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Mondini M, Costa S, Sponza S, Gugliesi F, Gariglio M, Landolfo S. The interferon inducible HIN-200 gene family in apoptosis and inflammation: implication for autoimmunity. Autoimmunity. 2010;43:226–231. doi: 10.3109/08916930903510922. [DOI] [PubMed] [Google Scholar]
- 19.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. PNAS. 2012;109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Orzalli MH, DeLuca NA, Knipe DM. HSV-1 ICP0 redistributes the nuclear IFI16 pathogen sensor and promotes its degradation. PNAS. 2012 In press. [Google Scholar]
- 21.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 Acts as a Nuclear Pathogen Sensor to Induce the Inflammasome in Response to Kaposi Sarcoma-Associated Herpesvirus Infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois C, Jin T, Xiao T, Fitzgerald P, Paludan SR, Bowie AG. IFI16 is an innate immune sensor for intracellular DNA. Nat.Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang ZQ, Yuan B, Bao MS, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature Immunology. 2011;12:959–U62. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stockinger S, Kastner R, Kernbauer E, Pilz A, Westermayer S, Reutterer B, Soulat D, Stengl G, Vogl C, Frenz T, Waibler Z, Taniguchi T, Rulicke T, Kalinke U, Muller M, Decker T. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 2009;5:e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumagai Y, Takeuchi O, Kato H, Kumar H, Matsui K, Morii E, Aozasa K, Kawai T, Akira S. Alveolar macrophages are the primary interferon-alpha producer in pulmonary infection with RNA viruses. Immunity. 2007;27:240–252. doi: 10.1016/j.immuni.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Kis-Toth K, Szanto A, Thai TH, Tsokos GC. Cytosolic DNA-Activated Human Dendritic Cells Are Potent Activators of the Adaptive Immune Response. Journal of Immunology. 2011;187:1222–1234. doi: 10.4049/jimmunol.1100469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci.Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melchjorsen J, Rintahaka J, Søby S, Horan KA, Ostergaard L, Paludan SR, Matikainen S. Innate recognition of HSV in human primary macrophages is mediated via the MDA5/MAVS pathway and MDA5/MAVS/Pol III independent pathways. J.Virol. 2010;84:11350–11358. doi: 10.1128/JVI.01106-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez Dosal R, Horan KA, Rahbek SH, Ichijo H, Chen ZJ, Mieyal JJ, Hartmann R, Paludan SR. HSV infection induces production of ROS, which potentiate signaling from pattern recognition receptors: role for S-glutathionylation of TRAF3 and 6. PLoS Pathog. 2011;7:e1002250. doi: 10.1371/journal.ppat.1002250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa S, Borgogna C, Mondini M, De Andrea M, Meroni PL, Berti E, Gariglio M, Landolfo S. Redistribution of the nuclear protein IFI16 into the cytoplasm of ultraviolet B-exposed keratinocytes as a mechanism of autoantigen processing. British Journal of Dermatology. 2011;164:282–290. doi: 10.1111/j.1365-2133.2010.10097.x. [DOI] [PubMed] [Google Scholar]
- 33.Birbach A, Gold P, Binder BR, Hofer E, de Martin R, Schmid JA. Signaling molecules of the NF-kappa B pathway shuttle constitutively between cytoplasm and nucleus. Journal of Biological Chemistry. 2002;277:10842–10851. doi: 10.1074/jbc.M112475200. [DOI] [PubMed] [Google Scholar]
- 34.Paludan SR. Requirements for the induction of interleukin-6 by herpes simplex virus-infected leukocytes. J.Virol. 2001;75:8008–8015. doi: 10.1128/JVI.75.17.8008-8015.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malmgaard L, Melchjorsen J, Bowie AG, Mogensen SC, Paludan SR. Viral activation of macrophages through TLR-dependent and -independent pathways. J.Immunol. 2004;173:6890–6898. doi: 10.4049/jimmunol.173.11.6890. [DOI] [PubMed] [Google Scholar]
- 36.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J.Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan ZY, Zak R, Luxton GWG, Ritchie TC, Bantel-Schaal U, Engelhardt JF. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. Journal of Virology. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc.Natl.Acad.Sci.U.S.A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen SB, Horan KA, Holm CK, Stranks AJ, Mettenleiter TC, Simon AK, Jensen SB, Rixon FJ, He B, Paludan SR. Activation of Autophagy by alpha-Herpesviruses in Myeloid Cells Is Mediated by Cytoplasmic Viral DNA through a Mechanism Dependent on Stimulator of IFN Genes. Journal of Immunology. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grandvaux N, Servant MJ, tenOever B, Sen GC, Balachandran S, Barber GN, Lin RT, Hiscott J. Transcriptional profiling of interferon regulatory factor 3 target genes: Direct involvement in the regulation of interferon-stimulated genes. Journal of Virology. 2002;76:5532–5539. doi: 10.1128/JVI.76.11.5532-5539.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rathinam VA, Jiang Z, Waggoner SN, Sharma S, Cole LE, Waggoner L, Vanaja SK, Monks BG, Ganesan S, Latz E, Hornung V, Vogel SN, Szomolanyi-Tsuda E, Fitzgerald KA. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat.Immunol. 2010;11:395–402. doi: 10.1038/ni.1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, Ohba Y, Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 44.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, Segal D, Sancho-Shimizu V, Lorenzo L, Puel A, Picard C, Chapgier A, Plancoulaine S, Titeux M, Cognet C, von Bernuth H, Ku CL, Casrouge A, Zhang XX, Barreiro L, Leonard J, Hamilton C, Lebon P, Heron B, Vallee L, Quintana-Murci L, Hovnanian A, Rozenberg F, Vivier E, Geissmann F, Tardieu M, Abel L, Casanova JL. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 45.Conrady CD, Zheng M, Fitzgerald KA, Lui C, Carr DJ. Resistance to HSV-1 infection in the epithelium resides with the novel innate sensor, IFI-16. Mucosal Immunology. 2012;5:173–183. doi: 10.1038/mi.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. Mycobacterium Tuberculosis Activates the DNA-Dependent Cytosolic Surveillance Pathway within Macrophages. Cell Host Microbe. 2012;11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reszka N, Zhou CH, Song B, Sodroski JG, Knipe DM. Simian TRIM5 alpha proteins reduce replication of herpes simplex virus. Virology. 2010;398:243–250. doi: 10.1016/j.virol.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J.Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.