Abstract
Various populations of memory phenotype CD8+ T cells have been described over the last 15–20 years, all of which possess elevated effector functions relative to naïve phenotype cells. Using a technique for isolating antigen specific cells from unprimed hosts, we recently identified a new subset of cells, specific for nominal antigen, but phenotypically and functionally similar to memory cells arising as a result of homeostatic proliferation (HP). We show here that these “Virtual Memory” cells are independent of previously identified “innate memory” cells, arising as a result of their response to IL-15 trans-presentation by lymphoid tissue-resident CD8α+ DCs in the periphery. The absence of IL-15, CD8+ T cell expression of either CD122 or Eomes, or of CD8a+ DCs all lead to the loss of Virtual Memory cells in the host. Our results show that CD8+ T cell homeostatic expansion is an active process within the non-lymphopenic environment, is mediated by IL-15, and produces antigen inexperienced memory cells which retain the capacity to respond to nominal antigen with memory-like function. Preferential engagement of these “Virtual Memory” T cells into a vaccine response could dramatically enhance the rate by which immune protection develops.
Introduction
Memory phenotype cells arise in a host either as a result of antigenic stimulation or as a result of homeostatic proliferation (HP) (1). Depending on its context, antigen stimulation induces the formation of one of a number of memory cell subsets, each with unique properties with respect to proliferation, trafficking, and effector response (1–3). Similarly, conditions of extreme lymphopenia induce the formation of memory phenotype cells through HP induced by cytokines such as IL-7, IL-12 and IL-15 (4). This form of proliferation results in the expression of many, though not all, memory activation markers and the acquisition of an increased degree of immune protective function relative to naïve phenotype cells (5–11). While HP requires TCR/MHC interactions (5, 12–14), it does not require or induce overt TCR mediated stimulation, as evidenced by the differential expression of activation markers such as CD49d (15). Until recently, the physiological relevance of HP outside of bone marrow transplantation was unclear, as was the representation of HP memory T cells within a normal, un-manipulated host.
In addition to antigen-mediated and HP memory cells, the loss of a variety of transcription factors results in the production of so called “innate memory” T cells within the thymus, largely in mice on the BALB/c background, but also to some degree in C57BL/6 animals (B6) (16). These cells are typically CD8+, bear a memory phenotype, and, like NKT cells, respond to stimulation by rapid production of IFNγ when in the periphery. It was recently determined that the development of these cells within the thymus requires IL-4 production by PLZF+ iNKT cells (17). The production of innate memory T cells is amplified in mice deficient in itk (17–23), slp76 (24, 25), Id3 (24, 26–29) or klf2 (17, 30). A lack of these transcription factors allows an increase in innate memory cell expansion within the thymus as a result of increased local production of IL-4 (16). It is currently unclear what repertoire of antigen specificities these innate memory cells might contain or what the precise functional impact of these cells, in the thymus or periphery, might be in regards to the development of protective immunity.
We and our collaborators recently described a novel subset of memory phenotype CD8+ T cells that exist in the periphery of normal, lymphoreplete hosts (15). These cells are phenotypically similar to both innate memory cells and HP memory cells. Further investigation of these memory phenotype cells revealed that they included cells specific for nominal antigen even in the absence of previous antigen exposure. Indeed, their phenotype (CD49dlo) was consistent with their having undergone HP, not with their having responded to antigenic stimulation. These Virtual Memory (VM) cells (memory phenotype cells specific for nominal antigen within an antigen-inexperienced host) bore all of the phenotypic and functional hallmarks of HP memory cells (4) with the notable exception that they were not derived from a lymphopenic environment.
These initial observations raised the question whether VM cells were induced by the same thymic processes that produced innate memory cells or rather by some form of HP in the periphery. Recent observations by our collaborators showed that VM cells were slightly, but statistically significantly reduced in number in B6 mice lacking IL-4 (31). This suggested that at least a portion of VM cells might be similar to innate memory cells. However, Akue et al. also showed that most VM cells acquired their properties in the periphery and not the thymus, with the suggestion that the response of recent thymic emigrants to the lymphopenic neonatal environment may be responsible for promoting HP and the production of VM cells in the normal B6 host.
That being said, the precise mechanism by which VM cells experience HP, as well as the capacity of VM cells to participate in normal immune responses in the periphery remains largely unexplored. Cells having undergone HP possess both phenotypic and functional characteristics of true antigen experienced memory cells (4, 5). Given that VM cells essentially represent a population of preexisting memory phenotype cells specific for nominal antigen in the antigen inexperienced host, their development and function must be considered in order for us to understand completely the nature of the “primary” immune response.
Here we confirm that the production of VM cells is independent of innate memory cell development in the thymus. Further, we show that VM cells develop as a result of CD122 stimulation on the T cells mediated by IL-15 encounter in the periphery. This occurs largely as a result of IL-15 trans-presentation by CD8α+ DCs despite substantial production of IL-15 by tissue derived DCs expressing or lacking CD103. These data indicate that HP is an active process during normal immune development (not just lymphopenia), and is mediated by IL-15 in the unmanipulated host. VM cells therefore represent a cell type within the unprimed repertoire that is one step closer to the formation of immunologic memory without the host ever having seen antigen.
Materials and Methods
Mice and Reagents
6–12 week old female C57BL6 mice were purchased from NCI (Bethesda, MD). IL-4−/− and IL-15Ra−/− mice were purchased from Jackson Laboratories (Bar Harbor, MA). RAG−/−, RAG−/− gBT1 TCR transgenic, OT-1 TCR transgenic, Ja18−/−, CD1−/−, and IL-4−/−, Tbet−/−, IL-15Ra−/−, CD122−/− (a kind gift from Charlie Surh at the Scripps Institute, La Jolla CA), and B6/129 BatF3−/− and B6 BatF3−/− (kind gifts from Ken Murphy at Washington University, St.Louis MO) mice were bred in the National Jewish Biological Resource Center. Spleen cells from i) Eomes conditional knockout mice (Eomesfl/fl × CD4-cre) were obtained from E. John Wherry (University of Pennsylvania, PA), ii) from PLZFlu/lu B6 mice were obtained from Harald Von Boehmer (Dana-Farber Cancer Institute, Boston, MA), and iii) id3−/− mice were obtained from Barbara Kee (University of Chicago, IL). Peptides were ordered from Global Peptide (Ft. Collins, CO). Class I MHC tetramers were produced as previously described (32) or purchased from Beckman Coulter (Immunomics, San Diego, CA). Fluorochrome conjugated antibodies against CD8, CD44, B220, CD4, CD69, CD49d (a4-integrin), CD122, were purchased from Beckton Dickinson (San Diego, CA) or eBioscience (San Diego, CA). Vaccinia Virus (WR strain) was produced by infection of Vero cells as previously described (32). Listeria expressing whole ovalbumin (LM-ova) protein under the control of the hyl promoter was a gift from Michael Bevan at the University of Washington (33). IL-15 (Peprotech, Rocky Hill, NJ) and IL-15Rα-Fc (R&D Systems, Minneapolis, MN) complexes were made as previously described (34). All mouse protocols were approved by the Institutional Animal Use and Care Committees at National Jewish Health.
Tetramer staining and magnetic bead sorting
Tissues from mice (Spleen, LN, ovary) were removed and collagenase digested for 45 minutes as previously described (32). In the absence of collagenase digestion, antigen specific T cells are poorly isolated from antigen-challenged hosts (35). Therefore, collagenase treatment was typically used for isolating cells from both naïve and antigen stimulated mice. However, qualitatively similar recovery of naïve and VM populations were observed with or without inclusion of collagenase in the isolation procedure. Cells were resuspended in 500ul sorting buffer (consisted of 250 ml 24G2 supernatant and 250ml HBSS containing 2% FCS and 0.2% azide, to prevent internalization of the tetramer) per spleen. Cells were stained with tetramer for 30–60 minutes at 37°C. Cells were then washed, resuspended in HBSS plus azide, and stained with anti-PE coupled MACS MicroBeads (Miltenyi Biotec, Auburn, CA) for 30 minutes rotating at 4°. Cells were again washed, resuspended in HBSS plus azide and the tetramer+ cells isolated on a magnetized MACS column (Miltenyi Biotec, Auburn, CA) according to manufacturers instructions. Following elution, cells were centrifuged and resuspended in FACS buffer and stained for markers used to enhance gating of tetramer+ CD8+ T cells (CD8 and CD3 as a positive gate; B220 and, in some experiments, CD4 or CD11b as a dump gate), as well as with antibodies to determine activation status (CD44, α4-integrin, CD122, LFA-1, Ly6C, CD69, CD25). Cells were analyzed on a Cyan ADP (Dako Cytomation) Data were analyzed using FlowJo software, following the gating strategy described in Figure 1. Statistical analysis was performed using GraphPad Prism (La Jolla, CA)
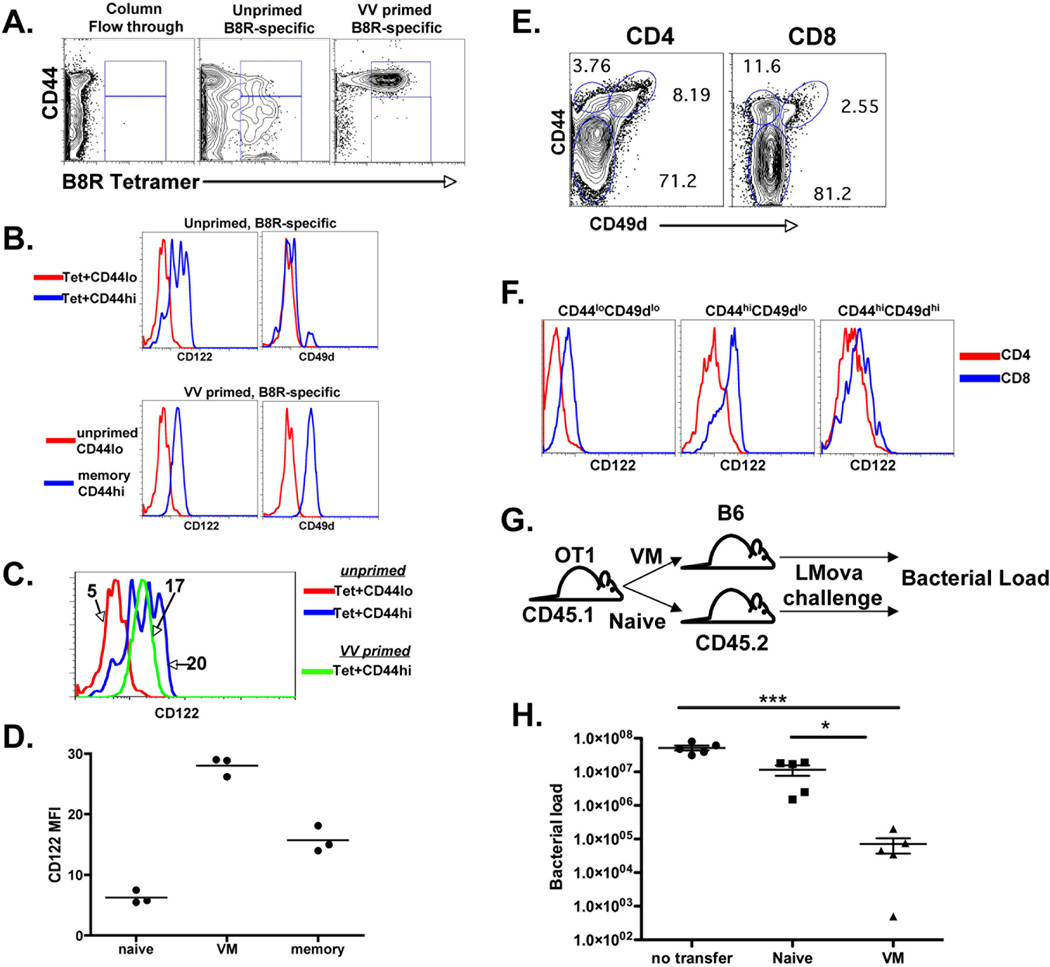
Figure 1.
VM cells are a distinct subset of CD8+ T cells with memory-like phenotype and function. A) B8R specific T cells were isolated from naive B6, and B6 mice challenged with VV 30 days previously, using magnetic enrichment of tetramer stained cells as described previously (15) and in the Materials and Methods. B) CD44hi and lo cells, from both naïve and VV-challenged hosts as shown in A, were analyzed by FACS for CD122 and CD49d expression. C, D) Overlay and graph of CD122 expression of the cells from the regions described in A and B. Data is shown as mean fluorescence intensity (MFI) of CD122 staining of each population. E) FACS analysis of spleen CD4 and CD8+ spleen cells for the distribution of CD122 and CD49d staining. F) Overlay of CD122 expression from the regions shown in E. G and H) Naïve and VM T cell transfer and immune protection study. CD44lo and CD44hiCD49dlo cells were sorted by FACS. And transferred into separate naïve hosts which were then challenged with LMova. Five days later, the bacterial load in the spleens was determined as previously described. All data are representative of 2–5 experiments performed. Data points are from 3–4 mice per group. Error bars represent standard deviation. All experiments shown were performed 2–5 times. * p value < 0.05, ** p value <0.001, *** p value <0.0001, unpaired t-test.
Immunizations
Where indicated, mice were injected with 50–100ug of B8R peptide together in combination with antiCD40 antibody (FGK45) and poly-I:C (GE Life Sciences) as previously described (36–39). For viral challenge, mice were injected i.p with 1×107 pfu of vaccinia virus strain WR as previously described (32). IL-15/Rα complexes were made by combining IL-15 and IL-15Rα-FC as previously described (34). Complexes were injected i.p. and the spleen cells analyzed 4–5 days later.
Immune protection
Spleen cells from CD45.1+ OT-I mice were sorted into CD44lo (naïve) and CD44hiCD49dlo (VM) CD8+ T cells by flow cytometry (15). 3×105 cells from each population were transferred into separate naïve B6 (CD45.2) mice which were then challenged i.v. with 1×105 CFU with LMova. Five days later, the bacterial load in the spleens were determined as previously described (40).
Bone Marrow Chimeras
Recipient mice were lethally irradiated with 900 rads followed by grafting via the tail vein with 4×106 total T cell-depleted donor bone marrow cells suspended in 200µL of PBS. Bone marrow was depleted of T cells using magnetic removal of CD3+ cells (Miltenyi Biotec, Auburn, CA). Bone marrow chimeras were grafted at a ratio of 1:1, representing 2×106 cells from CD45.1 WT and CD122−/− bone marrow. Chimeric mice were rested a minimum of 12 weeks before being analyzed or immunized for experiments. Chimeric mice were fed tirmethoprim-sulfamethoxazole-containing chow (Harlan Teklad 6596) for 6 weeks following reconstitution to reduce the risk of bacterial infection, but were switched to standard chow (Harlan Teklad 2919) well before immunization.
Generation and DC analysis of IL-15TE reporter mice
We generated an IL-15 translational reporter mouse by modifying a Bacterial Artificial Chromosome (BAC) and injecting it into pronuclei of C57BL/6J mice. 200 kb-long BAC clone RP23-331F16, which included the entire IL-15 genomic locus, was obtained from Children’s Hospital Oakland Research Institute in Oakland, California. Exon 8 of IL-15 was modified using a galK-based positive/negative selection system with reagents obtained from the National Cancer Institute Frederick campus (41). The modifications included addition of sequences coding for 2A-peptide from Thosea asigna virus (TaV) RAEGRGSLLTCGDVEENPGP, and Enhanced Green Fluorescence Protein (EGFP), to the end of Il-15. Our in vitro expression studies in COS-7 cells revealed that the wild type sequence of the TaV 2A peptide made the IL-15 biologically inactive (data not shown). We overcame this problem by mutating C to S within the 2A peptide, a change that most likely prevented disulfide bond formation of the IL-15 protein tagged with the 2A peptide (2A peptide remains attached to IL-15 after translation). The final construct contained a single open-reading-frame coding for IL-15, 2A from TaV(CtoS), and EGFP peptides, a fusion designated as IL-15TE. Sequence of the final BAC construct was verified by restriction digests and direct sequencing of the modified Exon 8 of IL-15. Next, IL-15 genomic sequences were separated from the vector sequences by restriction digest, purified on a Sephadex G-25 column, and injected into pronuclei of C57BL/6J mice. Founder mice were screened by Southern Blotting, and IL-15TE founder #20 containing 2 copies of the transgene was selected as the final reporter mouse. For the rationale behind creation of this translational reporter and the graphic representation of this approach, see Supplemental Figure 1 (42–45).
Dendritic cells were isolated from spleens in EHAA media (Invitrogen) containing DNAse (Worthington, Lakewood, NJ) and Collagenase D (Roche Diagnostics, Indianapolis, IN) as described (32). Fluorochrome conjugated antibodies against CD11c, CD11b, CD8, CD103, B220, CD40, Class II and CD326 were all purchased from eBioscience and DC stains and analysis were performed as previously described (46).
Results
VM cells are distinct from antigen experienced memory cells and are present in the CD8, not CD4 T cell subset
Using magnetic columns to enrich for tetramer stained T cells, we and others characterized the naïve T cell repertoire specific for different antigens within the unprimed host (15, 47, 48). This method revealed a unique population of memory phenotype T cells (VM cells) bearing the phenotype of cells having undergone HP (15). VM cells expressed intermediate levels of LFA1 and high levels of CD44, Ly6C, and CD122 ((15) and Fig 1A and B). However, in contrast to memory cells resulting from antigenic stimulation, VM cells expressed low levels of the α4 integrin, CD49d (Fig. 1A and B) (15). One quality of VM cells we previously noted was their high expression of CD122 (15). Indeed, closer examination revealed that VM cells expressed even higher levels of CD122 than antigen-experienced memory cells of the same specificity (Fig 1C and D). CD8+ T cells, both human and mouse, are well documented to have overall higher levels of CD122 expression than CD4+ T cells, and so it was little surprise to find that the VM subset was present only within the unprimed CD8+ and not the CD4+ T cell repertoire (Fig 1E). While there are CD122 expressing CD4+ T cells present in the unprimed host, they are essentially all CD49d+. These data suggest that the majority of CD122hi CD4+ T cells are the result of antigen stimulation and not the result of the cause(s) underlying CD8+ VM cell development. Consistent with this, the levels of CD122 expression on CD44hi CD4+ T cells was similar to that observed for CD49dhi CD8+ T cells but lower than that observed in the CD49dlo VM cell subset (Fig 1F).
HP memory cells derived from a lymphopenic environment have an identical phenotype to that of VM cells and were previously shown to mediate enhanced immunologic protection against infectious challenge (40). We used the same experimental system to determine whether or not VM cells, in the absence of any additional stimulation, could also mediated enhanced immunologic protection (Fig 1G). Naïve phenotype (CD44lo, CD49dlo) and VM phenotype CD44hi, CD49dlo) CD45.1+ OT1 cells were sorted and transferred into separate naïve CD45.2 B6 recipients. These recipients were then challenged with Listeria monocytogenes expressing ovalbumin (LMova) and four days later the bacterial load in the spleen was determined. Mice transferred with VM cells were better protected against LM challenge than either naïve OT1 transferred hosts or no transferred controls (Fig 1H). We conclude from these data (Fig 1) that VM cells represent a pool of T cells within the unprimed host that are i) unique to CD8+ T cells, ii) distinct from antigen-experienced memory cells, and iii) are phenotypically and functional similar to HP-memory cells.
VM cell development in B6 mice is largely independent of IL-4, NKT cells, PLZF, and Tbet
Memory phenotype CD8+ T cells arise in the thymus of a number of genetically modified mouse strains. Cells originally dubbed innate memory cells were found in elevated numbers in itk−/− (19, 20), id3−/− (24, 26–29), and klf2−/− mice (17, 30). These innate memory cells were recently shown to be dependent on IL-4 production by PLZF+ NKT cells within the thymus (17). Given the phenotypic similarity of our VM cells to innate memory cells, we first wished to examine whether VM cells might also arise via a similar NKT/IL-4/PLZF dependent mechanism.
We therefore examined the secondary lymphoid tissues of IL-4−/−, PLZF lu/lu (deficient in PLZF expression in lymphocytes) and NKT deficient mice (CD1−/−, Jα18−/−). Though all of these strains have reduced innate memory cells (16), a substantial pool of VM cells was still present (Fig 2A and B). Closer examination revealed that the IL-4−/−, CD1−/−, Jα18−/− and PLZF lu/lu strains did have modest, but statistically significant, reductions in VM phenotype cells as compared to the wild type (WT) controls (Fig 2C), consistent with recently published data (31). In addition, Tbet−/− mice displayed no loss of VM cells in the naïve T cell repertoire (Fig 2C), indicating that this transcription factor is also not required for the production of VM cells (49). Tetramer staining and magnetic bead pull down for each strain as described in Figure 1 revealed similar results for the B8R-specific T cell pool as compared to the bulk T cell pool (data not shown).
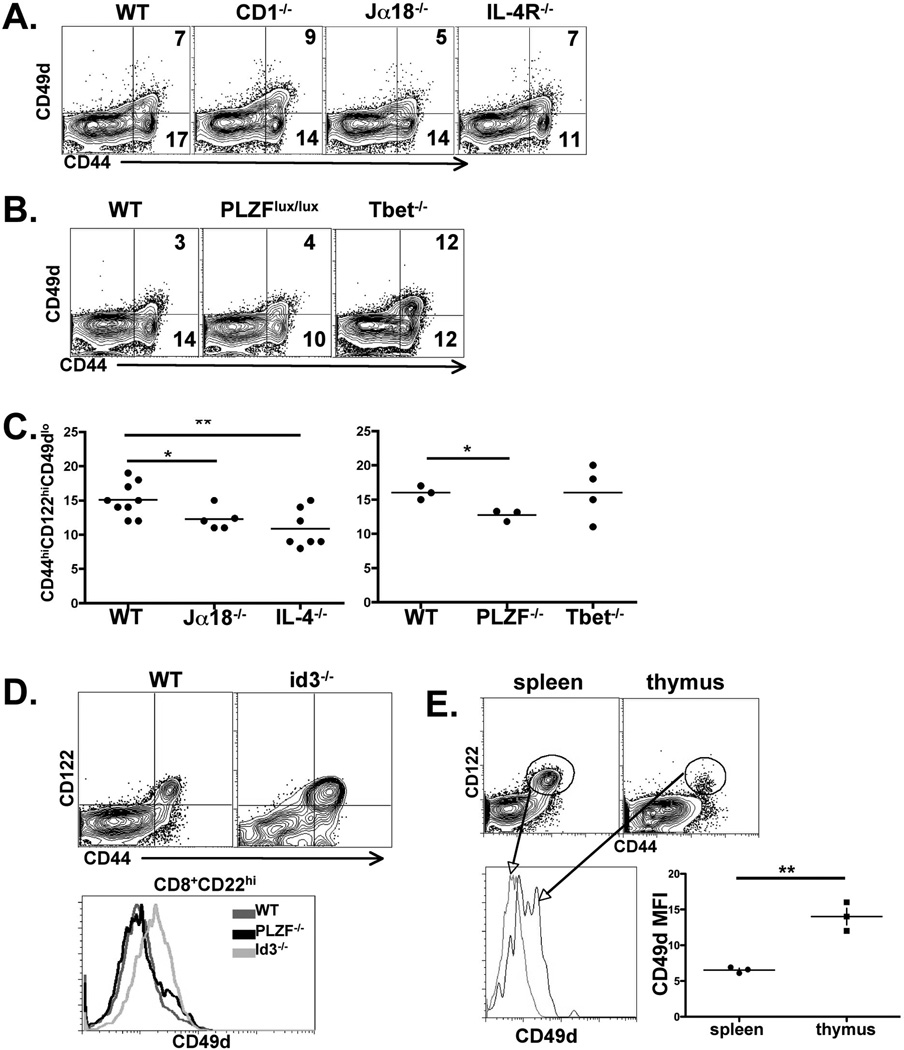
Figure 2.
VM cells are a population independent from Innate Memory cells. CD8+ spleen cells from the indicated strains of mice were isolated and stained for CD44, CD122 and CD49d expression. A, B) data shown were gated on all live, B220−, CD8+ events. Numbers indicate percent of CD8+ T cells in each quadrant. C) Graph of data gated as in A, each data point representing an individual, age matched mouse. Data are shown as percent of VM cells out of total CD8+ T cells from each strain. D) Dot plots and histograms of CD8+ T cells from WT and id3−/− mice. Histograms are of all CD44hi cells shown in the dot plots. Data shown are representative of 4 independent id3−/− spleens. E) Spleen and thymus from B6 hosts were isolated and CD49d expression was determined on all B220−CD4−CD8+CD44hiCD122hi cells in each tissue. Gating strategy is shown in the dot plots and histogram and mean fluorescent intensity of CD49d staining is shown in the graph, each data point representing an individual. All data are representative of 2–5 experiments performed. Error bars represent standard deviation. * p value < 0.05, ** p value <0.001, unpaired t-test.
id3−/− mice have elevated numbers of innate memory cells in the periphery (28, 29). Our examination of the CD8+ T cells in the secondary lymphoid tissue of id3−/− hosts was consistent with these reports, showing an elevated percentage of CD44hi, CD122hi CD8+ T cells in the periphery (Fig 2D). However, these cells in the id3−/− expressed higher levels of CD49d (Fig 2D), in contrast to VM cells. These data suggested that one potential means by which innate memory and VM cells might be distinguished could be their level of CD49d expression. We therefore examined the phenotype of innate memory cells in the thymus of normal B6 mice and compared their CD49d expression to that of VM cells found in the periphery. Though B6 mice have far fewer innate memory cells in the thymus as compared to BALBc mice (16, 17), they are detectable as a small population of CD8+CD44hiCD122hi cells (Fig 2E). Similar to the innate memory cells found in the id3−/− host, innate memory cells in the thymus express a statistically significant higher level of CD49d as compared to VM cells or naïve T cells in the periphery (Fig 2E). Collectively, our results are consistent with those recently reported (31) and support the conclusion that VM cells represent a subset of memory phenotype CD8+ T cells that are distinct from the innate memory cells described previously. Further, our data indicate that CD49d expression is a distinguishing marker between VM cells and innate memory cells.
VM cell development is CD122 and IL-15 dependent
We next wished to determine what mediators might be responsible for the appearance of VM cells in the unprimed host. We focused our attention on IL-15 and the IL-2Rβ/IL-15Rβ chain, CD122, for three reasons. First, as noted above, VM cells express a distinctly higher level of CD122, than even antigen-driven memory cells do (Fig. 1). Secondly, while resting naïve phenotype CD8+ T cells express readily detectable CD122, CD122 is only expressed on CD4+ T cells after antigen stimulus. This suggested a possible connection as to why VM cells are largely absent from the pool of CD4+ T cells (Figure 2). Thirdly, a previous report described a unique form of HP, primarily affecting CD8 and not CD4 cells, that is mediated by IL-2/IL-15 (50).
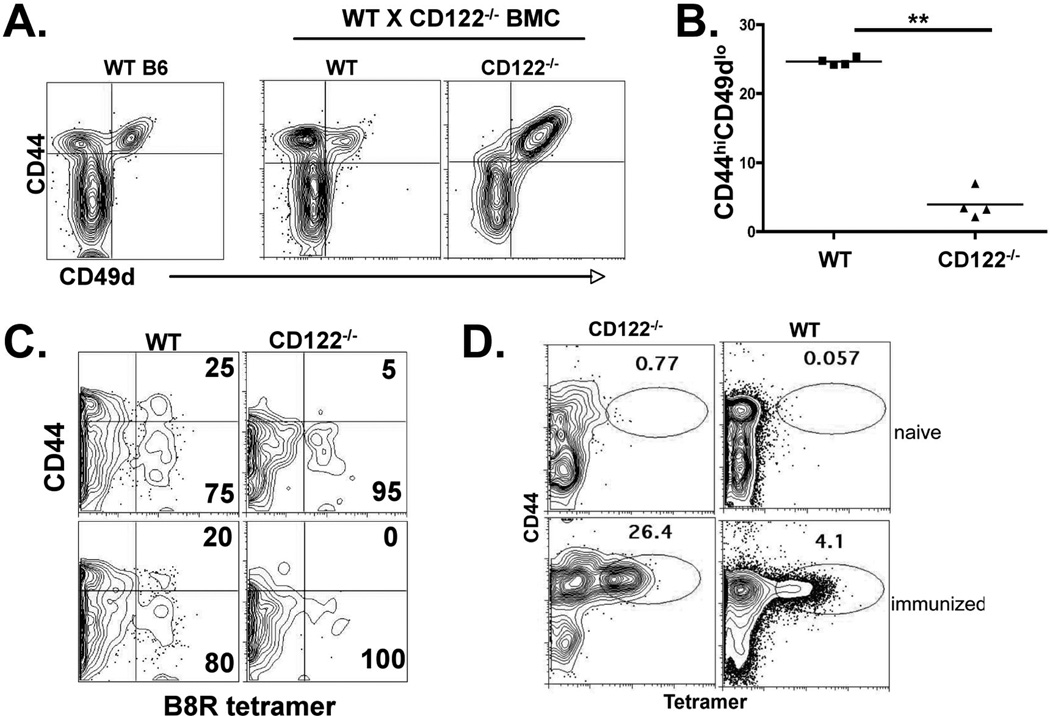
CD122−/− mice are marked by extreme lymphoproliferative syndrome similar to that observed in CD25−/− and scurffy/FoxP3−/− hosts (51). To avoid these issues associated with the intact CD122−/− mice, we made mixed bone marrow chimeras by transferring a mixture of bone marrow from CD122−/− (CD45.2) and WT (CD45.1) mice into irradiated Rag2−/− recipients. We assessed the unprimed bulk and antigen specific CD8+ T cell repertoire in the reconstituted mice for the presence or absence of VM phenotype cells derived from the WT and CD122−/− backgrounds. Not surprisingly, while VM cells derived from the WT bone marrow were abundant, essentially no VM cells derived from the CD122−/− bone marrow were observed (Fig 3A and B). Furthermore, tetramer staining and magnetic column enrichment of all B8R-specific T cells (dominant antigen derived from vaccinia virus (52)) from the chimeric hosts revealed a similar deficit in the B8R-specific VM cells derived from the CD122−/− background (Fig 3C). Interestingly, CD49d+CD44hi cells were readily observed in the bulk CD8+ T cells derived from the CD122−/− background (Fig 3A), suggesting no defects in the capacity of CD122−/− cells toward antigen-experienced memory formation, presumably against endogenous gut or food related antigens. Indeed, immunization of WT:CD122−/− chimeric mice resulted in the expansion of antigen specific CD122−/− cells that was 5–6 times that of the WT antigen specific T cells (Fig. 3D). This suggests that CD122−/− cells may have an increased sensitivity to TCR mediated expansion, a feature of CD122−/− cells previously overshadowed by the larger Treg defect in these mice. Regardless, these data confirm that VM cell formation in the periphery requires CD8+ T cell CD122 expression.
Figure 3.
VM cell development is CD122-dependent. WT × CD122−/− bone marrow chimeras were made as previously described and as in the Material Methods. Twelve weeks after reconstitution, spleen CD8+ T cells were analyzed for CD44 and CD49d expression. B) Percent of VM phenotype cells out of total CD8+ T cells from each background, WT or CD122−/− in the bone marrow chimeric mice shown in A. Each data point represents a separate chimeric host. Data are representative of 3 independent mixed chimera experiments, each with 3–5 chimeras. C) B8R specific T cells were isolated from the chimeric mice using magnetic enrichment of tetramer stained cells as described previously (15) and in the Materials and Methods. The column bound CD8+ cells were gated into CD45.1 (WT) and CD45.2 (CD122−/−) backgrounds and analyzed for CD44 expression and tetramer staining. Two representative mice are shown (top and bottom plots). D) Peripheral blood from WT×CD122−/− chimera’s before and after immunization with B8R peptide in conjunction with polyIC and antiCD40 as previously described (ref). Data shown are gated on all B220− CD8+CD45.1+ (WT) and B220−CD8+CD45.2+(CD122−/−) events. Data shown are representative of 4 immunized chimeric mice.
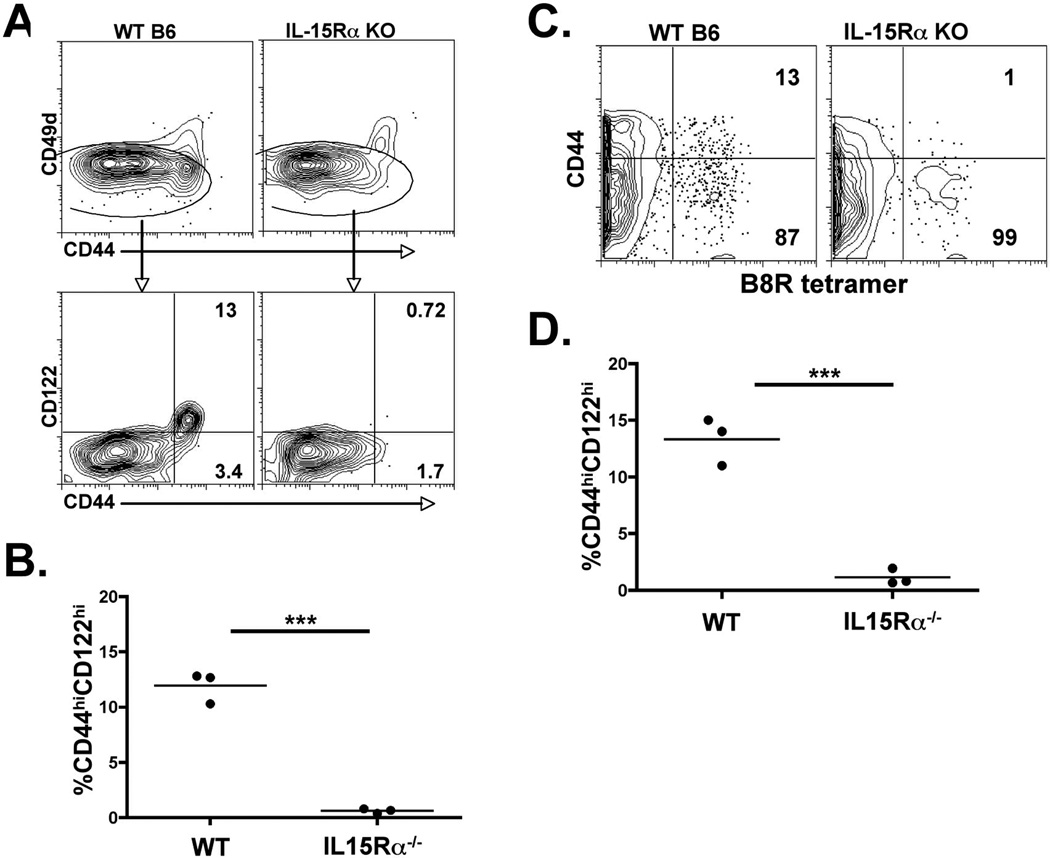
We next examined the potential role of IL-15 in VM cell development. IL-15−/− and IL-15Rα−/− mice have a deficit in their capacity to maintain memory CD8+ T cells over time (53–56). However, the T cell repertoire in these mice had never been analyzed for the presence of cells bearing a VM phenotype. We therefore examined IL-15Rα−/− mice for the presence or absence of VM phenotype cells, both in the bulk CD8+ T cell pool as well as within antigen specific T cells enriched from the unprimed repertoire. As previously described, the bulk CD8+ T cell pool had a reduction in the percentage of CD44hi cells (Fig 4A). However, closer examination revealed that CD44hiVLA4hi cells were present in frequencies similar to those in WT hosts, and that the major deficit in CD44hi cells was due to a loss of the CD44hiCD122hiVLA4lo VM cells (Fig 4A and B). We observed this loss of VM phenotype cells both in the bulk CD8+ T cell pool (Figure 4B) as well as in the collection of antigen specific T cells, in this case either for B8R (Figure 4C and D) or for HSVgB (not shown). Two conclusions can be made from these data. First, the development of VM cells within the normal unprimed host is largely IL-15 dependent. Second, though VM cells can be found at some of the earliest time points in neonates (31), the lack of VM cells in the IL-15Rα−/− host indicates that homeostatic stimuli typically associated with lymphopenia (ie. IL-7) do not contribute substantially to VM cell production.
Figure 4.
VM cell development is IL-15 dependent. Total CD8+ spleen cells (A and B), or B8R-specific spleen cells (C and D) from IL-15Rα−/− mice were analyzed for the percentage of VM cells. A and B) Total CD8+ T cells from WT and IL-15Rα−/− mice were analyzed for CD44 and CD49d expression (top plots). CD49dlo cells were gated on and analyzed for the frequency of CD44hiCD122hi cells (lower plots and B). C and D) B8R specific T cells were isolated from WT and IL-15Rα−/− mice as described previously (15) and in the Materials and Methods. Column bound CD8+ T cells were analyzed for CD44 expression of tetramer+ cells. Each data point is from an individual mouse. The data shown are representative of 3 independent experiments.
VM cells expand in vivo to IL-15Rα/IL-15 complexes
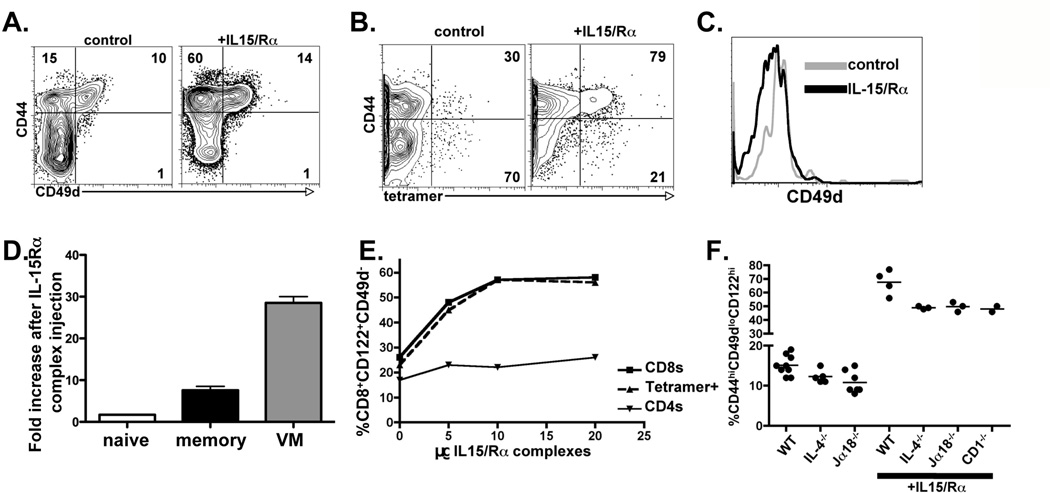
IL-15 stimulates cells by binding to IL-15Rα in APCs, and possibly other non-hematopoetic cells, to stimulate CD122/CD132 expressing T cells in trans (57–61). Given this unique feature of IL-15 mediated stimulation, others have utilized IL-15/Rα complexes to augment the response and survival of memory CD8+ T cells (34, 62). We predicted that if VM cells arose via an IL-15-dependent mechanism, then VM cells could be expected to respond preferentially to IL-15/Rα complexes. We injected WT hosts with IL-15/Rα complexes (34) and examined the resulting impact on bulk and antigen specific CD8+ T cells in the unprimed host. IL-15/Rα injection into B6 mice resulted in a dramatic expansion of CD8+ T cells with the largest expansion manifest by CD44hiCD122hiVLA4lo VM cells (Figure 5A). This effect was observed when examining either bulk CD8+ T cells (Figure 5A) or the antigen specific T cells isolated by magnetic column enrichment (Figure 5B). Importantly, the antigen specific cells expanded by IL-15/Rα treatment maintained low expression of CD49d (Figure 5C), consistent with HP devoid of overt TCR stimulation (15). Examining the total numbers of CD8+ T cells within each subset before and after IL-15 injection revealed a substantial fold increase in both memory and VM cells without any corresponding decrease in naïve phenotype cells (Figure 5D). These data suggest that the increase in VM cells occurred largely as a result of the expansion of the pre-existing VM cell pool. VM cell expansion occurred over a broad range of IL-15/Rα treatment and almost exclusively affected the CD8+ T cells and not the CD4+ T cells (Figure 5E). The small increase in the percentage of CD44hi CD4+ T cells that occurred at higher doses of IL-15/Rα complexes was due to an increase in frequency of CD44hiCD49dhi cells (not shown), consistent with the CD122 expression profile of these cells. In addition, even the strains of mice with a modest reduction in VM cells (see Fig 2C) responded to IL-15/Rα complex injection with a dramatic expansion of VM-phenotype cells (Fig 5F). These data show that IL-15-mediated stimulation of the CD8+ T cells in the unprimed host dramatically increases the frequency of VM cells. Further, the proliferation these cells experience does not influence their expression of CD49d, consistent with an absence of antigen/TCR stimulation in this process.
Figure 5.
IL-15 stimulation induces the expansion of VM cells in vivo. A) B6 mice were injected with 5ug of IL-15/Rα complexes i.p.. Spleen CD8+ T cells were analyzed 5 days later for the percent of cells with naïve (lower left quadrant), VM (upper left quadrant) and antigen memory (upper right quadrant) phenotypes. B) B8R specific T cells were isolated as described above from mice injected with IL-15/Rα complexes as in A. Column-bound cells were analyzed for the percent of CD44hi tetramer+ cells. C) Tetramer+ cells from B were further analyzed for their expression of CD49d. D) The absolute number of CD8+ T cells within the naïve (CD44lo), memory (CD44hi, CD49dhi) and VM (CD44hi, CD49dlo) CD8+ T cell pools were determined in control and IL-15/Rα complex injected mice. The fold expansion of cells in each subset was then calculated by dividing the cell number in the IL-15/Rα complex injected hosts by the number of cells (derived by averaging the number from 3 independent mice) in the controls. E) B6 mice were injected with increasing doses of IL-15/Rα complexes and 5 days later were analyzed as in A for the percent CD44hi cells out of the total CD4 and CD8 T cells (CD4s, CD8s) and as in B for the percent B8R+CD44hi cells out of the total B8R tetramer+ cells (tetramer+). F) IL-15/Rα complexes were injected as in A into the indicated strains. Five days later, the percent CD44hiCD49dloCD122hi cells out of total CD8+ T cells was determined as described above. Uninjected controls for each strain are the same data as shown in Figure 3, shown here for the purposes of comparison to the IL-15/Rα complex injected hosts.
VM cell development is compromised in Eomes−/− hosts
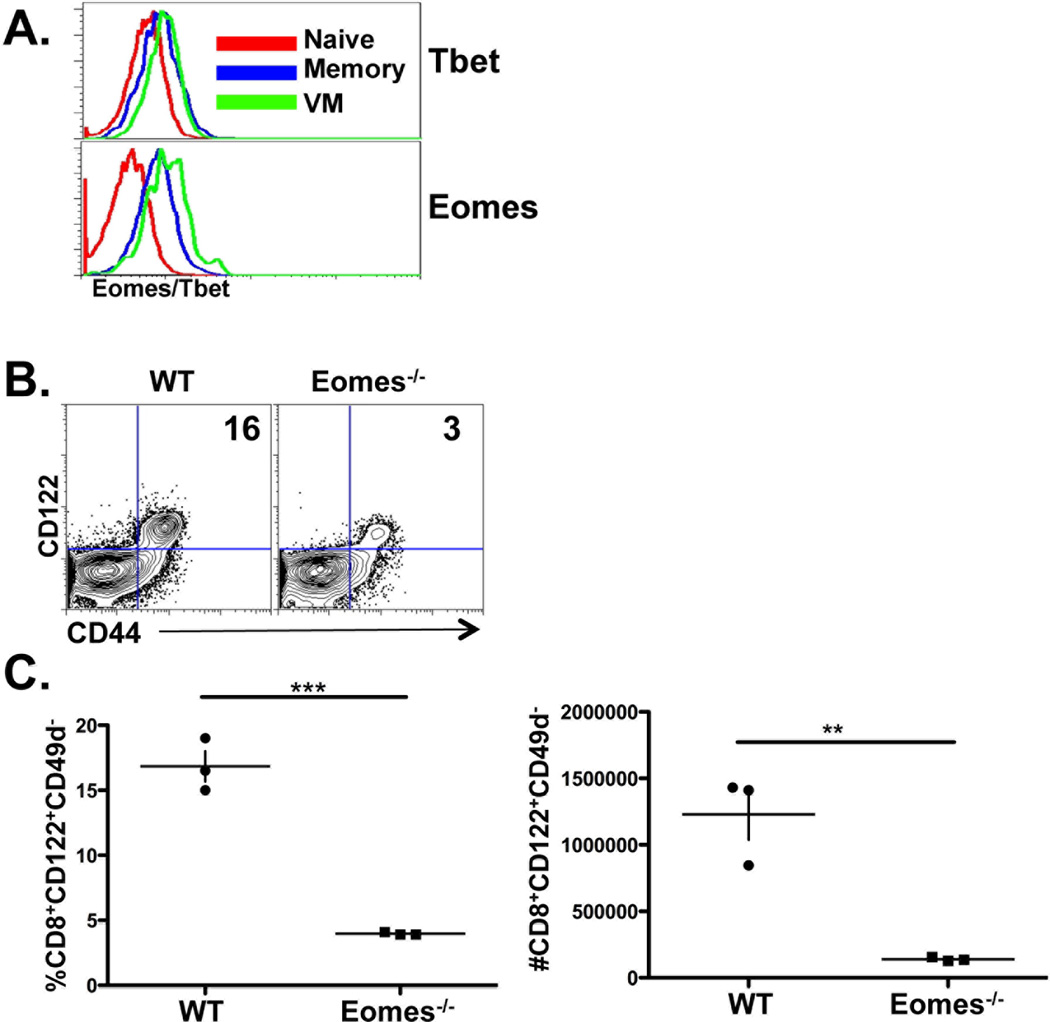
The maintenance of long-lived antigen-experienced memory cells requires IL-15 mediated stimulation through CD122/CD132 (55, 63), which leads to induction of the transcription factor, eomesodermin (Eomes). Interestingly, CD122 expression itself is largely dependent upon Eomes (64) which VM cells over-express compared to naïve cells (Fig 6A). Analysis of the different subsets of CD8+ T cells in the unprimed host revealed that, while CD49dhi and lo CD8+ T cells expressed roughly equivalent levels of Tbet, VM cells expressed elevated Eomes even compared to antigen experienced memory cells (Fig 6A). We speculated that this Eomes expression may be important in the development of VM cells such that Eomes deficiency would result in a loss of VM cells from the unprimed host. Indeed, CD8+ T cells in a T cell-specific conditional Eomes−/− host (CD4-Cre × Eomesfl/fl) displayed a substantial loss of VM cells from the periphery (Fig 6B and C) in both percentage and total numbers (Fig 6C). Further, the few VM phenotype cells present in these mice had lower expression of CD122 as compared to VM cells from WT mice, as might be expected. While a decrease in CD44hi CD8+ T cells in the Eomes conditional knockout, as well as the Eomes heterozygous mice, was already well established (64–66), we show here that the major deficit in these mice (similar to the IL-15−/− hosts) is the loss of VM cells.
Figure 6.
VM cells express high levels of Eomes and their development is Eomes-dependent. A) naïve, VM and antigen memory CD8+ T cells were analyzed for Eomes and Tbet expression as described above and in the Materials and Methods. Histograms are from CD8+ splenic T cells gated on CD44lo (naïve), CD44hiCD49dlo (VM) or CD44hiCD49dhi (antigen memory) cells. B and C) spleen CD8+ cells from WT and conditional Eomesfl/flCD4-cre knockout mice, gated on all CD49dlo events, were analyzed for CD44 and CD122 expression. C) Data is shown as both the percentage and total number of splenic VM cells from each host. Each data point in C represents an individual mouse. Data shown are representative of 3 experiments performed. ** p value <0.001, ***p value <0.0001, unpaired t-test.
Steady state IL-15 reporter expression in vivo by CD8α+ and tissue derived DCs
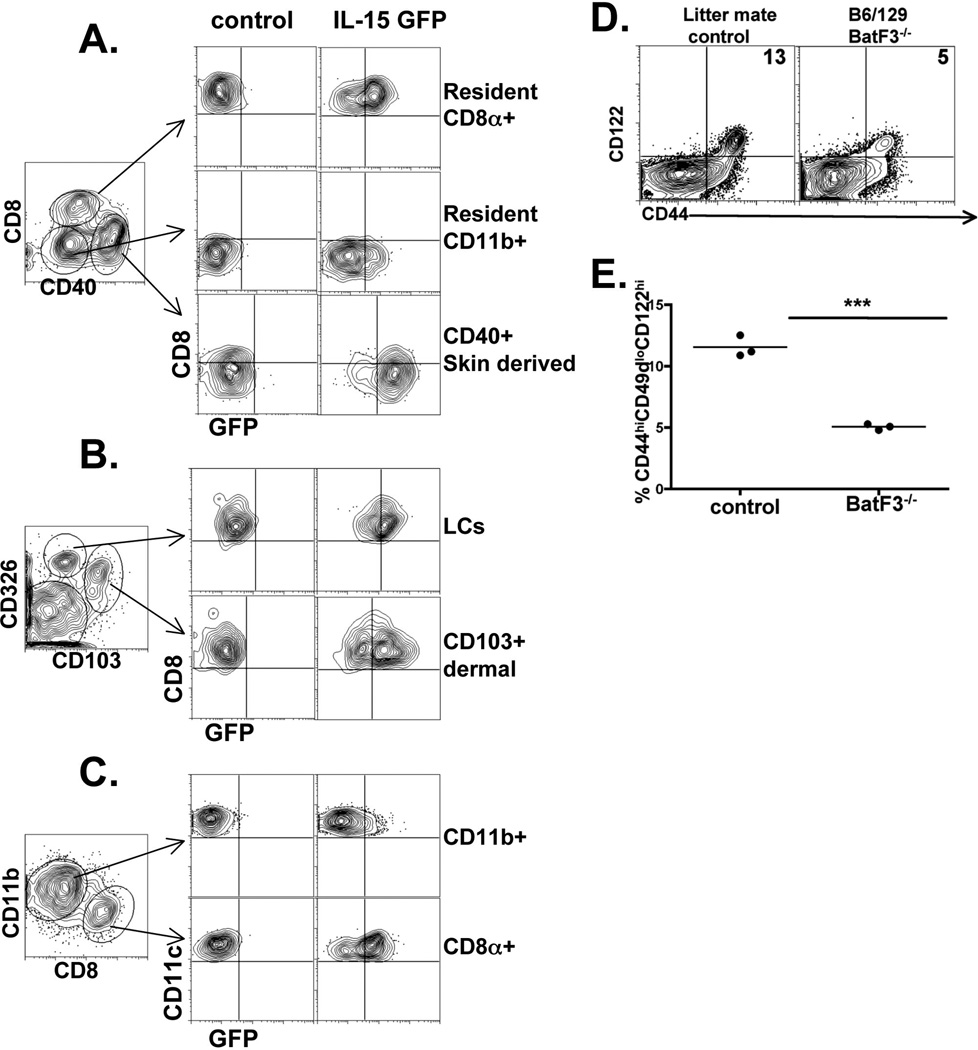
We next wished to determine the source of IL-15 expression/presentation that was responsible for generating VM cells from the pool of naïve phenotype cells. IL-15 expression has been described in a variety of DC and macrophage/monocyte subsets (57, 67, 68). Recent data using a transgenic IL-15 reporter system showed substantial expression/presentation of IL-15 in the CD8α+ DC subset (69). We also recently generated an IL-15 reporter system using a co-translational reporter mouse for IL-15 by introducing, as a transgene, a modified Bacterial Artificial Chromosome (BAC) containing the IL-15 genomic locus in which IL-15 is linked via a 2A peptide to green fluorescent protein (GFP) (see Supplemental Fig 1 and Materials and Methods). We isolated cells from the peripheral lymph nodes (LN) and spleens of these mice and analyzed the various DC populations for expression of the IL-15 reporter. Similar to the recently published data (69), minimal expression of GFP was noted in the resident CD11b+ DC subset in LN or spleen (Fig 7A, C). In contrast, the resident CD8α+ DCs as well as the tissue derived DC subsets displayed high levels of GFP expression (Fig 7A and B). In particular, Langerhan cells (LCs) as well as the skin-derived CD103+ DCs showed some of the highest GFP expression (Fig 7B). Analysis of splenic DC subsets mirrored that seen in the LN, with the dominant GFP+ DC subset being the CD8α+ DCs (Fig 7C). Thus, skin/tissue derived DCs as well as lymphoid derived CD8α+ DCs demonstrated substantial IL-15 expression/presentation in the resting host. These results are again fully consistent with published data using a different IL-15 reporter system (69).
Figure 7.
IL-15 is produced by tissue-derived and CD8+ lymphoid derived DC subsets which mediate VM cell formation in the periphery. IL-15 BAC transgenic mice expressing GFP under the control of the IL-15 locus were made as described in the Materials and Methods. The lymphoid tissues were isolated, collagenase digested as previously described, and the indicted DC subsets analyzed for their expression of IL-15 (GFP). A and B) DCs isolated from inguinal, axillary and brachial lymph nodes. Left most dot plot was gated on all B220−CD11c+ClassII+ events. The cells in the regions, representing the indicated DC subsets, were assessed for their expression of GFP. C) DCs isolated from the spleen and analyzed as in A and B. D and E) Analysis of spleen CD8+ T cells from B6/129 BatF3−/− mice and wild type littermate controls. The percent of CD44hiCD49dloCD122hi cells out of total CD8+ cells is shown in the upper right quadrant (dot plots) and in the graph. Each data point represents an individual mouse. *** p value <0.0001, unpaired t-test.
VM cell development is compromised in CD8α+ DC deficient hosts
Our observation of IL-15 expression in the CD8α+ and CD103+ DCs suggested that these subsets might play an important role in presenting IL-15 to support VM cell development. To explore the importance of these DC subsets we utilized the BatF3−/− mouse which have a significant defect in the development of both CD8α+ and CD103+ DCs (70–72). Even though IL-15 is still present within these hosts, we reasoned that if these DC subsets were important in IL-15 presentation to the naïve T cell pool, then we might expect to see a reduction in VM cells. Consistent with this expectation, we found a loss of VM cells (Fig 7D) roughly equivalent to that seen in the Eomes−/− hosts (Fig 6C and Fig 7E). This is again despite the fact that all other DC subsets and non-DC cell types capable of IL-15 expression (67, 68) are still present in these hosts.
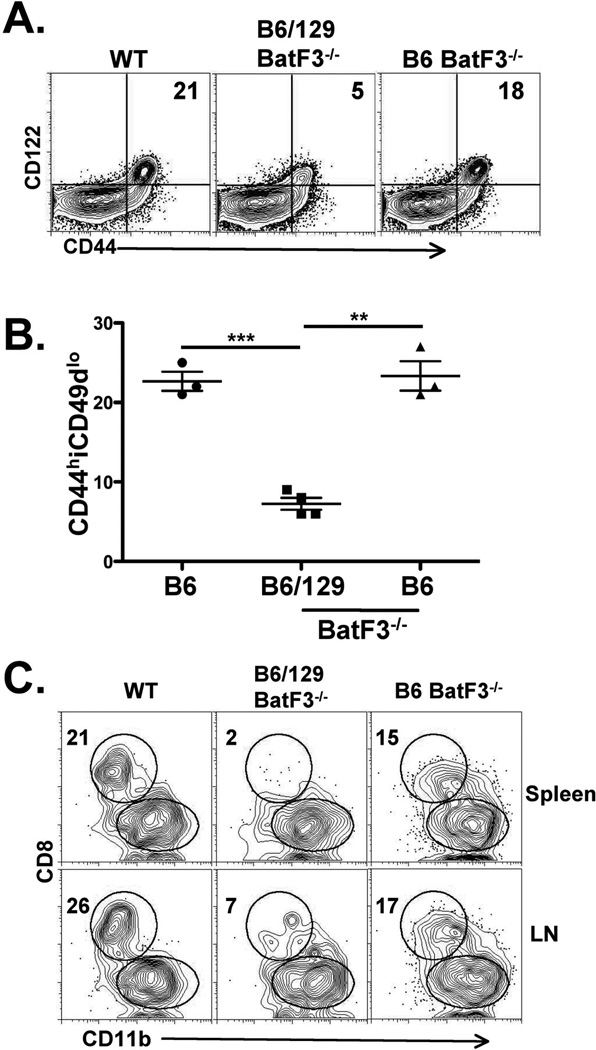
The BatF3−/− mice we initially utilized were on a B6/129 hybrid background, resulting from the intercrossing of F1–F2 B6/129 mice. We therefore repeated the experiment using BatF3−/− mice backcrossed to B6 for at least 10 generations (a kind gift from Dr. Ken Murphy, Washington University, St Louis, MO). To our surprise we found that the B6 BatF3−/− mice had a full complement of VM cells relative to WT B6 hosts (Fig 8A and B). However, we also observed that the lymphoid resident CD8α+ DCs were also present in almost normal numbers in these mice (Fig 8C). Of note, the CD103+DCs were absent in BatF3−/− mice on either B6 or B6/129 backgrounds (not shown). While it is unclear how the loss of BatF3 has such different effects on the presence of CD8α+ DCs in each background, the fact that the recovery of this DC subset corresponds with the full recovery of VM cell development indicates that VM cell development occurs largely as a result of the presentation of IL-15 by the CD8α+ DC subset, and does not require IL-15 production by the CD103+DCs.
Figure 8.
B6 BatF3−/− mice recover both CD8+DC cells and VM cells. A and B) Analysis of spleen CD8+ T cells from B6 WT, BatF3−/− crossed only 1–2 generations onto the B6 background (B6/129 BatF3−/−) and BatF3−/− mice crossed at least 10 generations onto the B6 background (B6 BatF3−/−). The percent of CD44hiCD49dlo cells out of total CD8+ cells is shown in the upper right quadrant (dot plots) and in the graph. Each data point represents an age matched, individual mouse. C) Analysis of lymphoid resident CD8+ and CD11b+ DCs from LN and spleens of the WT and Batf3−/− on the indicated backgrounds. DCs were isolated and analyzed as described in Figure 7. Numbers indicate the percent of DCs in that quadrant. Data are representative of 5–7 mice from 2 independent experiments. *** p value <0.0001, unpaired t-test
Discussion
Our data provide mechanistic insight into the influence of IL-15 in mediating a form of HP that is active during normal immune development, is independent of lymphopenia, and is responsible for the generation of a unique pool of CD8+ T cells capable of enhanced responses to antigen. This subset exists only within the CD8+ T cell pool, probably because CD8+ T cells express higher levels of CD122 than naïve CD4+ T cells (Fig 1) and are therefore capable of responding to IL-15 trans-presentation in the absence of overt antigen stimulation. Our data also confirm recently published data (69) showing steady state IL-15 expression by CD8+ DCs and we extend those findings to identify a novel function for this IL-15 trans-presentation, namely that of facilitating VM cell formation in the periphery. Collectively our data suggest a model in which elevated Eomes expression in a subset of naïve phenotype cells leads to greater expression of CD122, and therefore greater sensitivity to IL-15 presented by CD8α+ DCs, resulting in subsequent proliferation of the T cell and conversion to VM phenotype cells.
It remains an outstanding question as to what mediates the cellular decision of any given naïve phenotype CD8+ T cell to increase CD122 expression and respond to the IL-15 trans-presentation by the CD8+ DCs. In addition to cytokine mediated signals, there is evidence that increased so-called “tonic” TCR stimulation has an influence on the degree of HP experienced by T cells in a lymphopenic environment (5, 12–14). Indeed, more recent data indicates that the T cell response specifically to IL-15 is heavily influenced by TCR/MHC affinity (73). These data suggest that the VM repertoire may have subtle differences in TCR usage that confer an increase in TCR/self peptide/MHC interaction sufficient to contribute to VM cell formation. Cursory analysis using the available antibodies for TCR Vβ and Vα showed no difference between naïve and VM cells of the same specificity (RMK, unpublished data). However, the resolution of this method is almost surely inadequate to resolve these differences and deep sequencing is currently underway in pursuit of the full characterization of the naïve and VM cell repertoires.
Independent of repertoire issues, we can at least conclusively say that the decision to convert from naïve phenotype to VM phenotype largely requires the expression of Eomes. Though our and others’ (31) data firmly establish the derivation of VM cells in the periphery as unrelated to the generation of innate memory cells in the thymus, a requirement for Eomes expression is a shared feature for all CD8 memory cells, regardless of means by which they are created (1, 16). IL-15 mediated stimulation through CD122 leads to the induction of Eomes (55, 63), providing an effective link to all of the factors required for VM cell development. However, CD122 is itself dependent upon Eomes expression (64), calling into the question as what comes first, increased Eomes expression or elevated CD122? To resolve this dilemma, it is tempting to speculate on a potential role for the Wnt pathway. Eomes expression in antigen driven memory CD8+ T cells is at least partially maintained by the HMG-box transcription factor, TCF-1 (tcf7) (74–76). Two major isoforms of TCF-1, p45 and p33, are expressed in naïve T cells with both positive (p45) and negative (p33) regulatory functions based on their association with either coactivators (β-catenin) or repressors (groucho) respectively (77–83). In cooperation with β-catenin, the p45 isoform contributes to Eomes expression, which induces the subsequent expression of CD122 and sensitivity to IL-15 stimulation (76). It is interesting to note that ~90% of all CD8+ T cells in the TCF-1−/− host are VM phenotype (data not shown). The largely memory phenotype of the T cells in this host is well documented (77, 84) but is generally attributed to the lymphopenic periphery caused by aberrant thymic selection. However, a variety of other peripheral lymphopenic strains have typically 40–50% VM phenotype CD8+ T cells in the periphery (RMK unpublished results) suggesting a more direct connection between the loss of this transcription factors and VM cell formation.
Curiously, the TCF family member encoded by tcf7l2 (also known as TCF-4) similarly utilizes β-catenin as a coactivator, targets a very similar DNA binding motif, and is already known to associate tightly with the Eomes promoter (85). Taken together, these data support the hypothesis that the loss of TCF-1 results in the loss of both activating and repressor elements within the naïve T cell, allowing the cells to utilize other β-catenin responsive TCF transcription factor(s), such as TCF-4, to produce elevated Eomes expression, facilitating the cellular response to IL-15 and subsequent acquisition of the observed VM phenotype. It is again noteworthy that β-catenin−/− mice do not have any substantial aberrations in their T cell development (86), but they have not been analyzed for the presence or absence of VM cells. Ongoing experiments are currently examining TCF transcription factors and β-catenin in VM cell development.
Perhaps the most significant application of our data is the fact that we have identified a physiologically relevant, lymphopenia-independent form of T cell homeostatic expansion. While the reality of HP in extreme lymphopenia is well established (5–11), the physiological relevance of HP outside of bone marrow transplantation is unclear, as is the representation of HP memory T cells within a normal, un-manipulated host. Our data address both of these gaps in the knowledge base first by identifying nominal antigen-specific T cells within the pool of memory phenotype cells in the unprimed host, and second by clarifying the mechanism by which these cells come into existence. We can now say that HP, at least for a subset of CD8+ T cells, is an active process during normal immune development and not requiring a state of lymphopenia. Furthermore, this HP is largely mediated by IL-15 presentation by CD8α+ DCs in the unmanipulated host. The result of this HP is the generation of a pool of CD8+ T cells which can participate in de novo responses to nominal antigens but which have a functional capacity more similar to antigen-experienced memory cells.
Given that VM cells can mediate protection against infectious challenge (Fig 1) it is possible that they could be manipulated to provide increased immune protection, even in an antigen-inexperienced host. As we previously demonstrated that VM cells, at least in vitro, initiate proliferation more rapidly naïve phenotype cells (15), the decedents of VM precursors may be over represented in the pool of cells responding to antigenic challenge, ultimately shaping (if not mediating) such familiar features of the memory response as immunodominance (87) and population affinity maturation (88–90). Additionally, VM cells may access peripheral sites of inflammation even before their phenotypically naïve counterparts have had the opportunity to respond within the draining lymphoid tissue. This trafficking capacity might be expected to restrict the growth rate of the infectious organism in situ, subsequently affecting the overall antigen load and the inflammatory environment which ultimately controls the response of the naïve phenotype cells. Thus, the function of VM cells must be considered to completely understand the nature of the “primary” immune response.
Finally, these data confront our basic assumptions of what constitutes the naïve and memory T cell pools. While it is well agreed upon that the memory cell pool has a number of functionally distinct subsets (1), the pool of antigen inexperienced (naïve) T cells has always been considered relatively homogeneous. Most studies assume this functional and phenotypic homogeneity and focus instead on elucidating the mechanisms underlying the functional advantages within the various subsets of antigen-experienced memory cells. The data we present here are in stark contrast to this assumption and indicate that the primary CD8+ T cell response is actually the collective response of both naïve and memory phenotype T cells coexisting within the same, unprimed host. Further work must be done to more carefully clarify the functional characteristics of VM cells, their participation in primary and secondary responses, their role in population affinity maturation and immunodominance, and their ultimate role in promoting protective immunity.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. E. John Wherry, Barbara Kee, Harald VonBoehmer, Charlie Suhr, and Ken Murphy for their generosity in sharing the various knockout mice, and/or spleens cells, that were crucial to this project. Thanks to Sonia at the University of Colorado Cancer Center Flow Cytometry Facility. Finally, we thank Drs. Marc K. Jenkins and Steve Jameson from the Department of Microbiology and the Center for Immunology at the University of Minnesota, without whom our development of the tetramer staining and bead sorting methodology, or our identification of VM cells, would not have been possible.
This work was supported by grants from the NIH (AI06877, AI066121, AI18785) and the Department of Defense (W81XWH-07-1-0550). DoD support was associated with funding for the Center for Respiratory Biodefense at National Jewish Health.
References
- 1.Jameson SC, Masopust D. Diversity in T cell memory: an embarrassment of riches. Immunity. 2009;31:859–871. doi: 10.1016/j.immuni.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masopust D, Kaech SM, Wherry EJ, Ahmed R. The role of programming in memory T-cell development. Curr Opin Immunol. 2004;16:217–225. doi: 10.1016/j.coi.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Jameson SC. T cell homeostasis: keeping useful T cells alive and live T cells useful. Semin Immunol. 2005;17:231–237. doi: 10.1016/j.smim.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Le Campion A, Bourgeois C, Lambolez F, Martin B, Leaument S, Dautigny N, Tanchot C, Penit C, Lucas B. Naive T cells proliferate strongly in neonatal mice in response to self-peptide/self-MHC complexes. Proc Natl Acad Sci U S A. 2002;99:4538–4543. doi: 10.1073/pnas.062621699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marleau AM, Sarvetnick N. T cell homeostasis in tolerance and immunity. J Leukoc Biol. 2005;78:575–584. doi: 10.1189/jlb.0105050. [DOI] [PubMed] [Google Scholar]
- 8.Min B, McHugh R, Sempowski GD, Mackall C, Foucras G, Paul WE. Neonates support lymphopenia-induced proliferation. Immunity. 2003;18:131–140. doi: 10.1016/s1074-7613(02)00508-3. [DOI] [PubMed] [Google Scholar]
- 9.Min B, Sempowski GD, Paul WE. Neonates support "homeostatic" proliferation. Adv Exp Med Biol. 2002;512:91–95. [PubMed] [Google Scholar]
- 10.Schuler T, Hammerling GJ, Arnold B. Cutting edge: IL-7-dependent homeostatic proliferation of CD8+ T cells in neonatal mice allows the generation of long-lived natural memory T cells. J Immunol. 2004;172:15–19. doi: 10.4049/jimmunol.172.1.15. [DOI] [PubMed] [Google Scholar]
- 11.Surh CD, Sprent J. Regulation of mature T cell homeostasis. Semin Immunol. 2005;17:183–191. doi: 10.1016/j.smim.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Ernst B, Lee DS, Chang JM, Sprent J, Surh CD. The peptide ligands mediating positive selection in the thymus control T cell survival and homeostatic proliferation in the periphery. Immunity. 1999;11:173–181. doi: 10.1016/s1074-7613(00)80092-8. [DOI] [PubMed] [Google Scholar]
- 13.Kieper WC, Burghardt JT, Surh CD. A role for TCR affinity in regulating naive T cell homeostasis. J Immunol. 2004;172:40–44. doi: 10.4049/jimmunol.172.1.40. [DOI] [PubMed] [Google Scholar]
- 14.Seddon B, Zamoyska R. TCR and IL-7 receptor signals can operate independently or synergize to promote lymphopenia-induced expansion of naive T cells. J Immunol. 2002;169:3752–3759. doi: 10.4049/jimmunol.169.7.3752. [DOI] [PubMed] [Google Scholar]
- 15.Haluszczak C, Akue AD, Hamilton SE, Johnson LD, Pujanauski L, Teodorovic L, Jameson SC, Kedl RM. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206:435–448. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32:50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinreich MA, Odumade OA, Jameson SC, Hogquist KA. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11:709–716. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atherly LO, Brehm MA, Welsh RM, Berg LJ. Tec kinases Itk and Rlk are required for CD8+ T cell responses to virus infection independent of their role in CD4+ T cell help. J Immunol. 2006;176:1571–1581. doi: 10.4049/jimmunol.176.3.1571. [DOI] [PubMed] [Google Scholar]
- 19.Atherly LO, Lucas JA, Felices M, Yin CC, Reiner SL, Berg LJ. The Tec family tyrosine kinases Itk and Rlk regulate the development of conventional CD8+ T cells. Immunity. 2006;25:79–91. doi: 10.1016/j.immuni.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Broussard C, Fleischacker C, Horai R, Chetana M, Venegas AM, Sharp LL, Hedrick SM, Fowlkes BJ, Schwartzberg PL. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 21.Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 22.Felices M, Yin CC, Kosaka Y, Kang J, Berg LJ. Tec kinase Itk in gammadeltaT cells is pivotal for controlling IgE production in vivo. Proc Natl Acad Sci U S A. 2009;106:8308–8313. doi: 10.1073/pnas.0808459106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horai R, Mueller KL, Handon RA, Cannons JL, Anderson SM, Kirby MR, Schwartzberg PL. Requirements for selection of conventional and innate T lymphocyte lineages. Immunity. 2007;27:775–785. doi: 10.1016/j.immuni.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonzo ES, Gottschalk RA, Das J, Egawa T, Hobbs RM, Pandolfi PP, Pereira P, Nichols KE, Koretzky GA, Jordan MS, Sant'Angelo DB. Development of promyelocytic zinc finger and ThPOK-expressing innate gamma delta T cells is controlled by strength of TCR signaling and Id3. J Immunol. 2010;184:1268–1279. doi: 10.4049/jimmunol.0903218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan MS, Smith JE, Burns JC, Austin JE, Nichols KE, Aschenbrenner AC, Koretzky GA. Complementation in trans of altered thymocyte development in mice expressing mutant forms of the adaptor molecule SLP76. Immunity. 2008;28:359–369. doi: 10.1016/j.immuni.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lauritsen JP, Wong GW, Lee SY, Lefebvre JM, Ciofani M, Rhodes M, Kappes DJ, Zuniga-Pflucker JC, Wiest DL. Marked induction of the helix-loop-helix protein Id3 promotes the gammadelta T cell fate and renders their functional maturation Notch independent. Immunity. 2009;31:565–575. doi: 10.1016/j.immuni.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ueda-Hayakawa I, Mahlios J, Zhuang Y. Id3 restricts the developmental potential of gamma delta lineage during thymopoiesis. J Immunol. 2009;182:5306–5316. doi: 10.4049/jimmunol.0804249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verykokakis M, Boos MD, Bendelac A, Adams EJ, Pereira P, Kee BL. Inhibitor of DNA binding 3 limits development of murine slam-associated adaptor protein-dependent "innate" gammadelta T cells. PloS one. 2010;5:e9303. doi: 10.1371/journal.pone.0009303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verykokakis M, Boos MD, Bendelac A, Kee BL. SAP protein-dependent natural killer T-like cells regulate the development of CD8(+) T cells with innate lymphocyte characteristics. Immunity. 2010;33:203–215. doi: 10.1016/j.immuni.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31:122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akue AD, Lee JY, Jameson SC. Derivation and Maintenance of Virtual Memory CD8 T Cells. J Immunol. 2012 doi: 10.4049/jimmunol.1102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedl RM, Rees WA, Hildeman DA, Schaefer B, Mitchell T, Kappler J, Marrack P. T Cells Compete for Access to Antigen-bearing Antigen-presenting Cells. J. Exp. Med. 2000;192:1105–1114. doi: 10.1084/jem.192.8.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pope C, Kim SK, Marzo A, Masopust D, Williams K, Jiang J, Shen H, Lefrancois L. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- 34.Rubinstein MP, Lind NA, Purton JF, Filippou P, Best JA, McGhee PA, Surh CD, Goldrath AW. IL-7 and IL-15 differentially regulate CD8+ T-cell subsets during contraction of the immune response. Blood. 2008;112:3704–3712. doi: 10.1182/blood-2008-06-160945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jabbari A, Legge KL, Harty JT. T cell conditioning explains early disappearance of the memory CD8 T cell response to infection. J Immunol. 2006;177:3012–3018. doi: 10.4049/jimmunol.177.5.3012. [DOI] [PubMed] [Google Scholar]
- 36.Ahonen CL, Doxsee CL, McGurran SM, Riter TR, Wade WF, Barth RJ, Vasilakos JP, Noelle RJ, Kedl RM. Combined TLR and CD40 triggering induces potent CD8+ T cell expansion with variable dependence on type I IFN. J Exp Med. 2004;199:775–784. doi: 10.1084/jem.20031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahonen CL, Wasiuk A, Fuse S, Turk MJ, Ernstoff MS, Suriawinata AA, Gorham JD, Kedl RM, Usherwood EJ, Noelle RJ. Enhanced efficacy and reduced toxicity of multifactorial adjuvants compared with unitary adjuvants as cancer vaccines. Blood. 2008;111:3116–3125. doi: 10.1182/blood-2007-09-114371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanchez PJ, McWilliams JA, Haluszczak C, Yagita H, Kedl RM. Combined TLR/CD40 stimulation mediates potent cellular immunity by regulating dendritic cell expression of CD70 in vivo. J Immunol. 2007;178:1564–1572. doi: 10.4049/jimmunol.178.3.1564. [DOI] [PubMed] [Google Scholar]
- 39.Soares H, Waechter H, Glaichenhaus N, Mougneau E, Yagita H, Mizenina O, Dudziak D, Nussenzweig MC, Steinman RM. A subset of dendritic cells induces CD4+ T cells to produce IFN-gamma by an IL-12-independent but CD70-dependent mechanism in vivo. J Exp Med. 2007;204:1095–1106. doi: 10.1084/jem.20070176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamilton SE, Wolkers MC, Schoenberger SP, Jameson SC. The generation of protective memory-like CD8+ T cells during homeostatic proliferation requires CD4+ T cells. Nat Immunol. 2006;7:475–481. doi: 10.1038/ni1326. [DOI] [PubMed] [Google Scholar]
- 41.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic acids research. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bamford RN, DeFilippis AP, Azimi N, Kurys G, Waldmann TA. The 5' untranslated region, signal peptide, and the coding sequence of the carboxyl terminus of IL-15 participate in its multifaceted translational control. J Immunol. 1998;160:4418–4426. [PubMed] [Google Scholar]
- 43.de Felipe P, Hughes LE, Ryan MD, Brown JD. Co-translational, intraribosomal cleavage of polypeptides by the foot-and-mouth disease virus 2A peptide. The Journal of biological chemistry. 2003;278:11441–11448. doi: 10.1074/jbc.M211644200. [DOI] [PubMed] [Google Scholar]
- 44.Donnelly ML, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D, Ryan MD. The 'cleavage' activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring '2A-like' sequences. The Journal of general virology. 2001;82:1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 45.Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. The Journal of general virology. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- 46.Oh JZ, Kurche JS, Burchill MA, Kedl RM. TLR7 enables cross-presentation by multiple dendritic cell subsets through a Type I IFN-dependent pathway. Blood. 2011 doi: 10.1182/blood-2011-04-348839. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, Jenkins MK. Naive CD4(+) T Cell Frequency Varies for Different Epitopes and Predicts Repertoire Diversity and Response Magnitude. Immunity. 2007;27:203–213. doi: 10.1016/j.immuni.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Obar JJ, Khanna KM, Lefrancois L. Endogenous naive CD8+ T cell precursor frequency regulates primary and memory responses to infection. Immunity. 2008;28:859–869. doi: 10.1016/j.immuni.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q, Rao RR, Araki K, Pollizzi K, Odunsi K, Powell JD, Shrikant PA. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–553. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho JH, Boyman O, Kim HO, Hahm B, Rubinstein MP, Ramsey C, Kim DM, Surh CD, Sprent J. An intense form of homeostatic proliferation of naive CD8+ cells driven by IL-2. J Exp Med. 2007;204:1787–1801. doi: 10.1084/jem.20070740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard JJ, Ohashi PS, Griesser H, et al. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 1995;268:1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- 52.Tscharke DC, Karupiah G, Zhou J, Palmore T, Irvine KR, Haeryfar SM, Williams S, Sidney J, Sette A, Bennink JR, Yewdell JW. Identification of poxvirus CD8+ T cell determinants to enable rational design and characterization of smallpox vaccines. J Exp Med. 2005;201:95–104. doi: 10.1084/jem.20041912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–676. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 54.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JC, Joyce S, Peschon JJ. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, Ahmed R. Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J Exp Med. 2002;195:1541–1548. doi: 10.1084/jem.20020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan JT, Ernst B, Kieper WC, LeRoy E, Sprent J, Surh CD. Interleukin (IL)-15 and IL-7 jointly regulate homeostatic proliferation of memory phenotype CD8+ cells but are not required for memory phenotype CD4+ cells. J Exp Med. 2002;195:1523–1532. doi: 10.1084/jem.20020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Castillo EF, Schluns KS. Regulating the immune system via IL-15 transpresentation. Cytokine. 2012;59:479–490. doi: 10.1016/j.cyto.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dubois S, Mariner J, Waldmann TA, Tagaya Y. IL-15Ralpha recycles and presents IL-15 In trans to neighboring cells. Immunity. 2002;17:537–547. doi: 10.1016/s1074-7613(02)00429-6. [DOI] [PubMed] [Google Scholar]
- 59.Musso T, Calosso L, Zucca M, Millesimo M, Ravarino D, Giovarelli M, Malavasi F, Ponzi AN, Paus R, Bulfone-Paus S. Human monocytes constitutively express membrane-bound, biologically active, and interferon-gamma-upregulated interleukin-15. Blood. 1999;93:3531–3539. [PubMed] [Google Scholar]
- 60.Sandau MM, Schluns KS, Lefrancois L, Jameson SC. Cutting edge: transpresentation of IL-15 by bone marrow-derived cells necessitates expression of IL-15 and IL-15R alpha by the same cells. J Immunol. 2004;173:6537–6541. doi: 10.4049/jimmunol.173.11.6537. [DOI] [PubMed] [Google Scholar]
- 61.Stonier SW, Ma LJ, Castillo EF, Schluns KS. Dendritic cells drive memory CD8 T-cell homeostasis via IL-15 transpresentation. Blood. 2008;112:4546–4554. doi: 10.1182/blood-2008-05-156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rubinstein MP, Kovar M, Purton JF, Cho JH, Boyman O, Surh CD, Sprent J. Converting IL-15 to a superagonist by binding to soluble IL-15R{alpha} Proc Natl Acad Sci U S A. 2006;103:9166–9171. doi: 10.1073/pnas.0600240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyman O, Purton JF, Surh CD, Sprent J. Cytokines and T-cell homeostasis. Curr Opin Immunol. 2007;19:320–326. doi: 10.1016/j.coi.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 64.Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 65.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 66.Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma LJ, Acero LF, Zal T, Schluns KS. Trans-presentation of IL-15 by intestinal epithelial cells drives development of CD8alphaalpha IELs. J Immunol. 2009;183:1044–1054. doi: 10.4049/jimmunol.0900420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frasca L, Stonier SW, Overwijk WW, Schluns KS. Differential mechanisms of memory CD8 T cell maintenance by individual myeloid cell types. J Leukoc Biol. 2010;88:69–78. doi: 10.1189/jlb.1209816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Colpitts SL, Stoklasek TA, Plumlee CR, Obar JJ, Guo C, Lefrancois L. Cutting edge: the role of IFN-alpha receptor and MyD88 signaling in induction of IL-15 expression in vivo. J Immunol. 2012;188:2483–2487. doi: 10.4049/jimmunol.1103609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, Schreiber RD, Murphy TL, Murphy KM. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edelson BT, Kc W, Juang R, Kohyama M, Benoit LA, Klekotka PA, Moon C, Albring JC, Ise W, Michael DG, Bhattacharya D, Stappenbeck TS, Holtzman MJ, Sung SS, Murphy TL, Hildner K, Murphy KM. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8alpha+ conventional dendritic cells. J Exp Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Desch AN, Randolph GJ, Murphy K, Gautier EL, Kedl RM, Lahoud MH, Caminschi I, Shortman K, Henson PM, Jakubzick CV. CD103+ pulmonary dendritic cells preferentially acquire and present apoptotic cell-associated antigen. J Exp Med. 2011;208:1789–1797. doi: 10.1084/jem.20110538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stoklasek TA, Colpitts SL, Smilowitz HM, Lefrancois L. MHC class I and TCR avidity control the CD8 T cell response to IL-15/IL-15Ralpha complex. J Immunol. 2010;185:6857–6865. doi: 10.4049/jimmunol.1001601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao DM, Yu S, Zhou X, Haring JS, Held W, Badovinac VP, Harty JT, Xue HH. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J Immunol. 2010;184:1191–1199. doi: 10.4049/jimmunol.0901199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X, Yu S, Zhao DM, Harty JT, Badovinac VP, Xue HH. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat Immunol. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 78.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 79.Van de Wetering M, Castrop J, Korinek V, Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol Cell Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roose J, Clevers H. TCF transcription factors: molecular switches in carcinogenesis. Biochim Biophys Acta. 1999;1424:M23–M37. doi: 10.1016/s0304-419x(99)00026-8. [DOI] [PubMed] [Google Scholar]
- 81.Roose J, Molenaar M, Peterson J, Hurenkamp J, Brantjes H, Moerer P, van de Wetering M, Destree O, Clevers H. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–612. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 82.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science. 1999;285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 83.Oosterwegel M, van de Wetering M, Dooijes D, Klomp L, Winoto A, Georgopoulos K, Meijlink F, Clevers H. Cloning of murine TCF-1, a T cell-specific transcription factor interacting with functional motifs in the CD3-epsilon and T cell receptor alpha enhancers. J Exp Med. 1991;173:1133–1142. doi: 10.1084/jem.173.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Okamura RM, Sigvardsson M, Galceran J, Verbeek S, Clevers H, Grosschedl R. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]
- 85.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, Welboren W, Versteeg R, Cuppen E, van de Wetering M, Clevers H, Stunnenberg HG. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol Cell Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, Huelsken J, Held W. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 87.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 88.Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J Exp Med. 1999;189:701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rees W, Bender J, Teague TK, Kedl RM, Crawford F, Marrack P, Kappler J. An inverse relationship between T cell receptor affinity and antigen dose during CD4(+) T cell responses in vivo and in vitro. Proc Natl Acad Sci U S A. 1999;96:9781–9786. doi: 10.1073/pnas.96.17.9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10:485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.