Abstract
Calcium-calmodulin-dependent protein kinase II (CaMKII) has been implicated in a wide variety of cellular processes, which include a critical regulatory role in actin cytoskeletal assembly. CaMKII is ubiquitous in cells, expressed as one of four isoforms referred to as α, β, γ, and δ. Characterization of the CaMKII-actin interaction has been mainly focused on the β isoform, which has been shown to bundle actin filaments and sequester actin monomers in an activity dependent manner. Much less is known about the interactions of other CaMKII isoforms with actin. In the present report, isoform specific interactions of CaMKII with actin are described and reveal that the δ isoform of CaMKII bundles F-actin filaments similarly to the β isoform while the γ isoform induces a novel layered structure to filaments. Using electron tomography, CaMKII holoenzymes are clearly identified in the complexes bridging the actin filaments, allowing direct visualization of the interactions between CaMKII isoforms and actin. In addition, we determined the isoform specificity of CaMKII-mediated inhibition of actin polymerization and discovered that all isoforms inhibit polymerization to varying degrees with β > γ ≈ δ > α (from most to least effective). Ca2+/CaM activation of all kinase isoforms produced a robust increase in actin polymerization that surpassed the rates of polymerization in the absence of kinase inhibition. These results indicate that diversity exists between the types of CaMKII-actin interactions mediated by the different isoforms and that CaMKII isoform composition differentially impacts the formation and maintenance of the actin cytoskeleton.
Properly regulated assembly and disassembly of actin filaments coupled with stabilization of the actin cytoskeleton are fundamental processes necessary for the proper functioning of eukaryotic cells (1-3). Actin filaments serve a central role in molecular transport, providing tracks for motor protein based cargo delivery and participate in establishing the cellular cytoskeleton (1). Polymerization of actin from the monomeric, G-actin, form to the filamentous, F-actin, form is asymmetric, where the growth is more rapid at the barbed end than the pointed end. This asymmetric growth is responsible for mechanical force generation in cells (4, 5). Filament stability is controlled by capping proteins, such as gelsolin and tropomodulin, that interact with the filament ends to regulate polymerization rates (1, 2, 6). There are also G-actin binding proteins, for example thymosin and profilin, which offer additional regulation of filament growth by sequestering the monomeric form of actin. New filament formation depends on interactions between monomers which are initially unstable and consequently, any factors that reduce actin availability can have a major impact on whether polymerization of filaments is initiated or not (1). As a result, the nucleation of new filaments typically starts slowly, but once initiated, polymerization proceeds rapidly to allow for efficient response to cellular signaling (2). Once polymerized, filaments can be stabilized by interactions with multivalent actin binding proteins such as α-actinin, filamin and spectrin, to form the structural network of the cellular cytoskeleton (1).
Among actin regulatory proteins, βCaMKII has been shown to play multiple unique roles in actin assembly. Not only does it organize and stabilize F-actin into higher order assemblies (7-11), it also controls the rate and extent of actin polymerization through binding to and sequestering G-actin (11). Through such interactions, CaMKII can play the dual role of both an F-actin cross-linking protein and a sequestering protein through its ability to bind G-actin. These multivalent interactions with actin are made possible by the architecture of CaMKII holoenzymes which are composed of twelve subunits that oligomerize into a dodecameric complex. Each subunit is the product of one of four genes, α, β, γ and δ (12) that are assembled in random stoichiometry, forming pure or mixed holoenzymes depending on the level of expression of each subunit (13-15). The oligomeric complex formed by each isoform is similar in 3D structure (15), each with the potential to form multivalent interactions with multiple proteins. In addition to actin, the list of CaMKII binding partners is constantly expanding (e.g., α-actinin, the NMDA receptor, voltage-activated Ca2+-channels, to name a few (for reviews see (12, 16)) leading to the proposal that the kinase plays a role as a structural scaffold protein in addition to its role as a protein kinase. Expression of CaMKII isoforms is tissue specific with α and β isoforms highly enriched in nervous tissue and γ and δ found more broadly throughout the body (12). The four genes are alternatively spliced to form almost 30 protein products, the roles of which are poorly defined (12, 17). From deletion and comparative studies of CaMKII isoforms (largely focused on the β isoform) (9, 10, 18, 19), it has been suggested that the variable domain residing just C-terminal to the autoregulatory and CaM-binding domain of CaMKII (Fig. 1A), is responsible for targeting the enzyme to the actin cytoskeleton and therefore contains the site for actin-binding. However, one study examining deletion mutants and naturally occurring splice variants of the δ isoform of CaMKII concluded that the actin binding site was not in the variable domain, but resides in a sequence further C-terminal to the variable domain in an area largely conserved amongst all the CaMKII isoforms (20). Interestingly, interactions with actin were dependent on formation of the oligomeric state of the kinase; however, activity was not required (8, 9, 18). It was shown that β and not αCaMKII bundles actin filaments (9-11), implying that structural differences exist between isoforms to mediate actin cross-linking. Furthermore differences in inhibition of actin polymerization are observed between β and α isoforms (11), suggesting potential isoform differences in the affinity for G-actin. Despite some ambiguity in the literature concerning the exact domain of CaMKII responsible for actin binding, converging data indicates that Ca2+/CaM binding to the enzyme causes the enzyme to dissociate from actin (7-9, 11). These studies indicate that CaMKII serves to sequester G-actin and to cross-link actin filaments, processes which occur in a Ca2+-regulated manner and are mediated by the reversible binding of Ca2+/CaM.
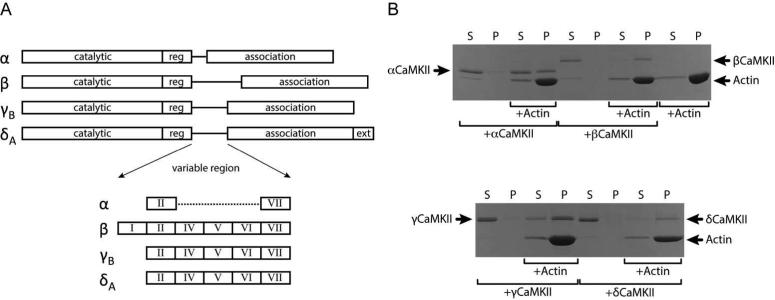
Figure 1. Domain map of CaMKII isoforms and their cosedimentation with F-actin.
A) The domain structures of all CaMKII isoforms used in these studies are highly conserved in sequence in the catalytic, regulatory and association domains. The only noteworthy differences are found in the variable linker region where alternative splicing results in inclusion or omission of various exons denoted by Roman numerals. Additionally the δA isoform contains a 21 amino acid C-terminal extension unique to this isoform. B) F-actin was incubated with CaMKII isoforms for 1 h and sedimented at 50,000 × g at RT for 1 h. The supernatant was collected and made 1X in SDS-sample buffer (125 μL total volume) and the pellet was resuspended in 125 μL of 1X SDS-sample buffer. Equivalent volumes of supernatant (S) and pellet (P) fractions from each reaction were then analyzed by SDS-PAGE and the gel was Coomassie stained. Control samples (reactions without actin) show that each kinase isoform remains in the supernatant (S) under these conditions. While all four kinase isoforms can be found in the pellet (P) with actin, there is a rank order of binding with β, δ, γ and α going from strongest to weakest.
Much less is known about the influence of the γ and δ isoforms of CaMKII on actin cross-linking or polymerization, prompting us to investigate their potential roles in the maintenance of actin structure. In addition, we examined whether activation by Ca2+/CaM-binding would reverse G-actin association with CaMKII, potentially leading to a burst in actin polymerization as has been proposed (11). In this report we present novel structural and kinetic data providing insight into the isoform specificity of the CaMKII-actin interaction. We show for the first time that the δ isoform of CaMKII bundles F-actin into rod-like assemblies similarly to the β isoform and that the γ isoform interaction with F-actin results in a unique layered structural assembly. We describe the isoform dependence on CaMKII-mediated inhibition of actin polymerization and rank their capacity for inhibition as (β > γ ≈ δ > α). We also provide experimental evidence that Ca2+/CaM-activation of each CaMKII isoform, pre-complexed with actin, does result in a rapid burst of actin polymerization. Finally, contrary to previous observations, we show that the α isoform of CaMKII interacts with actin, although weakly, and reveal its potential role in the regulation of the actin cytoskeleton.
Experimental Procedures
Protein Preparation
Purification of actin was carried out as previously described (11). The final purified protein was dialyzed into 5 mM Tris, 0.2 mM CaCl2, 0.2 mM ATP, and 0.5 mM dithiothreitol (DTT), pH 8.0 and stored at -80°C until needed. Preparations were shown to be more than 90% pure by SDS-PAGE analysis and protein concentration was determined by BCA assay (Pierce Chemical Co.). The actin depolymerization reaction consisted of 1 mg/mL G-actin, 5 mM Tris, 0.2 mM CaCl2, 0.2 mM Na2ATP, 0.5 mM DTT, 1 mM NaN3, pH 8.0. The reaction was incubated on ice for 1 hour then was allowed to undergo nearly complete depolymerization for ~16 hours at 4°C. From this pool of G-actin, F-action was produced by adding 10× polymerization buffer for 3 hours at room temperature (RT) with final concentrations of 1 mg/mL G-actin, 5 mM Tris, 50 mM KCl, 2 mM MgCl2, 1 mM Na2ATP, 0.2 mM CaCl2, 0.5 mM DTT, 1 mM NaN3, pH 8.0. Expression of all CaMKII isoforms was completed using a baculovirus system exactly as previously described (11, 21) and purification was carried out using CaM-sepharose affinity chromatography (for complete sequence comparison of isoforms used in this study refer to supplemental Fig. 1). Purified protein was dialyzed into 20 mM HEPES, 0.5 M NaCl, and 10% glycerol, pH 7.4 and aliquots frozen at -80°C. CaMKII protein was quantified using theoretical extinction coefficients of 1.226, 1.032, 1.068, and 1.104 respectively for the α, β, γ, and δ isoforms. A codon-optimized cDNA of calmodulin was cloned into a pET3a vector (Novagen) containing a T7 promoter. Protein expression was carried out in E. coli BL21(DE3) cells with a 0.4 mM IPTG induction at 32°C for 4 hours. Harvested cell pellets were lysed using an initial freeze-thaw cycle and then by sonication in 50 mM MOPS, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 0.1 mM PMSF, pH 7.5. 5 mM CaCl2 was added to lysis supernatant and applied to a High Trap Phenyl HP column (GE Healthcare) equilibrated with 50 mM Tris–HCl, 1 mM CaCl2, pH 7.5. Protein bound on the column was washed with 50 mM Tris–HCl, 1 mM CaCl2, 0.5 M NaCl, pH 7.5, and bound CaM was eluted with 10 mM Tris–HCl, 10 mM EDTA, pH 7.5. Purified CaM was then desalted into 50 mM HEPES, 100 mM NaCl, 2.5 mM EDTA, pH 7.5 to remove Ca2+, then finally desalted into 50 mM MOPS, pH 7.2.
Cosedimentation Assay
100 μL binding reactions were completed for 1 hour at RT with 1 mg/mL F-actin (polymerized as described above) and 0.1 mg/mL CaMKII α, β, γ, or δ in the following buffer: 5 mM Tris, 50 mM KCl, 2 mM MgCl2, 1 mM Na2ATP, 0.2 mM CaCl2, 0.5 mM DTT, 1 mM NaN3, pH 8.0. Control samples containing either no actin or no kinase were prepared similarly. Reactions were centrifuged at 50,000 × g at 24°C for 1 hour in a Beckman tabletop TL-1 ultracentrifuge. Supernatants were collected and were made to 1X SDS-sample buffer and pellets were suspended in an equivalent volume of 1X SDS-sample buffer and the same volume of each sample was subsequently analyzed with 12% SDS-PAGE. The distributions of proteins between supernatant and pellet fractions were visualized with Coomassie Blue stain.
Polymerization Assay
Actin polymerization was assessed using fluorescently labeled pyrene-actin (pyr-actin) purchased as part of a kit from Cytoskeleton Inc. Depolymerization of pyr-actin was carried out for 1 hour on ice then >16 hours at 4°C in general actin buffer + ATP: 0.4 mg/mL pyr-actin, 5 mM Tris, 0.2 mM CaCl2, 0.2 mM Na2ATP, pH 8.0. Polymerization of 0.25 μM actin was then assessed in the presence or absence of 2-fold excess CaMKII subunit concentration (0.5 μM) or BSA (to control for non-specific protein effects) at RT for 0.75-1.5 hours in 5 mM Tris, 50 mM KCl, 2 mM MgCl2, 1 mM Na2ATP, 0.2 mM CaCl2, pH 8.0. To observe the effects of CaMKII activation, a 1:1 molar ratio of CaM:CaMKII (subunits) was added to experiments at the time of adding polymerization buffer, or at 30 minutes after the start of polymerization. Fluorescence was monitored during the polymerization reaction in a PTI fluorimeter with 350 nm excitation and 405 nm emission. Excitation and emission slit widths were 1 and 10 nm, respectively. To avoid photobleaching from constant exposure over the time course of the experiments, data points were collected for a 1 second duration every 30 seconds then rotated out of the path of the excitation beam.
Electron Microscopy
F-actin-CaMKII reactions for electron microscopy were prepared as described in the cosedimentation assays. After binding for 1 hr at RT, 5 μL of each reaction was placed on a glow-discharged formvar carbon coated copper grid (Ted Pella) and incubated for 1 min. Excess sample was wicked away and the grids were washed once with methylamine tungstate (Nanoprobes Nano-W), stained for 30 seconds in methylamine tungstate, blotted, and air dried. Electron micrographs were collected on a JEOL 1400 Transmission Electron Microscope running at 120kV with a 2000 × 1300 pixel Gatan Orius SC1000 camera. The same grids were used to collect tomographic tilt-series on a 300kV FEI Polara F30 electron microscope equipped with a 4000 × 4000 pixel Tietz CCD camera. Series were collected at 39,000x magnification with a -5 to -10 μm defocus under a dose of ~400 electrons/Å2. Images were collected at two degree tilt increments from -60° to +60° with 2X binning which generated a final pixel size of 4.6 Å. Tilt series were aligned and tomographic reconstructions produced using Etomo, part of the IMOD software package (22).
Results
Isoform dependence on CaMKII/F-actin interaction
For preliminary insight into CaMKII interaction with actin filaments, we examined the cosedimentation of F-actin with α, β, γB, and δA isoforms of CaMKII (Fig. 1). These conventional assays were designed such that the centrifugation parameters resulted in sedimentation of F-action while non-interacting CaMKII and monomeric G-actin remained in the supernatant (Fig. 1B). Any CaMKII found in pellet fractions was assumed to interact with F-actin. Consistent with previously reported results, SDS-PAGE analysis illustrates that αCaMKII is found in both the supernatant and pellet fractions indicating partial association with F-actin, while the vast majority of βCaMKII is found in the pellet suggesting robust binding to actin filaments (11). The majority of γCaMKII and δCaMKII were in association with F-actin, but the γ isoform also showed a minor band in the supernatant as well indicating incomplete association under these conditions. These results suggest that CaMKII interaction with F-actin is isoform dependent with β bound actin most avidly, followed by δ, γ and α.
δCaMKII bundles actin filaments similarly to βCaMKII
Motivated by the apparent differences in F-actin association, EM grids were prepared from CaMKII binding reactions and negatively stained for imaging. Electron micrographs of CaMKII isoforms with F-actin indeed illustrate isoform-dependent differences in F-actin/CaMKII complexes (Figs. 2 and 3). As previously reported, βCaMKII binds to F-actin to produce actin bundles, parallel assemblies of filaments interspersed with bound CaMKII holoenzymes (9, 11). We observed that bundling by δCaMKII was very similar to βCaMKII in that bundles had comparable sizes and structures. Actin filaments were well organized in a mostly parallel fashion, with a degree of twist to the overall bundle (Fig. 2A). The configuration of holoenzyme molecules within the bundle did not show any apparent pattern regarding location or orientation. Bundle diameters on the order of 100 nm were consistently observed and the width did not vary significantly though out the length of the bundle, giving rise to relatively linear rod-like structures. Binding of either β or δ isoforms resulted in bundles with considerable variability in length, ranging to up to 15 μm. We observed that much of the F-actin on grids was incorporated into bundles and that bundles were discrete structures, seemingly unrelated to stray unbundled filaments in the same vicinity.
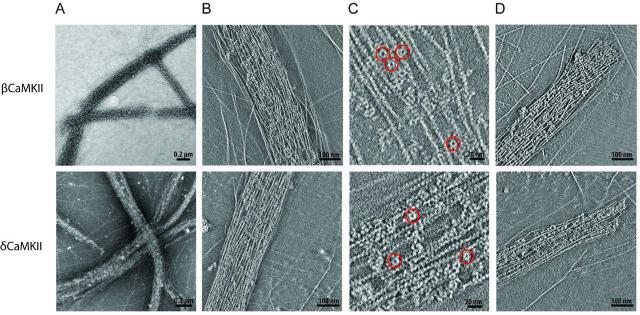
Figure 2. Structural analysis of actin bundles formed in the presence of the β and δ isoforms of CaMKII.
A) Left panels illustrate representative 2D projections of low power electron micrographs of negative stained F-actin in the presence of β (upper) and δ (lower) isoforms (n > 5). B) Panels show ~10 nm thick slices of two tomographic reconstructions to illustrate packing of β and δ CaMKII holoenzyme molecules within bundles. C) Several CaMKII holoenzyme molecules are highlighted by red circles in a zoomed in region of a representative slice from the same reconstructions shown in Panel B. D) The right panels show ~ 10 nm slices through two different tomographic reconstructions that illustrate examples of blunt ended bundles formed with β or δ CaMKII.
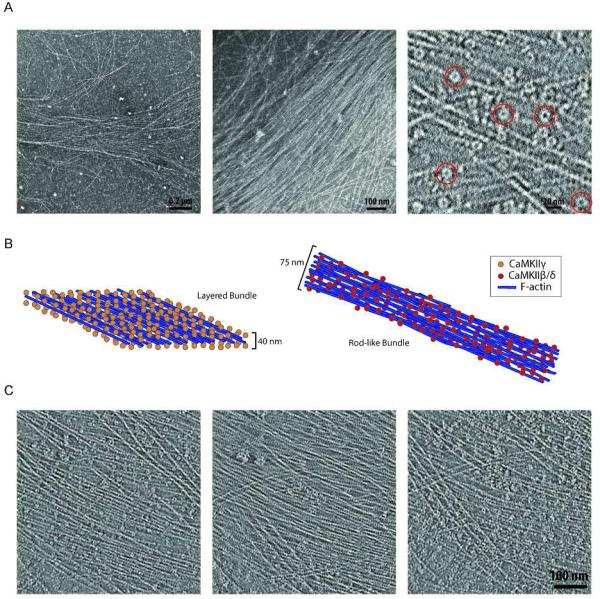
Figure 3. Structure of layered γCaMKII bundle.
A) Left and center panels show representative electron micrographs of F-actin bundles in the presence of γCaMKII (n = 6). The right panel illustrates a ~10 nm slice from a tomographic reconstruction where several CaMKII holoenzyme molecules are highlighted in red circles. B) A cartoon illustrates structural differences between the layered bundles observed in the presence of the γ isoform (orange spheres) and the rod-like bundles observed in the presence of the β or δ isoforms (red spheres). C) ~ 10 nm slices from tomograms illustrate the top, center, and bottom z sections of the layered bundle in the left, center, and right panels, respectively.
In addition to 2D micrographs, we also compiled 3D tomographic reconstructions from series of tilt images of negatively stained samples to provide additional insight into the 3D structures of bundles. 10 nm thick slices from these series are shown in Fig. 2B. In these images the packing of the bundle can be appreciated, where the small ring-like structures of individual holoenzyme molecules can be clearly identified among actin filaments. An enlargement of representative slices for each isoform revealed several CaMKII holoenzymes identified among actin filaments (Fig. 2C, highlighted in red circles). Actin bundles were tightly packed with a Z-axis thickness on the order of ~50-100 nm. One noteworthy observation was that bundle ends formed with β and γ often did not taper off but ended abruptly (Fig. 2D). Rarely did individual filaments extended beyond the length of the bundle. We wondered if this squared end may suggest a role for β and δ isoforms of CaMKII in capping of filaments, but could never discern holoenzyme molecules bound to the end of individual filaments. Overall the bundle structures resulting from the β and δ isoforms were indistinguishable, suggesting that their mode of interaction with F-actin is similar and that they presumably play similar roles in the maintenance of intracellular actin structure.
γCaMKII binding to F-actin generates an assembly with a layered structure
Remarkably, the γ isoform interaction with F-actin resulted in a completely different organization of filaments. In contrast to the discrete structures produced by β and δ isoforms, γCaMKII binding to F-actin resulted in wide and long filament bundles. These bundles contained a roughly parallel assembly of actin filaments, which appeared to be non-homogeneously linked to a loose meshwork of filaments with multiple branching sites in the x-y plane (Fig. 3A). Diameters were variable and although their lack of boundary made it difficult to determine bundle boundaries, they were, in general, 2- to 3-fold larger than the bundles formed by β and δ isoforms. 3D tomographic reconstructions of these bundles revealed that while the organization of filaments in the x-y plane was somewhat non-uniform, a layered structure was detected along the z axis, with actin filaments sandwiched between layers of kinase holoenzymes. The heights of these complexes are approximately 40 nm compared to the 50-100 nm range for the rod-like bundles. Fig. 3B illustrates a comparison of this layered bundle to rod-like bundles. In the left panel, a cartoon representing a segment of a layered bundle is depicted with γCaMKII holoenzymes represented as orange spheres and actin filaments as blue rods. The rod-like bundle is shown to the right with red spheres representing the β or δ isoform of CaMKII. The z-axis height of the rod-like bundles allows for more stacking of filaments while the layered bundle is dominated by one central sheet of roughly parallel filaments. Fig. 3C shows three tomographic slices from the top, center, and bottom of a representative layered bundle (left, center, and right panels, respectively). In the center panel representing the center of the bundle, mainly actin filaments are observed with only a few holoenzyme molecules seen, indicating the center of the bundle is largely made of filaments. In contrast, the left and right panels of Fig. 3C illustrate the outer most slices of the bundle where flat planes of holoenzyme molecules are positioned.
These results indicate that although all CaMKII isoform holoenzymes are structurally similar at low resolution (15), their capacity to bundle F-actin and their organization of filaments is isoform dependent. The nature of CaMKII dependent bundling of F-actin seen in EM micrographs and 3D tomographic reconstructions is congruous with the findings in the cosedimentation assay suggesting there is a relationship between isoform binding capacity and type of bundling. β and δ isoforms showed nearly complete association with F-actin which resulted in rod-like bundling while the γ isoform was not as completely associated with F-actin resulting in layered bundles. Despite extensive evaluation, under the present assay conditions, αCaMKII did not show evidence of bundling filaments analyzed using EM perhaps due to a weak overall interaction with F-actin.
Extent of CaMKII mediated inhibition of actin filament polymerization is isoform dependent
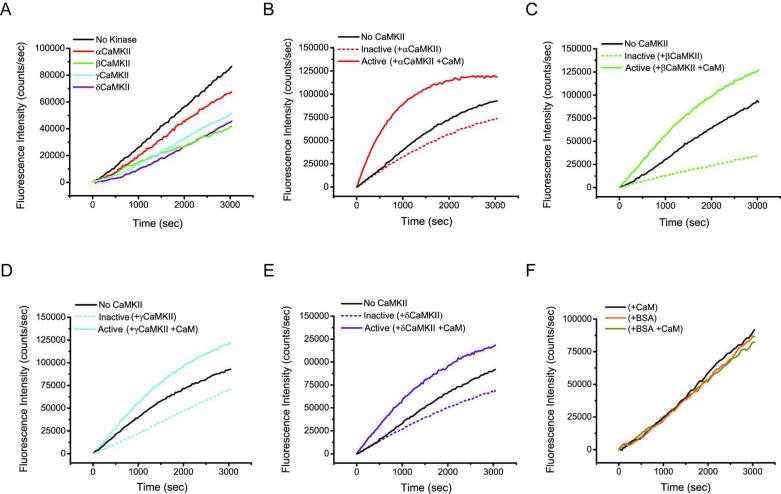
βCaMKII is able to bind both G- and F-actin (11) and decreases the actin polymerization rate while the α isoform yielded a modest reduction (9, 11). To examine how the γ and δ isoforms of CaMKII influence actin filament formation, actin polymerization was assessed in the presence of the various CaMKII isoforms. Polymerization was probed with fluorescent pyrene-actin, an environmentally sensitive molecule that increases in fluorescence as monomers assemble into filaments. An initial depolymerization reaction was carried out in an effort to maximize starting material that was largely monomeric. Polymerization was then observed over a 50-100 min time course in the presence (or absence) of various CaMKII isoforms, where CaMKII subunits were in two-fold excess of actin. Fig. 4A is a representative plot of this experiment illustrating that all CaMKII isoforms decreased the rate of actin polymerization. Consistent with previously reported data (9, 11), we observed a robust reduction in polymerization rate in the presence of the β isoform, while αCaMKII resulted in the least amount of inhibition. The rates of actin polymerization in the presence of the γ and δ isoforms (Fig. 4A, blue and purple traces) lie between α and β (red and green traces) suggesting they inhibit polymerization with an intermediate capacity. We discovered that the amount of inhibition was highly dependent on the G-actin/CaMKII subunit ratio that varied somewhat depending on the effectiveness of the initial actin depolymerization reaction. However, the trend of inhibition by the various isoforms (β > γ ≈ δ > α) was reproducible throughout multiple experiments (n = 4), clearly illustrating a robust isoform dependence on the CaMKII capacity to inhibit actin polymerization. The inhibition detected in these experiments was specific, as inclusion of BSA at the same concentration as CaMKII, had no discernible effect on actin polymerization (Fig. 4F).
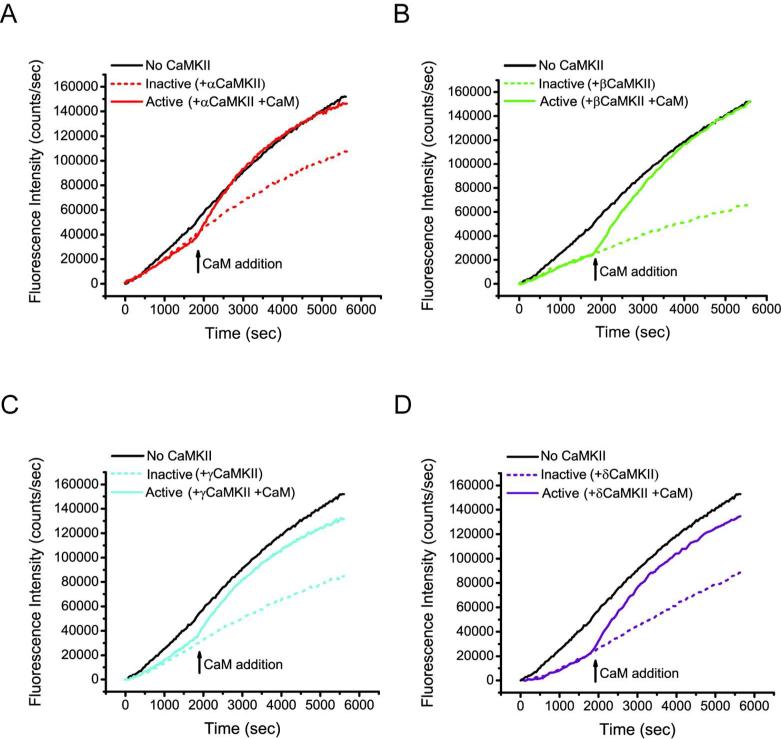
Figure 4. Impact of CaMKII binding on actin polymerization rates.
A) A representative plot of fluorescence intensity of pyr-actin measured in the presence or absence of each CaMKII isoform (n = 4). B) Actin polymerization kinetics in the presence of αCaMKII where kinase (two-fold molar excess over actin) was pre-incubated with G-actin, and then activated with the addition of equimolar CaM (to CaMKII subunits) at the same time polymerization was started. C-E) Identical experiments to that described in B, however the panels are reactions with βCaMKII, γCaMKII and δCaMKII, respectively. F) Control experiment showing a plot of the impact of BSA and Ca2+/CaM, at the same concentration as in Panel A added at the time of initiation of actin polymerization. Note that pre-incubation of kinase with G-actin followed by addition of Ca2+/CaM concurrent with actin polymerization produces a robust burst in actin polymerization with all four CaMKII isoforms.
CaMKII activation releases inhibition of actin polymerization and promotes rapid growth of filaments
We previously demonstrated that βCaMKII binds G-actin and that binding was reduced upon Ca2+/CaM activation of the kinase (11). Others have reported that the F-actin/βCaMKII interaction was also diminished in the presence of Ca2+/CaM (7, 9, 10, 19). These data led us to propose a dynamic mechanism of actin reorganization where exploitation of the multivalent architecture of the twelve subunit holoenzyme provides a kinetic advantage for rapid filament growth (11). In the basal state, our model predicts holoenzyme subunits serve to sequester monomeric G-actin and that Ca2+/CaM activation of CaMKII/actin complexes would facilitate a burst of actin polymerization due to a local release of G-actin. To examine this possibility, we first assessed whether Ca2+/CaM addition to reactions where CaMKII was pre-incubated with G-actin prior to initiating polymerization, impacted the rate of actin polymerization. Fig. 4B, illustrates the time course of actin polymerization when G-actin was pre-incubated with αCaMKII (or not) and then the reaction was initiated with polymerization buffer with or without a 1:1 molar ratio of Ca2+/CaM to CaMKII subunits. While the level of inhibition is modest with αCaMKII (dotted red trace) compared with no kinase (black trace), there is a significant increase in the rate of actin polymerization in reactions that also contained Ca2+/CaM (solid red trace). Importantly, inclusion of BSA, to test for non-specific protein effects, with or without the same concentration of added Ca2+/CaM, produced no significant change in the polymerization rate of actin (Fig 4F). In an analogous design, pre-incubation of the β, γ or δ isoforms of CaMKII with actin similarly showed enhanced rates of actin polymerization when Ca2+/CaM was added coincident with the actin polymerization buffer (Fig. 4, panels C, D and E, respectively). We conclude that the rate of actin polymerization associated with CaMKII when CaM is added is significantly increased compared to what is observed for the no CaMKII condition. Thus, the burst of polymerization we observe appears due to release of G-actin localized by CaMKII since otherwise, polymerization would have resembled that observed in the no CaMKII condition.
We next assessed whether addition of Ca2+/CaM to reactions where actin polymerization had progressed significantly in the presence of CaMKII would also have an impact on the rate of actin polymerization. In these reactions, we presume there is a mixture of polymerized F-actin and G-actin and that at least a portion of the CaMKII subunits in each reaction are interacting with the F-actin filaments. Fig. 5A illustrates a time course in which actin polymerization was initially monitored in the absence of CaM to establish αCaMKII mediated inhibition of polymerization (dotted red trace). After 30 min, αCaMKII was activated by addition of CaM (indicated by black arrow; Ca2+ is already present in the polymerization reactions). As controls, the no kinase condition (black trace) reports polymerization in the absence of αCaMKII. We observed that activation of αCaMKII (solid red trace) resulted in a rapid increase in fluorescence compared to the inactive condition (red dotted line) indicating a dramatic increase in the rate of actin polymerization. We also analyzed the impact of CaM addition and Ca2+/CaM binding to complexes formed between β, γ and δ CaMKII and actin (Fig 5., panels B, C and D, respectively) that were each permitted to polymerize before activation. Addition of CaM to each preformed complex led to a surge of actin polymerization similar to that observed with αCaMKII and actin (Fig. 5, panel A).
Figure 5. Impact of Ca2+/CaM binding on CaMKII inhibited actin polymerization rates.
A) Actin polymerization kinetics in the presence of αCaMKII where kinase was activated upon addition of CaM following 30 min of actin polymerization (saturating Ca2+ is already present in the reactions). CaMKII was added to the reaction in two-fold molar excess (subunit concentration) over actin and CaM was added at an equimolar concentration to CaMKII subunits. B-D) Identical experiments to that described in (A), however the panels display fluorescence traces showing the impact of CaM addition (black arrows) to actin polymerization reactions with added βCaMKII (B), γCaMKII (C) and δCaMKII (D), respectively.
Together, we interpret these results as evidence for rapid nucleation of localized actin monomers through Ca2+/CaM mediated release from CaMKII, supporting the model where release of G-actin, whether the kinase is bound to filaments or not, promotes an increased probability of new filament formation and/or increased rate of actin addition to existing filaments. These data indicate that all four of the isoforms of CaMKII can bind to and slow the polymerization rate of actin, including αCaMKII, and that Ca2+/CaM-binding leads to an apparent dissociation of G-actin with subsequent increased rates of polymerization.
Discussion
In this report we have shown that the products of each of the four mammalian genes of CaMKII, α, β, γ and δ interact with, and influence, aspects of actin polymerization and actin filament organization. CaMKII has been termed a multifunctional protein kinase, largely based on the many proteins that serve as substrates for the enzyme. In addition to this aspect of multifunctionality, CaMKII also serves a role as a structural scaffold, interacting with a number of proteins distinct from those that interact with the substrate-binding pocket (12, 16). Importantly one of these interacting proteins is actin, the assembly and bundling of which are impacted by binding of CaMKII to both G- and F-actin. This earlier work was dominated by analysis of the α and β isoforms of CaMKII with the consensus being that β interacts with the actin cytoskeleton, while α does not (19, 23) and that a domain unique to β is therefore responsible for actin-dependent interactions. The majority of this work also relied on co-localization experiments inside cells, or photobleaching recovery and was not derived from direct in vitro tests. The interaction of the γ and δ isoforms with actin or their influence on the actin cytoskeleton was largely unknown. The γ and δ isoforms have been shown to differentially localize to cellular actin rich structures (20), with δ showing stronger targeting to the actin cytoskeleton, but a direct interaction with G- or F-actin in vitro had not been analyzed.
We showed previously that compared to βCaMKII, αCaMKII weakly interacted with F-actin and could modestly impact the rate of G-actin polymerization (11). We reproduced that experimental finding in the present report and showed that αCaMKII has a modest, but significant impact on actin polymerization and that it binds, albeit weakly, to F-actin in pull-down experiments. This latter result is actually quite similar to the pull-down experiments described by (23), who showed that αCaMKII bound to purified F-actin, although less effectively than βCaMKII. These results suggest that while less efficient at binding to actin, αCaMKII does possess a site for interaction, complicating previous conclusions identifying the unique domain of βCaMKII as containing the actin-binding site that is entirely lacking in αCaMKII. Caran et al, (20) reached a similar conclusion from results evaluating distinct actin interactions of truncation mutants or naturally occurring splice variants of δCaMKII expressed inside cells. They showed that a domain beginning at amino acid 351, including the C-terminal oligomerization domain but lacking the variable domain, targeted as efficiently to actin as did full-length δCaMKII. Further, they showed that the δc splice variant lacking the variable domain sequences also targets to the actin cytoskeleton (20). We also evaluated the interactions of the δ and γ isoforms of CaMKII with actin in our panel of in vitro assays. Both bound to F-actin filaments, although γ appeared to bind with a lower apparent affinity than δ in pull-down assays. Both also inhibited actin polymerization to similar extents, similar to βCaMKII but more effectively than αCaMKII. These results indicate that all four isoforms of CaMKII can interact with G- and F-actin, but do so to different extents. Our results suggest there is no clear correlation between CaMKII linker sequence and actin remodeling and therefore we predict that alternative splicing of the linker region is only one aspect of a multifaceted regulatory mechanism responsible for the differences in isoform association with actin. Although it has been suggested that the additional variable domains present in the linker region of the β isoform could recognize actin (refer to Fig. 1A), our present results demand that a binding site must exist within the primary sequence of the α isoform. It is plausible to suggest then that while all isoforms contain the actin-binding site, it is the length of the linker region responsible for influencing access to actin binding resulting in variable affinities for each isoform. Variations in the length of the linker has been proposed to govern the frequency of conformational exchange within the enzyme (24, 25), which may serve to influence exposure of the actin binding site to alter affinity. This notion of altered affinities might explain previous results reported by O'leary et al (10) who showed that splice variants in the variable domain of βCaMKII exhibited altered actin binding. In addition, Fink et al. (19) showed that inserting the variable domain of β into αCaMKII increased the interaction of the chimeric construct with actin, but that it did not reconstitute the same magnitude of actin interaction as that found with βCaMKII.
We (11) and others (7, 9, 10) showed using EM approaches that βCaMKII interacts with F-actin to produce bundles of filaments. Using EM tomography, βCaMKII holoenzymes could be clearly identified as cross-linking the actin filaments (11). In this report, we extended this analysis to include higher-order actin filament structures assembled with the γ and δ holoenzymes of CaMKII. We showed previously using single particle EM reconstructions that the four CaMKII isoforms used in the present study assemble into dodecameric complexes that appear largely similar in overall architecture (15). Here we showed δCaMKII formed bundles of actin filaments that were indistinguishable from those formed by βCaMKII. These bundles were somewhat heterogeneous in length and could extend for many μm, sometimes crossing entire grid squares. Interestingly, the actin filaments within these complexes often terminated in a nearly identical fashion that produced distinct blunt ends to the bundles. Since we cannot distinguish CaMKII holoenzymes bound to the ends of filaments, we can only speculate why we observe blunt ends in our experimental system. Perhaps the structure of the bundle provides stability of individual actin filaments, where filaments that extend beyond the length of the bundle are subject to turnover or alternatively are susceptible to sheer force when samples are disturbed during application on the grid or staining. The CaMKII holoenzymes were clearly visible within the bundles and were relatively randomly distributed around the width and length of the bundle axes. However, the distance between filaments within the bundles was less variable and it appeared that CaMKII holoenzymes were helping to maintain a relatively consistent (20-30 nm) spacing between individual filaments within the bundles. This spacing is best appreciated in the movies of the tomographic reconstructions (Supplementary movies 1 and 2). A cartoon model summarizing observations of the relative orientation of CaMKII within the bundles is presented in Fig. 3, middle panel. In contrast to the tightly packed 3D actin bundles formed by β and δCaMKII, γCaMKII produced distinct higher-order, loosely associated large sheet-like structures. In these complexes, the γCaMKII holoenzymes could be identified in layers sandwiching a layer of actin filaments in between. Note that we did not detect multiple layers of actin filaments with kinase molecules sandwiched in between. This would indicate that the growth of these complexes is less restricted in the X-Y axis, but is capped in the Z-axis. The 3D arrangement of these layers and the holoenzymes bracketing them were again best appreciated in tomographic reconstructions (Supplementary movie 3). It is presently unclear why γCaMKII would produce different types of higher-order actin complexes than β or δCaMKII. Perhaps the rigidity in the linker region distinguishes the type of interactions formed with actin-filaments, giving a constraint preference or requirement for the binding interaction. Alternatively, other domains of CaMKII could interface with actin to modify the orientation of binding or serve to sterically restrict the types of interactions that can be formed with actin filaments. A thorough understanding of isoform dependent orientation of binding awaits definitive biochemical identification of the actin binding site(s) in CaMKII. Regardless of the underlying mechanism, to form the type of structures found with γCaMKII suggests an asymmetry in the orientation in which the kinase interacts with the filaments. While it is unclear whether such bundles or layers form inside cells with the different CaMKII isoforms, the data clearly indicates that the β, γ and δ isoforms can form multivalent interactions with actin filaments leading to the potential for organizing the actin cytoskeleton and in unique ways.
Binding of βCaMKII to actin filaments has been documented in a number of reports to be reversed by Ca2+/CaM-binding (7-10). We extended these observations by showing previously that Ca2+/CaM-binding also causes βCaMKII to release G-actin (11) and proposed that CaMKII might serve as a local reservoir for actin monomers that could be released following a Ca2+-influx. In the present study, we tested this hypothesis by evaluating the impact of CaM addition to actin polymerization assays that had been incubated with CaMKII to inhibit the rate of polymerization. As predicted, CaM addition produced an increased rate of actin polymerization that exceeded the rate of polymerization in the absence of CaMKII. The general observation was consistent for all four CaMKII isoforms, where CaM addition to the inhibited reactions led to a burst of actin polymerization. There are a number of possible explanations for the rate of polymerization to increase beyond that found in the absence of CaMKII. First, CaMKII is a dodecamer and we previously showed that 10-12 actin molecules could bind to one holoenzyme (11). This provides the opportunity that, upon addition of an excess of CaM, the release of localized G-actin would increase the probability for nucleation events, initializing multiple new filaments. The localized pool of G-actin then can facilitate growth of these new filaments as well as already formed actin filaments. Incubation with CaMKII might additionally lead to a change in the conformation of actin due either to phosphorylation, which was reported previously (10)], or due to some stabilization of actin caused by its cycle through binding to the kinase that increases its probability for interactions with G- or F-actin. Finally, it is possible that actin remains bound to CaMKII, while in competition for Ca2+/CaM-binding, and this bound actin might have a greater probability for initiating new filament formation. While these possibilities remain to be investigated, it is clear that CaMKII binding to actin slows its polymerization and that the addition of CaM leads to a burst in new filament formation and/or existing filament growth supporting the idea that CaMKII can link increases in Ca2+ concentration (through CaM activation) to alterations in polymerization of the actin cytoskeleton. We suggest that this model is applicable for all four isoforms, each able to bind both G- and F-actin, although we reiterate that the α isoform was not shown to assemble filaments into higher order structures.
Numerous studies have reported an impact of CaMKII on the cellular actin cytoskeleton and interestingly, many of them do not depend on the activity of the kinase. Perhaps most studied example is that of the activity-dependent reorganization of the actin cytoskeleton in neurons. βCaMKII was shown to impact neurite extension and synapse formation in primary cultures and this activity remained evident with catalytically inactive mutants, but required oligomerization of the subunits (9, 19). Similar conclusions were reached for a role of βCaMKII in formation of microspikes in primary cortical cultures (18). In addition to βCaMKII, a role for δCaMKII binding to the actin cytoskeleton and in neurite outgrowth has been proposed (26, 27). CaMKII has been shown to regulate the reorganization of the actin cytoskeleton at synapses, leading to activity-dependent increases in synaptic efficacy (9, 28). In this scenario, Ca2+ increases lead to CaM activation and binding to CaMKII. Based on previous and present results, we propose a model in which Ca2+/CaM-binding leads to dissociation of CaMKII from the actin cytoskeleton and also increases the local pool of G-actin which results in a local increase in actin filament formation and growth. In addition, the release of CaMKII from actin is permissive for its recruitment and binding to other molecules with the synapse, for example in the post-synaptic density (16, 29). When Ca2+ returns to baseline levels, CaM dissociates from the kinase and CaMKII again binds to the actin cytoskeleton to form higher order complexes and to sequester available free actin monomers. Sequestration of free actin would act to further stabilize the actin cytoskeleton by decreasing the probability of treadmilling. While this model was largely developed from results examining actin interactions with βCaMKII, based on the results in the present manuscript we can extend this model to cells that express the γ and δ isoforms of CaMKII as well.
We have shown that each isoform has a unique ability to promote variation in structural assemblies and actin polymerization, which is undoubtedly significant for reorganization of the actin cytoskeleton. Our results contribute evidence to the emerging picture where CaMKII decodes Ca2+-mediated reorganization of the actin cytoskeleton, and that the exploitation of its isoforms provide for a tunable response. We have shown this multifaceted regulation of actin is controlled at the structural level through isoform specific bundling and polymerization of F-action. The CaMKII interaction with actin is also regulated by activation of the kinase (7, 9-11, 19), where Ca2+/CaM affinity (15) and response to Ca2+ oscillations (24) are isoform specific. Tuning is further accomplished through the hetero-oligomerization of CaMKII into holoenzymes that are composed of varying ratios of different isoforms (12, 13). Thus these multifaceted levels of regulation are elegantly controlled by the selectivity and efficiency of isoforms as well as their composition within cells, offering the ability to tune Ca2+-mediated actin reorganization through CaMKII.
Supplementary Material
Supplementary Figure 1. Complete amino acid sequence alignment of CaMKII isozymes used in these studies, rat α, rat β, human γB, and rat δA. The domains in the variable linker region are identified in black boxes as annotated by Tombes, R.M. and Krystal, G.W. (1997) Identification of novel human tumor cell-specific CaMK-II variants. Biochim Biophys Acta 1355: 281-92. An asterisk denotes residues that are identical between isoforms while colons and periods indicate highly conserved and less conserved residues respectively.
Supplementary Video 1. Movie of a tomographic reconstruction of an actin bundle formed through interactions with βCaMKII. The movie shows images scrolling through the Z-axis of the tomogram first at a lower magnification and then at a higher magnification. Scale bar represents 50 nm
Supplementary Video 2. Movie of a tomographic reconstruction of an actin bundle formed through interactions with δCaMKII. The movie shows images scrolling through the Z-axis of the tomogram first at a lower magnification and then at a higher magnification. Scale bar represents 50 nm
Supplementary Video 3. Movie of a tomographic reconstruction of an actin bundle formed through interactions with γCaMKII. The movie shows images scrolling through the Z-axis of the tomogram first at a lower magnification and then at a higher magnification. Scale bar represents 50 nm
Acknowledgements
The authors would like to thank Dr. Matt Swulius and Dr. Jun Liu for on-going discussions concerning the automated collection of high-quality electron tomographic data. They would also like to thank Dr. Andy Bean for critically reviewing the manuscript.
Funding Source Statement: This work was supported by a grant from the NIH/NINDS R01NS026086. MNW also acknowledges an endowment from the William Wheless III Professorship. MMF was supported by NIH Training Grant 5T32GM008280. The Polara electron microscope was supported, in part, through the Structural Biology Center at UTHSC-Houston.
Abbreviations and Textual Footnotes
- CaM
calmodulin
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- G-actin
globular or monomeric actin
- F-actin
filamentous or polymerized actin
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- pyr-actin
pyrene labeled actin
- BSA
bovine serum albumin
- DTT
dithiothreitol
Footnotes
The authors declare no competing financial interest.
Supporting Information (supplemental materials may be accessed free of charge online at http://pubs.acs.org):
Contributor Information
Laurel Hoffman, The Department of Neurobiology and Anatomy, The University of Texas Medical School at Houston.
Madeline M. Farley, The Department of Neurobiology and Anatomy, The University of Texas Graduate School of Biomedical Sciences at Houston, The University of Texas Medical School at Houston
M. Neal Waxham, The Department of Neurobiology and Anatomy, The University of Texas Graduate School of Biomedical Sciences at Houston, The University of Texas Medical School at Houston, 6431 Fannin Street, Room 7.254 MSB Houston, TX 77030 713-500-5621 m.n.waxham@uth.tmc.edu.
References
- 1.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Current biology : CB. 2011;21:R560–569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoenenberger CA, Mannherz HG, Jockusch BM. Actin: from structural plasticity to functional diversity. European journal of cell biology. 2011;90:797–804. doi: 10.1016/j.ejcb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Rottner K, Stradal TE. Actin dynamics and turnover in cell motility. Current opinion in cell biology. 2011;23:569–578. doi: 10.1016/j.ceb.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annual review of biophysics and biomolecular structure. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 7.Ohta Y, Nishida E, Sakai H. Type II Ca2+/calmodulin-dependent protein kinase binds to actin filaments in a calmodulin-sensitive manner. FEBS letters. 1986;208:423–426. doi: 10.1016/0014-5793(86)81061-4. [DOI] [PubMed] [Google Scholar]
- 8.Shen K, Meyer T. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science. 1999;284:162–166. doi: 10.1126/science.284.5411.162. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto K, Narayanan R, Lee SH, Murata K, Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Leary H, Lasda E, Bayer KU. CaMKIIbeta association with the actin cytoskeleton is regulated by alternative splicing. Molecular biology of the cell. 2006;17:4656–4665. doi: 10.1091/mbc.E06-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanabria H, Swulius MT, Kolodziej SJ, Liu J, Waxham MN. {beta}CaMKII regulates actin assembly and structure. The Journal of biological chemistry. 2009;284:9770–9780. doi: 10.1074/jbc.M809518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hudmon A, Schulman H. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annual review of biochemistry. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- 13.Brocke L, Chiang LW, Wagner PD, Schulman H. Functional implications of the subunit composition of neuronal CaM kinase II. The Journal of biological chemistry. 1999;274:22713–22722. doi: 10.1074/jbc.274.32.22713. [DOI] [PubMed] [Google Scholar]
- 14.Kolb SJ, Hudmon A, Ginsberg TR, Waxham MN. Identification of domains essential for the assembly of calcium/calmodulin-dependent protein kinase II holoenzymes. The Journal of biological chemistry. 1998;273:31555–31564. doi: 10.1074/jbc.273.47.31555. [DOI] [PubMed] [Google Scholar]
- 15.Gaertner TR, Kolodziej SJ, Wang D, Kobayashi R, Koomen JM, Stoops JK, Waxham MN. Comparative analyses of the three-dimensional structures and enzymatic properties of alpha, beta, gamma and delta isoforms of Ca2+-calmodulin-dependent protein kinase II. The Journal of biological chemistry. 2004;279:12484–12494. doi: 10.1074/jbc.M313597200. [DOI] [PubMed] [Google Scholar]
- 16.Colbran RJ. Targeting of calcium/calmodulin-dependent protein kinase II. The Biochemical journal. 2004;378:1–16. doi: 10.1042/BJ20031547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tombes RM, Krystal GW. Identification of novel human tumor cell-specific CaMK-II variants. Biochimica et biophysica acta. 1997;1355:281–292. doi: 10.1016/s0167-4889(96)00141-3. [DOI] [PubMed] [Google Scholar]
- 18.Lin YC, Redmond L. CaMKIIbeta binding to stable F-actin in vivo regulates F-actin filament stability. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15791–15796. doi: 10.1073/pnas.0804399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fink CC, Meyer T. Molecular mechanisms of CaMKII activation in neuronal plasticity. Current opinion in neurobiology. 2002;12:293–299. doi: 10.1016/s0959-4388(02)00327-6. [DOI] [PubMed] [Google Scholar]
- 20.Caran N, Johnson LD, Jenkins KJ, Tombes RM. Cytosolic targeting domains of gamma and delta calmodulin-dependent protein kinase II. The Journal of biological chemistry. 2001;276:42514–42519. doi: 10.1074/jbc.M103013200. [DOI] [PubMed] [Google Scholar]
- 21.Putkey JA, Waxham MN. A peptide model for calmodulin trapping by calcium/calmodulin-dependent protein kinase II. The Journal of biological chemistry. 1996;271:29619–29623. doi: 10.1074/jbc.271.47.29619. [DOI] [PubMed] [Google Scholar]
- 22.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. Journal of structural biology. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 23.Shen K, Teruel MN, Subramanian K, Meyer T. CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron. 1998;21:593–606. doi: 10.1016/s0896-6273(00)80569-3. [DOI] [PubMed] [Google Scholar]
- 24.Bayer KU, De Koninck P, Schulman H. Alternative splicing modulates the frequency-dependent response of CaMKII to Ca(2+) oscillations. The EMBO journal. 2002;21:3590–3597. doi: 10.1093/emboj/cdf360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao LH, Stratton MM, Lee IH, Rosenberg OS, Levitz J, Mandell DJ, Kortemme T, Groves JT, Schulman H, Kuriyan J. A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin-dependent kinase II holoenzyme. Cell. 2011;146:732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson LD, Willoughby CA, Burke SH, Paik DS, Jenkins KJ, Tombes RM. delta Ca(2+)/Calmodulin-dependent protein kinase II isozyme-specific induction of neurite outgrowth in P19 embryonal carcinoma cells. Journal of neurochemistry. 2000;75:2380–2391. doi: 10.1046/j.1471-4159.2000.0752380.x. [DOI] [PubMed] [Google Scholar]
- 27.Donai H, Murakami T, Amano T, Sogawa Y, Yamauchi T. Induction and alternative splicing of delta isoform of Ca(2+)/calmodulin-dependent protein kinase II during neural differentiation of P19 embryonal carcinoma cells and during brain development. Brain research. Molecular brain research. 2000;85:189–199. doi: 10.1016/s0169-328x(00)00221-7. [DOI] [PubMed] [Google Scholar]
- 28.Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto K, Bosch M, Hayashi Y. The roles of CaMKII and F-actin in the structural plasticity of dendritic spines: a potential molecular identity of a synaptic tag? Physiology. 2009;24:357–366. doi: 10.1152/physiol.00029.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Complete amino acid sequence alignment of CaMKII isozymes used in these studies, rat α, rat β, human γB, and rat δA. The domains in the variable linker region are identified in black boxes as annotated by Tombes, R.M. and Krystal, G.W. (1997) Identification of novel human tumor cell-specific CaMK-II variants. Biochim Biophys Acta 1355: 281-92. An asterisk denotes residues that are identical between isoforms while colons and periods indicate highly conserved and less conserved residues respectively.
Supplementary Video 1. Movie of a tomographic reconstruction of an actin bundle formed through interactions with βCaMKII. The movie shows images scrolling through the Z-axis of the tomogram first at a lower magnification and then at a higher magnification. Scale bar represents 50 nm
Supplementary Video 2. Movie of a tomographic reconstruction of an actin bundle formed through interactions with δCaMKII. The movie shows images scrolling through the Z-axis of the tomogram first at a lower magnification and then at a higher magnification. Scale bar represents 50 nm
Supplementary Video 3. Movie of a tomographic reconstruction of an actin bundle formed through interactions with γCaMKII. The movie shows images scrolling through the Z-axis of the tomogram first at a lower magnification and then at a higher magnification. Scale bar represents 50 nm