Abstract
The development of mature B cells from hematopoietic stem cells is a strictly orchestrated process involving multiple regulatory genes. The transcription factor Sox4 is required for this process but its role has not been systematically studied, and the underlying mechanisms remain unknown. To determine when and how Sox4 functions in the stepwise process of B cell development, we used mice harboring conditional null alleles for Sox4 and a Cre transgene. Sox4 deletion in hematopoietic stem cells almost entirely eliminated pro-B cells in both fetal livers and adult bone marrow, resulting in a severe deficiency in later stage B cells including circulating mature B cells. Sox4-deficient pro-B cells, particularly those expressing the stem cell factor receptor c-Kit, readily underwent apoptosis, and even more so when c-Kit activity was inhibited by imatinib. C-Kit-expressing pro-B cells showed decreased activation of the c-Kit downstream protein Src upon Sox4 deletion. Likewise, the level of the anti-apoptotic Bcl2 protein was decreased in residual pro-B cells, and its restoration using a Bcl2 transgene not only allowed partial rescue of pro-B cell survival, but also B cell maturation in the absence of Sox4. Our findings indicate that Sox4 is required for the survival of pro-B cells and may functionally interact with c-Kit and Bcl2.
Introduction
B cells play pivotal roles in humoral immunity and are one of the key components of the immune system. Like other blood cell types, mature B cells arise from self-renewing pluripotent hematopoietic stem cells (HSCs) through a stepwise process involving coordinated cell proliferation along with progressive lineage commitment and differentiation. In the B cell lineage, HSCs first develop into lymphoid-primed multipotent progenitors (LMPPs), which have lost their self-renewal ability but remain multipotent, and then into common lymphocyte progenitors (CLPs), which in turn develop into B cells, T cells, natural killer cells, and dendritic cells (1). The first B cell specific progenitors arising from CLPs are pre-pro-B cells, which sequentially develop into pro-B, pre-B, immature, and ultimately mature B cells.
B cell development requires appropriate orchestration of a network of regulatory genes involved in cell survival, proliferation, and differentiation (2,3). Particularly in early B cell development, several key transcription factors act in a hierarchical order to precisely control the expression of critical genes (4,5). Pu.1 is involved in the hematopoietic lineage fate decision at the branchpoint of myeloerythroid and myelolymphoid progenitor populations (6). Ikaros is a key factor in B lineage specification and commitment. Ikaros deficient or hypomorphic mutant mice have severe defects in the development of the lymphoid system (7,8). Ebf1 and Pu.1 activate IL-7R, the first gene that distinguishes CLPs from LMPPs (9–11). E2A functions upstream of EBF and the two transcription factors collaborate to activate Pax5 expression (12,13). Pax5 is indispensable in B cell lineage commitment in that it activates B lineage specific genes and represses lineage inappropriate genes (14,15).
Programmed cell death frequently occurs during B cell development, but despite intensive studies, the molecular mechanisms that control this process are still elusive. Accumulating evidence has demonstrated that members of the Bcl2 protein family, including Bcl2 (16), Bcl-xL (17,18), Mcl1 (19), Bax (20), Bik (21), and Bim (22) are critical for B cell survival in vivo and in vitro. Other factors, such as Stat5 and Dicer1, are also involved in maintaining B cell survival. Stat5 has been shown to mediate the survival function of IL-7 signaling during B cell development (23,24). Conditional deletion of Dicer1 in the earliest stage of B cell development results in an almost complete absence of pre-B cells owing to a massive increase in apoptosis in pre-B cells (25,26). In adult mice, signaling by stem cell factor (SCF) and its receptor c-Kit is critical in pro-B and pro-T cell development (27). The downstream signaling pathways activated by c-Kit include activation of PI 3 and Src kinases. However, c-Kit mediated Src kinase activation but not PI 3 kinase activation is important for adult B cell development, likely through signaling events related to cell proliferation and cell survival (28).
Sox4 is a member of the SOX (SRY-related HMG box) transcription factor family and has been shown to play important roles in embryonic development and differentiation (29). Germline deletion of Sox4 is embryonically lethal in mice (30). Embryos with this deletion died at day 14 of development due to circulation failure as a result of malformation of the semilunar valves and ventricular septum. In vitro culture of liver cells from Sox4−/− embryos failed to generate B cells in presence of IL-7. Reconstitution of lethally irradiated adult mice with the Sox4−/− fetal liver cells showed a stringent arrest of donor B cell development at the pro-B cell stage. These findings indicated that Sox4 is indispensable for B cell development in the fetal liver. However, how Sox4 deficiency causes the fetal B cell developmental arrest and what role Sox4 plays in adult B cell development remain unknown. Sox4 has since then been shown to be critically required for cell survival and differentiation in many cell lineages other than B cells in embryonic development and postnatal life, and to act largely in redundancy with its close relatives Sox11 and Sox12 (31–35). In the study we report here, we used Sox4fl/fl mice and Vav-Cre recombinase to investigate the effect of Sox4 deletion at HSCs on B cell development. Our results showed that Sox4 was essential for pro-B cell survival and might functionally interact with c-Kit and Bcl2.
Materials and Methods
Mice
Mice with loxP sites flanking the entire coding region of the Sox4 gene were described previously (36). Vav-Cre mice were provided by Dr. Dimitris Kioussis at the National Institute for Medical Research, The Ridgeway, London (37). H2K-Bcl2 transgenic mice were provided by Dr. Irving Weissman at Stanford University, Stanford, CA (38). Genotyping was performed by PCR using genomic DNA extracted from mouse tails. All mice were bred and maintained in a specific pathogen-free animal facility at The University of Texas MD Anderson Cancer Center. All mouse experiments were performed in accordance with federal laws as well as guidelines of the National Institutes of Health, and protocols were approved by the MD Anderson Animal Care and Use Committee.
Imatinib treatment
To inhibit the c-Kit signaling pathway, 4- to 5-week-old mice were given intraperitoneal injections of 100 mg/kg imatinib (LC Laboratories, Woburn, MA) twice daily in a volume of 100 μL of PBS for 2, 3, or 7 consecutive days, as indicated. Mice were euthanized the day after the last injection, and samples were harvested for flow cytometric analysis.
Cell surface staining, flow cytometry, and cell sorting
Single-cell suspensions were prepared at the time of autopsy from fetal liver, bone marrow (femurs and tibias), and spleen. All samples were incubated with ice-cold red blood cell lysis buffer with ammonium chloride for 4 min in the dark before staining with a combination of mouse-specific antibodies at 4°C for 15 min in staining solution (PBS supplemented with 1% BSA). Blood was collected from the hearts of the mice in 15-mL Falcon tubes (BD Biosciences, San Jose, CA) containing 50 μL of 0.5 M EDTA in PBS. After two rounds of red blood cell lysis, the white blood cells were subjected to staining.
Antibodies used for flow cytometric analysis included the following: fluorescein isothiocyanate (FITC)–anti-IgM (Jackson ImmunoResearch, West Grove, PA); FITC-anti-CD21 (8D9), phycoerythrin (PE)–anti-CD11c (N418), PE-anti-CD25 (PC61,5), PE-anti-NK1.1 (PK136), allophycocyanin (APC)–anti-AA4.1 (AA4.1), PerCP-Cy5.5-anti-B220 (RA3-6B2), biotin–anti-BP1(FG35.4), and eFlour450-Streptavidin, all from eBioscience (San Diego, CA); FITC-anti-CD24 (M1-69), FITC-anti-IgD (11-26c.2a), PE-anti-CD43 (S7), PE-anti-Ter119 (Ter-119), PE-anti-c-kit (2B8), APC-anti-CD19 (1D3), biotin–anti-Cxcr4 (2B11), and biotin–anti-IL-7R (B12-1), all from BD Biosciences. Biotinylated antibodies were detected with FITC-, APC-, PerCP-Cy5.5-, eFlour450-, or PE-conjugated streptavidin (BD Biosciences). Samples were acquired on a FACSCalibur or Influx high speed sorter (BD), and data were analyzed with FlowJo software (Tree Star, Ashland, OR). The absolute number of each B cell population was derived from the total number of bone marrow cells per mouse and the percentage of the population in question.
Annexin-V and intracellular staining
For cell apoptosis analysis, the staining buffers containing surface antibodies were carefully removed before biotin-conjugated annexin-V was applied according to the manufacturer’s instructions (BD Biosciences). For intracellular detection of Bcl2 (2870), total Src (2109), or p-Src (2101, Cell Signaling Technology, Danvers, MA), cells were incubated with surface antibodies as indicated. After being washed with staining solution, cells were incubated in fixation solution (eBioscience 00-8222-49) for 20 min and subsequently washed twice with permeabilization buffer (eBioscience 00-8333-56). Cells were then incubated with specific primary antibodies for 30 min at room temperature and then incubated for 20 min on ice with PE-, APC-, or FITC-conjugated anti-rabbit second antibodies.
Semi-quantitative RT-PCR
Total RNA was isolated from freshly sorted bone marrow pre-pro-B or pro-B cells with TRIzol (Invitrogen, Carlsbad, CA). Fifty nanograms of total RNA were applied for first-strand cDNA synthesis by using the SuperScript II first-strand synthesis kit (Invitrogen). Serially diluted cDNA was used for 28–32 cycles of PCR with gene-specific primer pairs (table S1). PCR products were then separated in 1.5% agarose gel and visualized by a Dyversity imaging system (Syngene, Frederick, MD).
Statistical analysis
The Student t test, assuming unequal variances between the two samples, was applied to determine the significance of difference between experimental mice and their littermates. P values were considered significant when the probability of a difference was < 0.05.
Results
Sox4 is required for pro-B cell development
To determine at what step in normal B cell development Sox4 is first required, we generated mouse strains harboring the Sox4 conditional null allele and the Vav-Cre transgene. The Vav-Cre transgene is under the control of Vav1 gene promoter and is expressed in HSCs (39,40). Sox4fl/flVav-Cre mice were present at normal Mendelian ratios and externally indistinguishable from their littermate controls (Sox4fl/+Vav-Cre and Sox4+/+Vav-Cre) at birth and until at least 1 year of age.
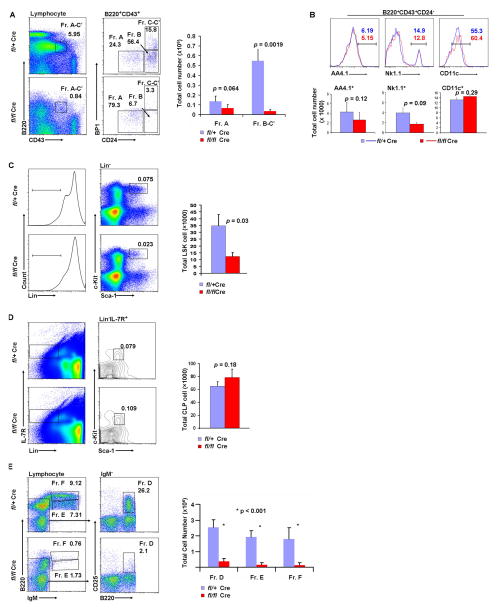
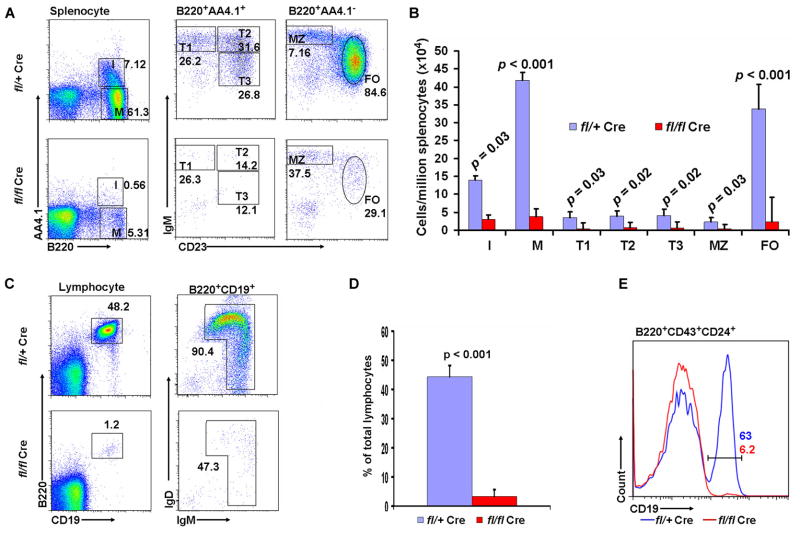
Flow cytometric analysis revealed that the number of pro-B cells, including fraction B (B220+CD43+CD24+BP1−) and fraction C-C′ (B220+CD43+CD24+BP1+), was much lower in the bone marrow of adult Sox4fl/flVav-Cre mice (Hardy’s nomenclature for subpopulations of developing B cells (41). While control littermates had 0.55 million proB cells, the Sox4fl/flVav-Cre mice had only 0.04 million (p = 0.0019; Fig. 1A). The number of fraction A cells (i.e., pre-pro-B, B220+CD43+CD24−BP1−) was slightly decreased (0.14 million in control mice versus 0.07 million in Sox4fl/flVav-Cre mice; p = 0.064). Fraction A cells are heterogeneous and can give rise to, in addition to B cells, natural killer (NK) and plasmacytoid dendritic cells (pDCs) (42–44). The AA4.1+ subset of fraction A cells contains B lineage precursors (45), the NK1.1+ subset produces NK cells and the CD11c+ subset generates pDCs (46,47). The numbers of AA4.1+, NK1.1+, and CD11c+ fraction A cells in Sox4fl/flVav-Cre bone marrow was comparable to those in the corresponding subsets of control mice (Fig. 1B). These data indicate that deletion of Sox4 significantly affected pro-B cells but only minimally affected pre-pro-B cells. Moreover, a moderate decrease in the number of Lin−c-Kit+Sca-1+ primitive HSCs (36 ×103 in control mice versus 12 × 103 in Sox4fl/flVav-Cre mice; p = 0.03; Fig. 1C) and a minimal increase in the number of CLPs (Lin−IL-7R+c-Kit+Sca-1+; 65 × 103 in control mice versus 78 × 103 in Sox4fl/flVav-Cre mice; p = 0.18; Fig. 1D) suggested that the marked reduction in pro-B cells was unlikely to be caused by changes in earlier stage cells (HSCs, CLPs, and pre-pro-B cells). As a consequence of the pro-B cell depletion, bone marrow fraction D (pre-B, IgM−B220+CD25+), fraction E (immature B, IgM+B220low), and fraction F (mature B, IgM+B220high) cells were almost completely eliminated (Fig. 1E). In the spleen, the percentages of immature (B220+AA4.1+, including transitional T1, T2, and T3) and mature (B220+AA4.1−, including marginal zone and follicular) B cells were severely reduced (7.12% and 61.3% in control mice versus 0.56% and 5.31% in Sox4fl/flVav-Cre mice, respectively; Fig. 2A–B). As shown in Fig. 2C–D, B220+CD19+ peripheral blood B cells were essentially absent. However, the peripheral blood residual B cells in Sox4fl/flVav-Cre mice expressed IgM and IgD on the surface, indicating that these cells were able to develop into mature B cells.
Figure 1. B cell development in the bone marrow of Sox4fl/flVav-Cre mice.
Flow cytometric analyses were performed to assess B cells at different developmental stages in Sox4fl/+Vav-Cre (fl/+Cre) and Sox4fl/flVav-Cre (fl/flCre) mice. The numbers in the plots are the percentages of the indicated cell groups. Bar graphs show the total number of cells per mouse presented as mean + SD with p values indicated. (A) Bone marrow B220+CD43+ lymphocytes were analyzed for fraction (Fr.) A (pre-pro-B; B220+CD43+Cd24−), fraction B (early pro-B; B220+CD43+Cd24+BP1−), and fraction C–C′ (late pro-B; B220+CD43+CD24+BP1+). Note that the percentage of fraction A cells was 24.3% in Sox4fl/+Vav-Cre mice and 79.3% in Sox4fl/flVav-Cre mice (flow cytometric plot). This was mostly due to the decrease in fractions B–C′ cells; the total number of fraction A cells was 0.14 million in Sox4fl/+Vav-Cre mice and 0.07 million in Sox4fl/flVav-Cre mice (bar graph). (B) Bone marrow fraction A cells were analyzed for the expression of NK1.1, AA4.1, and CD11c. (C) Bone marrow Lin− cells were analyzed for the expression of c-Kit and Sca-1 to assess the number of hematopoietic stem cells. Lin: lineage markers, including CD11b, B220, CD4, CD8, Ter119, NK1.1, Gr-1, and CD19. (D) Bone marrow Lin− and IL-7R+ cells were analyzed for the expression of c-Kit and Sca-1 to assess the number of common lymphocyte progenitors. (E) Bone marrow lymphocytes were analyzed for fractions D (pre-B; IgM−B220+CD25+), E (immature B; IgM+B220low), and F (mature B; IgM+B220high) B cells (n = 3–5 mice per group).
Figure 2. B cell development in the spleen, peripheral blood, and fetal liver of Sox4fl/flVav-Cre mice.
(A) Splenocytes were analyzed for immature (I, B220+AA4.1+) and mature (M, B220+AA4.1−) B cells. The immature B cells were further analyzed for transitional T1 (B220+AA4.1+IgM+CD23−), T2 (B220+AA4.1+IgM+CD23+), and T3 (B220+AA4.1+IgM−D23+) cells and the mature B cells were further analyzed for marginal zone (MZ, B220+AA4.1−IgM+CD23−) and follicular (FO, B220+AA4.1−IgM+CD23+) B cells. (B) The number of individual cells per million spleen lymphocytes. (C) Peripheral blood B cells (B220+CD19+) were analyzed for the expression of IgM and IgD. (D) Percentage of B cells in peripheral blood. (E) Fetal liver cells from day E13.5 embryos were analyzed for B220+CD43+CD24+CD19+ pro-B cells (data are representative of 2 independent experiments). Gating strategies and abbreviations are the same as in Figure 1 and n = 3 – 5 mice per group unless otherwise specified.
It was shown previously that Sox4−/− fetal liver hematopoietic cells fail to develop into mature B cells in recipient adult mice after transplantation (30). However, it was not known which early stage B cells were absent in the fetal livers of the Sox4-null embryos. We found that pro-B cells (B220+CD43+CD24+CD19+) were almost undetectable in the fetal livers of day 13.5 Sox4fl/flVav-Cre embryos (Fig. 2E). This result suggested that deletion of the Sox4 gene in embryonic HSCs resulted in the abrogation of fetal liver proB cells and that this abrogation was responsible for the B cell deficiency in fetal liver cell-reconstituted adult bone marrow and in vitro culture.
To exclude the possibility of incomplete or escape of Sox4 deletion in the residual B cells in Sox4fl/flVav-Cre mice, we sorted out the minute populations of pre-pro-B cells and pro-B cells and analyzed the Sox4 mRNA levels. Sox4 mRNA was not detectable in bone marrow fraction A cells (Fig. 3A) or residual pro-B cells (Fig. 3B) in Sox4fl/flVav-Cre mice by RT-PCR. Real-time RT-PCR showed that Sox4 mRNA in pro-B cells was decreased 133-fold in Sox4fl/flVav-Cre mice (Fig. 3C, left). We also crossed the Sox4fl/flVav-Cre mice with Rosa26–enhanced yellow fluorescent protein (EYFP) Cre reporter mice to assess Cre activity in residual B cells. More than 90% of bone marrow pre-pro-B (fraction A) cells and more than 95% of peripheral blood residual B220+CD19+ B cells were EYFP positive (Fig. 3D–E), implying that the floxed Sox4 allele had been deleted in these cells. The relative low percentages of EYFP+ cells in pro-B (fractions B-C′) and pre-B (fraction D) cells of Sox4fl/flVav-Cre mice compared with control mice were expected because of the high frequency of apoptosis in these cells (see below); EYFP+ (or GFP+) cells become EYFP− (or GFP−) cells when undergoing apoptosis (48). The presence of EYFP+ B cells in peripheral blood suggested that a minor portion of proB cells overcame the effect of Sox4 deficiency and continued to develop into mature B cells.
Figure 3. Sox4 deletion in various stage B cells in Sox4fl/flVav-Cre mice.
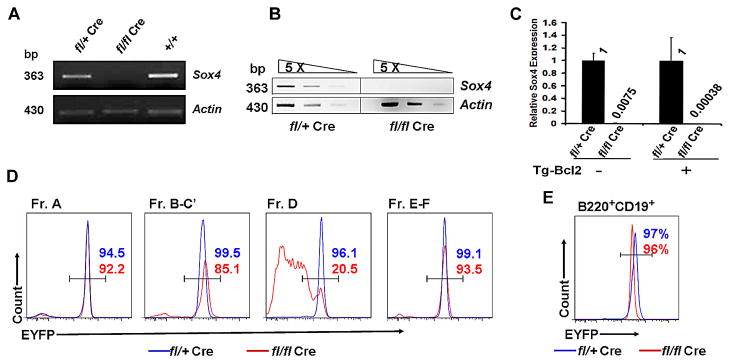
(A and B) Total RNA prepared from bone marrow FACS-sorted pre-pro-B cells (A) and pro-B cells (B) was subjected to RT-PCR to assess Sox4 expression. cDNA of pro-B cells was 5-times serially diluted for semiquantitative RT-PCR (B). (C) Quantitative RT-PCR analysis of Sox4 mRNA expression in pro-B cells of Sox4fl/+Vav-Cre and Sox4fl/flVav-Cre mice with or without Bcl2 transgenic overexpression. (D and E) Fractions A–F (D) and peripheral blood B cells (B220+CD19+; E) from Sox4f/+Vav-CreRosa26EYFP (blue line) and Sox4fl/flVav-CreRosa26EYFP (red line) mice were subjected to flow cytometric analysis to assess EYFP expression, which serves as an indicator for Sox4 deletion.
Sox4 is vital for the survival of pro-B and pre-B cells
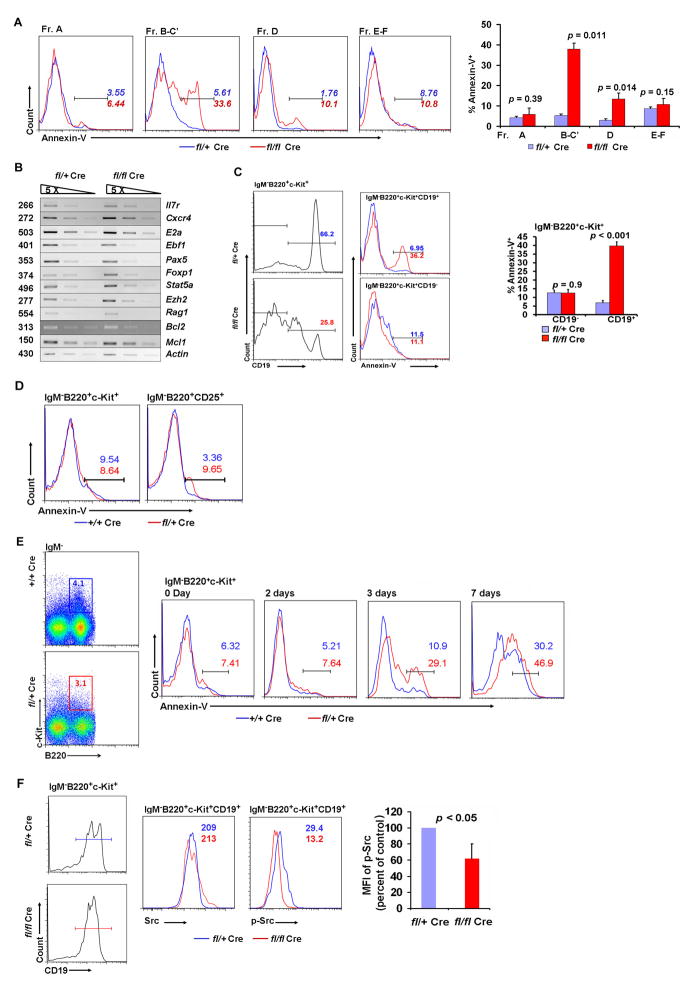
To measure apoptosis in minute populations of B cells, particularly in mice with B cell deficiency caused by Sox4 deletion, we used highly sensitive multi-color flow cytometry with annexin V staining (49,50). Annexin V staining may vary in B cells with different statuses of activation (51), but is a reliable marker for apoptosis when comparing cells at similar stages in development (52,53). As shown in Fig. 4A, a minimal increase in the frequency of apoptosis in pre-pro-B (fraction A) cells was detected in Sox4fl/flVav-Cre mice (3.55% versus 6.44%; p = 0.39), whereas the frequency of apoptosis in pro-B (fraction B–C′) and pre-B (fraction D) cells increased dramatically (5.61% versus 33.6%; p = 0.011; and 1.76% versus 10.1%; p = 0.014, respectively). In contrast, bone marrow immature and mature (fractions E–F) B cells of Sox4fl/flVav-Cre mice had a frequency of apoptosis comparable to controls. These results suggested that Sox4 was crucial for maintaining the survival of pro-B and pre-B cells but not required for the survival of later stage B cells.
Figure 4. Effect of Sox4 deletion and c-Kit inhibition on the apoptosis of B cells.
(A) Apoptotic frequency measured as the percentage of annexin V positive cells in fractions A to F in Sox4fl/+Vav-Cre (blue line) and Sox4fl/flVav-Cre (red line) mice. The histogram shows one of the representative experiments whereas the bar graph shows results (mean + S.D. with p values indicated) from 5 experiments. (B) The expression of critical B cell developmental genes in pro-B cells was determined by semiquantitative RT-PCR. Also included were two anti-apoptotic genes, Bcl2 and Mcl1. The actin cDNA was used as control. Three cDNA concentrations with 5-time serial dilution were used for each gene. The size of the PCR products is indicated in base pairs. (C) The frequency of apoptosis in c-Kit+ pro-B cells (IgM−B220+c-Kit+CD19+) was measured by the percentage of annexin V positive cells in Sox4fl/+Vav-Cre and Sox4fl/flVav-Cre mice. The apoptotic frequencies of c-Kit+ non-pro-B cells (IgM−B220+c-Kit+CD19−) are shown for comparison. The histogram shows one of the representative experiments, whereas the bar graph shows results (mean + SD with p values indicated) from 5 experiments. (D) Comparison of apoptosis in c-Kit+ pro-B cells and CD25+ pre-B cells between Sox4+/+Vav-Cre and Sox4fl/+Vav-Cre mice. (E) Frequency of apoptosis in bone marrow pro-B cells (IgM−B220+CD19+ c-Kit+) after imatinib treatment. Percentages of annexin V positive cells from mice that had received imatinib injections for the indicated number of days were determined by flow cytometry. Data are representative of 2 mice per group. (F) Flow cytometric analysis of total Src and phosphorylated Src (p-Y416) in c-Kit+ pro-B cells from Sox4fl/+Vav-Cre and Sox4fl/flVav-Cre mice. The numbers represent median fluorescence intensity (MFI). Gating strategies and abbreviations are the same as in Figures 1 and 2. The bar graph shows the percentage of p-Src in Sox4fl/flVav-Cre pro-B cells compared with that in Sox4fl/+Vav-Cre pro-B cells. n = 3–5 mice per group unless otherwise specified.
Sox4 cooperates with the c-Kit signaling pathway to regulate pro-B cell survival
To find out whether Sox4 exerts its function by regulating the genes known to be critical in early B cell development, we sorted out the small number of residual pro-B cells and characterized the expression of these genes by semiquantitative RT-PCR. Of the genes studied, including IL-7r, Cxcr4, E2a, Ebf1, Pax5, Foxp1, Stat5a, Ezh2, and Rag1, none showed a significant difference in expression between Sox4fl/flVav-Cre mice and their control littermates (Fig. 4B). Since Sox4 deficiency caused pro-B cell apoptosis, we also determined the mRNA levels of two anti-apoptotic genes, Bcl2 and Mcl1, which were also known to be involved in early B cell development, and found no significant difference in them, either (Fig. 4B). However, the possibility still existed that Sox4 regulates some of these gene products at the post-transcriptional level.
We analyzed apoptosis in residual c-Kit+ pro-B cells from the bone marrow of the Sox4fl/flVav-Cre mice. Lack of Sox4 expression significantly increased cell death in c-Kit+ pro-B cells (IgM−B220+c-Kit+CD19+; 6.95% versus 36.2%; p < 0.001) but not in c-Kit+ non-pro-B cells (IgM−B220+c-Kit+CD19−; 11.5% versus 11.1%; p = 0.9; Fig. 4C). Though deletion of one copy of the Sox4 gene (Sox4fl/+Vav-Cre) resulted in only minimal apoptosis of c-Kit+ pro-B cells (Fig. 4D, left), it caused nearly 3 times the frequency of apoptosis in pre-B cells compared with control (3.36% versus 9.65%; Fig. 4D, right), which did not express c-Kit. We hypothesized that c-Kit compensates for the deleterious effect of Sox4 deletion in pro-B cells. We administered 100 μg of the c-Kit inhibitor imatinib by intraperitoneal injection twice daily for 2, 3, or 7 days. Whereas the frequency of apoptosis in c-Kit+ pro-B cells was comparable between Sox4 fl/+Vav-Cre and Sox4+/+Vav-Cre mice after 0 and 2 days of injection, the difference in the frequency of apoptosis was 3-fold (29.1 % versus 10.9%) and 1.5-fold (46.9% versus 30.2%) after 3 and 7 days of injection, respectively (Fig. 4E). Since Src kinase is downstream of c-Kit and plays a critical role in pro-B cell development (28,54), we determined the levels of total Src and phosphorylated Src (p-Y416) in pro-B cells from Sox4fl/flVav-Cre and Sox4 fl/+Vav-Cre mice by flow cytometric analysis. We found that there was no overt difference in total Src, but phosphorylated Src (p-Y416) was significantly reduced upon Sox4 deletion (median fluorescence intensity [MFI] 29.4 versus 13.2; Fig. 4F).
B cell development in Sox4fl/flVav-Cre mice can be partially rescued by Bcl2
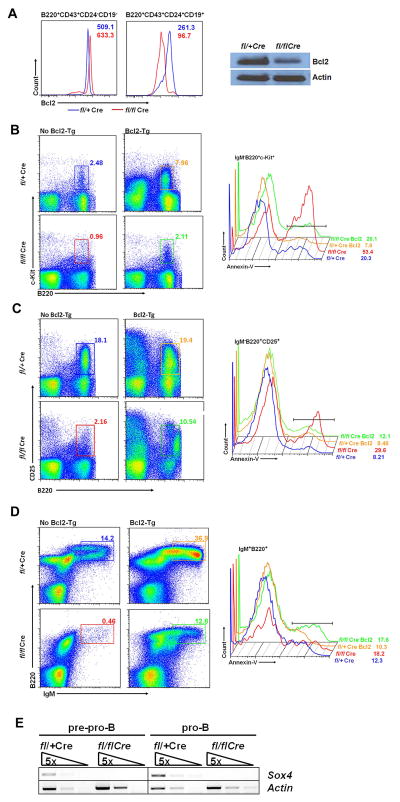
Since the frequency of apoptosis in Sox4-deficient pro-B cells was significantly increased and lack of the antiapoptosis protein Bcl2 was reported to increase the susceptibility of pro-B cells to proapoptotic agents (55,56), we analyzed the level of Bcl2 protein in Sox4-deficient pro-B cells. Although Bcl2 mRNA did not show significant change (Fig. 4B), Bcl2 protein was reduced in pro-B cells (MFI 261.3 versus 96.7), but not in pre-pro-B cells (MFI 509.1 versus 633.3; Fig. 5A, left) in the Sox4fl/flVav-Cre mice according to flow cytometric analysis. A reduction by 66% in Bcl2 protein was detected by Western blot in B220+ bone marrow B cells (Fig. 5A, right). These data indicated that the proapoptotic effect of Sox4 deficiency in pro-B cells was associated with reduction of Bcl2 at the protein level.
Figure 5. Effect of Bcl2 on B cell apoptosis induced by Sox4 deletion.
(A) Left pannel shows flow cytometry analysis of Bcl2 expression in pre-pro-B (B220+CD43+CD24−CD19−) and pro-B cells (B220+CD43+CD24+CD19+) from Sox4fl/+Vav-Cre (blue line) and Sox4fl/flVav-Cre (red line) mice. The numbers represent median fluorescence intensity. Data represent 3 independent experiments. Right panel shows Western blot results of the B220+ bone marrow cells from these mice. (B, C, and D) Frequency of apoptosis measured as the percentage of annexin V positive cells in bone marrow c-Kit+ pro-B cells (B), pre-B cells (C), and IgM+ (immature and mature) B cells (D) with or without forced Bcl2 expression. Data represent 3 mice per group. Gating strategies and abbreviations are the same as in Figures 1 and 2. (E) Semiquantitative RT-PCR of Sox4 expression in pre-pro-B (B220+CD43+CD24−CD19−) and pro-B (B220+CD43+CD24+CD19+) cells of the Sox4fl/+Vav-Cre and Sox4fl/flVav-Cre mice with forced Bcl2 expression.
Forced expression of Bcl2 in Bcl2-transgenic (Bcl2-Tg) mice has been shown to restore hematopoietic cell development in a number of gene-deficient mice (25). We crossed the Sox4fl/flVav-Cre mice with H2K-Bcl2-transgenic mice (38) to test whether forced Bcl2 expression could rescue the B cell deficiency caused by Sox4 deletion. Flow cytometric analysis showed that the number of B cells of various stages was increased in the Sox4fl/flVav-CreBcl2-Tg mice compared with the Sox4fl/flVav-Cre mice (Table 1). Comparison of the frequency of apoptosis in B cells of various stages from Sox4fl/+Vav-Cre and Sox4fl/flVav-Cre mice with or without Bcl2-Tg showed that the increase in the number of B cells in Bcl2-Tg mice was associated with a reduction in the frequency of apoptosis in pro-B cells (Fig. 5B) and pre-B cells (Fig. 5C) whereas the frequency of apoptosis in immature and mature B cells remained essentially the same (Fig. 5D). The rescued B cells did not have detectable Sox4 mRNA expression by semi-quantitative RT-PCR (Fig. 5E) and real-time RT-PCR showed a 2632-fold reduction in Sox4 mRNA levels in pro-B cells of Sox4fl/flVav-Cre mice (Fig. 3C, right), yet the rescued pro-B cells retained the normal expression pattern of critical B cell development genes listed in Fig. 4B. These results suggested that forced Bcl2 expression could partially restore B cell development by reducing apoptosis caused by Sox4 deficiency. Noticeably, bone marrow IgM+ late stage B cells (fractions E and F) and peripheral blood B220+CD19+ B cells were increased in number as well (Fig. 5D; Table 1), suggesting that the rescued pro-B cells could pass through the check point from pre-BCR+ pre-B cells to BCR+ immature and mature B cells without the function of Sox4.
Table 1.
Effect of forced Bcl2 expression on subpopulations of B cells
| Bone marrow B cells (millions)
|
Peripheral blood B cells (%) | ||||
|---|---|---|---|---|---|
| CD19+ pro-B | c-Kit+ pro-B | Pre-B | Immature + mature | ||
| fl/+ Vav-Cre | 0.912±0.087 | 0.354±0.045 | 3.544±0.285 | 2.576±0.336 | 45.85±9.67 |
| fl/fl Vav-Cre | 0.006±0.001 | 0.085±0.013 | 0.173±0.028 | 0.314±0.041 | 1.81±0.29 |
| fl/+ Vav-Cre Bcl2 | 0.766±0.103 | 0.823±0.147 | 2.256±0.198 | 8.674±1.201 | 59.36±10.83 |
| fl/fl Vav-Cre Bcl2 | 0.054±0.008 | 0.311±0.096 | 1.203±0.017 | 5.198±1.172 | 17.56±3.43 |
The numbers for bone marrow B cells are the total cells of each subpopulation from two femurs and two tibias. The numbers for peripheral blood B cells are the percentages of cells determined by flow cytometric analysis. fl/+: Sox4fl/+; fl/fl: Sox4fl/fl. Data represent the mean ± SD of three mice.
Discussion
In this study, we investigated the role of Sox4 in B cell development by conditionally knocking out the Sox4 gene in HSCs. We found that deletion of Sox4 markedly reduced the number of pro-B cells associated with an increased frequency of apoptosis, indicating that Sox4 was essential for the survival of pro-B cells. Our data also suggested that Sox4 functionally interacted with c-Kit and Bcl2 in protecting pro-B cells from apoptosis.
In the stepwise analysis of B cell development, depletion of fraction B pro-B cells is the earliest major change caused by Sox4 deficiency. In Sox4fl/flVav-Cre mice, though the number of Lin−c-Kit+Sca-1+ HSC-enriched cells was moderately decreased, there was no significant change in the number of CLPs and pre-pro-B cells. Thus, the nearly complete absence of pro-B cells was unlikely to have resulted from a decrease in the earlier stage cells. Nevertheless, to confirm that the heterogeneity in fraction A population did not obscure the B lineage subset, we measured the expression of various markers representing individual subsets within it, including AA4.1 for B cells (45), NK1.1 for NK cells, and CD11c for plasmacytoid dendritic cells (46, 47). We found that the size of each individual subset was not significantly altered in the absence of Sox4. Therefore, the marked reduction in the amount of pro-B cells is the major cause for the deficiency of downstream B cells. Our data also showed that deletion of Sox4 in embryonic HSCs caused fetal liver pro-B cell deficiency in Sox4fl/flVav-Cre mice and that deletion of Sox4 in adult bone marrow resulted in pro-B cell deficiency in Sox4fl/flVav-Cre mice. We conclude, therefore, that Sox4 is required for B cell development in both the fetal liver and adult bone marrow and that its deficiency causes pro-B cell abrogation.
Though controversy exists in literature, Sox4 appears to be a cell survival factor. This has been postulated for neural cells during spinal cord development (35) and for the development of late sympathetic ganglia (31, 33). In the current study, a significant increase in the frequency of apoptosis was observed in the residual pro-B and pre-B cells in Sox4fl/flVav-Cre mice. However, the frequency of apoptosis in pre-pro-B cells was not significantly different in the Sox4fl/flVav-Cre mice from that in the control mice. Likewise, the status of apoptosis of the immature and mature B cells was not affected by Sox4 deletion. Thus, during B cell development, Sox4 is required for the survival of proB and pre-B cells, but unlikely for other stage B cells.
As a transcription factor, Sox4 is expected to be part of the hierarchy of multiple transcription factors that control early B cell development, including Ebf1, E2A, and Pax5. However, the mRNA levels of these transcription factors and several other proteins such as CXCR4 and IL-7R that are critical in early B cell development were not changed in the Sox4 deficient pro-B cells, suggesting that the expression of these genes was not directly regulated by Sox4 at the transcription level. Nevertheless, it could not be ruled out that Sox4 might indirectly regulate these gene products at the protein level. While the Sox4 transcriptional program remains to be elucidated, our data show that c-Kit signaling and Bcl2 function are important in the control of pro-B cell survival supported by Sox4.
The SCF/c-Kit axis, through activation of Src, PI3K, and JAK/STAT, provides proliferation and survival signals to pro-B cells to ensure their proper development (57). C-Kit is a pro-B cell marker, and c-Kit-positive pro-B cells were diminished upon Sox4 deletion. We demonstrated that treatment with the tyrosine kinase inhibitor imatinib could sensitize the apoptotic effect of Sox4 deficiency in pro-B cells in vivo. Given the role of c-Kit in early B cell development, this sensitization most likely had resulted from the inhibition of c-Kit by imatinib and reflected functional cooperation of Sox4 and c-Kit in this process. This notion is supported by our findings that the level of Src phosphorylation (Y416) was lower in pro-B cells of the Sox4fl/flVav-Cre mice than in the Sox4 fl/+Vav-Cre mice. Src kinase is a downstream signaling component of the c-Kit pathway and its signaling is critical in pro-B cell development (28). Src phosphorylation (Y416) has been reported to provide survival signals and targeting Src activation could enhance apoptosis in multiple cell lines treated with pro-apoptotic agents (58,59). In our study, the lower Src phosphorylation (Y416) in the Sox4fl/flVav-Cre mice indicates that c-Kit/Src signaling is weakened in the absence of Sox4.
Bcl2 was reported to be highly expressed in normal bone marrow pro-B cells (17). Deficiency in B cell development caused by disruption of a number of genes can be completely or partially rescued by a Bcl2 transgene (24). We also detected a high level of Bcl2 expression in normal pre-pro-B and pro-B cells by flow cytometric analysis and the Bcl2 level was reduced in pro-B cells but not in pre-pro-B cells in Sox4fl/flVav-Cre mice. Since pro-B cells in the Sox4fl/flVav-Cre mice had an increased frequency of apoptosis, it is likely that Sox4 deletion caused pro-B cell apoptosis through reduction of the Bcl2 protein. We found that Bcl2 could partially restore the B cell development in Sox4fl/flVav-Cre mice without affecting the mRNA expression of genes critical for this process. It is noteworthy that the numbers not only of pro-B cells, but also of the later stage B cells, including pre-B and immature and mature B cells, were higher in Sox4fl/flVav-Cre mice in the presence of the Bcl2 transgene. This observation indicated that the rescued pro-B cells had undergone full differentiation and given rise to functional later stage B cells, suggesting that Bcl2, beyond its inhibition of apoptosis, compensates Sox4 in an as-yet-unknown way in B cell development.
To conclude, we report that deletion of the Sox4 gene in HSCs causes severe deficiencies of pro-B and later stage B cells. Sox4 promotes survival during early B cell development in bone morrow and may functionally interact with c-Kit and Bcl2 in this process. Inhibition of c-Kit with imatinib sensitizes the apoptotic effect of Sox4 deficiency, and forced Bcl2 expression could partially rescue B cell development in the absence of Sox4.
Supplementary Material
Acknowledgments
We would like to thank Drs. Dimitris Kioussis, and Irving Weissman for providing the Vav-Cre mice, and H2K-Bcl2 transgenic mice, respectively.
This work was supported by grant 5R03AI079779-02 to X.S. from National Institutes of Health, Physician Scientist Program Award to X.S. from MD Anderson Cancer Center, Research Scholar Grant 119645-RSG-10-131-01-DDC to X.S. from the American Cancer Society, and MD Anderson’s Cancer Center Support Grant CA016672.
Footnotes
The authors have no conflicting financial interest.
References
- 1.Kondo M, I, Weissman L, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 2.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 3.Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat Rev Immunol. 2006;6:107–116. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- 4.Busslinger M. Transcriptional control of early B cell development. Annu Rev Immunol. 2004;22:55–79. doi: 10.1146/annurev.immunol.22.012703.104807. [DOI] [PubMed] [Google Scholar]
- 5.Fuxa M, Skok JA. Transcriptional regulation in early B cell development. Curr Opin Immunol. 2007;19:129–136. doi: 10.1016/j.coi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Arinobu Y, Mizuno S, Chong Y, Shigematsu H, Iino T, Iwasaki H, Graf T, Mayfield R, Chan S, Kastner P, Akashi K. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1:416–427. doi: 10.1016/j.stem.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Kirstetter P, Thomas M, Dierich A, Kastner P, Chan S. Ikaros is critical for B cell differentiation and function. Eur J Immunol. 2002;32:720–730. doi: 10.1002/1521-4141(200203)32:3<720::AID-IMMU720>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 8.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 9.Kikuchi K, Lai AY, Hsu CL, Kondo M. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J Exp Med. 2005;201:1197–1203. doi: 10.1084/jem.20050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller JP, Izon D, DeMuth W, Gerstein R, Bhandoola A, Allman D. The earliest step in B lineage differentiation from common lymphoid progenitors is critically dependent upon interleukin 7. J Exp Med. 2002;196:705–711. doi: 10.1084/jem.20020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polli M, Dakic A, Light A, Wu L, Tarlinton DM, Nutt SL. The development of functional B lymphocytes in conditional PU.1 knock-out mice. Blood. 2005;106:2083–2090. doi: 10.1182/blood-2005-01-0283. [DOI] [PubMed] [Google Scholar]
- 12.O’Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 13.Sigvardsson M, O’Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 14.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 15.Thevenin C, Nutt SL, Busslinger M. Early function of Pax5 (BSAP) before the pre-B cell receptor stage of B lymphopoiesis. J Exp Med. 1998;188:735–744. doi: 10.1084/jem.188.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuzaki Y, Nakayama K, Tomita T, Isoda M, Loh DY, Nakauchi H. Role of bcl-2 in the development of lymphoid cells from the hematopoietic stem cell. Blood. 1997;89:853–862. [PubMed] [Google Scholar]
- 17.Fang W, Mueller DL, Pennell CA, Rivard JJ, Li YS, Hardy RR, Schlissel MS, Behrens TW. Frequent aberrant immunoglobulin gene rearrangements in pro-B cells revealed by a bcl-xL transgene. Immunity. 1996;4:291–299. doi: 10.1016/s1074-7613(00)80437-9. [DOI] [PubMed] [Google Scholar]
- 18.Kirman I, Zhao K, Wang Y, Szabo P, Telford W, Weksler ME. Increased apoptosis of bone marrow pre-B cells in old mice associated with their low number. Int Immunol. 1998;10:1385–1392. doi: 10.1093/intimm/10.9.1385. [DOI] [PubMed] [Google Scholar]
- 19.Vikstrom I, Carotta S, Luthje K, Peperzak V, Jost PJ, Glaser S, Busslinger M, Bouillet P, Strasser A, Nutt SL, Tarlinton DM. Mcl-1 is essential for germinal center formation and B cell memory. Science. 2010;330:1095–1099. doi: 10.1126/science.1191793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu L, Chaudhury P, Osmond DG. Regulation of cell survival during B lymphopoiesis: apoptosis and Bcl-2/Bax content of precursor B cells in bone marrow of mice with altered expression of IL-7 and recombinase-activating gene-2. J Immunol. 1999;162:1931–1940. [PubMed] [Google Scholar]
- 21.Jiang A, Clark EA. Involvement of Bik, a proapoptotic member of the Bcl-2 family, in surface IgM-mediated B cell apoptosis. J Immunol. 2001;166:6025–6033. doi: 10.4049/jimmunol.166.10.6025. [DOI] [PubMed] [Google Scholar]
- 22.Craxton A, Draves KE, Gruppi A, Clark EA. BAFF regulates B cell survival by downregulating the BH3-only family member Bim via the ERK pathway. J Exp Med. 2005;202:1363–1374. doi: 10.1084/jem.20051283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dai X, Chen Y, Di L, Podd A, Li G, Bunting KD, Hennighausen L, Wen R, Wang D. Stat5 is essential for early B cell development but not for B cell maturation and function. J Immunol. 2007;179:1068–1079. doi: 10.4049/jimmunol.179.2.1068. [DOI] [PubMed] [Google Scholar]
- 24.Malin S, McManus S, Cobaleda C, Novatchkova M, Delogu A, Bouillet P, Strasser A, Busslinger M. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat Immunol. 2010;11:171–179. doi: 10.1038/ni.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 26.Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn. 2011;240:808–819. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waskow C, Paul S, Haller C, Gassmann M, Rodewald HR. Viable c-Kit(W/W) mutants reveal pivotal role for c-kit in the maintenance of lymphopoiesis. Immunity. 2002;17:277–288. doi: 10.1016/s1074-7613(02)00386-2. [DOI] [PubMed] [Google Scholar]
- 28.Agosti V, Corbacioglu S, Ehlers I, Waskow C, Sommer G, Berrozpe G, Kissel H, Tucker CM, Manova K, Moore MA, Rodewald HR, Besmer P. Critical role for Kit-mediated Src kinase but not PI 3-kinase signaling in pro T and pro B cell development. J Exp Med. 2004;199:867–878. doi: 10.1084/jem.20031983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schepers GE, Teasdale RD, Koopman P. Twenty pairs of sox: extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167–170. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 30.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 31.Bhattaram P, Penzo-Mendez A, Sock E, Colmenares C, Kaneko KJ, Vassilev A, Depamphilis ML, Wegner M, Lefebvre V. Organogenesis relies on SoxC transcription factors for the survival of neural and mesenchymal progenitors. Nat Commun. 2010;1:9. doi: 10.1038/ncomms1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nissen-Meyer LS, Jemtland R, Gautvik VT, Pedersen ME, Paro R, Fortunati D, Pierroz DD, Stadelmann VA, Reppe S, Reinholt FP, Del Fattore A, Rucci N, Teti A, Ferrari S, Gautvik KM. Osteopenia, decreased bone formation and impaired osteoblast development in Sox4 heterozygous mice. J Cell Sci. 2007;120:2785–2795. doi: 10.1242/jcs.003855. [DOI] [PubMed] [Google Scholar]
- 33.Potzner MR, Tsarovina K, Binder E, Penzo-Mendez A, Lefebvre V, Rohrer H, Wegner M, Sock E. Sequential requirement of Sox4 and Sox11 during development of the sympathetic nervous system. Development. 2010;137:775–784. doi: 10.1242/dev.042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pramoonjago P, Baras AS, Moskaluk CA. Knockdown of Sox4 expression by RNAi induces apoptosis in ACC3 cells. Oncogene. 2006;25:5626–5639. doi: 10.1038/sj.onc.1209566. [DOI] [PubMed] [Google Scholar]
- 35.Thein DC, Thalhammer JM, Hartwig AC, Crenshaw EB, 3rd, Lefebvre V, Wegner M, Sock E. The closely related transcription factors Sox4 and Sox11 function as survival factors during spinal cord development. J Neurochem. 2010;115:131–141. doi: 10.1111/j.1471-4159.2010.06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penzo-Mendez A, Dy P, Pallavi B, Lefebvre V. Generation of mice harboring a Sox4 conditional null allele. Genesis. 2007;45:776–780. doi: 10.1002/dvg.20358. [DOI] [PubMed] [Google Scholar]
- 37.de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 38.Domen J, Gandy KL, Weissman IL. Systemic overexpression of BCL-2 in the hematopoietic system protects transgenic mice from the consequences of lethal irradiation. Blood. 1998;91:2272–2282. [PubMed] [Google Scholar]
- 39.Bustelo XR, Rubin SD, Suen KL, Carrasco D, Barbacid M. Developmental expression of the vav protooncogene. Cell Growth Differ. 1993;4:297–308. [PubMed] [Google Scholar]
- 40.Georgiades P, Ogilvy S, Duval H, Licence DR, Charnock-Jones DS, Smith SK, Print CG. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis. 2002;34:251–256. doi: 10.1002/gene.10161. [DOI] [PubMed] [Google Scholar]
- 41.Hardy RR, Hayakawa K. A developmental switch in B lymphopoiesis. Proc Natl Acad Sci U S A. 1991;88:11550–11554. doi: 10.1073/pnas.88.24.11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balciunaite G, Ceredig R, Massa S, Rolink AG. A B220+ CD117+ CD19- hematopoietic progenitor with potent lymphoid and myeloid developmental potential. Eur J Immunol. 2005;35:2019–2030. doi: 10.1002/eji.200526318. [DOI] [PubMed] [Google Scholar]
- 43.Rolink A, ten Boekel E, Melchers F, Fearon DT, Krop I, Andersson J. A subpopulation of B220+ cells in murine bone marrow does not express CD19 and contains natural killer cell progenitors. J Exp Med. 1996;183:187–194. doi: 10.1084/jem.183.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li YS, Wasserman R, Hayakawa K, Hardy RR. Identification of the earliest B lineage stage in mouse bone marrow. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 46.Diao J, Winter E, Chen W, Cantin C, Cattral MS. Characterization of distinct conventional and plasmacytoid dendritic cell-committed precursors in murine bone marrow. J Immunol. 2004;173:1826–1833. doi: 10.4049/jimmunol.173.3.1826. [DOI] [PubMed] [Google Scholar]
- 47.Pelayo R, Hirose J, Huang J, Garrett KP, Delogu A, Busslinger M, Kincade PW. Derivation of 2 categories of plasmacytoid dendritic cells in murine bone marrow. Blood. 2005;105:4407–4415. doi: 10.1182/blood-2004-07-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steff AM, Fortin M, Arguin C, Hugo P. Detection of a decrease in green fluorescent protein fluorescence for the monitoring of cell death: an assay amenable to high-throughput screening technologies. Cytometry. 2001;45:237–243. doi: 10.1002/1097-0320(20011201)45:4<237::aid-cyto10024>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 49.Jackson SM, Capra JD. IgH V-region sequence does not predict the survival fate of human germinal center B cells. J Immunol. 2005;174:2805–2813. doi: 10.4049/jimmunol.174.5.2805. [DOI] [PubMed] [Google Scholar]
- 50.van Engeland M, Nieland LJ, Ramaekers FC, Schutte B, Reutelingsperger CP. Annexin V-affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry. 1998;31:1–9. doi: 10.1002/(sici)1097-0320(19980101)31:1<1::aid-cyto1>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 51.Dillon SR, Mancini M, Rosen A, Schlissel MS. Annexin V binds to viable B cells and colocalizes with a marker of lipid rafts upon B cell receptor activation. J Immunol. 2000;164:1322–1332. doi: 10.4049/jimmunol.164.3.1322. [DOI] [PubMed] [Google Scholar]
- 52.Meffre E, Nussenzweig MC. Deletion of immunoglobulin beta in developing B cells leads to cell death. Proc Natl Acad Sci U S A. 2002;99:11334–11339. doi: 10.1073/pnas.172369999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reichlin A, Hu Y, Meffre E, Nagaoka H, Gong S, Kraus M, Rajewsky K, Nussenzweig MC. B cell development is arrested at the immature B cell stage in mice carrying a mutation in the cytoplasmic domain of immunoglobulin beta. J Exp Med. 2001;193:13–23. doi: 10.1084/jem.193.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matsuda K, Koguma M, Okuyama R, Nakazawa T, Matsuzaki Y, Nakauchi H, Yanai N, Terasaki T, Obinata M. A novel stromal cell-dependent B lymphoid stem-like cell line that induces immunoglobulin gene rearrangement. J Biochem. 1999;125:602–612. doi: 10.1093/oxfordjournals.jbchem.a022326. [DOI] [PubMed] [Google Scholar]
- 55.Merino R, Ding L, Veis DJ, Korsmeyer SJ, Nunez G. Developmental regulation of the Bcl-2 protein and susceptibility to cell death in B lymphocytes. Embo J. 1994;13:683–691. doi: 10.1002/j.1460-2075.1994.tb06307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakayama K, Negishi I, Kuida K, Sawa H, Loh DY. Targeted disruption of Bcl-2 alpha beta in mice: occurrence of gray hair, polycystic kidney disease, and lymphocytopenia. Proc Natl Acad Sci U S A. 1994;91:3700–3704. doi: 10.1073/pnas.91.9.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linnekin D. Early signaling pathways activated by c-Kit in hematopoietic cells. Int J Biochem Cell Biol. 1999;31:1053–1074. doi: 10.1016/s1357-2725(99)00078-3. [DOI] [PubMed] [Google Scholar]
- 58.Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z, Sahin AA, Esteva FJ, Hortobagyi GN, Yu D. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461–469. doi: 10.1038/nm.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang T, Liu S, Yang P, Han C, Wang J, Liu J, Han Y, Yu Y, Cao X. Fibronectin maintains survival of mouse natural killer (NK) cells via CD11b/Src/beta-catenin pathway. Blood. 2009;114:4081–4088. doi: 10.1182/blood-2009-05-219881. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.