Abstract
The advent of high throughput sequencing technologies has revealed that pervasive transcription generates RNAs from nearly all regions of eukaryotic genomes. Normally these transcripts undergo rapid degradation by a nuclear RNA surveillance system primarily featuring the RNA exosome. This multimeric protein complex plays a critical role in the efficient turnover and processing of a vast array of RNAs in the nucleus. Despite its initial discovery over a decade ago, important questions remain concerning the mechanisms that recruit and activate the nuclear exosome. Specificity and modulation of exosome activity requires additional protein cofactors, including the conserved TRAMP polyadenylation complex. Recent studies suggest that helicase and RNA binding subunits of TRAMP direct RNA substrates for polyadenylation, which enhances their degradation by Dis3/Rrp44 and Rrp6, the two exosome associated ribonucleases. These findings indicate that the exosome and TRAMP have evolved highly flexible functions that allow recognition of a wide range of RNA substrates. This flexibility provides the nuclear RNA surveillance system with the ability to regulate the levels of a broad range of coding and non-coding RNAs, which results in profound effects on gene expression, cellular development, gene silencing and heterochromatin formation. This review summarizes recent findings on the nuclear RNA surveillance complexes, and speculates upon possible mechanisms for TRAMP-mediated substrate recognition and exosome activation.
The RNA exosome, a highly conserved protein complex originally discovered in S. cerevisiae, has emerged as a major component of 3′-5′ ribonucleoytic RNA surveillance pathways in both archae and eukaryotes 1–5. The eukaryotic exosome is composed of a 9-subunit core, referred to as exo-9. Six of these proteins are arranged in a hexameric ring with a central channel, while the remaining 3 proteins form a trimeric cap and sit on top of this ring structure 6. In S. cerevisiae, catalytic activity resides in an additional component Dis3/Rrp44 that associates with the catalytically inactive core exosome to form the functional cytoplasmic and nuclear exo-10 complex 7–9. RNA substrates with sufficiently long single-stranded 3′ ends may thread through the core to reach the active site of Dis3; alternatively, RNA may bypass the exosome core channel and enter the active site of Dis3 directly 8, 9. In the nucleus, exo-10 can associate further with the nonessential 3′-5′ exoribonuclease Rrp6 to form the exo-11 complex 10. Rrp6 can also function independently of exo-10 on a subset of RNA targets, since mutants unable to interact with exo-10 still complement some functions of native Rrp6 11.
Remarkably, experiments have shown that the exosome functions in degradation of a wide range of substrates, including coding and noncoding RNAs produced by all 3 major RNA polymerases 12–15. Furthermore, the exosome functions in the 3′ end formation of a growing list of transcripts 10, 13, 16–20. Hence, the exosome is not only able to adapt to a multitude of RNA substrates, but its catalytic activities can be modulated, likely by protein cofactors such as TRAMP, to distinguish between processing and degradation. The exosome relies on multiple co-factors, such as the TRAMP complex, for recruitment of its vast repertoire of substrates, and possibly modulation of its activity. Originally discovered in yeast, the TRAMP complex is composed of a noncanonical poly(A) polymerase Trf4 or Trf5, a zinc-knuckle protein Air1 or Air2, and the RNA helicase Mtr4 21–24. TRAMP polyadenylates RNAs destined for Rrp6 and the core exosome, assisting in transcript recognition and exosome activation. In this way, TRAMP plays a critical role in ridding the cell of non-coding transcripts generated through pervasive Pol II transcription, as well as functioning in the biogenesis and turnover of functional coding and non-coding RNAs 16, 19, 25. Extensive research has revealed the critical role the TRAMP plays in determining nuclear RNA fate, though the mechanism used for recognition of RNA substrates and subsequent activation of the exosome remains elusive.
COMPOSITION AND STRUCTURE OF TRAMP
TRAMP4 and TRAMP5 are named for the presence of the non-canonical poly(A) polymerase Trf4 or Trf5, respectively. Despite considerable similarities to the catalytic region and central domain of the canonical poly(A) polymerase Pap1, Trf4 and Trf5 lack a recognizable RNA binding domain and require one of two RNA binding proteins, Air1 or Air2, for polyadenylation of substrates 24, 26. The related TRF and AIR genes likely arose from the whole-genome duplication of Saccharomyces cerevisiae 27; consistently, the polymerases share 58% identity while Air1 and Air2 are 45% identical. The majority of Air conservation is clustered within the first third of the proteins, which contain 5 zinc knuckle motifs (Figure 1a). The zinc knuckles appear to function in both protein-protein and protein-RNA interactions. Mutational analysis reveals that zinc knuckles one through four of both Air1 and Air2 are required for efficient polyadenylation and degradation of a variety of RNA substrates, suggesting that these zinc knuckles function in RNA binding 26, 28, 29. Indeed, the crystal structure of Trf4 in complex with a fragment of Air2 spanning the fourth and fifth zinc knuckles reveal that the critical RNA binding residues of zinc knuckle four are exposed, in agreement with the suggestion that this region binds RNA (Figure 1b) 28. In contrast, the critical residues of zinc knuckle five are buried against the surface of Trf4, indicating that the fifth zinc knuckle, along with residues located upstream, bind Trf4. Biochemical evidence also supports a role for this region in Air2-Trf4 binding, and suggests that it is necessary for Air1-Trf4 binding as well. Specifically, mutation of a conserved IWRxYxL motif located just downstream of zinc knuckle four disrupts interaction of Air1 and Air2 with Trf4 26, 29. Interestingly, mutations in zinc knuckle four of either Air1 or Air2 disrupt a small portion of Trf4-Air1/2 complexes, suggesting that zinc knuckle four does affect Trf4 binding 29. It is possible that mutations in here induce slight conformational changes in adjacent regions, including the critical IWRxYxL motif, explaining why mutations in this region affect binding despite lack of interaction between Trf4 and critical zinc knuckle amino acids.
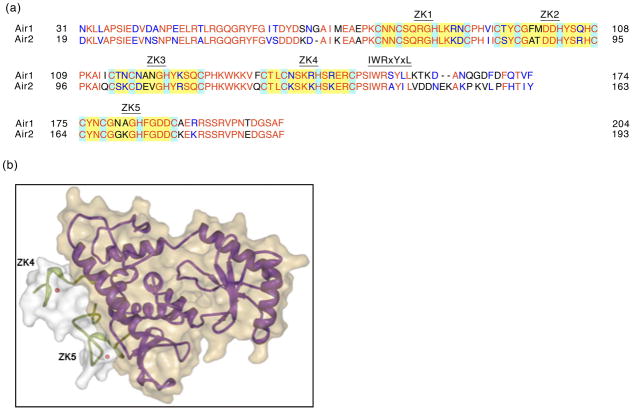
Figure 1.
Structural features of Air1 and Air2 and interaction with Trf4. (a). The protein sequences of Saccharomyces cerevisiae Air1 and Air2 were aligned using red font to indicate identical residues, and blue font to denote similar residues. Only the most highly conserved sequences are shown (amino acids 31–204 of Air1, and 19–193 of Air2). The numbered yellow boxes indicate the positions of the 5 zinc knuckle motifs, with the conserved CCHC residues highlighted in light blue boxes. The IWRxYxL motif is also specified. (b). Crystal structure of the catalytic and central domains of Trf4 (residues 161–481; purple) bound to zinc knuckles 4 (ZK4) and 5 (ZK5) of Air2 (residues 122–198; green)28. Zinc atoms are shown as red balls. The structure was modelled from Protein Data Base structure 3NYB using RCSB PDB Protein Workshop 4.0.
Air1, but not Air2, co-precipitates with Trf5, though no information is known about the residues necessary for this interaction 23. Based on the high level of conservation between Trf4 and Trf5 and Air1 and Air2, it is tempting to speculate that the face of interaction is conserved between the TRAMP4 and TRAMP5 complexes. Indeed, the Air2 binding pocket of Trf4 is highly conserved in Trf5 28. This, however, raises the question of why Air2 does not also interact with Trf5. The answer may be in the sequence: although the IWRxYxL motif and the fifth zinc knuckles are conserved between Air1 and Air2, the region in between varies greatly. In fact, Air2 contains two more residues in this linker region than Air1. Since this linker region directly contacts Trf4 it is possible that Air2 lacks residues necessary for interaction with Trf5 28. Alternatively, TRAMP4 and TRAMP5 composition may be dictated by differential localizations of Trf and Air proteins. Consistent with this, while all Trf and Air proteins are found in the nucleus, Trf5-GFP and Air1-GFP fusions exhibit slight nucleolar enrichment 29–31.
TRAMP4 and TRAMP5 also contain the nuclear 3′-5′ RNA helicase Mtr4 21–24. The crystal structure of Mtr4 reveals that it is composed of central ATPase core similar to those found in other DExH helicases, but also has a unique arch domain that resembles the KOW domain of ribosomal proteins 32, 33. Indeed, the novel arch/KOW domain is required for 5.8s rRNA processing, as well as for binding to tRNAiMet 32, 33. Although there is no crystal structure for Mtr4 in complex with TRAMP, it is known that the interaction with TRAMP occurs through conserved residues on the helicase core and does not rely on the unique arch of Mtr4 32. Furthermore, Mtr4 likely interacts directly with Trf4 and Trf5 independently of the Air proteins, as Trf5 and Mtr4 co-precipitate in the absence of Air1 or Air2 21. In this regard, the ability of Mtr4 to bind poly(A) RNA suggests that oligoadenylation by Trf polymerases may facilitate RNA unwinding in the absence of Air proteins 34, 35. Since Trf4 and Mtr4 appear to exist in excess over Air1, Air2 and Trf5, complexes between Mtr4 and Trf4 could have such a role in vivo 36.
OVERLAPPING AND DISTINCT FUNCTIONS OF MTR4 AND TRAMP
The requirement of Mtr4 for efficient processing or degradation by the exosome varies greatly and appears to depend on the RNA substrate. First, Mtr4 can function as an exosome cofactor independently of the TRAMP complex in the processing of structured RNAs such as the 5.8s rRNA 13, 21, 37, 38. Indeed, there is a vast cellular excess of Mtr4 compared to other TRAMP components, supporting the claim that Mtr4 can function independently of TRAMP 21. The crystal structure of Mtr4 suggests why it is required for efficient processing of pre-rRNAs. An archless version of Mtr4 results in the accumulation of a 5.8s rRNA species 30 nucleotides longer than the mature species, similar to the extended transcript seen in the absence of Rrp6 33. Interestingly, it is predicted that the distance from the top of the exosome, where the RNA enters the core, to the active site of Dis3 encompasses approximately 30 nucleotides 8. It seems likely, therefore, that exo-10 can degrade the 7s precursor to within 30 nucleotides of the mature end, at which point Rrp6 and Mtr4 ensure efficient trimming of the last nucleotides 10, 33. It is possible that the arch of Mtr4 acts as an arm to secure the RNA, and that hydrolysis of ATP induces a conformational change in Mtr4 and transfers the 5.8s + 30 extended transcript from Dis3 to Rrp6 33.
Alternatively, Mtr4 interacts with TRAMP, functionally and physically, for processing and/or degradation of a different set of transcripts. Both Mtr4 and TRAMP are required for efficient degradation of hypomodified precursor tRNAiMet, a highly structured substrate 39. It appears that Mtr4 functions downstream of TRAMP polyadenylation for this transcript, as mutation or absence of Mtr4 results in defects in degradation but not polyadenylation 24, 39. It has also been suggested that both Mtr4 and Trf4 function in termination of snoRNA transcription at one of two possible sites, possibly through recruitment of the Nrd1/Nab3 termination complex 19. Interestingly, a related pathway may function in the 3′ end formation of CTH2 mRNA 16. NAB2 is a unique example where TRAMP and Mtr4 share a substrate, but not a function. Trf4 and the core exosome are required for trimming a 3′ extended version of NAB2 to its mature end, at which point Mtr4 and Rrp6 likely function to degrade the transcript if it is not stabilized via canonical polyadenylation by Pap1 17. Thus, Trf4 and Mtr4 serve opposing roles in the processing and degradation, respectively, of a single transcript. Further work may reveal additional substrates where TRAMP and exosome components display differential functions within a single transcript.
Finally, a third group of substrates may require TRAMP, but not Mtr4, to effectively stimulate degradation by the exosome. In vitro assays with a portion of the 7S pre-rRNA reveal a requirement for TRAMP, but not the ATP dependent helicase activity of Mtr4 40. The observed enhancement of degradation was also not structurally dependent on Mtr4, as TRAMP without Mtr4 stimulated degradation as effectively as TRAMP + Mtr4 21, 24. Additional experiments are necessary to determine if TRAMP can stimulate degradation by the exosome in the absence of Mtr4 in vivo.
MECHAMISM OF TRAMP’S EXOSOMAL ENHANCEMENT
It has long been known that mutations in exosome components cause an accumulation of poly(A)+ RNA 13, 20, 41, 42. Thus, when TRAMP and its poly(A) polymerase activity was first discovered, it was generally believed that it enhanced exosome degradation or processing through successive rounds of polyadenylation and subsequent recruitment of the exosome. For this to be true, RNAs polyadenylated by Trf4 and Trf5 must be distinguishable from stable mRNAs polyadenylated by the canonical poly(A) polymerase Pap1. Indeed, initial experiments suggested that poly(A) tails added in vivo by Trf4 and Trf5 are 10–50 nts, while Pap1 adds tails 60–80 nucleotides 15, 21, 23, 43–45. More recent studies showed that the Mtr4 helicase can modulate the activity of Trf4, suppressing polymerization after addition of 3–5 adenosines 34. This is in agreement with a concurrent study reporting that the distribution of poly(A) tails added by TRAMP peaks around 4–5 adenosines 14. The fact that early studies used RNase H/olido(dT) cleavage or poly(A) selection, which requires poly(A) tails of ~15 nucleotides, for identification of exosome and TRAMP substrates likely explains why only longer poly(A) tails were initially identified 13, 19, 21.
Despite the fact that both Trf4 and Trf5 polyadenylate RNAs destined for the exosome, the role that this polyadenylation plays in RNA degradation remains unclear. The addition of a single stranded poly(A) tail to the ends of transcripts destined for the exosome seems an elegant way to modify highly structured RNAs, making them a more favourable substrate for the sterically restricted exosome core. This mechanism is reminiscent of polyadenylation of mRNA in bacteria, which likewise facilitates degradation of RNAs by the PNPase subunit of the degradosome 46. Consistently, a catalytically inactive Trf4 is insufficient to stimulate degradation of unmodified pre-tRNAiMet in vitro and in vivo 24, 38. However, multiple studies have also found that the polyadenylation activity of Trf4 is not necessary for the activation and/or recruitment of the exosome 21, 22, 40, 44, 47, 48. Furthermore, a catalytically inactive form of Trf4 is able to restore ~90% of elevated transcripts in trf4-Δ to wild-type levels and suppresses the synthetic lethality of trf4-Δ trf5-Δ 22, 47, 49.
The fact that polyadenylation by Trf4 is not universally required for all TRAMP substrates may be explained, in part, by the presence of Mtr4. The 3′-5′ helicase activity of Mtr4 can unwind highly structured RNA substrates in vitro, and is required for degradation of some tRNAs in vivo 24, 39. It is likely that the helicase is necessary to produce a single stranded end for capture by the exosome. Until recently, however, it was unclear if the helicase activity of Mtr4 is active while in complex with TRAMP, and if so, how this activity coordinates with the 5′-3′ polyadenylation activity of Trf4/5. Remarkably, in vitro experiments suggest that Trf4/Air2 stimulates the helicase activity of Mtr4, specifically the ATP affinity and rate of strand separation 50, consistent with the finding that Mtr4 preferentially binds to strings of adenosines 35. Furthermore, although unwinding by Mtr4 does not require polyadenylation by Trf4, the addition of adenosines to short 3′ overhangs can gradually increase the extension to the minimum 5 nucleotides required for Mtr4 unwinding. Thus, if a transcript contains a natural 3′ overhang of sufficient length for Mtr4/exosome recruitment, polyadenylation by Trf4 may not be required. Combining this with earlier findings, it appears that Mtr4 and TRAMP activities are coordinated threefold: 1.) Trf4 can prime a substrate for Mtr4 by adenylating the 3′ end, 2.) Mtr4 modulates Trf4 activity, slowing polyadenylation after addition of 3–4 adenosines, and 3.) Trf4 stimulates the RNA helicase activity of Mtr4 directly 34, 50. Although it is unclear if these in vitro findings represent in vivo mechanisms, it is of note that Mtr4 preferentially unwinds substrates containing overhangs of 5–6 nucleotides, correlating with the average length of tails found on TRAMP substrates in vivo 14, 50.
Together, these recent studies suggest a mechanism by which TRAMP coordinates its intrinsic polyadenylation and helicase activities to generate substrates with short, single stranded 3′ overhangs. TRAMP therefore functions in what may be thought of as a “priming” pathway, generating substrates suitable for the exosome (Figure 2(a)). But what purpose do these tails serve mechanistically? It is possible that the tails generated by TRAMP remain exposed for capture by the exosome or Rrp6, if these enzymes function as cellular scavengers of exposed 3′ ends. Consistently, Pab1 poly(A) binding protein, part of the complex responsible for stabilization of 3′ ends of mRNAs, requires 12 nucleotide overhangs for efficient binding 34, 51, while a related PABP (poly(A) binding protein) Nab2 may bind to adenosine tails of about 20 nucleotides 52. Due to this requirement any transcript containing poly(A) shorter than ~20 nucleotides will not be bound by PABPs and exported to the cytoplasm for translation 53, 54. Moreover, inefficient canonical polyadenylation has been directly linked to increased degradation by the exosome 42, 55, 56. TRAMP substrates that do not show a dependency on polyadenylation or helicase activity likely have 3′ overhangs already well suited for degradation or processing by the exosome, but this does not explain why TRAMP is still required for activation or recruitment of the exosome. One explanation may involve the kinetics of RNA maturation (Figure 2(b)). If RNA matures with rapid kinetics, the end remains bound by 3′ end formation machinery in an mRNP (messenger ribonucleoprotein) complex. Defects in the RNA, however, will stall the processing machinery, likely leading to eventual dissociation of the mRNP complex and exposure of the 3′ overhang. Binding of TRAMP to the exposed 3′ ends would prevent re-association of the 3′ end processing. TRAMP is not a processive enzyme, and the constant dissociation/reassociation may allow for the exosome to capture the exposed end 21, 34. Three key pieces of evidence support this idea. First, decreased canonical polyadenylation on transcripts with stalled 3′ processing leads to Trf4 and Rrp6 dependent RNA degradation by the exosome 55. Second, recent evidence reveals that Trf4 can act on poly(A) tails, suggesting that TRAMP can mediate degradation of transcripts already carrying a canonical poly(A) tail 57. Third, the canonical polyadenylation pathway involving Pab2, required for maturation of snoRNAs in S. pombe, functions in an antagonistic manner to TRAMP-mediated RNA decay of snoRNAs 58. Together, these studies suggest a “kinetic” or “competitive” pathway between 3′ end maturation processes and the TRAMP/exosome degradation pathway.
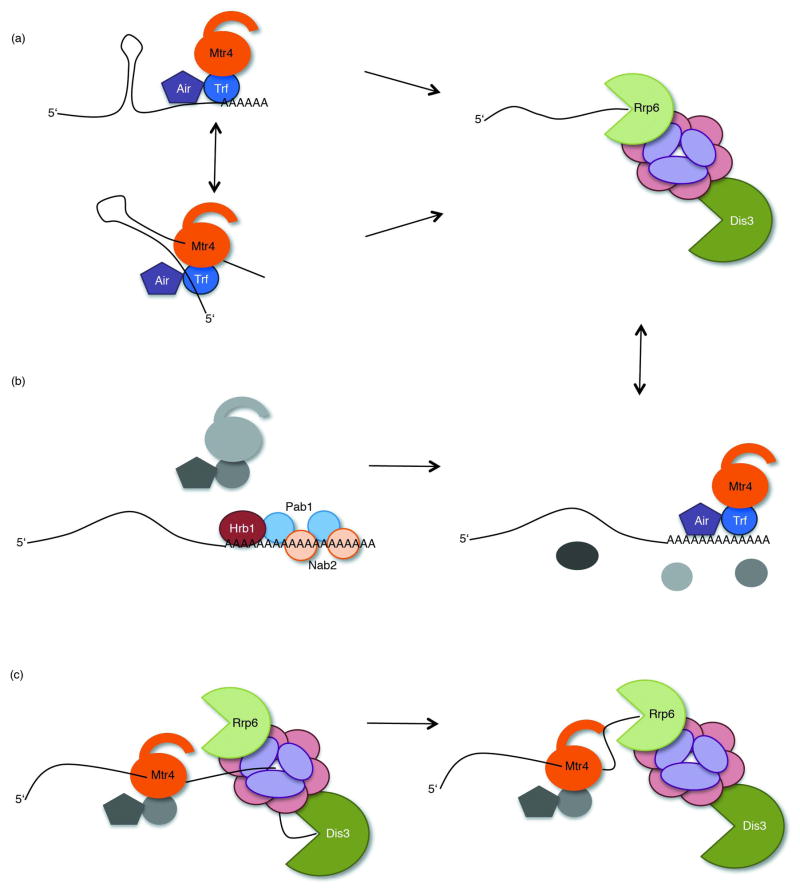
Figure 2.
Proposed mechanisms of TRAMP enhancement of exosomal degradation. (a) The priming model. Highly structured transcripts are modified by TRAMP, producing single stranded ends that are available for capture and subsequent degradation by Rrp6 and/or Dis3. These single stranded extensions are generated by the poly(A) polymerase activity of Trf4 or Trf5 (top), and/or by the 3′-5′ helicase activity of Mtr4 (bottom). (b) The kinetic or competitive model. Rapidly maturing RNAs are normally bound by 3′ end processing machinery. For simplicity, only the poly(A) binding proteins Hrb1, Nab2, and Pab1 are represented. For a complete review of 3′ end processing, see 117. If the processing machinery encounters defects, however, the processing machinery will pause and eventually disassociate, exposing the 3′ end of the transcript for capture by TRAMP and the exosome. The inactive and unassociated TRAMP and 3′ end processing components are shown in grey. (c) The scaffold model. The 7s rRNA precursor is trimmed by Dis3 to within 30 nucleotides of it′s mature 3′ end. The Mtr4 arch domain then acts as an arm to transfer the 5.8s + 30 extended transcript to Rrp6, which trims the transcript to it’s final length. It is unclear if the Air proteins are required for this process and are therefore shown in grey.
As previously mentioned, it has also been suggested that Mtr4, specifically the arch domain, bridges TRAMP and exosome functions by handing off a transcript to the exosome or Rrp6 (Figure 2(c)). 32, 33 In this model TRAMP may be thought of as a scaffold, functioning only as an intermediate to ensure presentation of transcripts to the degradation machinery. This hypothesis is particularly appealing for transcripts where polyadenylation and helicase activity of TRAMP are not required for exosome activation. It remains to be determined if the “scaffold” model is used for transcripts other than rRNAs.
A fourth, but not exclusive, possibility is that the TRAMP complex functions to retain aberrant transcripts in the nucleus (Figure 3). A few key pieces of evidence support this model. First, the Air proteins were originally identified as in vitro inhibitors of the arginine methylation function of Hmt1 and were named accordingly (arginine methyltransferase-interacting RING finger protein) 59. Hmt1 methyltransferase activity is required to modify Nab2 and a shuttling protein Npl3, which can then transport RNPs from the nucleus to the cytoplasm 60. If the Air proteins also inhibit Hmt1 function in vivo, the presence of TRAMP at aberrant or immature transcripts may inhibit the activation (i.e. methylation) of Nab2 and Npl3 and export of these RNPs. Retention of these transcripts in the nucleus likely ensures degradation by the nuclear surveillance machinery. It is of note that over-expressed Air1 co-purifies with Hrb1, an mRNA export protein related to Npl3 61–63. It is tempting to speculate that TRAMP may also serve to somehow regulate shuttling of Hrb1 and associated transcripts, though an interaction between endogenously expressed proteins and possible functional implications of such an interaction remain to be explored. In addition to the functional connection between TRAMP and nuclear export, polyadenylated RNAs accumulate in the nucleus of air1-Δ air2-Δ cells, but not in the absence of only one of the Air proteins 59. Although the role of Air1 and Air2 in context of the TRAMP complex was unknown at the time of the study, it is unlikely that this increase in poly(A)+ species is related to TRAMP polyadenylation. Since the Air proteins are required for polyadenylation by Trf4 and Trf5, an absence of Air proteins would decrease, not increase, polyadenylation of TRAMP substrates. Therefore, the accumulation of nuclear poly(A)+ RNA in air1-Δ air2-Δ is likely a result of defective nuclear mRNA turnover and/or export.
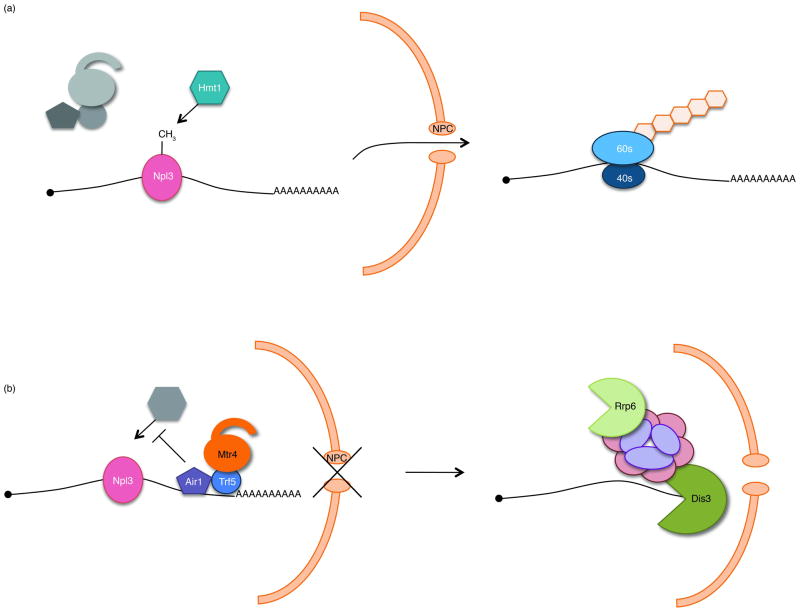
Figure 3.
Model of Air mediated inhibition of mRNP export. (a) Following accurate transcription, 3′ end processing, and polyadenylation, Npl3 binds properly processed mRNPs. Following methylation by Hmt1, Npl3 mediates export of the mRNP to the cytoplasm for translation. (b) If an mRNP is not properly processed, Air (likely in context of the TRAMP context) is recruited to the aberrant mRNP and inhibits export by blocking Hmt1 mediated methylation of Npl3. TRAMP recruits the exosome for degradation of mRNPs retained in the nucleus.
Despite extensive efforts to determine the exact mechanism behind TRAMP’s ability to enhance the function of the exosome, the molecular details of the connection between these complexes remain elusive. Many intriguing possibilities and pathways have been proposed thus far. Given the wide variety of substrates recognized by TRAMP and the exosome, it is not surprising that multiple proteins have evolved to function cooperatively to ensure correct targeting of transcripts to the nuclear RNA surveillance system. The possible pathways for delivery to the surveillance system outlined here are likely not mutually exclusive, allowing the system to adapt to a wide array of transcripts.
SPECIFICITY OF TRAMP4 AND TRAMP5
Despite significant similarities between TRAMP4 and TRAMP5, early experiments suggested specificity between the complexes. Specifically, Trf4 is required for 3′ end formation of CTH2 and NAB2 and the degradation of internal genomic spacer (IGS) region of rRNAs, while Trf5 appears to preferentially enhance the degradation of aberrant rRNA precursors 16, 17, 23, 44. Furthermore, while Trf4 and Trf5 both function in the polyadenylation of sn- and snoRNAs intermediates, they display slight differences in substrate preference 43. A subsequent microarray analysis of total RNA revealed that deletion of TRF4 and TRF5 affects different subsets of transcripts, confirming substrate specificity of the proteins 47. However, these studies assumed that the other distinct TRAMP protein, Air1 or Air2, functioned interchangeably in the TRAMP complex. Indeed, early studies on relatively few RNA substrates failed to identify substrate specificity between Air1 and Air2 21, 24, 44. However, the Air protein subunits are the likely source of TRAMP RNA binding, suggesting they regulate the specificity of nuclear RNA surveillance 26, 29. Accordingly, TRAMP4 preferentially contains Air2, while TRAMP5 appears to only contain Air1 21–24. Recent work from our lab combined phenotypic, genetic and deep sequencing techniques to confirm that Air1 and Air2 serve as critical determinants of TRAMP specificity in S. cerevisiae 64. Air2 functions in the polyadenylation and subsequent degradation of many snoRNAs, and plays a role in regulation of transcripts encoding proteins involved in iron and carbon metabolism. Air1, along with the ribonuclease Rrp6, is required for stable copy number of the endogenous 2μ plasmid. Together, these findings reveal that Air1 and Air2 function uniquely, and direct the specificity of TRAMP4 and TRAMP5 complexes. The high degree of similarity between the two proteins begs the question of what protein characteristics determine the observed specificity. There are several possible determinants of Air specificity, and it is likely that they are not mutually exclusive.
First, it is possible that the zinc knuckles of Air1 and Air2 bind to specific and distinct sequence motifs located on target transcripts, as do zinc knuckles of other RNA binding proteins 65, 66. Computational methods such as MEME analysis can be used to identify sequence motifs found on a subset of transcripts, such as those bound by a protein of interest 67. Unfortunately, MEME analysis of subsets of transcripts exhibiting differential expression in the absence of either AIR1 or AIR2 has not yet identified conserved sequence motifs (K. Schmidt, J. S. Butler, unpublished) 64. This may be a consequence of inclusion of transcripts that exhibit differential gene expression due to indirect effects of AIR1 or AIR2 deletion. Regardless, despite the current lack of evidence, target sequence recognition by Air1 and Air2 has not been ruled out as a possible method for TRAMP recruitment.
Additionally, certain structural characteristics of transcripts may result in either recruitment or exclusion of particular RNA binding proteins (RBPs). First, efficient binding of RBPs to sequence motifs requires a level of structural accessibility on target transcripts 68. Irregular RNA folds, such as those likely found on aberrant transcripts, may sequester sequence motifs with regions of dsRNA, rendering them inaccessible to proteins necessary for 3′ end processing, transcript maturation or nuclear export. In the absence of normally bound proteins, the 3′ ends are available for Air1 or Air2 binding and degradation by the exosome. Alternatively, transcript structure may contribute directly to recruitment of RBPs. Binding of an RBP to a particular structure may occur with sequence specificity, such as is the case of the yeast sequence-specific RBP Vts1 69, or may occur independently of structure sequence. For example, K-turns are 60° kinks that occur in double stranded portions of many functional RNAs, including ribosomal RNAs, snRNAs, mRNAs and riboswitches 70. This unique structural feature serves as a binding platform for a diverse range of RBPs, and the varied sequences that have thus far been found within K-turn motifs suggest that RBP specificity is achieved independently of sequence. It is therefore possible that Air1 and Air2 (or associated proteins) recognize distinct structural characteristics on target RNAs.
Differences in Air protein function may also be related to uniquely associated proteins. For example, if Air1 or Air2 interacts with other RBPs, the second protein may mediate recruitment of TRAMP to target transcripts. Indeed, Air2 co-purifies with Nrd1 and Nab3, RNA binding proteins that direct transcription termination of non-polyadenylated RNAs including sn-/snoRNAs and CUTs 71–74. The physical interaction between Air2 and Nrd1 appears to be functionally relevant, as Trf4 and/or Air2 are linked to Nrd1/Nab3 directed transcription termination of a variety of transcripts 16, 19, 64, 74–76, and Air2 is required for turnover of Nrd1/Nab3 terminated snoRNAs 64. The observed Air2 and Nrd1 interaction may result from close association of these proteins on shared transcript targets, or Air2 and Nrd1 may interact independently of RNA and be co-recruited to RNA substrates. Regardless of the mechanism of interaction, it seems likely that the presence of TRAMP at regions of transcription termination allows for active surveillance of newly synthesized transcript.
In contrast, over-expressed Air1 was shown to co-purify with Hrb1, a poly(A) binding protein involved in mRNA export 61–63. Hrb1 is methylated by the arginine methyltransferase Hmt1, which also methylates and regulates the export of the hnRNP proteins Nab2, Hrp1 and Npl3 60, 77–79. The function of Hrb1 methylation, however, remains unclear 77. As previously mentioned, Air1 and Air2 inhibit Hmt1 mediated methylation of Npl3 59. Therefore, it is possible that Hmt1 mediates the physical interaction between Hrb1 and Air1. It may be interesting to determine if endogenous Air1 and Hrb1 interact, if this interaction if mediated through the Hmt1 methyltransferase, and finally if this interaction is unique from the interaction of Air2 and Hmt1/Npl3. Identification and characterization of unique protein interactors of Air1 and Air2 may reveal mechanisms of TRAMP recruitment and/or specificity.
Finally, it has been proposed that unique sub-nuclear localizations of TRAMP components contribute to observed substrate specificities. In S. cerevisiae and other eukaryotes, nuclear proteins may be located throughout the nucleus, or remain concentrated within the denser nucleolus, where ribosomal RNA is transcribed. As mentioned, Trf5-GFP and Air1-GFP fusions exhibit slight nucleolar enrichment compared to Trf4-GFP and Air2-GFP 29–31. Given this, it would be expected that Trf5 and Air1 are the preferred proteins involved in RNA surveillance of ribosomal RNAs. However, Trf4 is required for efficient polyadenylation and turnover of both precursor and mature ribosomal RNAs 21, 38, 80. Furthermore, Trf4-GFP shifts localization and accumulates in the nucleolus under conditions of nucleolar ribosomal RNA accumulation 80. This suggests that the observed sub-nuclear localizations of TRAMP components result from, rather than cause, inherent differences in substrate specificities. It is therefore unlikely that protein localization is the strongest determinant of TRAMP4 and TRAMP5 specificity.
CONSERVATION OF TRAMP: FROM YEAST TO HUMANS
Components of the TRAMP complex are conserved through a wide range of species, including yeast, flies and even higher eukaryotes. The fission yeast Schizosaccharomyces pombe’s TRAMP complex, consisting of Cid14, Air1 and Mtr4, is functionally homologous to budding yeast’s TRAMP 81. The nuclear Cid14 is required for polyadenylation and degradation of S. pombe rRNAs, and is able to complement lack of a function S. cerevisiae Trf4 protein 82. Drosophila melanogastor contains two Trf homologs, DmTrf4-1 and DmTrf4-2. DmTrf4-1 likely polyadenylates snRNAs and signals for their degradation by DmRrp6, analogous to the S. cerevisiaes Trf4 and Rrp6 83.
Mammals also contain TRAMP homologs. Murine Papd5, similar to Trf4, polyadenylates aberrant pre-rRNA transcripts and targets them for degradation by the exosome 84. It remains unknown if Papd5 functions independently, or in context of a murine TRAMP complex. Human hPAPD5/hTrf4-2 and hPAPD7/hTrf4-1 are 37% identical to the catalytic domain of S. cerevisiae Trf4, and hZCCHC7/hAir1 is 35% identical to Air1 across the 5 zinc knuckle motifs. The highest conserved TRAMP component, however, is hMtr4/SKIV2L2, exhibiting 51% overall identity to Mtr4. Consistent with S. cerevisiae’s Mtr4, hMtr4 interacts functionally and physically with the exosome for efficient 3′ end processing of 5.8S rRNA 85, 86. Although a functionally related TRAMP complex has yet to be characterized in humans, it is likely that such a complex exists. First, PROMPTs (PROMoter uPstream Transcripts) and some mRNAs destined for degradation by the exosome contain short, non-encoded poly(A) tails, suggesting that polyadenylation is likely involved in targeting transcripts to the human exosome 87, 88. Curiously, although hTrf4-2 adenylates 3′ ends of PROMPTs, this is not required for turnover of these transcripts 86, 88. Rather, the Nuclear EXosome Targeting (NEXT) complex, composed of hMtr4, a zinc knuckle protein ZCCHC8 and a putative RNA binding protein RBM7, likely target PROMPTs for degradation by the exosome 86. Second, co-IP experiments suggest that the TRAMP homologs hMtr4, hTrf4-1 and hTrf54-2 are recruited to the pre-mRNA spliceosome to degrade aberrantly spliced transcripts and/or introns 47, 86, 89. While it is unclear if these proteins are recruited to the pre-mRNA processing machinery as a complex, hMTR4 and hRRP6 precipitates contain hAir1 and hTrf4-2, suggesting structurally conservation of a human TRAMP complex 86. Lastly, the putative dimer hTrf4-2/hAir1 polyadenylates rRNA degradation products, while hMtr4 likely recruits the exosome for degradation, again analogous to functions of the yeast homologs 86. Together, these studies are suggestive of a functional human Trf/Air/Mtr4 complex.
GENE SILENCING AND THE EXOSOME
In eukaryotes, DNA is wrapped around histone octomers to form nucleosomes, which further condense to form chromatin. The structure and condensation of chromatin is regulated by chromatin remodeling factors and DNA- and histone- modifying enzymes, which alter the chromatin and allow for higher degrees of compaction. Heterochromatin, regions of highly condensed DNA commonly marked by methylation of H3K9, is necessary for stable inheritance of chromosomes and for proper regulation of gene expression. Historically, compact heterochromatin is believed to repress gene expression by occluding transcriptional machinery. Consistently, heterochromatin is often found in areas of repetitive DNA sequences, including telomeric, centromeric, and ribosomal DNA. Recently, however, the pervasive presence of, and transcription by, Pol II was shown to generate non-coding RNAs corresponding to regions of DNA previously believe to be transcriptionally silent, including heterochromatin 44, 76, 90, 91. This suggests that organisms are able to silence these regions post-transcriptionally, such as through degradation of transcripts by the RNAi or RNA surveillance machinery. Furthermore, multiple heterochromatin formation pathways have evolved, and it has recently been suggested that RNA surveillance machinery may contribute to heterochromatin formation directly.
Non-coding RNAs can initiate heterochromatin formation through either the RNAi machinery (RNAi-dependent) in organisms capable of RNAi, including the fission yeast Schizosaccharomyces pombe, or through RNA processing machinery (RNAi-independent). In the RNAi-dependent pathway, dicer processes dsRNAs generated from heterochromatic regions into small interfering RNAs (siRNAs), which are then bound by argonaute. The argonaute-siRNA complexes are subsequently loaded onto the RNA-induced transcriptional silencing (RITS) complex, which is related to the RISC posttranslational silencing complex of RNAi. Clr4/Suv39h targets RITS to heterochromatic regions through siRNA binding, as well as through binding of Chp1 to H3K9 methylated chromatin. RITS and other associated factors then act as a scaffold to recruit chromatin modifiers capable of generating regions of heterochromatin, including Clr4, which further methylates H3K9 and spreads heterochromatin 92. Mlo3, an RNA quality control and export factor, is one protein recruited to these regions of heterochromatin formation 93, 94. The recruitment of Mlo3 may be a critical determinate of RNA fate, as the protein interacts with the S. pombe’s TRAMP complex 93. TRAMP and the exosome can then degrade cryptic unstable transcripts originating from heterochromatic regions 95–97. In this way, even if RNA is transcribed from regions of heterochromatin, the cell is able to quickly eliminate it though recruitment of TRAMP and degradation by the exosome in this Co-Transcriptional Gene Silencing (CTGS) pathway.
Organisms are also capable of forming heterochromatin independently of RNAi machinery. In Saccharomyces cerevisiae, which lacks RNAi completely, heterochromatin is found at the silent mating type loci (HMLa and HMRα), in addition to rDNA, centromeric and sub-telomeric regions. Histone deacetylation by Sir2, along with the activities of other histone and chromatin modifying proteins, are required for the establishment and maintenance of heterochromatin 98. TRAMP and the exosome are also required for efficient silencing of some heterochromatic regions in Saccharomyces cerevisiae, as absence of these components results in an accumulation of transcripts originating from regions of repressed chromatin 44, 76, 99. Interestingly, it appears that ncRNA transcription is required for efficient silencing, as mutations in the Nrd1-Nab3-Sen1 3′ end-processing pathway also results in de-repression of silent heterochromatin 44, 76. Even in RNAi capable S. pombe, components of RNA surveillance act in a pathway parallel to the RNAi-dependent pathway to promote centromeric heterochromatin formation 100. Similar to S. cerevisiae, transcription of non-coding heterochromatic RNAs in fission yeast is required for efficient targeting of heterochromatin, though the exact mechanism of RNAi-independent heterochromatin nucleation at centromeres remains unknown 100.
Recently, the RNA degradation machinery of S. pombe was found to directly impact formation of heterochromatin. Heterochromatin ‘islands’ are located around meiotic genes, whose expression is repressed during vegetative growth 101. During vegetative growth, meiotic mRNAs are recognized by the RNA binding protein Mmi1, which interacts with the poly(A) binding protein Pab2 to promote post-transcriptional elimination of meiotic RNAs via the exosome 102–105. More recently, ChIP analysis revealed that loss of Mmi1 or Rrp6 abolished H3K9me at heterochromatin islands, directly implicating the RNA degradation factors in the assembly of heterochromatin at meiotic genes 101. The requirement of RNA degradation machinery for efficient silencing of heterochromatic DNA is two-fold; first, RNA degradation machinery degrades transcripts ‘escaping’ from silent heterochromatin, and second, the proteins are directly involved in the formation of heterochromatin, possibly by recruiting chromatin-modifying proteins. It therefore seems that cells have evolved an elegant system in which non-coding RNAs serve as a read-out of transcriptional activity, signalling for elimination of these RNAs through formation of heterochromatin and/or recruitment of RNA degradation machinery. Surprisingly, posttranscriptional elimination of meiotic RNAs is not dependent on Cid14, the S. pombe homolog of Trf4 102, 103, though it remains unclear if TRAMP is required alongside Rrp6 for formation of heterochromatin.
In S. cerevisiae, TRAMP is linked directly to gene silencing. In addition to heterochromatin, S. cerevisiae has evolved a number of other ways to efficiently silence genes. These pathways involve non-coding RNAs that are subject to TRAMP-mediated degradation 47, 64. First, in a transcriptional interference mechanism, non-coding RNA transcription can directly alter chromatin structure and histone modification of a region of DNA. These alterations can affect the ability of polymerases or activators to bind to the cognate sense strand, thereby modulating transcription of the coding sense transcript. Transcriptional interference has thus far been shown to repress expression of genes involved in such diverse processes as amino acid biosynthesis, mitochondrial function, and meiosis 106–109. Remarkably, transcription of an antisense non-coding RNA can also activate transcription of the sense transcript, as is the case for PHO5 110.
In addition to the cis regulatory pathway of transcriptional interference (where transcription of a non-coding RNA can affect transcription of a directly adjacent sense RNA), non-coding RNAs can also affect sense transcription in trans. In order for a non-coding RNA to function in trans, the transcript itself, and not the act of transcription, is able to direct site-specific modifications of its cognate sense transcript at separate DNA loci. Expression of PHO84 is regulated in trans by non-coding RNAs that direct RNA-dependent modifications of the Hda1/2/3 histone deacetylase complex 111, 112. Rrp6 is responsible for turnover of the PHO84 regulatory non-coding RNAs, as deletion of Rrp6 stabilizes these transcripts 111. Thus, the RNA degradation machinery can indirectly affect gene silencing of PHO84 by regulating expression levels of trans regulatory non-coding RNAs. Uncovering more genes subject to trans non-coding RNA regulation may expand the role of RNA surveillance factors in gene regulation.
Finally, it has been suggested that a major role of the RNAi pathway is to silence transposable elements 113, 114. In the absence of RNAi, S. cerevisiae has evolved to inhibit Ty1 retrotransposition through histone deacetylation by Set1, mediated in trans by an antisense Ty1 CUT 115, 116. Interestingly, although the cytoplasmic 5′ to 3′ exoribonuclease Xrn1 degrades the antisense Ty1 RNA, it does not appear that Rrp6, Trf4 or Trf5 are involved in turnover of the antisense RNA 47, 116. Nevertheless, deletion of trf4 or trf5 results in correlated fluctuations in the sense and antisense RNAs from Ty1, suggesting that TRAMP functions in retrotransposon silencing through an uncharacterized mechanism 47.
Conclusion
Research on nuclear RNA metabolism in eukaryotes over the last decade revealed a remarkably complex array of transcription products whose processing and degradation occur within the nucleus. In addition to mRNA, tRNA, rRNA and sn-snoRNAs, transcription produces a vast array of long and short non-coding RNAs whose potential for regulation of gene expression is just beginning to come to light. All of these RNAs undergo post-transcriptional processing events that determine their final structures and concentrations. Studies have revealed that the RNA exosome plays a central role in shaping the final products and, in many cases, destroying transcripts that fail to pass structural and/or kinetic transitions. These include barriers to splicing, polyadenylation and correct ribonucleoprotein complex formation. Understanding the detailed mechanisms that specifically target transcripts for processing or degradation by the RNA exosome constitutes an exciting challenge for the field of RNA surveillance.
The TRAMP complex provides one mechanism for control of RNA surveillance by the nuclear exosome. Its ability to recognize and target transcripts for interaction with the exosome suggests that it plays a critical role in substrate choice. Recent studies of TRAMP function indicate that it can function in pathways that lead to 3′ end formation or degradation of RNAs. Whether these distinctions reflect properties inherent to the components of TRAMP or its ability to interact with various adaptors remains an important area of ongoing research. Nevertheless, current evidence makes clear the roles that TRAMP and the exosome play in controlling the fate of RNAs in the nucleus, which results in significant and fascinating contributions to the control of gene expression in eukaryotes.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-0817324 to J.S.B.) and the National Institutes of Health (NIH T-32 GM068411-05 to K.S.).
Footnotes
The authors have no conflicts of interest.
Contributor Information
Karyn Schmidt, Department of Biochemistry and Biophysics, Center for RNA Biology, University of Rochester Medical Center, Rochester, NY 14642.
J. Scott Butler, Email: Scott_Butler@urmc.rochester.edu, Department of Biochemistry and Biophysics, Center for RNA Biology, Department of Microbiology and Immunology, University of Rochester Medical Center, Rochester, NY 14642.
References
- 1.Fasken MB, Corbett AH. Mechanisms of nuclear mRNA quality control. RNA biology. 2009;6:237–241. doi: 10.4161/rna.6.3.8330. [DOI] [PubMed] [Google Scholar]
- 2.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nature reviews. Molecular cell biology. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 3.Lykke-Andersen S, Brodersen DE, Jensen TH. Origins and activities of the eukaryotic exosome. J Cell Sci. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- 4.Doma MK, Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 7.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- 8.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 9.Wang HW, Wang J, Ding F, Callahan K, Bratkowski MA, Butler JS, Nogales E, Ke A. Architecture of the yeast Rrp44 exosome complex suggests routes of RNA recruitment for 3′ end processing. Proc Natl Acad Sci U S A. 2007;104:16844–16849. doi: 10.1073/pnas.0705526104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briggs MW, Burkard KT, Butler JS. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem. 1998;273:13255–13263. doi: 10.1074/jbc.273.21.13255. [DOI] [PubMed] [Google Scholar]
- 11.Callahan KP, Butler JS. Evidence for core exosome independent function of the nuclear exoribonuclease Rrp6p. Nucleic Acids Res. 2008;36:6645–6655. doi: 10.1093/nar/gkn743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res. 2000;28:1684–1691. doi: 10.1093/nar/28.8.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Hoof A, Lennertz P, Parker R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol. 2000;20:441–452. doi: 10.1128/mcb.20.2.441-452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wlotzka W, Kudla G, Granneman S, Tollervey D. The nuclear RNA polymerase II surveillance system targets polymerase III transcripts. The EMBO journal. 2011;30:1790–1803. doi: 10.1038/emboj.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciais D, Bohnsack MT, Tollervey D. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 2008;36:3075–3084. doi: 10.1093/nar/gkn160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth KM, Byam J, Fang F, Butler JS. Regulation of NAB2 mRNA 3′-end formation requires the core exosome and the Trf4p component of the TRAMP complex. RNA. 2009;15:1045–1058. doi: 10.1261/rna.709609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemay JF, D’Amours A, Lemieux C, Lackner DH, St-Sauveur VG, Bahler J, Bachand F. The nuclear poly(A)-binding protein interacts with the exosome to promote synthesis of noncoding small nucleolar RNAs. Mol Cell. 2010;37:34–45. doi: 10.1016/j.molcel.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 19.Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell. 2008;32:247–258. doi: 10.1016/j.molcel.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 22.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 23.Houseley J, Tollervey D. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 2006;7:205–211. doi: 10.1038/sj.embor.7400612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson JT, Wang X. Nuclear RNA surveillance: no sign of substrates tailing off. Crit Rev Biochem Mol Biol. 2009;44:16–24. doi: 10.1080/10409230802640218. [DOI] [PubMed] [Google Scholar]
- 26.Holub P, Lalakova J, Cerna H, Pasulka J, Sarazova M, Hrazdilova K, Arce MS, Hobor F, Stefl R, Vanacova S. Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation. Nucleic Acids Res. 2012;40:5679–5693. doi: 10.1093/nar/gks223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 28.Hamill S, Wolin SL, Reinisch KM. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci U S A. 2010;107:15045–15050. doi: 10.1073/pnas.1003505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fasken MB, Leung SW, Banerjee A, Kodani MO, Chavez R, Bowman EA, Purohit MK, Rubinson ME, Rubinson EH, Corbett AH. Air1 zinc knuckles 4 and 5 and a conserved IWRxY motif are critical for the function and integrity of the TRAMP RNA quality control complex. J Biol Chem. 2011 doi: 10.1074/jbc.M111.271494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 31.Walowsky C, Fitzhugh DJ, Castano IB, Ju JY, Levin NA, Christman MF. The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J Biol Chem. 1999;274:7302–7308. doi: 10.1074/jbc.274.11.7302. [DOI] [PubMed] [Google Scholar]
- 32.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci U S A. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E. The RNA Helicase Mtr4p Modulates Polyadenylation in the TRAMP Complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernstein J, Patterson DN, Wilson GM, Toth EA. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, a 3′->5′ helicase partner of the nuclear exosome. J Biol Chem. 2008;283:4930–4942. doi: 10.1074/jbc.M706677200. [DOI] [PubMed] [Google Scholar]
- 36.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 37.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA. 2006;12:508–521. doi: 10.1261/rna.2305406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Jia H, Jankowsky E, Anderson JT. Degradation of hypomodified tRNA(iMet) in vivo involves RNA-dependent ATPase activity of the DExH helicase Mtr4p. RNA. 2008;14:107–116. doi: 10.1261/rna.808608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callahan KP, Butler JS. TRAMP complex enhances RNA degradation by the nuclear exosome component Rrp6. J Biol Chem. 2010;285:3540–3547. doi: 10.1074/jbc.M109.058396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:8581–8586. doi: 10.1073/pnas.0402888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burkard KT, Butler JS. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egecioglu DE, Henras AK, Chanfreau GF. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houseley J, Kotovic K, El Hage A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip Rev RNA. 2011;2:348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 46.Mohanty BK, Kushner SR. Bacterial/archaeal/organellar polyadenylation. Wiley Interdiscip Rev RNA. 2011;2:256–276. doi: 10.1002/wrna.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.San Paolo S, Vanacova S, Schenk L, Scherrer T, Blank D, Keller W, Gerber AP. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 2009;5:e1000555. doi: 10.1371/journal.pgen.1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rougemaille M, Gudipati RK, Olesen JR, Thomsen R, Seraphin B, Libri D, Jensen TH. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castano IB, Heath-Pagliuso S, Sadoff BU, Fitzhugh DJ, Christman MF. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 1996;24:2404–2410. doi: 10.1093/nar/24.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jia H, Wang X, Anderson JT, Jankowsky E. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc Natl Acad Sci U S A. 2012;109:7292–7297. doi: 10.1073/pnas.1201085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol. 1987;7:3268–3276. doi: 10.1128/mcb.7.9.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Viphakone N, Voisinet-Hakil F, Minvielle-Sebastia L. Molecular dissection of mRNA poly(A) tail length control in yeast. Nucleic Acids Res. 2008;36:2418–2433. doi: 10.1093/nar/gkn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soucek S, Corbett AH, Fasken MB. The long and the short of it: The role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Biochim Biophys Acta. 2012;1819:546–554. doi: 10.1016/j.bbagrm.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saguez C, Schmid M, Olesen JR, Ghazy MA, Qu X, Poulsen MB, Nasser T, Moore C, Jensen TH. Nuclear mRNA surveillance in THO/sub2 mutants is triggered by inefficient polyadenylation. Mol Cell. 2008;31:91–103. doi: 10.1016/j.molcel.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 56.Das B, Butler JS, Sherman F. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol Cell Biol. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schmid M, Poulsen MB, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz LM, Moore C, Jensen TH. Rrp6p controls mRNA poly(A) tail length and its decoration with poly(A) binding proteins. Mol Cell. 2012;47:267–280. doi: 10.1016/j.molcel.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Larochelle M, Lemay JF, Bachand F. The THO complex cooperates with the nuclear RNA surveillance machinery to control small nucleolar RNA expression. Nucleic Acids Res. 2012;40:10240–10253. doi: 10.1093/nar/gks838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue K, Mizuno T, Wada K, Hagiwara M. Novel RING finger proteins, Air1p and Air2p, interact with Hmt1p and inhibit the arginine methylation of Npl3p. J Biol Chem. 2000;275:32793–32799. doi: 10.1074/jbc.M004560200. [DOI] [PubMed] [Google Scholar]
- 60.McBride AE, Cook JT, Stemmler EA, Rutledge KL, McGrath KA, Rubens JA. Arginine methylation of yeast mRNA-binding protein Npl3 directly affects its function, nuclear export, and intranuclear protein interactions. J Biol Chem. 2005;280:30888–30898. doi: 10.1074/jbc.M505831200. [DOI] [PubMed] [Google Scholar]
- 61.Hacker S, Krebber H. Differential export requirements for shuttling serine/arginine-type mRNA-binding proteins. J Biol Chem. 2004;279:5049–5052. doi: 10.1074/jbc.C300522200. [DOI] [PubMed] [Google Scholar]
- 62.Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 63.Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A. 2004;101:1858–1862. doi: 10.1073/pnas.0308663100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt K, Xu Z, Mathews DH, Butler JS. Air proteins control differential TRAMP substrate specificity for nuclear RNA surveillance. RNA. 2012;18:1934–1945. doi: 10.1261/rna.033431.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J, Allain FH. Structural basis of pre-let-7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat Struct Mol Biol. 2012;19:84–89. doi: 10.1038/nsmb.2202. [DOI] [PubMed] [Google Scholar]
- 67.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li X, Quon G, Lipshitz HD, Morris Q. Predicting in vivo binding sites of RNA-binding proteins using mRNA secondary structure. RNA. 2010;16:1096–1107. doi: 10.1261/rna.2017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aviv T, Lin Z, Ben-Ari G, Smibert CA, Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol. 2006;13:168–176. doi: 10.1038/nsmb1053. [DOI] [PubMed] [Google Scholar]
- 70.Lilley DM. The structure and folding of kink turns in RNA. Wiley Interdiscip Rev RNA. 2012;3:797–805. doi: 10.1002/wrna.1136. [DOI] [PubMed] [Google Scholar]
- 71.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 72.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 73.Vasiljeva L, Buratowski S. Nrd1 interacts with the nuclear exosome for 3′ processing of RNA polymerase II transcripts. Mol Cell. 2006;21:239–248. doi: 10.1016/j.molcel.2005.11.028. [DOI] [PubMed] [Google Scholar]
- 74.Arigo JT, Eyler DE, Carroll KL, Corden JL. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell. 2006;23:841–851. doi: 10.1016/j.molcel.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 75.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 76.Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 77.Shen EC, Henry MF, Weiss VH, Valentini SR, Silver PA, Lee MS. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 1998;12:679–691. doi: 10.1101/gad.12.5.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng X, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J Biol Chem. 2002;277:7752–7760. doi: 10.1074/jbc.M110053200. [DOI] [PubMed] [Google Scholar]
- 79.Xu C, Henry MF. Nuclear export of hnRNP Hrp1p and nuclear export of hnRNP Npl3p are linked and influenced by the methylation state of Npl3p. Mol Cell Biol. 2004;24:10742–10756. doi: 10.1128/MCB.24.24.10742-10756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dez C, Houseley J, Tollervey D. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 2006;25:1534–1546. doi: 10.1038/sj.emboj.7601035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Keller C, Woolcock K, Hess D, Buhler M. Proteomic and functional analysis of the noncanonical poly(A) polymerase Cid14. RNA. 2010;16:1124–1129. doi: 10.1261/rna.2053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol. 2006;26:1710–1721. doi: 10.1128/MCB.26.5.1710-1721.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakamura R, Takeuchi R, Takata K, Shimanouchi K, Abe Y, Kanai Y, Ruike T, Ihara A, Sakaguchi K. TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster. Mol Cell Biol. 2008;28:6620–6631. doi: 10.1128/MCB.00448-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–111. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schilders G, van Dijk E, Pruijn GJ. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lubas M, Christensen MS, Kristiansen MS, Domanski M, Falkenby LG, Lykke-Andersen S, Andersen JS, Dziembowski A, Jensen TH. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011;43:624–637. doi: 10.1016/j.molcel.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 87.West S, Gromak N, Norbury CJ, Proudfoot NJ. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell. 2006;21:437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 88.Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A, Jensen TH. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179–7193. doi: 10.1093/nar/gkr370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nag A, Steitz JA. Tri-snRNP-associated proteins interact with subunits of the TRAMP and nuclear exosome complexes, linking RNA decay and pre-mRNA splicing. RNA Biol. 2012;9:334–342. doi: 10.4161/rna.19431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 91.Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 92.Reyes-Turcu FE, Grewal SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–163. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang K, Fischer T, Porter RL, Dhakshnamoorthy J, Zofall M, Zhou M, Veenstra T, Grewal SI. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331:1624–1627. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thakurta AG, Gopal G, Yoon JH, Kozak L, Dhar R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–2523. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang SW, Stevenson AL, Kearsey SE, Watt S, Bahler J. Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol. 2008;28:656–665. doi: 10.1128/MCB.01531-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Buhler M, Haas W, Gygi SP, Moazed D. RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell. 2007;129:707–721. doi: 10.1016/j.cell.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 97.Murakami H, Goto DB, Toda T, Chen ES, Grewal SI, Martienssen RA, Yanagida M. Ribonuclease activity of Dis3 is required for mitotic progression and provides a possible link between heterochromatin and kinetochore function. PLoS One. 2007;2:e317. doi: 10.1371/journal.pone.0000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bi X. Functions of chromatin remodeling factors in heterochromatin formation and maintenance. Sci China Life Sci. 2012;55:89–96. doi: 10.1007/s11427-012-4267-1. [DOI] [PubMed] [Google Scholar]
- 99.Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reyes-Turcu FE, Zhang K, Zofall M, Chen E, Grewal SI. Defects in RNA quality control factors reveal RNAi-independent nucleation of heterochromatin. Nat Struct Mol Biol. 2011;18:1132–1138. doi: 10.1038/nsmb.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, Grewal SI. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science. 2012;335:96–100. doi: 10.1126/science.1211651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen HM, Futcher B, Leatherwood J. The fission yeast RNA binding protein Mmi1 regulates meiotic genes by controlling intron specific splicing and polyadenylation coupled RNA turnover. PLoS One. 2011;6:e26804. doi: 10.1371/journal.pone.0026804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.St-Andre O, Lemieux C, Perreault A, Lackner DH, Bahler J, Bachand F. Negative regulation of meiotic gene expression by the nuclear poly(a)-binding protein in fission yeast. J Biol Chem. 2010;285:27859–27868. doi: 10.1074/jbc.M110.150748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 105.Yamanaka S, Yamashita A, Harigaya Y, Iwata R, Yamamoto M. Importance of polyadenylation in the selective elimination of meiotic mRNAs in growing S. pombe cells. EMBO J. 2010;29:2173–2181. doi: 10.1038/emboj.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bird AJ, Gordon M, Eide DJ, Winge DR. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 2006;25:5726–5734. doi: 10.1038/sj.emboj.7601453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 108.Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- 109.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci U S A. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 112.Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev. 2009;23:1534–1545. doi: 10.1101/gad.522509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Coy S, Vasiljeva L. The exosome and heterochromatin: multilevel regulation of gene silencing. Adv Exp Med Biol. 2010;702:105–121. [PubMed] [Google Scholar]
- 114.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci U S A. 2009;106:15657–15662. doi: 10.1073/pnas.0908305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chan S, Choi EA, Shi Y. Pre-mRNA 3′-end processing complex assembly and function. Wiley Interdiscip Rev RNA. 2011;2:321–335. doi: 10.1002/wrna.54. [DOI] [PMC free article] [PubMed] [Google Scholar]