Abstract
Because abnormal development of striatal neurons is thought to be part of pathology underlying major psychiatric illnesses, we studied the expression pattern of genes involved in striatal development and of genes comprising key striatal-specific pathways, during an active striatal maturation period, the first two postnatal weeks in rat. This period parallels human striatal development during the second trimester, when prenatal stress is though to lead to increased risk for neuropsychiatric disorders.
In order to identify genes involved in this developmental process, we used subtractive hybridization, followed by quantitative real-time PCR, which allowed us to characterize the developmental expression of over 60 genes, many not previously known to play a role in neuromaturation. Of these 12 were novel transcripts, which did not match known genes, but which showed strict developmental expression and may play a role in striatal neurodevelopment.
We show that during the first two postnatal weeks in rat, an early gene expression network, still lacking key striatal-specific signaling pathways, is downregulated and replaced by a mature gene expression network, containing key striatal-specific genes including the dopamine D1 and D2 receptors, conferring to these neurons their functional identity. Therefore, before this developmental switch, striatal neurons lack many of their key phenotypic characteristics. This maturation process is followed by a striking rise in expression of myelination genes, indicating a striatal-specific myelination event.
Such strictly controlled developmental program has the potential to be a point of susceptibility to disruption by external factors. Indeed, this period is known to be a susceptibility period in both humans and rats.
Keywords: neurodevelopment, schizophrenia, dopamine, striatum, medium spiny neurons, gene expression
Introduction
The goal of this study was to analyze the developmental gene expression within the striatum during an active neuromaturation period. Evidence, showing that this is a susceptibility period in both rats and humans, suggested that there is an important developmental process in progress during this time, the classification of which may advance our understanding of striatal neuromaturation and of diseases resulting from its abnormal completion.
The striatum is an important signaling hub of the brain, collecting information and distributing it to a number of targets. It receives extensive glutamatergic inputs from the cortex, the thalamus, the hippocampus, and the amygdala. It integrates this information and conveys it to its targets through basal ganglia output nuclei: the globus pallidus, subthalamic nucleus, substantia nigra, and the ventral tegmental area (VTA) (reviewed in (Bolam et al., 2000; Kreitzer and Malenka, 2008)). Over 95% of striatal neurons are GABAergic medium spiny neurons (MSNs) (Matamales et al., 2009). The remainder of the neuronal population is composed of GABAergic and cholinergic interneurons, which modulate MSN function (Tepper and Bolam, 2004).
There are two types of MSNs, each forming one of two major output pathways of the striatum. MSNs which express dopamine receptor type 1 (D1R) and dynorphin form the direct pathway to substantia nigra, while MSNs expressing dopamine receptor type 2(D2R), enkephalin and substance P project to globus pallidus, before reaching substantia nigra, and form the indirect pathway (Gerfen, 1992). These exert opposing effects on one of the major MSN signalling cascades involving adenylyl cyclase (AC), cAMP and cAMP-dependent protein kinase (PKA), which is an important regulator of neuronal function (Sibley, 1995). D2Rs also activate the Gβγ-IP3 pathway and generate Ca2+ influx (Hernandez-Lopez et al., 2000). In addition to their differential dopamine receptor expression, the striatonigral and striatopallidal neurons differ significantly in their expression of other signalling pathways, in particular in expression of NMDA (Jocoy et al., 2011), AMPA (Deng et al., 2007) and adenosine receptor subunits (Schiffmann et al., 1991), as well as neuropeptides (Gerfen and Young, 1988) and cannabinoids (Laviolette and Grace, 2006). Because of their very different properties (Alexander et al., 1986), imbalance in these two pathways has been implicated in diseases such as Huntington’s, Parkinson’s, bipolar disorder, depression and schizophrenia (Dujardin and Laurent, 2003; Simpson et al., 2010; Thomas, 2006). Specifically, abnormal expression of the D2R, which is the primary site of action for all antipsychotics, has been strongly linked to the etiology of schizophrenia (Seeman, 2011). Hence, the establishment of proper balance of D1R and D2R expression during striatal neurodevelopment is essential. Furthermore, evidence supports the hypothesis that dysregulation of MSN function observed in neuropsychiatric disease may result from an alteration in an early neurodevelopmental process (reviewed in (Crittenden and Graybiel, 2011; Welham et al., 2009)). A number of variables including stress, nutritional deficiencies (Bao et al., 2012) and immune challenge (Meyer, 2011; Meyer et al., 2011) during the second trimester of human development (Mednick et al., 1994; Mednick et al., 1988), a period when the striatum is undergoing active neurodevelopment, are thought to be able to induce such alterations (reviewed in (Brown and Patterson, 2011; Palomo et al., 2002)).
Because the striatum has many pathways connecting to many brain regions, and because its dysfunction is associated with a number of neuropsychiatric disorders (Dujardin and Laurent, 2003; Meyer-Lindenberg et al., 2002; Simpson et al., 2010; Thomas, 2006), the neuromaturation period of the striatum warranted a detailed study. Since the equivalent stage of striatal development in rats occurs during the second postnatal week of life (Clancy et al., 2001), we were able to analyze this critical progression of striatal maturation in a postnatal animal model.
The striatal expression of candidate genes, such as D1 and D2 dopamine receptors, and genes known to be involved in striatal development, was examined during the second postnatal week in rat. We then used the subtractive hybridization method to identify additional genes involved in this process. In total we have classified the developmental expression of over 60 genes, many previously not known to participate in the developmental process. This includes several novel transcripts, which do not match the sequence of any known genes, but which are strictly developmentally regulated.
We show that striatal neurons undergo a controlled transformative neuromaturation program during the second postnatal week in rat. During this period, an entire network of early gene transcripts became down-regulated and replaced by a mature transcriptional network. This mature network included the D1 and D2 dopamine receptors, which exhibited a steep rise in expression during the second postnatal week. It also included many genes used as MSN markers, which represent key MSN-specific pathways. This maturation process was followed by a striking myelination event, with a steep upregulation of myelin-related genes, which may represent another point of susceptibility, as expression of these genes was shown to be altered in response to immune system activation (Fatemi et al., 2008; Leitner and Connor, 2012; Tatar et al., 2010).
In summary, we show that immature striatal neurons do not express many genes essential for their MSN-specific function until the end of the second postnatal week. Because they gain their full projection neuron identity during this neurodevelopmental process, it’s abnormal completion may be of high relevance to neuropsychiatric diseases thought to result from abnormal striatal neuron development. This is further supported by observations that this period is a known susceptibility period for the later development of psychiatric illness (Boksa and El-Khodor, 2003; Brown and Patterson, 2011; Egan and Weinberger, 1997; Fredriksson et al., 2004; Hansen et al., 2004; Keshavan and Murray, 1997; Mednick et al., 1994; Palomo et al., 2002). Hence, disruption of this process by environmental insults may play a significant role in the etiology of neuropsychiatric disease.
Methods
Animals
All procedures with animals were performed in accordance with guidelines from the Canadian Council on Animal Care and were approved by the University of Toronto Animal Care Committee. Sprague Dawley rats (Charles River, St. Constant, Quebec, Canada) were housed one litter per cage and maintained on a 12h:12h light:dark schedule with free access to food and water.
Tissues
Animals were euthanized by carbon dioxide overdose. Tissues were dissected and quickly frozen in precooled tubes on dry ice. Tissues were collected at 2pm,starting at postnatal day 1 (P1) through P25 and in adulthood, at 14 time points, using three animals per each time point per each of three experiments. Rat brain tissue for P1, P4 and P6 samples were pooled from 7, 4, and 3 animals, respectively. Several litters were used to avoid variations.
RNA extraction
Total RNA was extracted using the Aurum fatty tissue RNA extraction kit (BioRad, cat. # 732–6820) and treated with DNase. The RNA concentration was determined through absorption at 260 nm using the Nano Drop instrument (Fisher Scientific).
cDNA synthesis
The SUPERSCRIPT IIITM First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) was used to prepare cDNA, using oligo (dT)20 and 2 μg of total RNA as per manufacturer’s instructions. The cDNA was treated with 1 μl RNase H and then stored at −20°C.
QRT-PCR
Primers were designed using Primer Express 3.0 software and synthesized by SIGMA-Genosys (Oakville, Ontario). Standard templates of 90–150bp in length were generated by PCR, purified using Invitrogen Pure Link PCR Micro Kit (K310050) and their concentration determined using Nano Drop Spectrophotometer. The cDNA levels within samples were determined using quantitative real-time PCR (QRT-PCR) on an ABI ViiA™ 7 Real-Time PCR System using the Maxima® SYBR Green/ROX qPCR Master Mix (2X) (cat. # K0223). 2 μl of 1/10 cDNA dilution (or 2 μl of standard) were used per PCR reaction. The primers used are listed in Table SIV. Dilutions were made to flank the concentration of the template within the sample at least 100 times above and below their concentration, with slope of the standard curve near or at 3.3. The accuracy and precision was measured as % deviation from known concentration of a standard within concentration range of the samples and a SD of triplicate repeats, respectively. The values were standardized per total RNA and per β-glucuronidase levels.
Subtractive hybridization
Total RNA was extracted as described. The mRNA was isolated (Sigma Gen Elute mRNA kit) and subtracted using the Suppression Subtractive Hybridization (SSH) cDNA Subtraction Kit (Clontech) to selectively amplify differentially expressed genes. This method is optimal for identification of rare transcript target cDNAs. P4 (driver) and P21 (tester) were used to identify candidate genes of the mature transcriptional network. The tissues were used in reverse order (P21 driver, P4 tester) to identify candidate genes of the early transcriptional network. The resulting cDNA templates were run on 1% agarose gel, extracted, cloned into a vector (Clone Jet, Fermentas), transfected into E. coli and amplified directly from bacterial colonies. The PCR was cleaned using Invitrogen Pure Link PCR Micro Kit (K310050) and the template sequenced by The Centre for Applied Genomics (TCAG, Toronto, Ontario) using primers complementary to the vector. Transcripts were matched to rat reference sequences in the NCBI database using BLAST. Developmental striatal expression was determined by QRT-PCR.
Results
Developmental expression
Developmental expression of DRs, associated factors and striatal-specific genes (Crittenden and Graybiel, 2011; Gerfen, 1992; Gerfen and Young, 1988; Lobo et al., 2006) was determined starting at postnatal day 1 (P1) through P25, at 14 time points, using quantitative real-time PCR (QRT-PCR) (Figure 1) For detailed information see Supplement Figure S1 and Table SI. Each time point is represented by three independent striatal samples (three animals, Figure S2-A). The data were standardized to total RNA, as well as to β-glucuronidase, which we have shown to be a housekeeping gene with a stable expression in both the early postnatal and adult brain (Novak and Tallerico, 2006). Both standardization methods yielded the same expression pattern. Therefore, there was no variation due to RNA degradation or cDNA synthesis (expression per total RNA is shown). Error bars represent standard error of the mean (SE) (Figure S2-B).
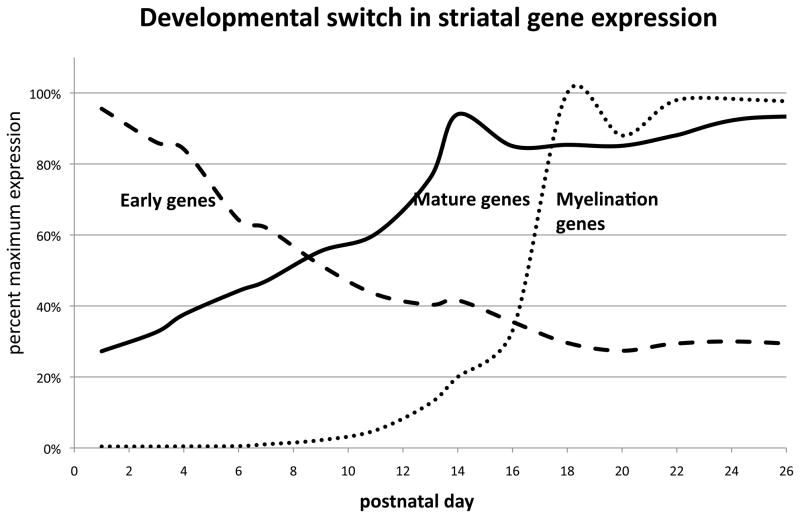
Figure 1. Developmental switch in gene expression networks.
This figure shows the average expression genes, which are part of the early gene expression network, genes that are part of the mature gene expression network, and of myelination genes. The expression is shown as percent of maximal expression during this time period.
Early genes. This group of genes became downregulated within the first week after birth. This graph is based on the expression of genes: Early B-cell factor (Ebf1), Ikaros (IKZF1), retinoid X receptor G (RXRg), mediator of ARE-directed mRNA degradation (Auf1, HNRNPD), chicken ovalbumin upstream promoter transcription factor-interacting protein 1 (Ctip2, B-cell CLL/lymphoma 11B, BCL11B), Brain-4 (Brn4, POU3F4), retinoic acid receptor B splice variant 1 (RARbV1), cocaine-and amphetamine-regulated transcript (CART) (Table SI, Figure S1).
Mature genes. These genes became upregulated during the second week of life and peaked at P14, after which their expression approached adult levels. This group includes many genes essential for MSN-specific functions, also used as MSN markers. The graph is based on the expression of genes: dopamine receptor 1 (DRD1), dopamine receptor 2 (DRD2), calcium/calmodulin-dependent protein kinase II alpha subunit (CaMKIIα) and beta subunit (CaMKIIβ), Enkephalin, Dynorphin, dopamine-and cAMP-regulated phosphoprotein 32-KD (DARPP-32, protein phosphatase 1 regulatory subunit 1B, PPP1R1B, neurokinin B (NKB, tachykinin 3 TAC3), downstream regulatory element (DRE)-antagonist modulator (DREAM, potassium channel-interacting protein, KCNIP3, calsenilin) (Table SI, Figure S1).
Myelination genes. These include myelin basic protein (MBP), myelin-associated glycoprotein (MAG), myelin-oligodendrocyte glycoprotein (MOG), phospholipid protein 1 (Plp1), and transferrin (Tf). The expression of these myelination genes peaks at P17, hence 3 days after a peak in the expression of DRs and other genes of the mature MSN gene expression network (Table SI).
The results indicate that two distinct and mutually exclusive transcriptional networks were present within the rat striatum during the first two postnatal weeks. As expected, each of these transcriptional networks contains genes which fulfill specific requirements of the particular developmental stage (Table I) (For full names and brief description of function, see Supplement Table SI). Expression in pg cDNA is considered equivalent to expression in pg mRNA, as cDNA synthesis is expected to produce cDNA in 1:1 ratio to mRNA, hence values are listed as pg mRNA per mg total RNA.
The early transcriptional network
This group of genes became downregulated within the first week after birth and include early transcription factors (TFs) essential for MSN differentiation (Figure 1, Figure S1). However, some persisted longer throughout this period than others, likely indicating their position in the signaling/transcriptional pathway hierarchy (Ang, 2006)(Figure S1). Figure S1 shows the expression of genes as % of the maximum expression during this period in order to account for varying scales of expression. This group of genes includes early transcription factors (TFs) essential for MSN differentiation, such as Ebf-1 andCtip2, factors which regulate striatal D1R gene expression including Brn4, RARBV1 and RXRg, and transcription factors such as AUF1 and Ikaros, which regulate expression of MSN-specific genes such as enkephalin, an opioid peptide expressed by the striatopallidal MSNs (Table SI).
The mature transcriptional network
These genes became upregulated during the second week of life and peaked at P14, after which they leveled off to adult levels (Figure1)(Table SI, Figure S1). This group includes many genes essential for MSN-specific functions, also used as MSN markers. For example, the D1R neuron-associated opioid peptide dynorphin, which acts to inhibit dopamine release, and the opioid peptide enkephalin expressed by D2R neurons, as well as neurokinin B (NKB), at achykin in which modulates AMPA and GABA signaling in MSNs. Another member of this group is CaMK II (calcium/calmodulin-dependent protein kinase II), a key downstream effector which bridges the DA and Ca2+ signalling pathways, dysregulation of which has been linked to the etiology of schizophrenia (Greenstein et al., 2007; Novak and Seeman, 2008; Novak et al., 2006).
Of note is that genes known to be expressed by the striatonigral neurons (such as D1R, dynorphin) peaked in exact synchrony with genes expressed by the striatopallidal neurons (D2R, enkephalin, Adora2a), indicating that both types of projection neurons matured along the same timeline.
Transitional genes
These genes are expressed only during the period between the early and the mature MSN gene expression networks and may play an important role in the transition between the two transcriptional networks. The dopamine receptor regulating factor, DRRF, a regulator of expression of the D1R and D2R genes, closely paralleled the expression of dopamine receptors (Figure S2-C). It was upregulated during the second postnatal week, peaked at P14, along with the D1R and D2R transcripts, and was sharply downregulated thereafter to the adult level, similar to its pre-P7 expression.
Frontal cortex
Frontal cortex tissues from the same animals were processed simultaneously with the striatal tissues. The gene expression of DRs within the frontal cortex indicated that the switch in gene expression networks that occurred during the first two postnatal weeks in striatum was not present in the frontal cortex (Figure S2-B), likely because frontal cortex in rat develops earlier than the striatum.
Subtractive hybridization
Subtractive hybridization of mRNA from P4 and P21 striatal tissues was performed to identify genes of the mature network, i.e. expressed at P21 but not P4. Genes of the early transcriptional network were identified through a subtraction using these tissue sources in the reverse order. Transcripts were matched to NCBI rat genomic sequence and rat mRNA reference sequences. In case of no available rat mRNA reference sequence match, a search for homology to non-reference rat mRNA, Ests and mouse mRNA sequences was performed (Table SI). The developmental expression was confirmed in striatal tissues, showing that the transcripts are developmentally expressed during the first two postnatal weeks. Expression was determined using primers specific to the cloned sequence. Over 95% of templates analyzed showed the predicted developmental pattern according to the type of subtraction performed, which indicated that the subtraction procedure was successful. All novel transcripts were numbered S01 through S11, the cloned sequence and primers used are provided in Supplemental Table SIII.
S01
A correlation in expression was noted for the novel template S01 and FoxP1, which showed not only nearly identical expression patterns, but were expressed at an uncommonly similar mRNA levels (Figure S3).
S07
Template S07 showed a developmental expression pattern highly similar to that of D2R and DRRF (Figure S3).
New splice variant of Rapgef5
We also identified a new splice variant of the Rap guanine nucleotide exchange factor 5 (Rapgef5). Rapgef5 was expressed at high level at P1, decreasing to about half of its P1 expression by P18. The cloned sequence contained additional five exons, 5′ of the currently known first exon and an alternative splice site in the original exon 1 (Table SIII, Figure S4). To confirm the expression of this splice variant in P2 tissues, we have designed primers specific to this splice variant. Amplification yielded a single band of the correct size, indicating that this splice variant is expressed in early postnatal striatum. The developmental expression of this splice variant has been confirmed and follows the same pattern as was determined using original primers designed to the 5′ end of the template, which is common to both the new splice variant and the reference sequence published by NCBI.
Additional candidate templates
Templates obtained through subtractive hybridization, but for which developmental expression is yet to be confirmed are listed in Table SII. In total, this group contains 89 additional templates.
Myelination event
Between P14 to P17 myelination-related genes underwent a dramatic increase in expression, indicating that a myelination event was under way (Figure 1), (Table SI). The expression of myelin basic protein (MBP), myelin-associated glycoprotein (MAG), myelin-oligodendrocyte glycoprotein (MOG), phospholipid protein 1 (Plp1), and transferrin (Tf) surged at the mRNA level between P11and P17by10, 400%, 1700%, 1900%, 9,400% and 4,800%, respectively. (i.e. the expression increased 104, 17, 19, 94, and 48 fold from P11 to P17, with most of the increase occurring between P14 and P17). After the peak at P17, their expression plateaued. four days after the expression of genes belonging to the “mature MSN” transcriptional network has leveled off (Figure 1).
Discussion
The objective of this study was to analyze striatal neurodevelopment in the rat during the first two postnatal weeks, a period which corresponds to the second trimester in humans (Clancy et al., 2001), when environmental factors such as infection or stress are known to lead to an increase in susceptibility to later development of neuropsychiatric disease, including schizophrenia (Boksa and El-Khodor, 2003; Egan and Weinberger, 1997; Fredriksson et al., 2004; Hansen et al., 2004; Mednick et al., 1994)(reviewed in (Brown and Patterson, 2011; Keshavan and Murray, 1997; Palomo et al., 2002)). Specifically, the human day 116 post conception (PC) (birth at 270 PC, hence 3.9 gestational month) in limbic development corresponds to rat P7 in striatal development, and 152 PC days in human limbic development (5.1 months) corresponds to P14 in rat striatal development (Clancy et al., 2001).
However, until now the only indication that striatal neurons are undergoing changes during these first two postnatal weeks was evidence that the GABA and the NMDA receptors undergo alteration in their subunit composition (Lau et al., 2003). We show that striatal neurons undergo changes that are far more extensive. Our data show that there is a strictly timed maturation program under way within the striatum during the first two postnatal weeks in rat, which results in a complete metamorphosis of gene expression. An entire transcriptional network, responsible for early postnatal striatal development and still lacking genes that define mature striatal neurons, became downregulated and replaced by a transcriptional network characteristic of mature MSN neurons. Therefore, the striatum, a structure known to be involved in the etiology of diseases such as Huntington’s, Parkinson’s, bipolar disorder, depression and schizophrenia (Dujardin and Laurent, 2003; Simpson et al., 2010; Thomas, 2006)(Simpson et al., 2010), undergoes an intensive developmental program during this key period. Understanding this process and how it may be altered will likely provide us with an important insight into the pathological changes that may predispose an individual to the later development of disease.
The early transcriptome, which became downregulated during the first postnatal week, contained a range of early transcription factors, many of which are known to be involved in striatal neurodevelopment. These include factors associated with the expression of genes of the striatonigral neurons, such as Brn4, CART and Ebf, or genes expressed by the striatopallidal neurons, such as Auf1 and Ikaros, or factors involved in general MSN differentiation, including Ctip2, RARbV1 and RXRg (see Table 1).
Table 1. Grouping of genes analyzed by this study according to their function.
Genes expressed early in development are underlined. Mature genes, which become expressed during the maturation period, are in bold. DRRF, a transitionally expressed gene, is in italic font.
| Proliferation, differentiation | Neural migration | Myelination | Signaling | D1R, D2R - related | G-protein-related | Glutamate |
|---|---|---|---|---|---|---|
| Ccrn4 | Tubb2b | MBP | Akap8 | AUF1 | NM23B | SLC1A2 |
| Ctip2 | Tubb3 | Plp1 | Clk4 | Brn-4 | Gnb4 | Grina |
| Dpysl3 | Epha4 | Mobp | CaMKIIα | CART | Adora2a | Gpr158 |
| Ablim3 | Cdh4 | Tf | CaMKIIβ | Ikaros | DARPP-32 | |
| Ube2v1 | Mag | RARbV1 | D1R | |||
| Ebf1 | Aqp4 | DRRF | D2R | |||
| Epha4 | Mog | DREAM | ||||
| FoxP1 | dynorphin | |||||
| Rapgef5 | enkephalin | |||||
| RARbV1 | ||||||
| RXRg | ||||||
| Trim 28 | ||||||
| Fbxl16 |
Before the second postnatal week, striatal neurons lacked their MSN phenotype as defined by the expression of MSN markers, which represent essential MSN-specific pathways. It was during the second postnatal week that genes of the mature transcriptome became upregulated within the striatum, giving the neurons their MSN functional identity, as well as a means of communication with each other and with their interneurons. These included the D1 and D2 dopamine receptor genes, as well as CaMKIIα, CaMKIIβ and DARPP-32, which are an important part of the dopamine receptor downstream signaling pathway. BothD2R and CaMKIIβ have been linked to schizophrenia (Greenstein et al., 2007; Novak and Seeman, 2008; Novak et al., 2006). The transcription factor Dream and its target, dynorphin, an endogenous opioid which is coexpressed with D1R and tonically inhibits the mesolimbic dopamine system, also became upregulated during this period (Buxbaum, 2004; Schwarzer, 2009). Enkephalin, another endogenous opioid which provides inhibitory neuromodulation (Harkany et al., 2008) and is coexpressed with the D2R, as well as neurokinin B and its receptor (NKB-R)(Bodnar, 2007), also came online during the second postnatal week. NKB modulates both DAergic and GABAergic signaling in the striatum (Preston et al., 2000; Zhang et al., 2008). D1R & D2R, CaMK II, DARPP-32 and NKB (Furuta et al., 2000) are genes specifically only expressed by MSNs and not present in striatal interneurons. We observed that genes known to be expressed by the striatonigral (D1R-expressing) neurons peaked at the same time as genes expressed by the striatopallidal (D2R-expressing) neurons, indicating that the maturation of both lineages is synchronized.
The dopamine receptor regulating factor, DRRF, a transcription factor that regulates the expression of D1 and D2 dopamine receptors, became transiently expressed during this maturation process. It perfectly paralleled the expression of the D1 and D2 receptor genes and was quickly downregulated after the peak at P14. Therefore, it likely participates in the neurodevelopmental upregulation of DR expression. However, it is possible that DRRF also participates in controlling the expression of many other genes during this period (Hwang et al., 2001; Lee et al., 2008).
In order to more fully classify this neurodevelopmental process, we performed a subtractive hybridization of tissues before and after the second postnatal week. This allowed us to identify additional genes which participate in the neurodevelopmental switch. The transcripts identified were strictly developmentally expressed and each belonged to either the early or the late transcriptomes, but not both (genes of early network are underlined, genes of the mature network appear in bold below). Some of these genes were previously known to play a role in neurodevelopmental disease, but most were not known to play a role in the striatal neurodevelopmental process (Table I, Table S1). Among these were genes involved in signaling, such as Clk4, Akap8 and Rapgef5, as well as transcription factors including Ccrn4, Trim28, Nflb and FoxP1. We also isolated genes involved in the glutamatergic pathway, such as Slc1A2 and Grina, or in G-protein signaling, such as NM23B, Gnb4, Adora2a and Gpr158. Genes known to be involved in outgrowth and development also formed a large group including Ablim3, Dpysl3, Cdh4, Ube2v1 and genes involved specifically in neural migration such as Tubb2b, Tubb3, Epha4 and Fbxl16. One significantly large group was formed by myelination-related genes, these include MBP, Plp1, Mobp, Tf, Mag, and Mog.
In addition to transcripts of known genes, we have identified 11 novel templates (S01–S11) (the DNA sequences of these clones are listed in Table S III). These are developmentally expressed and match rat genomic DNA sequence, with most also matching expressed sequence tags (Ests) within the NCBI database. There was no sequence homology among these templates. Some code for novel, hypothetical, proteins.
Of special interest is theS01transcript (Figure S3). In most cases the expression pattern of individual genes is very unique. An exception occurs when, as noted by others (Bithell et al., 2003), two genes participate in the same process or are controlled by the same mechanism. We observed such correlation for the adenosine 2A receptor (Adora2A) and the D2R. Adora 2A receptors are coexpressed with D2R, form heterodimers and modulate D2R signaling (Schiffmann et al., 2007). High similarity in expression patterns is also observed between the dopamine receptor regulating factor, DRRF, and the D2R. It is, therefore, of note that the pattern and even the magnitude of expression of S01 is identical to the expression of FoxP1 (S3A). Hence S01 may be a novel factor participating in the same developmental process governed by FoxP1, which is thought to play a role in the development and formation of a circuit in the basal ganglia necessary for sensorimotor integration (Teramitsu et al., 2004) involving the matrix neurons (Tamura et al., 2004). Another template of interest, S07, showed a high homology to the developmental expression of the D2R and the DRRF.
A number of the pathways involved in this neuromaturation process participate in guiding neuronal migration. Striatal neurons form a patch and matrix pattern. The patch neurons send projections to the substantia nigra embryonically, while the matrix neurons form these projections during the early postnatal period (Fishell and van der Kooy, 1987; Fishell and van der Kooy, 1989). The D1R promotes and D2R inhibits migration of GABAergic neurons (Araki et al., 2007; Crandall et al., 2007), with GABARs also being critical for neural migration (Ben-Ari et al., 2004; Goetz et al., 2007; Odeon et al., 2010). Hence, alterations in this neurodevelopmental program, leading to abnormal DR expression, could not only alter the signaling properties of MSN neurons, but also have the capacity to affect neuronal migration of striatal matrix neurons. This would lead to an imbalance in the number of patch and matrix projections and abnormal innervation of substantia nigra. Such imbalance is thought to play a role in basal ganglia disease (reviewed by (Crittenden and Graybiel, 2011)).
An additional 89 templates were identified, but their developmental expression has not yet been determined (listed in Table SII). Given that over 95% of templates for which we have determined expression were confirmed to be strictly developmentally regulated, this group contains genes which are strong candidates as participants in the developmental process.
In addition to the switch in gene expression networks during the second postnatal week, we have identified yet another developmental milestone within the striatum. About four days after the conclusion of the switch in gene expression networks, a steep rise in expression of myelination-related genes began, involving MBP, MAG, MOG, Plp1 and Tf. Tf in particular plays an important role in early postnatal oligodendroglial cell differentiation (Connor and Fine, 1987) and mediates the expression of other myelination genes including MBP and Plp, and, hence, the level of myelination (Escobar Cabrera et al., 1994). The levels of Tf can be altered through the activation of the immune system, such as in response to human influenza virus infection, and lead to reduced Tf expression (Fatemi et al., 2008). This leads to a decreases in the expression of Plp and MBP, hence affecting both the composition of myelin and the level of myelination (Bartzokis et al., 2007; Connor and Fine, 1987)(reviewed in (Leitner and Connor, 2012)). Therefore, an infection may lead to a defect in myelination and, potentially, be a contributing factor to the etiology of neurological disease (Dusek et al., 2012).
The myelination process is also extremely sensitive to the precise level of Plp expression (Readhead et al., 1994). Proteolipid protein, PLP1, or lipophilin, the primary constituent of myelin (Popot et al., 1991), plays an important role in brain-immune system interaction. Inflammation during increased Plp levels can cause massive microglial cell activation and axonal degeneration, with the upregulation of inflammatory markers including TNF-α (tumor necrosis factor-α) and IL-6 (Tatar et al., 2010), known to be upregulated in schizophrenia (reviewed in (Watanabe et al., 2010)). Therefore, infection resulting in neuroinflammation during this key myelination event, when Plp levels are very high, may lead to neural degeneration and/or hypomyelination. MOG could also play a role in abnormal neurodevelopment. It is not only a mediator of interactions between the myelin and the immune system, but is also a regulator of oligodendrocyte microtubule stability, providing a link between activation of the immune system and oligodendrocyte function (reviewed in (Johns and Bernard, 1999)). This is in line with observations that infection during the end of the second trimester and beginning of third trimester may result in neuropathology (Brown and Patterson, 2011; Egan and Weinberger, 1997; Mednick et al., 1994; Mednick et al., 1988; Meyer, 2011; Meyer et al., 2011). Furthermore, both expression (Hakak et al., 2001; Katsel et al., 2005; McCullumsmith et al., 2007) and association studies (Ayalew et al., 2012) implicate genes involved in myelination, in particular MBP, MAG, MOG and Tf, in neuropsychiatric disease (Ayalew et al., 2012; Davis et al., 2003; Felsky et al., 2012; Hakak et al., 2001; Katsel et al., 2005; Martins-de-Souza et al., 2010; McCullumsmith et al., 2007; Sokolov, 2007)(reviewed in (Davis et al., 2003)).
In summary, our results show that the striatum is undergoing a previously unknown, strictly timed switch in transcription networks, followed by a wave of myelination, which may predispose it to environmental insults during this period, leading to increased likelihood for the development of neuropsychiatric disease. Therefore, the classification of this developmental process may yield significant advances in our understanding of the neurodevelopmental basis of neuropsychiatric disease.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute on Drug Abuse (to SRG and BFO, #007223) and a Canadian Institutes on Health Research Postdoctoral Fellowship (to GN, #213845). SRG holds a Canada Research Chair in Molecular Neuroscience. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors thank Dr. Philip Seeman for reading this manuscript and for helpful feedback. We would like to thank Dr. John Vincent for the use of QRT-PCR equipment. We would like to acknowledge the contribution of Jace Jones Tabahin processing samples for sequencing listed in Table II.
Footnotes
Conflict of Interest
Authors declare that there are no competing financial interests in relation to the work described.
References
- Alexander GE, DeLong MR, Strick PL. Parallel organization of functionally segregated circuits linking basal ganglia and cortex. Annu Rev Neurosci. 1986;9:357–381. doi: 10.1146/annurev.ne.09.030186.002041. [DOI] [PubMed] [Google Scholar]
- Ang SL. Transcriptional control of midbrain dopaminergic neuron development. Development. 2006;133(18):3499–3506. doi: 10.1242/dev.02501. [DOI] [PubMed] [Google Scholar]
- Araki KY, Sims JR, Bhide PG. Dopamine receptor mRNA and protein expression in the mouse corpus striatum and cerebral cortex during pre-and postnatal development. Brain Res. 2007;1156:31–45. doi: 10.1016/j.brainres.2007.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O’Donovan MC, Niculescu AB. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Molecular psychiatry. 2012 doi: 10.1038/mp.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y, Ibram G, Blaner WS, Quesenberry CP, Shen L, McKeague IW, Schaefer CA, Susser ES, Brown AS. Low maternal retinol as a risk factor for schizophrenia in adult offspring. Schizophrenia research. 2012 doi: 10.1016/j.schres.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartzokis G, Lu PH, Tishler TA, Fong SM, Oluwadara B, Finn JP, Huang D, Bordelon Y, Mintz J, Perlman S. Myelin breakdown and iron changes in Huntington’s disease: pathogenesis and treatment implications. Neurochem Res. 2007;32(10):1655–1664. doi: 10.1007/s11064-007-9352-7. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, Khalilov I, Represa A, Gozlan H. Interneurons set the tune of developing networks. Trends Neurosci. 2004;27(7):422–427. doi: 10.1016/j.tins.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Bithell A, Alberta J, Hornby F, Stiles CD, Williams BP. Expression of the guanine nucleotide exchange factor, mr-gef, is regulated during the differentiation of specific subsets of telencephalic neurons. Brain Res Dev Brain Res. 2003;146(1–2):107–118. doi: 10.1016/j.devbrainres.2003.09.017. [DOI] [PubMed] [Google Scholar]
- Bodnar RJ. Endogenous opiates and behavior: 2006. Peptides. 2007;28(12):2435–2513. doi: 10.1016/j.peptides.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boksa P, El-Khodor BF. Birth insult interacts with stress at adulthood to alter dopaminergic function in animal models: possible implications for schizophrenia and other disorders. Neuroscience and biobehavioral reviews. 2003;27(1–2):91–101. doi: 10.1016/s0149-7634(03)00012-5. [DOI] [PubMed] [Google Scholar]
- Bolam JP, Hanley JJ, Booth PA, Bevan MD. Synaptic organisation of the basal ganglia. Journal of anatomy. 2000;196 (Pt 4):527–542. doi: 10.1046/j.1469-7580.2000.19640527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AS, Patterson PH. Maternal infection and schizophrenia: implications for prevention. Schizophrenia bulletin. 2011;37(2):284–290. doi: 10.1093/schbul/sbq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum JD. A role for calsenilin and related proteins in multiple aspects of neuronal function. Biochem Biophys Res Commun. 2004;322(4):1140–1144. doi: 10.1016/j.bbrc.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Clancy B, Darlington RB, Finlay BL. Translating developmental time across mammalian species. Neuroscience. 2001;105(1):7–17. doi: 10.1016/s0306-4522(01)00171-3. [DOI] [PubMed] [Google Scholar]
- Connor JR, Fine RE. Development of transferrin-positive oligodendrocytes in the rat central nervous system. J Neurosci Res. 1987;17(1):51–59. doi: 10.1002/jnr.490170108. [DOI] [PubMed] [Google Scholar]
- Crandall JE, McCarthy DM, Araki KY, Sims JR, Ren JQ, Bhide PG. Dopamine receptor activation modulates GABA neuron migration from the basal forebrain to the cerebral cortex. J Neurosci. 2007;27(14):3813–3822. doi: 10.1523/JNEUROSCI.5124-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden JR, Graybiel AM. Basal Ganglia disorders associated with imbalances in the striatal striosome and matrix compartments. Frontiers in neuroanatomy. 2011;5:59. doi: 10.3389/fnana.2011.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KL, Stewart DG, Friedman JI, Buchsbaum M, Harvey PD, Hof PR, Buxbaum J, Haroutunian V. White matter changes in schizophrenia: evidence for myelin-related dysfunction. Arch Gen Psychiatry. 2003;60(5):443–456. doi: 10.1001/archpsyc.60.5.443. [DOI] [PubMed] [Google Scholar]
- Deng YP, Xie JP, Wang HB, Lei WL, Chen Q, Reiner A. Differential localization of the GluR1 and GluR2 subunits of the AMPA-type glutamate receptor among striatal neuron types in rats. Journal of chemical neuroanatomy. 2007;33(4):167–192. doi: 10.1016/j.jchemneu.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin K, Laurent B. Dysfunction of the human memory systems: role of the dopaminergic transmission. Current opinion in neurology. 2003;16(Suppl 2):S11–16. doi: 10.1097/00019052-200312002-00003. [DOI] [PubMed] [Google Scholar]
- Dusek P, Jankovic J, Le W. Iron dysregulation in movement disorders. Neurobiol Dis. 2012;46(1):1–18. doi: 10.1016/j.nbd.2011.12.054. [DOI] [PubMed] [Google Scholar]
- Egan MF, Weinberger DR. Neurobiology of schizophrenia. Curr Opin Neurobiol. 1997;7(5):701–707. doi: 10.1016/s0959-4388(97)80092-x. [DOI] [PubMed] [Google Scholar]
- Escobar Cabrera OE, Bongarzone ER, Soto EF, Pasquini JM. Single intracerebral injection of apotransferrin in young rats induces increased myelination. Developmental neuroscience. 1994;16(5–6):248–254. doi: 10.1159/000112116. [DOI] [PubMed] [Google Scholar]
- Fatemi SH, Reutiman TJ, Folsom TD, Huang H, Oishi K, Mori S, Smee DF, Pearce DA, Winter C, Sohr R, Juckel G. Maternal infection leads to abnormal gene regulation and brain atrophy in mouse offspring: implications for genesis of neurodevelopmental disorders. Schizophrenia research. 2008;99(1–3):56–70. doi: 10.1016/j.schres.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsky D, Voineskos AN, Lerch JP, Nazeri A, Shaikh SA, Rajji TK, Mulsant BH, Kennedy JL. Myelin-associated glycoprotein gene and brain morphometry in schizophrenia. Frontiers in psychiatry/Frontiers Research Foundation. 2012;3:40. doi: 10.3389/fpsyt.2012.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, van der Kooy D. Pattern formation in the striatum: developmental changes in the distribution of striatonigral neurons. J Neurosci. 1987;7(7):1969–1978. doi: 10.1523/JNEUROSCI.07-07-01969.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishell G, van der Kooy D. Pattern formation in the striatum: developmental changes in the distribution of striatonigral projections. Brain Res Dev Brain Res. 1989;45(2):239–255. doi: 10.1016/0165-3806(89)90042-4. [DOI] [PubMed] [Google Scholar]
- Fredriksson A, Archer T, Alm H, Gordh T, Eriksson P. Neurofunctional deficits and potentiated apoptosis by neonatal NMDA antagonist administration. Behavioural brain research. 2004;153(2):367–376. doi: 10.1016/j.bbr.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Furuta T, Mori T, Lee T, Kaneko T. Third group of neostriatofugal neurons: neurokinin B-producing neurons that send axons predominantly to the substantia innominata. J Comp Neurol. 2000;426(2):279–296. doi: 10.1002/1096-9861(20001016)426:2<279::aid-cne9>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Annu Rev Neurosci. 1992;15:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Young WS., 3rd Distribution of striatonigral and striatopallidal peptidergic neurons in both patch and matrix compartments: an in situ hybridization histochemistry and fluorescent retrograde tracing study. Brain Res. 1988;460(1):161–167. doi: 10.1016/0006-8993(88)91217-6. [DOI] [PubMed] [Google Scholar]
- Goetz T, Arslan A, Wisden W, Wulff P. GABA(A) receptors: structure and function in the basal ganglia. Prog Brain Res. 2007;160:21–41. doi: 10.1016/S0079-6123(06)60003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenstein R, Novak G, Seeman P. Amphetamine sensitization elevates CaMKIIbeta mRNA. Synapse. 2007;61(10):827–834. doi: 10.1002/syn.20429. [DOI] [PubMed] [Google Scholar]
- Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, Haroutunian V, Fienberg AA. Genome-wide expression analysis reveals dysregulation of myelination-related genes in chronic schizophrenia. Proc Natl Acad Sci U S A. 2001;98(8):4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HH, Briem T, Dzietko M, Sifringer M, Voss A, Rzeski W, Zdzisinska B, Thor F, Heumann R, Stepulak A, Bittigau P, Ikonomidou C. Mechanisms leading to disseminated apoptosis following NMDA receptor blockade in the developing rat brain. Neurobiol Dis. 2004;16(2):440–453. doi: 10.1016/j.nbd.2004.03.013. [DOI] [PubMed] [Google Scholar]
- Harkany T, Mackie K, Doherty P. Wiring and firing neuronal networks: endocannabinoids take center stage. Curr Opin Neurobiol. 2008;18(3):338–345. doi: 10.1016/j.conb.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Lopez S, Tkatch T, Perez-Garci E, Galarraga E, Bargas J, Hamm H, Surmeier DJ. D2 dopamine receptors in striatal medium spiny neurons reduce L-type Ca2+ currents and excitability via a novel PLC[beta]1-IP3-calcineurin-signaling cascade. J Neurosci. 2000;20(24):8987–8995. doi: 10.1523/JNEUROSCI.20-24-08987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CK, D’Souza UM, Eisch AJ, Yajima S, Lammers CH, Yang Y, Lee SH, Kim YM, Nestler EJ, Mouradian MM. Dopamine receptor regulating factor, DRRF: a zinc finger transcription factor. Proc Natl AcadSci U S A. 2001;98(13):7558–7563. doi: 10.1073/pnas.121635798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jocoy EL, Andre VM, Cummings DM, Rao SP, Wu N, Ramsey AJ, Caron MG, Cepeda C, Levine MS. Dissecting the contribution of individual receptor subunits to the enhancement of N-methyl-d-aspartate currents by dopamine D1 receptor activation in striatum. Frontiers in systems neuroscience. 2011;5:28. doi: 10.3389/fnsys.2011.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns TG, Bernard CC. The structure and function of myelin oligodendrocyte glycoprotein. J Neurochem. 1999;72(1):1–9. doi: 10.1046/j.1471-4159.1999.0720001.x. [DOI] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Haroutunian V. Variations in myelin and oligodendrocyte-related gene expression across multiple brain regions in schizophrenia: a gene ontology study. Schizophrenia research. 2005;79(2–3):157–173. doi: 10.1016/j.schres.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Keshavan MS, Murray R. Neurodevelopment & adult psychopathology. xvi. Cambridge; New York: Cambridge University Press; 1997. p. 282. [Google Scholar]
- Kreitzer AC, Malenka RC. Striatal plasticity and basal ganglia circuit function. Neuron. 2008;60(4):543–554. doi: 10.1016/j.neuron.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau WK, Lui PW, Wong CK, Chan YS, Yung KK. Differential expression of N-methyl-D-aspartate receptor subunit messenger ribonucleic acids and immunoreactivity in the rat neostriatum during postnatal development. Neurochemistry international. 2003;43(1):47–65. doi: 10.1016/s0197-0186(02)00191-2. [DOI] [PubMed] [Google Scholar]
- Laviolette SR, Grace AA. The roles of cannabinoid and dopamine receptor systems in neural emotional learning circuits: implications for schizophrenia and addiction. Cellular and molecular life sciences: CMLS. 2006;63(14):1597–1613. doi: 10.1007/s00018-006-6027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Jang MK, Lee OH, Kim OS, Kim YM, Yajima S, Lee YC, Mouradian MM. Transcriptional auto-regulation of the dopamine receptor regulating factor (DRRF) gene. Molecular and cellular endocrinology. 2008;289(1–2):23–28. doi: 10.1016/j.mce.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Leitner DF, Connor JR. Functional roles of transferrin in the brain. Biochim Biophys Acta. 2012;1820(3):393–402. doi: 10.1016/j.bbagen.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9(3):443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Maccarrone G, Wobrock T, Zerr I, Gormanns P, Reckow S, Falkai P, Schmitt A, Turck CW. Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. Journal of psychiatric research. 2010;44(16):1176–1189. doi: 10.1016/j.jpsychires.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Matamales M, Bertran-Gonzalez J, Salomon L, Degos B, Deniau JM, Valjent E, Herve D, Girault JA. Striatal medium-sized spiny neurons: identification by nuclear staining and study of neuronal subpopulations in BAC transgenic mice. PloS one. 2009;4(3):e4770. doi: 10.1371/journal.pone.0004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullumsmith RE, Gupta D, Beneyto M, Kreger E, Haroutunian V, Davis KL, Meador-Woodruff JH. Expression of transcripts for myelination-related genes in the anterior cingulate cortex in schizophrenia. Schizophrenia research. 2007;90(1–3):15–27. doi: 10.1016/j.schres.2006.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mednick SA, Huttunen MO, Machon RA. Prenatal influenza infections and adult schizophrenia. Schizophrenia bulletin. 1994;20(2):263–267. doi: 10.1093/schbul/20.2.263. [DOI] [PubMed] [Google Scholar]
- Mednick SA, Machon RA, Huttunen MO, Bonett D. Adult schizophrenia following prenatal exposure to an influenza epidemic. Arch Gen Psychiatry. 1988;45(2):189–192. doi: 10.1001/archpsyc.1988.01800260109013. [DOI] [PubMed] [Google Scholar]
- Meyer U. Progress in neuro-psychopharmacology & biological psychiatry. 2011. Developmental neuroinflammation and schizophrenia. [DOI] [PubMed] [Google Scholar]
- Meyer U, Weiner I, McAlonan GM, Feldon J. The neuropathological contribution of prenatal inflammation to schizophrenia. Expert review of neurotherapeutics. 2011;11(1):29–32. doi: 10.1586/ern.10.169. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Miletich RS, Kohn PD, Esposito G, Carson RE, Quarantelli M, Weinberger DR, Berman KF. Reduced prefrontal activity predicts exaggerated striatal dopaminergic function in schizophrenia. Nat Neurosci. 2002;5(3):267–271. doi: 10.1038/nn804. [DOI] [PubMed] [Google Scholar]
- Novak G, Seeman P. Hyperactive mice show elevated D2High receptors, a model for schizophrenia: calcium/calmodulin-dependent kinase II alpha knockouts. 2008. [DOI] [PubMed] [Google Scholar]
- Novak G, Seeman P, Tallerico T. Increased expression of calcium/calmodulin-dependent protein kinase II beta in frontal cortex in schizophrenia and depression. Synapse. 2006;59(1):61–68. doi: 10.1002/syn.20211. [DOI] [PubMed] [Google Scholar]
- Novak G, Tallerico T. Nogo A, B and C expression in schizophrenia, depression and bipolar frontal cortex, and correlation of Nogo expression with CAA/TATC polymorphism in 3′-UTR. Brain Res. 2006;1120(1):161–171. doi: 10.1016/j.brainres.2006.08.071. [DOI] [PubMed] [Google Scholar]
- Odeon MM, Salatino AE, Rodriguez CB, Scolari MJ, Acosta GB. The response to postnatal stress: amino acids transporters and PKC activity. Neurochem Res. 2010;35(7):967–975. doi: 10.1007/s11064-010-0153-z. [DOI] [PubMed] [Google Scholar]
- Palomo T, Kostrzewa RM, Archer T, Beninger RJ. Neurodevelopmental liabilities in schizophrenia and affective disorders. Neurotox Res. 2002;4(5–6):397–408. doi: 10.1080/1029842021000022061. [DOI] [PubMed] [Google Scholar]
- Popot JL, Pham Dinh D, Dautigny A. Major myelin proteolipid: the 4-alpha-helix topology. The Journal of membrane biology. 1991;123(3):278. doi: 10.1007/BF01870411. [DOI] [PubMed] [Google Scholar]
- Preston Z, Richardson PJ, Pinnock RD, Lee K. NK-3 receptors are expressed on mouse striatal gamma-aminobutyric acid-ergic interneurones and evoke [(3)H] gamma-aminobutyric acid release. Neuroscience letters. 2000;284(1–2):89–92. doi: 10.1016/s0304-3940(00)00968-x. [DOI] [PubMed] [Google Scholar]
- Readhead C, Schneider A, Griffiths I, Nave KA. Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron. 1994;12(3):583–595. doi: 10.1016/0896-6273(94)90214-3. [DOI] [PubMed] [Google Scholar]
- Schiffmann SN, Fisone G, Moresco R, Cunha RA, Ferre S. Adenosine A2A receptors and basal ganglia physiology. Prog Neurobiol. 2007;83(5):277–292. doi: 10.1016/j.pneurobio.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann SN, Jacobs O, Vanderhaeghen JJ. Striatal restricted adenosine A2 receptor (RDC8) is expressed by enkephalin but not by substance P neurons: an in situ hybridization histochemistry study. J Neurochem. 1991;57(3):1062–1067. doi: 10.1111/j.1471-4159.1991.tb08257.x. [DOI] [PubMed] [Google Scholar]
- Schwarzer C. 30 years of dynorphins--new insights on their functions in neuropsychiatric diseases. Pharmacol Ther. 2009;123(3):353–370. doi: 10.1016/j.pharmthera.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P. All roads to schizophrenia lead to dopamine super sensitivity and elevated dopamine D2(high) receptors. CNS neuroscience & therapeutics. 2011;17(2):118–132. doi: 10.1111/j.1755-5949.2010.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley DR. Molecular biology of dopamine receptors. In: Ariano MA, Surmeier DJ, editors. Molecular and cellular mechanisms of neostriatal function. New York Austin: Springer; R.G. Landes Co; 1995. pp. 255–272. [Google Scholar]
- Simpson EH, Kellendonk C, Kandel E. A possible role for the striatum in the pathogenesis of the cognitive symptoms of schizophrenia. Neuron. 2010;65(5):585–596. doi: 10.1016/j.neuron.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov BP. Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse. Comorbidity, shared traits, or molecular phenocopies? Int J Neuropsychopharmacol. 2007;10(4):547–555. doi: 10.1017/S1461145706007322. [DOI] [PubMed] [Google Scholar]
- Tamura S, Morikawa Y, Iwanishi H, Hisaoka T, Senba E. Foxp1 gene expression in projection neurons of the mouse striatum. Neuroscience. 2004;124(2):261–267. doi: 10.1016/j.neuroscience.2003.11.036. [DOI] [PubMed] [Google Scholar]
- Tatar CL, Appikatla S, Bessert DA, Paintlia AS, Singh I, Skoff RP. Increased Plp1 gene expression leads to massive microglial cell activation and inflammation throughout the brain. ASN neuro. 2010;2(4):e00043. doi: 10.1042/AN20100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper JM, Bolam JP. Functional diversity and specificity of neostriatal interneurons. Curr Opin Neurobiol. 2004;14(6):685–692. doi: 10.1016/j.conb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Teramitsu I, Kudo LC, London SE, Geschwind DH, White SA. Parallel FoxP1 and FoxP2 expression in songbird and human brain predicts functional interaction. J Neurosci. 2004;24(13):3152–3163. doi: 10.1523/JNEUROSCI.5589-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA. Striatal specificity of gene expression dysregulation in Huntington’s disease. J Neurosci Res. 2006;84(6):1151–1164. doi: 10.1002/jnr.21046. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Someya T, Nawa H. Cytokine hypothesis of schizophrenia pathogenesis: evidence from human studies and animal models. Psychiatry and clinical neurosciences. 2010;64(3):217–230. doi: 10.1111/j.1440-1819.2010.02094.x. [DOI] [PubMed] [Google Scholar]
- Welham J, Isohanni M, Jones P, McGrath J. The antecedents of schizophrenia: a review of birth cohort studies. Schizophrenia bulletin. 2009;35(3):603–623. doi: 10.1093/schbul/sbn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Andren PE, Chergui K, Svenningsson P. Neurokinin B/NK3 receptors exert feedback inhibition on L-DOPA actions in the 6-OHDA lesion rat model of Parkinson’s disease. Neuropharmacology. 2008;54(7):1143–1152. doi: 10.1016/j.neuropharm.2008.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.