Abstract
Specialized DNA polymerases participate in replication stress responses and in DNA repair pathways that function as barriers against cellular senescence and genomic instability. These events can be co-opted by tumor cells as a mechanism to survive chemotherapeutic and ionizing radiation treatments, and as such, represent potential targets for adjuvant therapies. Previously, a high-throughput screen of ~16,000 compounds identified several first generation proof-of-principle inhibitors of human DNA polymerase kappa (hpol κ). The indole-derived inhibitor of 5-lipoxygenase activating protein (FLAP), MK886, was one of the most potent inhibitors of hpol κ discovered in that screen. However, the specificity and mechanism of inhibition remained largely undefined. In the current study, the specificity of MK886 against human Y-family DNA polymerases and a model B-family DNA polymerase was investigated. MK886 was found to inhibit the activity of all DNA polymerases tested with similar IC50 values, the exception being a six- to eight-fold increase in the potency of inhibition against human DNA polymerase iota (hpol ι), a highly error-prone enzyme that uses Hoogsteen base-pairing modes during catalysis. The specificity against hpol ι was partially abrogated by inclusion of the recently annotated 25 a.a. N-terminal extension. Based on Michaelis-Menten kinetic analyses and DNA binding assays the mechanism of inhibition by MK886 appears to be mixed. In silico docking studies were used to produce a series of models for MK886 binding to Y-family members. The docking results indicate that two binding pockets are conserved between Y-family polymerases, while a third pocket near the thumb domain appears to be unique to hpol ι. Overall, these results provide insight into the general mechanism of DNA polymerase inhibition by MK886.
Keywords: DNA replication, polymerase, translesion DNA synthesis, small-molecule inhibitor
INTRODUCTION
Genomic replication is fundamental to all biological systems,1 with impediments to accurate replication arising from both exogenous and endogenous sources.2 Aberrant activation of DNA damage responses and increased replication stress contribute to some cancers, including highly invasive brain tumors.3 These biological responses are generally regarded as protective in nature under normal conditions but their up-regulation in tumor cells may contribute to challenges in the design and implementation of effective therapies.4 Evolutionary pressure has retained DNA repair pathways as a means of eliminating DNA damage prior to replication. The Y-family DNA polymerases are specialized enzymes that are recruited to stalled replication forks in the event that DNA adducts or specialized DNA structures are encountered by the replication machinery.5 In this manner, the Y-family members allow cells to “tolerate” DNA damage by facilitating progression of the replication machinery past blocking lesions that may arise following exposure to ionizing radiation and/or reactive chemicals, such as activated polycyclic aromatic hydrocarbons, reactive oxygen and nitrogen species, aflatoxins, aromatic amines and many other DNA damaging agents.6 Mis-regulation of Y-family DNA polymerase members have been documented at the transcript and protein level in several tumor types.7–9 As such, they represent targets for sensitizing tumor cells to genotoxic agents, such as cisplatin, doxorubicin or temozolomide.6, 10–13
The ability to target individual members of any class of enzymes requires sufficient structural and/or functional diversity to achieve specific inhibition. DNA polymerases present a unique opportunity for targeted inhibition. While the active site residues involved in nucleic acid synthesis are generally conserved in different DNA and RNA polymerases from a variety of organisms14, considerable variation exists for both the structural and functional attributes of polymerase sub-families. Of note here, are the translesion DNA synthesis properties of Y-family DNA polymerases.15, 16 The structure of Y-family DNA polymerases exhibit several general features that distinguish them from their high-fidelity counterparts (e.g. spacious active sites and the lack of proofreading exonuclease activity).16 Y-family DNA polymerases can accommodate cumbersome DNA adducts in their active site in a manner that remains conducive to polymerization.17–20 The finger domain of Y-family DNA polymerases is composed of shorter α-helices than those present in A-, B- or C-family polymerases, a trend that can be found in all Y-family members studied to date. Additionally, the polymerase-associated or “little finger” domain is only observed in Y-family DNA polymerases. Yet even within the Y-family there is substantial diversity of structure and function, especially near the extreme N-terminus of the four mammalian enzymes. Based on the structural and functional properties of the Y-family, it is not unreasonable to suppose that these specialized polymerases could be targets of specific inhibition by small molecules, yielding interesting biological results. However, finding small molecule inhibitors of specialized polymerases has proven challenging. Although, recent reports have identified natural product inhibitors of Y-family DNA polymerases,21, 22 synthetic routes to obtain these compounds remain to be determined and specificity to individual Y-family members has yet to be achieved.
Recently, along with additional collaborators, we identified several inhibitors of human DNA polymerase kappa (hpol κ).23 Among the top hits for inhibition of hpol κ was the indole derivative 3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isoproylindol-2-yl]-2,2-dimethylpropanoic acid, MK886. MK886 was originally discovered as an inhibitor of leukotriene biosynthesis.24, 25 The compound has been studied extensively and is readily available from multiple commercial sources. MK886 is an inhibitor of both 5-lipoxygenase activator protein (FLAP) and cyclooxgenase-1 (COX-1) activities.26–28 At concentrations above 10 μM, MK886 can induce apoptosis through FLAP-independent inhibition of peroxisome proliferator activated receptor alpha (PPARα).29 An early study showed that DNA replication was inhibited at concentrations of MK886 as low as 1 μM and that DNA fragmentation occurred at sub-micromolar MK886 concentrations.30 However, direct inhibition of DNA polymerase activity was never tested until our recent study with hpol κ.23 The identification of MK886 as an inhibitor of hpol κ led us to investigate the specificity and mechanism of polymerase inhibition by this indole-derived compound. Here we report that MK886 appears to be a general inhibitor of DNA polymerase activity across the Y-family and that the compound also inhibits a B-family DNA polymerase. The mechanism of inhibition appears to be mixed, based on Michaelis-Menten kinetic analyses. Finally, some specificity for MK886 inhibition of hpol ι was observed and the structural rationale for the increased potency against this highly error-prone enzyme was examined biochemically and using molecular modeling tools. The results provide a framework for understanding DNA polymerase inhibition by MK886.
EXPERIMENTAL PROCEDURES
All chemicals were molecular biology grade or better. All dNTPs were purchased from GE Healthcare Life Sciences (Piscataway, NJ). The oligonucleotides used in this work were synthesized by either Integrated DNA technologies (Coralville, IA) or Biosearch Technologies, Inc. (Novato, CA) and purified by the manufacturer using HPLC, with analysis by matrix-assisted laser desorption time-of-flight MS. The primer sequence used in the extension assays, inhibition assays, single-nucleotide kinetics assays and the binding assays was 5′-(FAM-TTT)-GGGGGAAGGATTC-3′. The template DNA sequence used in the extension assays, inhibition assays and the binding assays was 5′-TCACGGAATCCTTCCCCC-3′. MK886 was purchased from either Sigma-Aldrich (St. Louis, MO) or EMD Millipore Corp. (Billerica, MA).
Expression and purification of recombinant proteins
The pBG101 plasmid was used to prepare constructs encoding human DNA polymerases η (a.a. 1–437), ι (a.a. 1–446 and 26–446) and κ (a.a. 19–526). The pBG101 vector encodes a 6X-histidine tag and a glutathione transferase (GST) fusion protein upstream of the polymerase-encoding region. A protease recognition sequence (LEVLFQGP) just upstream of the polymerase insert allows cleavage of the N-terminal affinity tags during purification. All the human polymerases used in the study were expressed in Escherichia coli (strain BL21 DE3) and purified in an identical manner. Briefly, pBG101 vector encoding the polymerases just downstream of 6X-Histidine and GST-tags was transformed into E. coli cells (BL21 (DE3) strain). Cells were grown at 37 °C and 250 rpm for three hours (OD600 = 0.5–0.6), followed by induction for three hours (37 °C and 250 rpm) by addition of isopropyl β-D-1-thiogalactopyranoside (1 mM), and finally harvested by centrifugation. Buffer containing 50 mM Tris-HCl (pH 7.4), 0.5 M NaCl, 10% glycerol (v/v), 5 mM β-mercaptoethanol (β-ME), lysozyme (1 mg/ml), and a protease inhibitor cocktail (Roche, Basel, Switzerland) was added to the harvested pellet. The suspension was sonicated and supernatant recovered from an ultracentrifugation step (35,000 g, 1 h, 4 °C). After the removal of cellular debris by ultracentrifugation, the resulting clear lysate was loaded onto a 5 mL HisTrap column (GE Healthcare Life Sciences) followed by washing the column sequentially with 50 mM Tris-HCl (pH 7.3 at 22 °C) buffer containing 0.5 M NaCl, 5 mM β-ME, 10% glycerol and 20 mM imidazole to remove non-specifically bound proteins. The remaining bound proteins were then eluted using a linear gradient from 60 mM to 400 mM imidazole. The eluted proteins were loaded onto a 2 mL GSTrap column (GE Healthcare Life Sciences) in 25 mM HEPES (pH 7.5) buffer containing 0.1 M NaCl, 5 mM β-ME, and 10% glycerol. Cleavage of the GST tag was performed on the bound proteins by injecting a solution containing the PreScission protease (GE Healthcare Life Sciences) onto the column and allowing it to incubate overnight at 4 °C. The GST-tag-free proteins were eluted in the GSTrap running buffer and concentrated using an Amicon spin concentrator (MilliPore). The purity of each polymerase was analyzed by SDS-polyacrylamide gel electrophoresis. The highly pure proteins were stored at −80 °C in the HEPES buffer (pH 7.5) containing 0.1 M NaCl, 5 mM β-ME, and 30% glycerol. The model B-family DNA polymerase, Dpo1, from Sulfolobus solfataricus was expressed and purified as described previously.31
Determination of IC50 values using polymerase assays
To obtain an estimate of the IC50 for the inhibition of DNA polymerase activity by MK886, polymerase-catalyzed DNA synthesis was measured in the presence of all four dNTPs at a single-time point with increasing concentrations of inhibitor. The MK886 concentration range used for the enzymes hpol η1–437, hpol κ19–526 and Dpo1 was 1–200 μM, while for hpol ι26–446 and hpol ι1–446 it was 0.1–100 μM. For DNA substrate preparation, the FAM-labeled primer was annealed to a complementary template oligonucleotide (1:2, primer:template molar ratio) by incubating at 95 °C for five minutes followed by slow cooling to room temperature. The full-length extension assays were performed in 50 mM HEPES (pH 7.5) buffer containing 50 mM NaCl, 1 mM DTT, 0.1 mg/mL BSA and 10% (v/v) glycerol. The DNA was pre-incubated with the enzyme for five minutes prior to starting the polymerase reaction. Next, inhibitor was added and allowed to incubate with the polymerase•DNA solution for five minutes. In the control reaction, DMSO was added instead of the inhibitor solution. Each reaction was initiated by the addition of the dNTP•Mg2+ (0.25 mM of each dNTP and 5 mM MgCl2) solution to a pre-incubated enzyme•DNA complex (20 nM enzyme and 200 nM DNA). The reactions were performed in a final volume of 15 μL at 37 °C. Reactions were terminated by adding quench solution (20 mM EDTA and 95% (v/v) formamide) to each reaction at different time intervals, and then heating the samples at 95 °C for five minutes. The samples were separated using a 15% (w/v) polyacrylamide/7M urea gel and the products were analyzed using a Typhoon imager and ImageQuant™ software (GE Healthcare Life Sciences). Total product formed in each reaction was calculated by adding the band intensities of the +1, +2, +3, +4 and +5 products. The fraction of substrate converted to product was calculated by dividing the above by the total amount in each reaction (summation of +0, +1, +2, +3, +4 and +5 product bands). The percent product formed at each MK886 concentration was calculated by normalizing to the DMSO control, which was considered as 100%. This percent product formed was plotted as a function of MK886 concentration and fit to a four-parameter logistic model (equation 1) using Prism software (GraphPad, San Diego, CA):
| Equation 1 |
The experiments were performed using at least 11 concentrations of MK886 and were performed at different time points to verify the results.
Fluorescence anisotropy experiments to determine the effect of MK886 upon polymerase binding to DNA
Fluorescein-labeled DNA substrate (2 nM) was incubated with varying concentrations of protein and fluorescence polarization was measured in a Biotek SynergyH4 plate reader using the appropriate filter sets (λex = 485 ± 20 nm and λem = 525 ± 20 nm). All titrations were performed at 25 °C in 50 mM HEPES buffer (pH 7.5) containing 10 mM KOAc, 10 mM MgCl2, 0.1 mM ZnCl2, 0.1 mM EDTA, 2 mM β-E, 10% (v/v) DMSO and 0.1 mg/mL BSA. The experiments were performed in the presence of 0, 1, 10, 25, 50, 100, 150 and 250 μM MK886. DMSO was substituted in the no MK886 control experiment. Polarization was determined using equation 2:
| Equation 2 |
where F∥ equals fluorescence intensity parallel to the excitation plane and F⊥ equals the fluorescence intensity perpendicular to the excitation plane. The resulting change in polarization units was plotted against protein concentration and fit to a quadratic equation (equation 3).
| Equation 3 |
Where P = the measured change in fluorescence polarization, Etotal = the concentration of enzyme, DNAtotal = the concentration of fluorescein-labeled DNA and Kd is the measured equilibrium dissociation constant for enzyme binding to DNA. The binding curves were fit such that DNAtotal was fixed to be constant between curves.
Kinetic analysis of polymerase activity in the presence of MK886
In order to determine the mechanism of inhibition of Y-family DNA polymerases by MK886, kinetic analyses were performed using hpol ι26–446 under steady-state conditions using two approaches. In the first approach, single nucleotide insertion by hpol ι26–446 was studied on a fluorescein-labeled primer-template, in the presence of varying concentrations of MK886. The dsDNA substrate, in which the incoming correct nucleotide is dCTP, was generated as described earlier. Polymerase assay conditions were also identical to those described previously. The final concentrations of enzyme and dsDNA were 5 nM and 100 nM, respectively. The rate of product formation was quantified at 10 different dCTP concentrations between 1–1500 μM. Reactions were repeated in the presence of DMSO (control), or at different concentrations of the inhibitor MK886 (20 nM, 50 nM, 1 μM and 10 μM). Reactions were initiated by adding dCTP:Mg2+ (Mg2+ at 2 mM; dCTP at concentrations between 1–1500 μM), and were terminated at different time points by the addition of quench solution containing 95% (v/v) formamide and 20 mM EDTA. Products were separated through a 16% (w/v) acrylamide/7M urea gel and the bands were visualized and quantified as described earlier. At each dCTP concentration, in each set of experiments at different MK886 concentrations, velocities were calculated by plotting the amount of product formed as a function of time. The resulting velocities were then plotted as a function of dCTP concentration and fit to the Michealis-Menten equation. After correcting for enzyme concentration, the steady-state kinetic parameters of turnover number (kcat) and Michaelis constant (KM,dNTP) were obtained.
In the second approach, hpol ι26–446 -catalyzed primer extension was studied using the fluorogenic substrate-based assay described by Dorjsuren et al.32 All three oligonucleotides for this assay were purchased from Biosearch Technologies, Inc. (Novato, CA). Briefly, unlabeled 19-mer primer (5′TCACCCTCGTACGACTCTT-3′), 3'-carboxytetramethylrhodamine (TAMRA) – labeled 10-mer reporter oligonucleotide (5′-TTTTTTTTGC-6-TAMRA-3′) and 5'-Black Hole Quencher 2 (BHQ-2) - labeled 29-mer template oligonucleotide (5′-BHQ2-GCAAAAAAAAAAGAGTCGTACGAGGGTGA-3′) were annealed in a molar ratio of 1.4:1:1.5 to generate a tripartite dsDNA substrate. Assays were performed with 5 nM hpol ι26–446 and 50 nM of the tripartite DNA substrate in a buffer containing 50 mM Tris-HCl (pH 8.0 at 22 °C), 0.01% Tween-20, 2 mM dithiothreitol, 40 mM KCl and 0.25 mM MnCl2 at 25°C. The reporter strand displacement assay was performed in a 200 μL reaction mixture, in a 96-well plate. Initially, 170 μL of a master mix containing the enzyme, inhibitor at appropriate concentration (or DMSO in control), and assay buffer was pipetted into each well of the 96-well plate. This was followed by the addition of 10 μL of stock solution of dTTP to each well, to obtain final concentrations of dTTP between 1–200 μM. Reactions were initiated by the addition of 20 μL of the DNA substrate to each well with a multi-channel pipette. After mixing well, the fluorescence was continuously measured in a Biotek SynergyH4 plate reader using the appropriate filter sets (λex = 525 nm and λem = 598 nm) for up to 60 minutes. The measured relative fluorescence units (RFUs) were converted to a nanomolar quantity by calculating the total change in fluorescence observed between the start of the reaction and the time point at which the fluorescence change was maximal and considering that change to be 100% of substrate converted to product. The percentage of substrate converted to product was multiplied by the concentration of dsDNA in the reaction mixture. Product formation was then plotted as a function of time, and by considering only the linear portion of each curve, velocities were calculated for each dTTP concentration. These were then plotted as a function of dTTP concentration, and fit to a hyperbola. After correcting for enzyme concentration, the steady-state kinetic parameters were obtained as described previously.
In silico docking of MK886 in Y-family DNA polymerase structures
Prior to performing molecular docking, the protein PDB files were prepared for docking using the Dock Prep tool available in the free software package UCSF Chimera.33 This involved the addition of hydrogens, removal of solvent (water) molecules, and assigning partial charges (using the AMBER99 force field). Additionally, some extra molecules/ligands such as DNA, dNTP and metal ions, if present, were removed manually from the coordinates file. The 3-dimensional coordinates for the MK886 molecule were generated using the Marvin Sketch free software tool in the ChemAxon package. Automated in silico molecular docking was performed using the web-based docking server SwissDock (http://www.swissdock.ch/) that is based on the docking algorithm EADock DSS.34 The processed coordinates file (as described above) for each of the proteins and for the ligand MK886 were uploaded and docking runs were performed using the “Accurate” parameters option, which is the most exhaustive in terms of number of binding modes sampled. Docking runs were performed as `blind', covering the entire protein surface, and not defining any specific region of the protein as the binding pocket, in order to avoid sampling bias. Output `clusters' were obtained after each docking run and were ranked according to the FullFitness (FF) scoring function specified by the SwissDock algorithm (cluster 0 being the cluster with the best FullFitness score). A greater negative FF score indicates a more favorable binding mode with a better fit. Within each cluster, the individual binding poses were further arranged and ranked based on their FF score.
Docking runs were performed using the PDB files 4EBC (hpol ι), 3MR2 (hpol η), and 2OH2 (hpol κ) downloaded from the RCSB public database, for target Y-family DNA polymerases, either with the DNA coordinates (binary) in place, or after removing the DNA atoms (apoenzyme). Docking with each protein PDB target was repeated five times. The best binding mode hits obtained after the five docking runs (based on FF score) were displayed using UCSF Chimera. All figures with molecular representations of the docking results were generated using UCSF Chimera and PyMOL. A series of `control' docking runs were also performed using the structure coordinates of two other proteins, viz. HIV-1 protease (PDB file 1HPV) and bovine serum albumin (BSA; PDB file 4F5S), to test the validity of our docking results with the Y-family DNA polymerases and MK886. The PDB file 1HPV, which is a crystal structure of the HIV-1 protease dimer in complex with a small-molecule inhibitor VX478, was processed as described above. The atoms of the inhibitor VX478 were also removed. Five independent docking runs were performed by uploading the coordinates of the protease dimer and VX478 separately in SwissDock. Similar multiple docking runs were performed with the BSA crystal structure coordinates, with both MK886 and VX478. Results of docking runs of MK886 with BSA were compared to similar results with the Y-family DNA polymerases, while those with VX478 and BSA were compared with those of HIV-1 protease.
RESULTS
Inhibition of human Y-family DNA polymerases by MK886
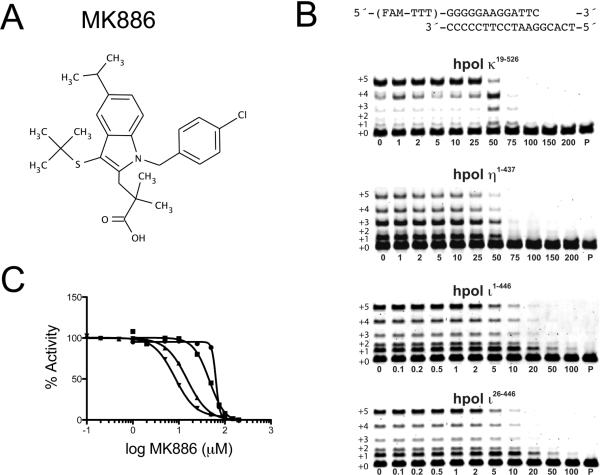
The small molecule MK886 was previously identified as an inhibitor of hpol κ utilizing a high-throughput fluorescent-based screen.23 Our first goal was to determine the specificity of MK886 against hpol κ. DNA synthesis was measured in the presence of increasing concentrations of MK886. To calculate polymerase activity, the total amount of product was first quantified for our DMSO control. Then the product formed in the presence of MK886 was divided by the control to obtain a % activity. Under the conditions tested here, which used a five-minute time point, hpol κ19–526 was inhibited by MK886 with an IC50 value of 63.8 ± 1.7 μM (Figure 1). Similar to hpol κ, the core polymerase domain of hpol η1–437 was inhibited with an IC50 value of 45.8 ± 8.4 μM, indicating that catalysis by hpol η and hpol κ is perturbed to a similar extent by MK886. Similar results were obtained when these experiments were repeated using different time points (Figure S1). The IC50 values reported here are slightly different from those reported by our groups previously, likely due to variations in the experimental design (e.g. enzyme concentrations and DNA substrates).23
Figure 1.
MK886 inhibits DNA synthesis by the Y-family DNA polymerases. A. The chemical structure of MK886 is shown. B. DNA polymerase-catalyzed primer extension assays were performed in the presence of increasing concentrations of MK886 with hpol κ19–526, hpol η1–437, hpol ι1–446 and hpol ι26–446 in the presence of all four dNTPs (0.25 mM) and MgCl2 (5 mM). The primer-template DNA used in the assays is shown above the gels. The concentration of MK886 (μM) is indicated underneath each lane. The un-reacted primer (P) was run as a control for each set of experiments. Extended polymerase products (+1 through +5) are indicated to the left of the gels. C. The IC50 values for MK886 inhibition of DNA polymerase activity were 63.8 ± 1.7 μM, 45.8 ± 8.4 μM, 15.1 ± 2.1 μM and 8.2 ± 0.6 μM for hpol κ19–526 (●), hpol η1–437 (■), hpol ι1–446 (▲) and hpol ι26–446 (▼), respectively.
Additionally, the inhibitory effects of MK886 were measured using two recombinant versions of hpol ι (Figure 1B, bottom two gels). The two versions of hpol ι were used because the original annotation of the hpol ι-coding region erroneously started downstream of the actual translational start site. The first version of hpol ι was predicted to be 715 amino acids in length and the newly annotated protein has an additional 25 amino acids at the N-terminus (740 a.a.). Thus, the polymerase core that has been utilized for crystallization purposes on many occasions by our group and others was originally denoted as being residues 1–420. The initial hpol ι construct contained residues 26–446 (hpol ι26–446) and the construct that resulted from the updated annotation, with the extended N-terminus, is comprised of residues 1–446 (hpol ι1–446).
DNA synthesis by the two recombinant versions of hpol ι was tested in the presence of MK886 (Figure 1). The measured IC50 values were 8.2 ± 0.6 μM and 15.1 ± 2.1 μM for hpol ι26–446 and hpol ι1–446, respectively. Analyses of these data revealed that the IC50 is lower for both enzymes relative to what was observed with hpol η and hpol κ and that there was a greater inhibition against the shorter version of hpol ι26–446, which lacks the N-terminal 25 a.a. extension (IC50 = 8.2 ± 0.6 μM). These assays were repeated using different time points to further verify these results (Figure S1). The more potent inhibition of the hpol ι construct that lacks the N-terminal extension was suggestive of potential site of interaction between the small-molecule inhibitor MK886 and hpol ι26–446. These results are interesting when one considers the relative importance of N-terminal extensions in the determination of Y-family polymerase mechanisms of action. For example, hpol κ lacking the N-terminal 70 amino acids is inactive and Rev1 requires an arginine residue in the N-digit for the protein-template directed mechanism of nucleotide selection. The stronger inhibition of hpol ι26–446 by MK886 is discussed later with regards to structural features that are only observed for this Y-family member. Perhaps most importantly, the IC50 experiments show that, of the Y-family members tested here, hpol ι is most strongly inhibited by MK886 with the shorter version, hpol ι26–446, exhibiting the lowest IC50 value.
MK886 inhibits a model B-family DNA polymerase
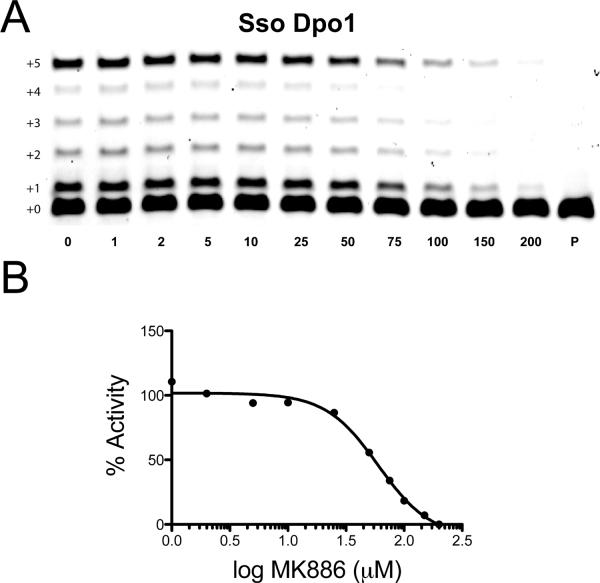
The small molecule MK886 was clearly able to inhibit all of the Y-family DNA polymerases tested, exhibiting some specificity against hpol ι. To extend these analyses, inhibition of a model B-family DNA polymerase, Dpo1 from Sulfolobus solfataricus was tested (Figure 2). B-family DNA polymerases are typically high-fidelity enzymes involved in processive replication events, though there are well-studied examples of B-family polymerases, such as pol ζ, that participate in DNA repair/translesion synthesis.35–37 Dpo1 is a high-fidelity enzyme that appears to function as the primary replicative enzyme in the crenarchaeote S. solfataricus.31 It is a robust enzyme that consists of a single sub-unit and serves as an excellent model system for understanding the more complex eukaryotic B-family members. An exonuclease defective mutant form of Dpo1 was utilized for our inhibition assays. Dpo1 was inhibited by MK886 with an IC50 value (59.8 ± 18.5 M) that is similar to that observed for the Y-family members hpol η1–437 and hpol κ19–526 (Figure 2). These results show that MK886 can inhibit DNA synthesis by enzymes from different families but that it shows the most potency against hpol ι. Based on these results, experimental strategies were designed to understand the mechanism of polymerase inhibition by MK886.
Figure 2.
MK886 inhibits DNA synthesis by the B-family DNA polymerase Dpo1 from Sulfolobus solfataricus. A. Dpo1-catalyzed primer extension assays were performed in the presence of increasing concentrations of MK886. The concentration of MK886 (μM) is indicated underneath each lane. The un-reacted primer (P) was run as a control for each set of experiments. Extended polymerase products (+1 through +5) are indicated to the left of the gels. B. The IC50 for MK886 inhibition of Dpo1 polymerase activity was determined to be 59.8 ± 18.5 μM.
Effect of MK886 upon DNA binding by hpol ι and hpol κ
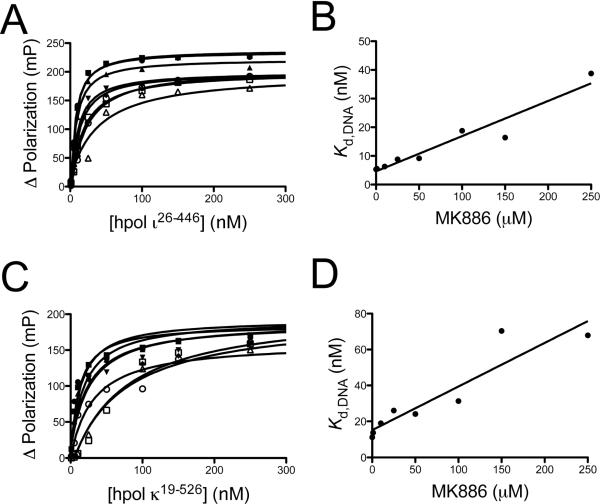
The molecular rationale for increased potency against hpol ι was investigated further in order to determine the mechanism of action. The ability of MK886 to interfere with polymerase binding to primer-template DNA was determined by measuring the equilibrium dissociation constant (Kd,DNA) for hpol ι26–446 in the presence of increasing concentrations of MK886 (Figure 3A and 3B). For hpol ι26–446, we obtained Kd,DNA values of 5.3 ± 0.7 nM, 5.5 ± 0.7 nM, 6.3 ± 0.9 nM, 8.8 ± 1.3 nM, 9.1 ± 1.6 nM, 18.8 ± 2.7 nM, 16.4 ± 2.1 nM and 38.8 ± 17.1 nM at MK886 concentrations of 0, 1, 10, 25, 50, 100, 150 and 250 μM, respectively. There was no observed change in the affinity of hpol ι26–446 for DNA at 10 μM MK886 relative to the DMSO control, which is near the IC50 value for inhibition of hpol ι26–446. There appears to be a linear relationship between the amount of MK886 present and the observed binding constant for hpol ι26–446, with the Kd,DNA increasing ~0.12 nM for every 1 μM MK886 in solution. Only at MK886 concentrations at or above 100 μM was there a substantial decrease in affinity for DNA binding (i.e. >2-fold increase in the Kd,DNA), suggesting that while high-concentrations of MK886 may inhibit hpol ι26–445 activity by interfering with binding to primer-template DNA, this is not the primary mode of polymerase inhibition observed at lower concentrations of the inhibitor.
Figure 3.
The effect of MK886 upon DNA binding by Y-family DNA polymerases is slightly more pronounced for hpol κ19–526 than hpol ι26–446. A. Fluorescein-labeled primer-template DNA (FAM-13/18mer; 2 nM) was incubated with increasing concentrations of hpol ι26–446 and the change in fluorescence polarization was measured. The experiment was repeated in the presence of increasing concentrations of MK886 (1 μM to 250 μM). B. The measured equilibrium dissociation constants (Kd,DNA) determined in panel A were re-plotted as a function of MK886 concentration. The data was fit to a linear equation and the slope used as a measure of MK886 inhibition of polymerase binding to DNA. C. Fluorescein-labeled primer-template DNA (FAM-13/18mer; 2 nM) was incubated with increasing concentrations of hpol κ19–526 and the change in fluorescence polarization was measured. The experiment was repeated in the presence of increasing concentrations of MK886 (1 μM to 250 μM). D. The measured equilibrium dissociation constants (Kd,DNA) deterimined in panel C were re-plotted as a function of MK886 concentration. The data was fit to a linear equation and the slope used as a measure of MK886 inhibition of polymerase binding to DNA.
The ability of MK886 to inhibit DNA binding by hpol κ was also tested (Figure 3C and 3D). Like hpol ι26–446, a moderate increase in the equilibrium dissociation constant was observed as the concentration of MK886 was increased. For hpol κ19–526, Kd,DNA values of 11.1 ± 5.1 nM, 13.7 ± 5.1 nM, 19.0 ± 5.6 nM, 26.0 ± 9.1 nM, 24.1 ± 9.4 nM, 31.3 ± 12.1 nM, 70.4 ± 15.7 nM and 67.9 ± 17.1 nM were obtained at MK886 concentrations of 0, 1, 10, 25, 50, 100, 150 and 250 μM, respectively. Again, the trend appeared to be fairly linear in nature and linear regression analysis indicated that the Kd,DNA for hpol κ19–526 binding to primer-template DNA increased ~0.24 nM for every 1 μM increase in MK886 concentration. The small molecule inhibitor produces a change in binding affinity for hpol κ that was ~2-fold greater than that observed for hpol ι26–446. The Kd,DNA for hpol κ binding to DNA was increased ~1.8-fold at concentrations near the measured IC50 value for inhibition of polymerase activity, which would indicate that this is an important feature in the mechanism of MK886 inhibition of hpol κ19–526. These data are interpreted as evidence of increased specificity for interactions between MK886 and hpol ι26–446 that occur at sites on the protein that are not involved in DNA binding.
Kinetic analysis of the mechanism of polymerase inhibition by MK886
In order to further assess the mechanism of polymerase inhibition by MK886, single-nucleotide insertion kinetic analyses of hpol ι26–446 activity were performed in the presence of increasing concentrations of inhibitor. The concentration of nucleotide triphosphate was varied (1 μM to 1.5 mM) and the initial velocity of dNTP insertion was measured. These experiments were repeated in the presence of 20 nM, 50 nM, 1 μM and 10 μM MK886. Steady-state kinetic analyses showed that both the turnover number (kcat) and the Michealis constant (Km,dNTP) were altered by the inclusion of MK886 (Table 1), a result that was consistent with a mixed-model of inhibition of DNA polymerase activity by MK886. A fluorescence-based assay was also used to determine the effect of MK886 upon the steady-state kinetic parameters that define hpol ι26–446-catalyzed insertion of dNTPs in the presence of increasing amounts of inhibitor (Figure S2). Again, a diminished kcat and increased KM,dNTP were observed as the concentration of MK886 was increased in the reaction mixture (Table 2), a result that is indicative of mixed-type inhibition by MK886.
Table 1.
Steady-state kinetic parameters for hpol ι26–446-catalyzed single-nucleotide insertion in the presence of MK886.
| [MK886] (μM) | k cat | K M,dNTP | kcat/KM,dNTP | |
|---|---|---|---|---|
| (min−1) | (μM) | (min−1 μM−1) | Fold decrease in kcat/KM,dNTP | |
| 0 | 2.2 ± 0.1 | 28.4 ± 7.6 | 0.077 | - |
| 0.02 | 1.4 ± 0.1 | 27.1 ± 12.3 | 0.052 | 1.5 |
| 0.05 | 1.2 ± 0.1 | 33.1 ± 11.8 | 0.036 | 2.1 |
| 1 | 1.6 ± 0.1 | 44.1 ± 16.4 | 0.036 | 2.1 |
| 10 | 1.5 ± 0.1 | 56.0 ± 19.4 | 0.027 | 2.8 |
Table 2.
Steady-state kinetic parameters for hpol ι26–446-catalyzed primer extension in the presence of MK886 as determined using a fluorescence-based assay.
| [MK886] (μM) | k cat | K M,dNTP | kcat/KM,dNTP | |
|---|---|---|---|---|
| (min−1) | (μM) | (min−1 μM−1) | Fold decrease in kcat/KM,dNTP | |
| 0 | 5.6 ± 0.1 | 0.8 ± 0.2 | 7.0 | - |
| 20 | 4.4 ± 0.1 | 1.0 ± 0.2 | 4.4 | 1.6 |
| 40 | 4.3 ± 0.1 | 1.3 ± 0.2 | 3.3 | 2.1 |
| 60 | 4.1 ± 0.2 | 1.4 ± 0.3 | 2.9 | 2.4 |
Results of molecular docking of MK-886 to the Y-family DNA polymerases
We attempted to crystallize hpol ι26–446 in the presence of MK886 in order to assess the structural rationale for polymerase inhibition. Diffracting crystals were obtained of both binary and ternary complexes of hpol ι26–446. MK886 was either co-crystallized or soaked in after the crystals were formed. However, none of the diffraction patterns revealed clear density for the small molecule, even after multiple rounds of refinement (data not shown). After several unsuccessful attempts to obtain data sets with density for MK886 we turned to in silico docking with the web server-based SwissDock in an effort to better understand the molecular features associated with polymerase inhibition.
Prior to performing docking analyses with MK886 and the Y-family polymerases, a series of control docking experiments were performed to establish that the algorithms would provide reasonable models for potential small-molecule binding sites. For a “positive control”, the previously solved crystal structure for HIV-1 protease in complex with the small-molecule inhibitor VX-478 was used.38 For “negative controls” either VX-478 or MK886 were docked onto bovine serum albumin (BSA). Importantly, SwissDock correctly predicted the binding mode observed for VX-478 in the HIV-1 protease crystal structure (Figure S3). Both VX-478 and MK886 molecules were distributed across multiple sites on BSA (Figures S3 and S4). Notably, docking runs with BSA produced variable results, with no consistently identified binding pockets (Figure S4). Thus, it was confirmed that SwissDock could accurately identify a small-molecule binding site for a previously solved structure and that the program did not identify binding pockets for either VX-478 or MK886 on BSA.
Docking submissions were next performed with the apo-forms of three Y-family DNA polymerases: hpol ι26–446 (4EBC), hpol η1–437 (3MR2), and hpol κ19–526 (2OH2). An average of 34 clusters were obtained for hpol ι, 32 clusters for hpol η, and 32 clusters for hpol κ from five docking runs of MK-886 with each of the three Y-family polymerases (Table S1). The term “cluster” refers a group of MK886 molecules with binding modes occurring at one site on the target protein. The binding modes within a cluster differ only by small rotational changes or rotations around bonds and are ranked according to the predicted binding energy.
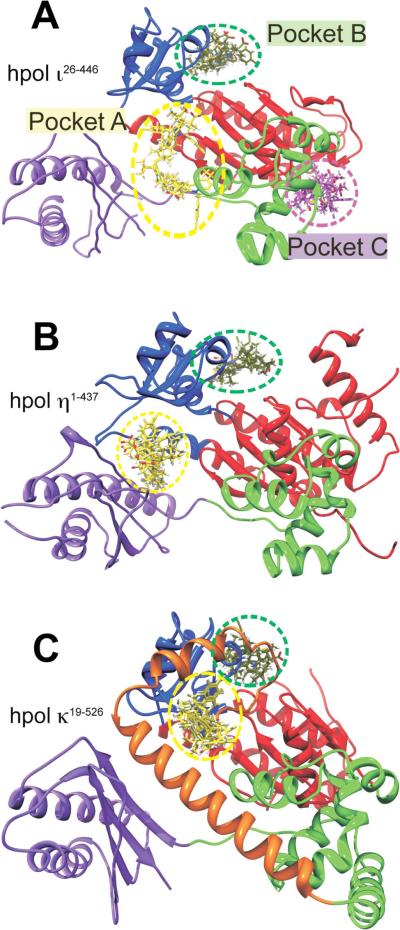
In docking analyses, three MK886 binding pockets for hpol ι26–446 were consistently identified (Figure 4A) and only two MK886 binding pockets for hpol η1–437 (Figure 4B) and hpol κ19–526 (Figure 4C). For all three polymerases, the highest number of clusters was found to localize at the interface between the DNA-binding cleft and the active-site of each Y-family member, which we refer to as pocket A (Figure 4). In addition to this pocket, two more distinct binding pockets were observed for hpol ι. The first of these, which we call pocket B, lies at the junction between the finger and palm sub-domains of hpol ι (Figure 4A). Pocket B was also identified in docking analyses with hpol η1–437 and hpol κ19–526. The final binding pocket for MK886 on hpol ι26–446, which we refer to as pocket C, lies at the junction between the palm and thumb sub-domains (Figure 4A). Consistently it was observed that at least one cluster from the top 5 (top 10 in the case of pocket C) localized to these three pockets on hpol ι26–446. It is interesting to note that nearly all of the binding modes identified in our docking analyses for the polymerases localized to one of these three pockets, with only an occasional outlier cluster, which localized to a completely different region of the polymerase. The consistent identification of MK886 binding pockets on the DNA polymerases is in stark contrast to what was observed with BSA where the top 10 binding modes changed positions dramatically between docking experiments (Figure S4).
Figure 4.
Docking results for MK886 reveal potential binding sites, including a pocket specific to hpol ι26–446. In silico docking runs were performed as described in Experimental Procedures. A. The crystal structure of hpol ι26–446 (PDB ID 4EBC) is shown as a cartoon representation, with the sub-domains colored as follows: Finger, blue; Thumb, green; Palm, red; little finger (or palm-associated domain), purple. The MK886 binding modes with the highest FF scores from each of the five docking runs are shown as sticks, localized to each of the three binding pockets (A, B and C, indicated by dashed yellow, green and magenta circles respectively). MK886 clusters binding to the pocket A are shown in yellow, pocket B clusters (junction of fingers and palm sub-domains) are in green, while those at pocket C are shown in magenta. Panels B and C show crystal structures of hpol η1–437 (PDB ID 3MR2) and hpol κ19–526 (PDB ID 2OH2), respectively, in the same relative orientation with respect to hpol ι26–446 in panel A, and with the domains colored identically. The `N-clasp' sub-domain in hpol κ19–526 is shown in orange in panel C. Note the absence of any MK886 clusters in hpol η1–437 and hpol κ19–526, in the region corresponding to pocket C of hpol ι26–446. All crystal structure representations shown were generated using the software PyMol (DeLano Scientific, San Carlos, CA).
Docking runs were performed with two versions of the target polymerase PDB files, one in which the coordinates for the DNA atoms were left in place (binary form), and the other in which these were removed to give an “apo” form. The only difference observed in the results obtained with these two versions was in the number of clusters observed in pocket A. As expected, fewer clusters were found localized to pocket A when the binary form of the target PDB was used, as compared to when we used the apo form PDB coordinates since the DNA occludes much of pocket A for each enzyme. However, the number and positions of clusters obtained in the pockets B and C using these two versions did not vary significantly, and hence, in subsequent discussion, only the results with the apo form are considered, since this gave the maximum number of hits.
DISCUSSION
The number of polymerases encoded by the human genome includes at least fifteen enzymes.16 It has been recognized that some of the more recently identified polymerases appear to play minimal roles in genomic replication (i.e. processive/high-fidelity events) but rather participate in specialized replication events, such as DNA repair, somatic hypermutation and replication stress response.39, 40 The Y-family DNA polymerases represent a class of enzymes that can bypass DNA adducts because of a relatively spacious active site and lack of proofreading activity.41, 42 Prokaryotic Y-family enzymes constitute the central polymerase activity in the so- called “S.O.S” response.43, 44 To a large extent, eukaryotes also rely upon the Y-family polymerases to bypass unrepaired DNA adducts.5 The relevance of Y-family polymerases to human disease is best illustrated by studies with the skin cancer-prone disease xeroderma pigmentosum variant (XPV), a severe cancer-prone disease that results from inactivating mutations in the gene that codes for hpol η and leads to inaccurate replication of UV-induced cyclobutane pyrimidine dimers.45–47 Perhaps not surprisingly, altered activity/expression of Y-family members has been observed in multiple cancer types.7–9 The importance of these enzymes to mechanisms of mutagenesis and a potential role for them in cancer initiation/progression led us to identify small molecule inhibitors against one of the human enzymes, hpol κ, which is thought to play important roles in the relatively accurate bypass of bulky N2-guanyl DNA adducts and in the mutagenic bypass of the common oxidative lesion 7,8-dihydro-8-oxo-2′- deoxyguanosine (8-oxo-dG or oxoG).48–50 These investigations identified the small molecule MK886 that was originally pursued as an inhibitor of leukotriene biosynthesis.24, 25 The current study was an attempt to determine the in vitro specificity and mode of polymerase inhibition by MK886.
The first conclusion that can be drawn from our results is that MK886 inhibits hpol ι26–446 more potently than the other polymerases tested here. The IC50 values indicate that MK886 inhibits the truncated version of hpol ι six- to eight-fold more effectively than either hpol η1–437 or hpol κ19–526 (Figure 1 and Figure S1). The increased potency observed for hpol ι could be due to a number of factors. Interestingly, hpol ι is the only DNA polymerase that preferentially utilizes Hoogsteen base pairing modes during nucleotide selection opposite undamaged templates.51 This unusual mechanism of nucleotide selection is due in large part to a narrowed active site, that constrains the distance between C1′-C1′ atoms to around 9 Å, and induces purines to adopt a syn-orientation about the glycosyl bond in order to accommodate the purine:pyrimidine base pair.
Based on our analyses, it would appear that MK886 only interferes with hpol ι26–446 DNA binding at concentrations above the measured IC50 values (Figure 3A and 3B). This indicates that the small-molecule acts through a mechanism that is not dependent upon direct interference with polymerase·primer-template DNA interactions. MK886 has a more pronounced effect upon hpol κ19–526 DNA binding affinity, increasing the Kd,DNA about 2-fold at concentrations of MK886 near the IC50 value (Figure 3C and 3D). Steady-state results with hpol ι26–446 in which the dNTP concentration was varied point to a mixed-mode of inhibition by MK886 (i.e. an increased Km,dNTP is accompanied by a diminished turnover number; see Tables 1 and 2). Thus, at least for hpol ι26–446, MK886 presumably acts through a mechanism that interferes with the ternary complex in some fashion.
We attempted to reconcile our functional results with structural determinants associated with polymerase inhibition by MK886. Our attempts to crystallize hpol ι26–446 were successful in that they produced X-ray diffraction data sets but we did not observe convincing electron density for MK886 in the resulting maps (data not shown). As an alternative strategy, we chose to utilize a freely available docking program (SwissDock) to analyze potential binding pockets for MK886 using coordinates from previously reported crystal structures of three Y-family DNA polymerases tested in our functional assays. The web server (http://www.swissdock.ch) allows for automated docking to be performed with existing coordinate files.52 For our docking submissions we focused on the Y-family DNA polymerases. Multiple crystal structures have been reported for hpol η1–437, hpol κ19–526 and hpol ι26–446.17, 49, 51, 53–55 Both apo and binary coordinate files were submitted to SwissDock for each enzyme. For our discussion, we consider only the apo form since the only observed difference was in the binding pocket near the primer-template DNA/active site interface (pocket A). The resulting MK886 binding sites distant from the DNA/active site interface were the same whether DNA was present during the docking procedure or not.
In the docking runs performed with the hpol η1–437 and hpol κ19–526 PDB targets, clusters were obtained only in the pockets A and B (Figure 4). For hpol η1–437 and hpol κ19–526 there were no clusters that corresponded to pocket C, as observed with hpol ι26–446. Pockets A and B corresponded very well for all three polymerases. Thus, pocket C was found to be unique to hpol ι26–446. As noted above, there exist two versions of hpol ι that differ by 25 amino acids in the length at the N-terminus in the public database. These correspond to accession number NP_009126.2 (740 amino acids) and accession number AAD50381 (715 amino acids). All previous biochemical and structural studies were based on the shorter version of the enzyme (i.e. the protein lacking the newly annotated 25 amino acids at the N-terminus). The same shorter construct was also used in the crystal structure 4EBC that was utilized in our docking studies. Superimposition of three polymerase structures (4EBC, 3MR2 and 2OH2) clearly suggests that, in the case of hpols η and κ, the binding pocket C is not as well formed as in hpol ι (Figure S5). We can tentatively attribute the absence of any MK886 binding clusters in the pocket C with hpols η and κ to the structural changes at the palm-thumb junction produced by N-terminal amino acids in these structures. In the light of this, it is interesting to note that, when we performed biochemical experiments to calculate IC50 for MK886 with the two versions of hpol ι (Figure 1), we observed a lower IC50 for hpol ι26–446 (8.2 ± 0.6 μM) as compared to hpol ι1–446 (15.1 ± 2.1 μM). From the results obtained with our docking studies, we can explain this difference as caused due to the presence of the extra 25 amino acids at the N-terminus of hpol ι1–446. Pocket C is formed quite near the N-terminus of the previously crystallized hpol ι26–446. In the longer version of the protein, it can be envisaged that pocket C would be either occluded, or would not be formed at all, as seen with hpols η and κ (Figure S5). In the shorter protein (hpol ι26–446) on the other hand, there is no occlusion, which presents an additional binding for MK886 (pocket C). The presence of the additional pocket may contribute to more pronounced inhibition of hpol ι26–446 by MK886.
The negative impact upon catalysis if MK886 binds to pocket A is obvious for all three polymerases. In the hpol ι26–446 docking model, interactions between MK886 and the side-chains of Arg103 and Arg331 would disrupt key electrostatic interactions between the polymerase and the template strand. Similar effects upon DNA binding could be predicted for hpol η1–437 and hpol κ19–526. Consideration of inhibitor binding to pocket B in hpol ι26–446 shows that the electrostatic interaction between the docked MK886 molecule and Asn216 stations the inhibitor near the “gate” to the dNTP binding cleft, which could also reasonably be assumed to interfere with productive binding of the incoming nucleotide triphosphate. Likewise, binding of MK886 to pocket B of either hpol η1–437 or hpol κ19–526 could conceivably interfere with productive binding of dNTP. Finally, pocket C is only observed with hpol ι26–446 where MK886 is found to interact with residues in the thumb and palm domains. The potential inhibitory effect of pocket C is less obvious than either pocket A or pocket B. In pocket C, MK886 interacts with residues that are near the base of the αH helix (Gln227) in the thumb domain, not far removed from the binding site of a third metal ion. Transient coordination of a third metal ion was recently shown to play a role in catalysis by hpol η and metal ion coordination has been suggested to be the rate-limiting step in catalysis by hpol η.55, 56 Other studies have revealed that conformational changes in the thumb domain play a role in nucleotide selection by Y-family polymerases.57, 58 Thus, it would appear that binding of MK886 to pocket C near the thumb region of hpol ι26–446 may contribute to the more potent inhibitory effect by interfering with either conformational changes or metal ion dynamics, and that this effect is not observed for either hpol η1–437 and hpol κ19–526 since they do not possess a well-formed pocket C (Figure S5).
CONCLUSIONS
MK886 is an indole-derived compound that has been shown to perturb leukotriene biosynthesis.24, 25 MK886 inhibits DNA synthesis in cell culture at concentrations as low as 1 μM and can induce DNA fragmentation at 0.1 μM.30 At higher concentrations MK886 has antiproliferative properties, inducing apoptosis through inhibition of multiple pathways.26, 28, 29, 59–61 We recently discovered that MK886 is also an inhibitor of hpol κ.23 The initial discovery that MK886 can inhibit a translesion polymerase led us to study the in vitro specificity of the molecule as an inhibitor of DNA replication and to investigate the mode of action against these enzymes. Our results clearly show that MK886 can inhibit both the B-family polymerase Sso Dpo1 and the Y-family DNA polymerases hpol η and κ with similar efficiency. The relatively high IC50 values for MK886 inhibition observed for these enzymes makes it unlikely that it could be used to target translesion DNA synthesis properties in vivo. However, future studies with MK886 derivatives may identify more potent and/or specific inhibitors of translesion DNA polymerase activity.
MK886 exhibits the most potent inhibitory properties against the highly error-prone Y-family member, hpol ι. The mechanism of inhibition appears to be mixed in nature. The mixed-mode of inhibition is supported by steady-state kinetic results and is likely the result of MK886 binding to multiple pockets identified by our docking experiments. The docking experiments with hpol ι26–446 reveal a third binding pocket (pocket C) for MK886 near the N-terminus that does not exist for either hpol κ19–526 or hpol η1–437 (Figures 4 and S5). The existence of multiple binding pockets is supported by our results showing increased potency against a truncated version of hpol ι, which lacks the newly annotated N-terminal extension. Experiments with the extended version of hpol ι1–446 show that the addition of 25 amino acids at the N-terminus of the enzyme diminishes the effectiveness of MK886 as an inhibitor. Pocket C is located very near the N-terminal residues of hpol ι26–446. However, the longer version of the protein is still inhibited, indicative of a MK886 binding site distinct from pocket C.
In summary, our results show that MK886 exerts a previously unrecognized biochemical effect by acting as a polymerase inhibitor. It is difficult to make direct comparisons with the IC50 values determined in our in vitro polymerase assays and those values reported for MK886 inhibition of 5-lipoxygenase activating protein (FLAP) since most of the previous studies utilized cell culture assays or simple binding assays to measure inhibition of FLAP.25, 62 Still, it is worth noting that the IC50 value for MK886 inhibition of hpol ι is comparable to the IC50 value for COX1 inhibition (8 μM) by MK886 that was determined using in vitro activity assays.27 However, it remains unclear whether MK886 can provide targeted modulation of hpol ι translesion polymerase activity in vivo.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by National Institutes of Health Grants R00 GM084460 (R.L.E.), R01 GM055237 (M.E.) and R03 MH094179 (R.S.L.). This work was also supported by NRF grant 2012R1A1A2042391 from MEST Korea (to J.-Y.C.) and by a grant from the Arkansas Breast Cancer Research Program (R.L.E.), with additional support from the University of Arkansas for Medical Sciences Translational Research Institute (CTSA Grant Award UL1TR000039).
ABBREVIATIONS
- dNTP
deoxynucleoside triphosphate
- FAM
fluorescein
- HIV-1
human immunodeficiency virus type 1
- pol
polymerase
- Sso
Sulfolobus solfataricus
- TLS
translesion DNA synthesis
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information Available. The supporting information section contains a table summarizing the docking results (Table S1), as well as results from our kinetic analysis and docking studies (Figures S1–S5). This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- (1).Alberts B. Molecular biology of the cell. 4th ed. Garland Science; New York: 2002. [Google Scholar]
- (2).Geacintov N, Broyde S. The Chemical Biology of DNA Damage. Wiley-VCH; Weinheim: 2010. [Google Scholar]
- (3).Bartkova J, Hamerlik P, Stockhausen MT, Ehrmann J, Hlobilkova A, Laursen H, Kalita O, Kolar Z, Poulsen HS, Broholm H, Lukas J, Bartek J. Replication stress and oxidative damage contribute to aberrant constitutive activation of DNA damage signalling in human gliomas. Oncogene. 2010;29:5095–5102. doi: 10.1038/onc.2010.249. [DOI] [PubMed] [Google Scholar]
- (4).Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- (5).Chang DJ, Cimprich KA. DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yamanaka K, Lloyd RS. Functions of Translesion DNA polymerases: Implications for cancer risk and opportunities as therapeutic targets. In: Madhusudan S, Wilson DM 3rd, editors. DNA Repair and Cancer: Bench to Clinic. Science Publishers; Enfield, NH: 2012. [Google Scholar]
- (7).Albertella MR, Lau A, O'Connor MJ. The overexpression of specialized DNA polymerases in cancer. DNA Repair (Amst) 2005;4:583–593. doi: 10.1016/j.dnarep.2005.01.005. [DOI] [PubMed] [Google Scholar]
- (8).Lemée F, Bavoux C, Pillaire MJ, Bieth A, Machado CR, Pena SD, Guimbaud R, Selves J, Hoffmann JS, Cazaux C. Characterization of promoter regulatory elements involved in downexpression of the DNA polymerase kappa in colorectal cancer. Oncogene. 2007;26:3387–3394. doi: 10.1038/sj.onc.1210116. [DOI] [PubMed] [Google Scholar]
- (9).Wang H, Wu W, Wang HW, Wang S, Chen Y, Zhang X, Yang J, Zhao S, Ding HF, Lu D. Analysis of specialized DNA polymerases expression in human gliomas: association with prognostic significance. Neuro. Oncol. 2010;12:679–686. doi: 10.1093/neuonc/nop074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ceppi P, Novello S, Cambieri A, Longo M, Monica V, Lo Iacono M, Giaj-Levra M, Saviozzi S, Volante M, Papotti M, Scagliotti G. Polymerase eta mRNA expression predicts survival of non-small cell lung cancer patients treated with platinum-based chemotherapy. Clin. Cancer Res. 2009;15:1039–1045. doi: 10.1158/1078-0432.CCR-08-1227. [DOI] [PubMed] [Google Scholar]
- (11).Chen YW, Cleaver JE, Hanaoka F, Chang CF, Chou KM. A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Mol. Cancer Res. 2006;4:257–265. doi: 10.1158/1541-7786.MCR-05-0118. [DOI] [PubMed] [Google Scholar]
- (12).Makridakis NM, Reichardt JK. Translesion DNA polymerases and cancer. Front. Genet. 2012;3:174. doi: 10.3389/fgene.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Moraes MC, de Andrade AQ, Carvalho H, Guecheva T, Agnoletto MH, Henriques JA, Sarasin A, Stary A, Saffi J, Menck CF. Both XPA and DNA polymerase eta are necessary for the repair of doxorubicin-induced DNA lesions. Cancer Lett. 2012;314:108–118. doi: 10.1016/j.canlet.2011.09.019. [DOI] [PubMed] [Google Scholar]
- (14).Castro C, Smidansky ED, Arnold JJ, Maksimchuk KR, Moustafa I, Uchida A, Gotte M, Konigsberg W, Cameron CE. Nucleic acid polymerases use a general acid for nucleotidyl transfer. Nat. Struct. Mol. Biol. 2009;16:212–218. doi: 10.1038/nsmb.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- (16).Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. (U. S. A.) 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Eoff RL, Angel KC, Egli M, Guengerich FP. Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by Sulfolobus solfataricus DNA polymerase Dpo4: steady-state and pre-steady-state kinetics and x-ray crystallography of correct and incorrect pairing. J. Biol. Chem. 2007;282:13573–13584. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- (19).Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, Jerina DM, Yang W. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc. Natl. Acad. Sci. (U. S. A.) 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang H, Eoff RL, Kozekov ID, Rizzo CJ, Egli M, Guengerich FP. Versatility of Y-family Sulfolobus solfataricus DNA polymerase Dpo4 in translesion synthesis past bulky N2-alkylguanine adducts. J. Biol. Chem. 2009;284:3563–3576. doi: 10.1074/jbc.M807778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kimura T, Takeuchi T, Kumamoto-Yonezawa Y, Ohashi E, Ohmori H, Masutani C, Hanaoka F, Sugawara F, Yoshida H, Mizushina Y. Penicilliols A and B, novel inhibitors specific to mammalian Y-family DNA polymerases. Bioorg. Med. Chem. 2009;17:1811–1816. doi: 10.1016/j.bmc.2009.01.064. [DOI] [PubMed] [Google Scholar]
- (22).Mizushina Y, Motoshima H, Yamaguchi Y, Takeuchi T, Hirano K, Sugawara F, Yoshida H. 3-O-methylfunicone, a selective inhibitor of mammalian Y-family DNA polymerases from an Australian sea salt fungal strain. Mar. Drugs. 2009;7:624–639. doi: 10.3390/md7040624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Yamanaka K, Dorjsuren D, Eoff RL, Egli M, Maloney DJ, Jadhav A, Simeonov A, Lloyd RS. A comprehensive strategy to discover inhibitors of the translesion synthesis DNA polymerase κ. PLoS One. 2012;7:e45032. doi: 10.1371/journal.pone.0045032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gillard J, Ford-Hutchinson AW, Chan C, Charleson S, Denis D, Foster A, Fortin R, Leger S, McFarlane CS, Morton H, et al. L-663,536 (MK-886) (3-[1-(4-chlorobenzyl)-3-t-butyl-thio-5-isopropylindol-2-yl]-2,2 - dimethylpropanoic acid), a novel, orally active leukotriene biosynthesis inhibitor. Can. J. Physiol. Pharmacol. 1989;67:456–464. doi: 10.1139/y89-073. [DOI] [PubMed] [Google Scholar]
- (25).Rouzer CA, Ford-Hutchinson AW, Morton HE, Gillard JW. MK886, a potent and specific leukotriene biosynthesis inhibitor blocks and reverses the membrane association of 5-lipoxygenase in ionophore-challenged leukocytes. J. Biol. Chem. 1990;265:1436–1442. [PubMed] [Google Scholar]
- (26).Datta K, Biswal SS, Kehrer JP. The 5-lipoxygenase-activating protein (FLAP) inhibitor, MK886, induces apoptosis independently of FLAP. Biochem J. 1999;340(Pt 2):371–375. [PMC free article] [PubMed] [Google Scholar]
- (27).Koeberle A, Siemoneit U, Northoff H, Hofmann B, Schneider G, Werz O. MK-886, an inhibitor of the 5-lipoxygenase-activating protein, inhibits cyclooxygenase-1 activity and suppresses platelet aggregation. Eur. J. Pharmacol. 2009;608:84–90. doi: 10.1016/j.ejphar.2009.02.023. [DOI] [PubMed] [Google Scholar]
- (28).Lim JY, Oh JH, Jung JR, Kim SM, Ryu CH, Kim HT, Jeun SS. MK886-induced apoptosis depends on the 5-LO expression level in human malignant glioma cells. J. Neurooncol. 2009;97:339–346. doi: 10.1007/s11060-009-0036-9. [DOI] [PubMed] [Google Scholar]
- (29).Kehrer JP, Biswal SS, La E, Thuillier P, Datta K, Fischer SM, Vanden Heuvel JP. Inhibition of peroxisome-proliferator-activated receptor (PPAR)alpha by MK886. Biochem J. 2001;356:899–906. doi: 10.1042/0264-6021:3560899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Dittmann KH, Mayer C, Rodemann HP, Petrides PE, Denzlinger C. MK-886, a leukotriene biosynthesis inhibitor, induces antiproliferative effects and apoptosis in HL-60 cells. Leuk. Res. 1998;22:49–53. doi: 10.1016/s0145-2126(97)00132-x. [DOI] [PubMed] [Google Scholar]
- (31).Choi JY, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011;286:31180–31193. doi: 10.1074/jbc.M111.258038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Dorjsuren D, Wilson DM, 3rd, Beard WA, McDonald JP, Austin CP, Woodgate R, Wilson SH, Simeonov A. A real-time fluorescence method for enzymatic characterization of specialized human DNA polymerases. Nucleic Acids Res. 2009;37:e128. doi: 10.1093/nar/gkp641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lang PT, Brozell SR, Mukherjee S, Pettersen EF, Meng EC, Thomas V, Rizzo RC, Case DA, James TL, Kuntz ID. DOCK 6: combining techniques to model RNA-small molecule complexes. RNA. 2009;15:1219–1230. doi: 10.1261/rna.1563609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem. 2011 doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- (35).Haracska L, Unk I, Johnson RE, Johansson E, Burgers PM, Prakash S, Prakash L. Roles of yeast DNA polymerases delta and zeta and of Rev1 in the bypass of abasic sites. Genes Dev. 2001;15:945–954. doi: 10.1101/gad.882301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Sharma S, Canman CE. REV1 and DNA polymerase zeta in DNA interstrand crosslink repair. Environ. Mol. Mutagen. 2012;53:725–740. doi: 10.1002/em.21736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sharma S, Shah NA, Joiner AM, Roberts KH, Canman CE. DNA polymerase zeta is a major determinant of resistance to platinum-based chemotherapeutic agents. Mol. Pharmacol. 2012;81:778–787. doi: 10.1124/mol.111.076828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kim EE, Baker CT, Dwyer MD, Murcko MA, Rao BG, Tung RD, Navia MA. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 1995;117:1181–1182. [Google Scholar]
- (39).Seki M, Gearhart PJ, Wood RD. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 2005;6:1143–1148. doi: 10.1038/sj.embor.7400582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Front. Biosci. 2006;11:2496–2517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- (41).Friedberg EC, Wagner R, Radman M. Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science. 2002;296:1627–1630. doi: 10.1126/science.1070236. [DOI] [PubMed] [Google Scholar]
- (42).Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- (43).Radman M. SOS replication: a distinct DNA replication mechanism which is induced by DNA-damaging treatments? DNA Repair (Amst) 1970;4:732–738. [PubMed] [Google Scholar]
- (44).Sutton MD, Smith BT, Godoy VG, Walker GC. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 2000;34:479–497. doi: 10.1146/annurev.genet.34.1.479. [DOI] [PubMed] [Google Scholar]
- (45).Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Pol eta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- (46).Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- (47).Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Choi JY, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase kappa. J. Biol. Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- (49).Irimia A, Eoff RL, Guengerich FP, Egli M. Structural and functional elucidation of the mechanism promoting error-prone synthesis by human DNA polymerase kappa opposite the 7,8-dihydro-8-oxo-2'-deoxyguanosine adduct. J. Biol. Chem. 2009;284:22467–22480. doi: 10.1074/jbc.M109.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Ogi T, Shinkai Y, Tanaka K, Ohmori H. Pol kappa protects mammalian cells against the lethal and mutagenic effects of benzo[a]pyrene. Proc. Natl. Acad. Sci. (U. S. A.) 2002;99:15548–15553. doi: 10.1073/pnas.222377899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- (52).Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Ketkar A, Zafar MK, Banerjee S, Marquez VE, Egli M, Eoff RL. A Nucleotide-Analogue-Induced Gain of Function Corrects the Error-Prone Nature of Human DNA Polymerase iota. J. Am. Chem. Soc. 2012;134:10698–10705. doi: 10.1021/ja304176q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, Prakash S, Prakash L, Aggarwal AK. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Molecular cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- (55).Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012;487:196–201. doi: 10.1038/nature11181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Ummat A, Silverstein TD, Jain R, Buku A, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase eta is pre-aligned for dNTP binding and catalysis. J. Mol. Biol. 2011;415:627–634. doi: 10.1016/j.jmb.2011.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Beckman JW, Wang Q, Guengerich FP. Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence. J. Biol. Chem. 2008;283:36711–36723. doi: 10.1074/jbc.M806785200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Eoff RL, Sanchez-Ponce R, Guengerich FP. Conformational changes during nucleotide selection by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 2009;284:21090–21099. doi: 10.1074/jbc.M109.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Cianchi F, Cortesini C, Magnelli L, Fanti E, Papucci L, Schiavone N, Messerini L, Vannacci A, Capaccioli S, Perna F, Lulli M, Fabbroni V, Perigli G, Bechi P, Masini E. Inhibition of 5-lipoxygenase by MK886 augments the antitumor activity of celecoxib in human colon cancer cells. Mol. Cancer Ther. 2006;5:2716–2726. doi: 10.1158/1535-7163.MCT-06-0318. [DOI] [PubMed] [Google Scholar]
- (60).Li Y, Yin S, Nie D, Xie S, Ma L, Wang X, Wu Y, Xiao J. MK886 inhibits the proliferation of HL-60 leukemia cells by suppressing the expression of mPGES-1 and reducing prostaglandin E2 synthesis. Int. J. Hematol. 2011;94:472–478. doi: 10.1007/s12185-011-0954-0. [DOI] [PubMed] [Google Scholar]
- (61).Magro AM, Magro AD, Cunningham C, Miller MR. Down-regulation of vinculin upon MK886-induced apoptosis in LN18 glioblastoma cells. Neoplasma. 2007;54:517–526. [PMC free article] [PubMed] [Google Scholar]
- (62).Mancini JA, Abramovitz M, Cox ME, Wong E, Charleson S, Perrier H, Wang Z, Prasit P, Vickers PJ. 5-lipoxygenase-activating protein is an arachidonate binding protein. FEBS Lett. 1993;318:277–281. doi: 10.1016/0014-5793(93)80528-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.