Abstract
Retroviruses and retrotransposons package genomic RNA into virus-like particles (VLPs) in a poorly understood process. Expression of the budding yeast retrotransposon Ty3 results in the formation of cytoplasmic Ty3 VLP assembly foci comprised of Ty3 RNA and proteins, and cellular factors associated with RNA processing body (PB) components, which modulate translation and effect nonsense-mediated decay (NMD). A series of Ty3 RNA variants were tested to understand the effects of read-through translation via programmed frameshifting on RNA localization and packaging into VLPs, and to identify the roles of coding and non-coding sequences in those processes. These experiments showed that a low level of read-through translation of the downstream open reading frame (as opposed to no translation or translation without frameshifting) is important for localization of full-length Ty3 RNA to foci. Ty3 RNA variants associated with PB components via independent determinants in the native Ty3 untranslated regions (UTRs) and in GAG3-POL3 sequences flanked by UTRs adapted from non-Ty3 transcripts. However, despite localization, RNAs containing GAG3-POL3 but lacking Ty3 UTRs were not packaged efficiently. Surprisingly, sequences within Ty3 UTRs, which bind the initiator tRNAMet proposed to provide the dimerization interface, were not required for packaging of full-length Ty3 RNA into VLPs. In summary, our results demonstrate that Gag3 is sufficient and required for localization and packaging of RNAs containing Ty3 UTRs and support a role for POL3 sequences, translation of which is attenuated by programmed frameshifting, in both localization and packaging of the Ty3 full-length gRNA.
Keywords: yeast, retrotransposon, retrovirus, assembly, virus-like particles, packaging, RNA processing bodies, retrosome
1. Introduction
Trafficking and packaging of gRNA into VLPs is a central process in the assembly of retroviruses and LTR retrotransposons (Beckham & Parker, 2008; Cochrane et al., 2006; Johnson & Telesnitsky, 2010; Klein et al., 2007; Sandmeyer & Clemens, 2010; White & Lloyd, 2012). Among retroviruses, significant differences exist with respect to where VLPs form. Assembly is cytoplasmic for beta-retroviruses and primate foamy virus, and contrastingly occurs at the plasma membrane for alpha- and gamma-retroviruses and lentiviruses. Even among retroviruses that undergo final assembly at the membrane, identification of gRNA is not constrained to a common compartment. Saccharomyces cerevisiae copia-like Ty1 and gypsy-like Ty3 retroelements are among the most extensively studied for LTR retrotransposon VLP assembly (Sandmeyer, 2002; Voytas & Boeke, 2002). These Ty elements assemble intracellularly in concentrations of retrotransposon proteins, gRNA and host RNA binding proteins, which we refer to as retrosomes (Beliakova-Bethell et al., 2006; Checkley et al., 2010; Dutko et al., 2010; Malagon & Jensen, 2008; Sandmeyer & Clemens, 2010). The current study was undertaken in order to identify the cis-acting sequences in the Ty3 LTR retrotransposon gRNA that stipulate delivery to the retrosome assembly site and packaging into VLPs.
1.1 Yeast Ty gRNAs
S. cerevisiae LTR retrotransposons Ty1-5 are ancient retrovirus-like elements which have proliferated throughout metazoans (Sandmeyer, 2002; Voytas & Boeke, 2002). Ty elements share their overall genome and gRNA structure with retroviruses in that the U3RU5 segments composing each LTR and intervening sequences are transcribed into a gRNA with RU5-GAG-POL-U3R structure. Copia-like Ty1, 2, 4, and 5 and gypsy-like Ty3 are only distantly related to one another. Both retrotransposons, similar to retroviruses, utilize translational frameshifting to produce a Gag-Pol polyprotein precursor at a level less than one-twentieth that of free Gag (Farabaugh, 1995; Farabaugh et al., 1993; Kirchner et al., 1992). Ty1 Gag-p49 is processed into CA-p45 and Gag-p4, and Gag-Pol is processed into CA-p45, PR, IN and RT domains (Voytas & Boeke, 2002). The Gag-p4 sequence also serves as the N-terminal portion of PR. While Ty1 does not have a canonical, retroviral-like NC domain containing a zinc finger motif, other domains of Gag and Gag-Pol contribute NC-like functionalities: a basic region of CA-p45 has RNA binding capability (Cristofari et al., 2000), and the Gag-p4 portion of PR is necessary for reverse transcription (Lawler et al., 2002). In contrast, the genome organization of Ty3 is more closely related to animal retroviruses. Ty3 Gag3-p34 is processed into CA-p24, spacer-p3 and NC-p7. CA contains a MHR motif and NC contains one zinc-binding motif, CCHC, Gag3-Pol3 is processed into CA-p24, spacer-p3, NC-p9, PR, RT and IN domains (Hansen et al., 1992; Kirchner & Sandmeyer, 1993; Kuznetsov et al., 2005). Both Ty1 and Ty3 use initiator tRNAMet as the reverse transcription primer, and mutations introduced to the TΨC arm and acceptor stem of the primer tRNA disrupt transposition (Keeney et al., 1995). Ty1 has a bipartite PBS in the 5’ end of GAG (Friant et al., 1998); in contrast, Ty3 has a bipartite PBS split between the 5’ and 3’ UTRs (Gabus et al., 1998). Both Ty1 (Feng et al., 2000) and Ty3 (Nymark-McMahon et al., 2002) VLP gRNAs are dimeric, and Ty1 protease activity contributes to the stability of dimeric gRNA (Feng et al., 2000). Association of the initiator tRNAMet with gRNA is proposed, based on in vitro experiments, to provide the dimeric gRNA interface (Cristofari et al., 1999; Gabus et al., 1998).
Ty1 and Ty3 expression can be artificially regulated under the GAL1-10 UAS to induce high levels of retrotransposition (Boeke et al., 1986; Hansen et al., 1988). Cis-acting sequences required for the intracellular packaging of retrotransposon gRNA into VLPs are challenging to define since VLPs can only be isolated from cell lysates and are heavily contaminated with cytoplasmic RNA. However, transposition assays using donor Ty elements in which internal domains are substituted with genetic markers have delimited sequences sufficient in cis for retrotransposition within the terminal 200 nts of the gRNAs of both types of Ty elements (Kirchner et al., 1992; Xu & Boeke, 1990).
1.2 Ty assembly foci
Cells induced to express Ty3 protein visualized with anti-CA or GFP fused to Gag3-Pol3 and RNA tagged with a MS2-binding site and expressed in the presence of MS2-RFP show an early punctate staining pattern and within 1–2 h nuclear pore-associated foci containing Ty3 protein and RNA (Beliakova-Bethell et al., 2006; Hansen et al., 1992). A combination of observations indicates that these foci are centers of Ty3 assembly. First, electron microscopy shows that cells expressing Ty3 accumulate clusters of VLPs rather than isolated VLPs (Hansen et al., 1992); second, RNA and protein co-localize to foci shown by immuno-electron microscopy to correspond to VLP foci (Beliakova-Bethell et al., 2006); third, mutations in the CA or NC domain of GAG3 that are predicted to affect multimerization or RNA binding disrupt formation of foci (Larsen et al., 2008; Larsen et al., 2007); and fourth, deletions of some host genes required for transposition disrupt foci formation at a point after protein synthesis (Beliakova-Bethell et al., 2006; Irwin et al., 2005) and (manuscript in preparation).
Genome-wide screens for factors which support retrotransposition of Ty1 (Griffith et al., 2003; Nyswaner et al., 2008; Scholes et al., 2001) and Ty3 (Irwin et al., 2005) implicated PB components at steps downstream of transcription. Similar to animal cells, budding yeast PB foci contain components of the deadenylation-dependent 5’ to 3’ degradation apparatus, including decapping enhancers Edc3, Pat1, and DEAD-box helicase Dhh1 (hDDX6/p54); decapping factors Dcp1,2; and the 5’ to 3’ exonuclease Xrn1, among other proteins (Aizer et al., 2008; Anderson & Kedersha, 2009; Buchan et al., 2010; Kedersha & Anderson, 2007). PB composition overlaps that of SG. For example, Xrn1 and Dhh1 are found in both PB and SG foci (Swisher & Parker, 2010) and PB formation precedes and is physically associated with SG formation in yeast cells undergoing nutritional deprivation (Buchan et al., 2008; Hoyle et al., 2007).
Despite association of Ty proteins and RNAs with PB proteins, the relationship of Ty assembly foci to PBs is complex. Because Ty RNA and foci contain a combination of RNA granule components, we have adopted the term “retrosome” to refer to Ty assembly foci (Sandmeyer & Clemens, 2010). In the case of Ty1, PB proteins not only facilitate retrotransposition, and are required for cluster formation (Checkley et al., 2010; Malagon & Jensen, 2008). However, functionally Ty1 clusters appear to differ from PBs; for example, they do not enlarge upon glucose deprivation (Checkley et al., 2010). However, they can be adjacent to, or temporally overlapping with PB reporters Dhh1- and Dcp2-GFP (Dutko et al., 2010). Although Ty1 retrosomes are not congruent with PBs, deletion of Xrn1 slows turnover of a repressive Ty1 antisense RNA (Berretta et al., 2008; Matsuda & Garfinkel, 2009; Dijk et al., 2011) and decreases packaging of Ty1 RNA into VLPs (Dutko et al., 2010).
Examination of cells expressing Ty3 fused at the downstream end of POL3 to RFP-coding sequence together with a PB marker (Lsm1-, Dcp2-, Xrn1-, Dhh1-, or Edc3-GFP) showed that, in contrast to Ty1 retrosomes, Ty3 retrosomes contain PB components (Beliakova-Bethell et al., 2006) and at least one SG component (manuscript in preparation). Similar to Ty1, Ty3 requires PB proteins Dhh1 and Xrn1 for retrotransposition and deficiency in either factor affects Ty3 focus formation (Beliakova-Bethell et al., 2006; Irwin et al., 2005). We speculate that retrosomes promote Ty3 VLP assembly by sequestering gRNA from the translation apparatus, concentrating Ty3 proteins and host assembly factors, and mediating localization of large numbers of VLPs in the vicinity of the nuclear pore complex.
Intracellular assembly of LTR retrotransposon VLPs presents problems analogous to those encountered in retrovirus translation, packaging and Gag multimerization. However, Ty3 VLPs complete assembly and presumably some extent of reverse transcription, and become remodeled to enable nuclear entry within a single host cell. Results from previous studies have highlighted the role of Gag3 protein domains in packaging and assembly of Ty3 VLPs. Mutagenesis of Gag3 showed that the CA NTD is critical for assembly (Larsen et al., 2007). Gag3 with mutations in the MHR forms diffuse foci, and fails to assemble or package RNA efficiently (Larsen et al., 2007; Orlinsky et al., 1996). Neutralization of the acidic charge of the Gag3 spacer arrests assembly, suggesting that interactions between spacer and the basic NTD of NC help to drive condensation of the VLP (Clemens et al., 2011). Disruption of the NC zinc-binding motif results in loss of Ty3 RNA foci but does not completely abrogate Gag3-Pol3 foci and Gag3 aggregates form in the nucleus (Larsen et al., 2008). These observations suggest that cytoplasmic localization of Gag3 depends upon gRNA interaction. This raises the possibility that during the course of wt Ty3 induction, free Gag3 could exceed the available supply of gRNA, and redistribute to the nucleus.
Retrovirus gRNA
Experiments examining the effects of transcription or translation inhibition on retrovirus gRNA packaging versus virion production have demonstrated that retrovirus RNAs can be committed to packaging in the nucleus or in the cytoplasm, without or after translation. MoMLV gRNA forms a separate pool from translated RNA soon after synthesis (Levin et al., 1974; Levin & Rosenak, 1976) and RNAs which are co-transcribed are preferentially co-packaged (Flynn et al., 2004). RSV Gag shuttles to the nucleus and promotes gRNA export, consistent with gRNA designation in the nucleus (Parent, 2011). In contrast, for HIV-1 and HIV-2, inhibition of transcription or nuclear export decreases cytoplasmic RNA, which affects translation products and virion gRNA similarly, indicating that these gRNAs are drawn from a common pool (Dorman & Lever, 2000). Within the cytoplasmic pool, viruses may differ with respect to mode of genomic capture. HIV-2 encoding truncated Gag is not packaged efficiently in trans, indicating that assembly is potentially initiated co-translationally with a resulting cis bias (Griffin et al., 2001; Kaye & Lever, 1999). On the other hand, in the case of HIV-1, inhibition of translation resulted in rerouting of RNA to packaging, indicating that translation is not a pre-requisite (Butsch & Boris-Lawrie, 2000; Dorman & Lever, 2000) and reviewed in (Butsch & Boris-Lawrie, 2002).
Retrovirus gRNA packaging requires specific signals in the RNA, typically a cluster of stem-loops overlapping the 5’ UTR which are recognized by the NC domain of Gag [reviewed in (Cimarelli & Darlix, 2002; D'Souza & Summers, 2005; Darlix et al., 1995; Johnson & Telesnitsky, 2010)]. Retroviral genomes are duplex, and dimer formation prior to packaging could facilitate proper copy number by contributing to recognition of gRNA by Gag. For example, dimerization of MLV gRNA exposes a high-affinity NC binding site within psi (D'Souza & Summers, 2004). Mutations in this sequence disrupt packaging of gRNA in vivo, supporting a model in which Gag binds to two high affinity sites in psi that are exposed upon dimerization (Gherghe et al., 2010). Disruption of specific basic residues contained in MLV NC reduces the level of packaged dimers without increasing the level of packaged monomers, consistent with packaging of a dimeric species (Housset et al., 1993). A genetic analysis of HIV-1 gRNA encapsidation also supports a role for dimeric species in packaging. In vivo visualization of gRNA using fluorescently-tagged RNA-binding proteins indicates that the vast majority of VLPs contain RNA dimers (Chen et al., 2009; Chin et al., 2007). Strikingly, RNAs with complementary DIS are preferentially co-packaged (Moore et al., 2007; Ni et al., 2011). More recent NMR studies of HIV-1 RNA-Gag interactions suggest conformation-driven packaging could be similar to that proposed for MLV. The HIV-1 5’ leader sequence can exist in two conformations, one of which promotes dimerization and exposes high-affinity NC binding sites proposed to promote packaging (Heng et al., 2012; Lu et al., 2011). Thus, dimerization emerges a s a potentially general mechanism for selection of gRNAs for packaging.
1.2 Translation and gRNA capture
Selection of retroelement gRNAs from cytoplasmic pools implies coordination of translation template and genome functions. Evidence suggests that translation factors can associate with RNAs during nuclear export and affect downstream packaging fates. For example, in mouse cells, HIV-1 packaging is inefficient, but can be rescued by expression of human CBC, which binds RNA during export (Cochrane et al., 2006). Although HIV-1 gRNAs can be translated, translation is not necessary for packaging into virions (Butsch & Boris-Lawrie, 2000; Nikolaitchik et al., 2006). The proximity of psi to the site of Gag translation initiation suggests that local Gag accumulation could trigger assembly. This is supported by the observation that newly-translated HIV-1 Gag can feedback and eventually inhibit ongoing translation. This sequence of events would be consistent with the proposed model of co-translational assembly (Anderson & Lever, 2006).
In vivo retrovirus Gag association with gRNA has been monitored in several kinds of experiments, but with some consistent themes that relate to observations in retrotransposon systems. Pulse-chase analysis of HIV-1 Gag utilizing a cycloheximide block coupled with biarsenical tetracysteine staining showed an early diffuse pattern followed by collection in the perinuclear region and endosomal trafficking to the cell surface (Perlman & Resh, 2006). FRET analysis used to detect direct interactions between Gag and gRNA, as well as immunofluorescence (Poole et al., 2005) showed an early punctate cytoplasmic pattern, followed by concentration of Gag and gRNA in a pericentriolar region, and subsequently cell surface localization. Expression of psi-minus gRNA resulted in a diffuse Gag pattern, indicating that Gag-RNA interaction is required for normal trafficking and localization. Consistent with this, mutations in the HIV-1 Gag zinc-binding motif that disrupt high-affinity binding to psi caused Gag to be diffuse and cytoplasmic rather than punctate and plasma membrane-associated (Grigorov et al., 2007). Similarly, expression of psi-minus MLV RNA resulted in diffuse patterns of Gag (Basyuk et al., 2005; Basyuk et al., 2003), indicating that functional association of Gag with gRNA in the cytoplasm enables correct localization to the assembly site.
Thus, Gag is translated and in association with gRNA collects at the microtubule organizing complex, likely with newly-exported RNA for delivery via endosomal trafficking to the plasma membrane (Cochrane et al., 2006; Klein et al., 2007). Data are consistent with a model in which gRNAs undergo early cytoplasmic translation and potentially initiate assembly co-translationally (Anderson & Lever, 2006; Balvay et al., 2007; Griffin et al., 2001). For lenti-, gamma and some alpha retroviruses, assembly is completed at the plasma membrane, where a myristate moiety conjugated to the Gag amino-terminus and the basic region of Gag combine to stabilize Gag in the membrane, thereby allowing completion of multimerization and assembly (Cimarelli & Darlix, 2002).
1.3 Formation of HIV-1 gRNA ribonucleoprotein complexes
As discussed above, data from multiple systems supports co-trafficking of Gag and gRNA to the plasma membrane. Although Gag multimerizes upon RNA binding, there is less agreement regarding the level of this multimerization and nature of host factors within these RNPs. Some reports suggest that only low-level Gag multimers (dimers and trimers) are associated with gRNA prior to membrane delivery (Kutluay & Bieniasz, 2010). High-throughput siRNA screens (Brass et al., 2008; Bushman et al., 2009), mass spectrometric protein analysis (Mouland et al., 2001; Ott, 2008), biochemical assays (Bolinger et al., 2007) and fluorescence visualization of tagged candidate proteins have implicated host proteins in HIV-1 gRNA trafficking. In addition to CRM1 and CBC, HIV-1 RNA export requires DDX3 (Yedavalli et al., 2004) and hnRNP A2 (Levesque et al., 2006; Mouland et al., 2001). In vitro studies led to identification of the ATPase ABCE1 as a host co-factor required for HIV-1 preassembly through a series of 80S, 150S, 500S complexes (Dooher et al., 2007). The double-stranded RNA-binding protein Staufen (Abrahamyan et al., 2010) has also been identified as a component of the HIV-1 gRNA-protein granule. During HIV-1 infection, Staufen gRNA-protein granule formation coincides with a decrease in the abundance of stress granule and PB-related protein complexes (Abrahamyan et al., 2010).
Components of RNA PBs, in addition to being implicated in Ty VLP assembly, have recently been identified as both co-factors and restriction factors for HIV-1 assembly. Animal cell PBs, similar to S. cerevisiae, contain deadenylation, decapping, 5’ exonuclease and NMD- related proteins, but in contrast to S. cerevisiae, also contain eukaryotic RISC components (Anderson & Kedersha, 2009; Buchan et al., 2010; Moser & Fritzler, 2010; Thomas et al., 2011). Two recent investigations showed localization of HIV-1 RNA with PB components and identified miRNAs that antagonized HIV-1 replication. Consistent with host restriction activity, there were positive effects on HIV-1 replication of knockdown of RISC and PB components DDX6 (yeast Dhh1), Dcp2 and Ago3 (Chable-Bessia et al., 2009; Nathans et al., 2009). However, the role of PB-associated proteins is likely complex. The PB DEAD box helicase Mov10 associates with the RISC complex and is reported to modulate the activity of the subunit Ago2 in human cells (Liu et al., 2012). Overexpression decreased production of infectious exogenous retroviruses including HIV-1 (Burdick et al., 2010; Furtak et al., 2010; Wang et al., 2010) and LINE and Alu retrotransposons (Arjan-Odedra et al., 2012). Knockdown of endogenous levels of Mov10 has modest effects on exogenous retroviruses and reports are mixed as to the nature of those effects. However, consistent with the overexpression studies, knockdown of Mov10 increased retrotransposition of endogenous elements (Arjan-Odedra et al., 2012). Two other PB proteins, DEAD box helicase DDX6 and Dcp2, are associated with HIV-1 ABCE1 assembly complexes. DDX6 depletion reduces HIV-1 particle production, but not gRNA incorporation into assembled particles (Reed et al., 2012). DDX6 is also associated with primate foamy virus (PFV) Gag viral RNA and localizes with them at the peri-microtubule organizing complex assembly site (Yu et al., 2011). However, this case appears different from HIV-1 since PFV is not associated with the canonical PB marker Dcp1, and depletion of DDX6 reduced PFV gRNA packaging rather than particle production.
In the current study we set out to understand the roles of cis-acting, untranslated (UTRs) and translated (GAG3-POL3) regions in Ty3 RNA localization with PB reporters. Because PB proteins implicated in Ty3 retrotransposition act as modulators of translation and RNA stability, we also explored the relationship between translation and assembly foci. We found that Ty3 gRNA localization is sensitive to the state of gRNA translation and that RNAs with Ty3 UTRs and GAG3-POL3 flanked by heterologous UTRs (hUTRs) localize independently to foci with PB proteins but only the former is packaged into VLPs. Surprisingly, packaging was not sensitive to mutations deleting the region proposed to mediate dimerization.
2. Methods
2.1 Yeast and bacterial culture conditions
Yeast and bacterial culture methods were as previously described (Amberg, 2005; Ausubel, 2002) except where noted. Bacterial strains HB101 [F− hsd-20 (rB−mB−) recA13 leuB6 ara14 proA2 lacY1 galK2 rpsL20 (Smr) xyl-5 myl-1 supE44λ] or DH5α [F− Φ80lacZΔ M15Δ (lacZYA-argF) U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 thi-1 gyrA96 relA1 λ−] were used for plasmid preparations. Ty3 or lacZ RNAs tagged with the MS2 binding site and MS2-RFP (Beliakova-Bethell et al., 2006) were expressed in strains derived from BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), by fusion of the genomic copy of DHH1 or DCP2 in-frame to the sequence encoding GFP(S65T) (Huh et al., 2003). Strain BY4741 trp1Δ was used for experiments with donor and helper plasmids. For 6 h packaging assays, a Ty3 null strain (ygr109wΔ::loxP yil080wΔ::loxP) was used. This strain was derived from BY4741 and constructed using loxP knock-out cassette amplification (Fang et al., 2011) with primers F5’-TCTGCTGCGCTACCACTGCGCCATACGAGCTTGATTTTCTGAAAGCGACTCTAGAG GATC-3’ and R5’-TATAGTCATATTCCCATAAATAGAGCATCCAAAATGGAA-3’ for YGR109W and F5’-ACAGGGTTTGTAGCACTGGTATACGTTTTTCCATATCATGGAACACGACTCTAGAGGATC-3’ and R5’-GCCTTAACCAACTGGGCCAAGAGACCAGATATTTCATATGTTCTGATTCGAGCTCGGTAC-3’ for YIL080W. To select for cells with particular prototrophic markers, synthetic dextrose medium [0.67% yeast nitrogen base-2% dextrose], synthetic galactose medium [0.67% yeast nitrogen base-2% galactose], or synthetic raffinose medium [0.67% yeast nitrogen base-1% raffinose-2% (vol/vol) glycerol-2% (vol/vol) lactic acid] containing complete amino acids, inositol and adenine sulfate lacking selection nutrients was used (Amberg, 2005).
2.2 Recombinant DNA constructions
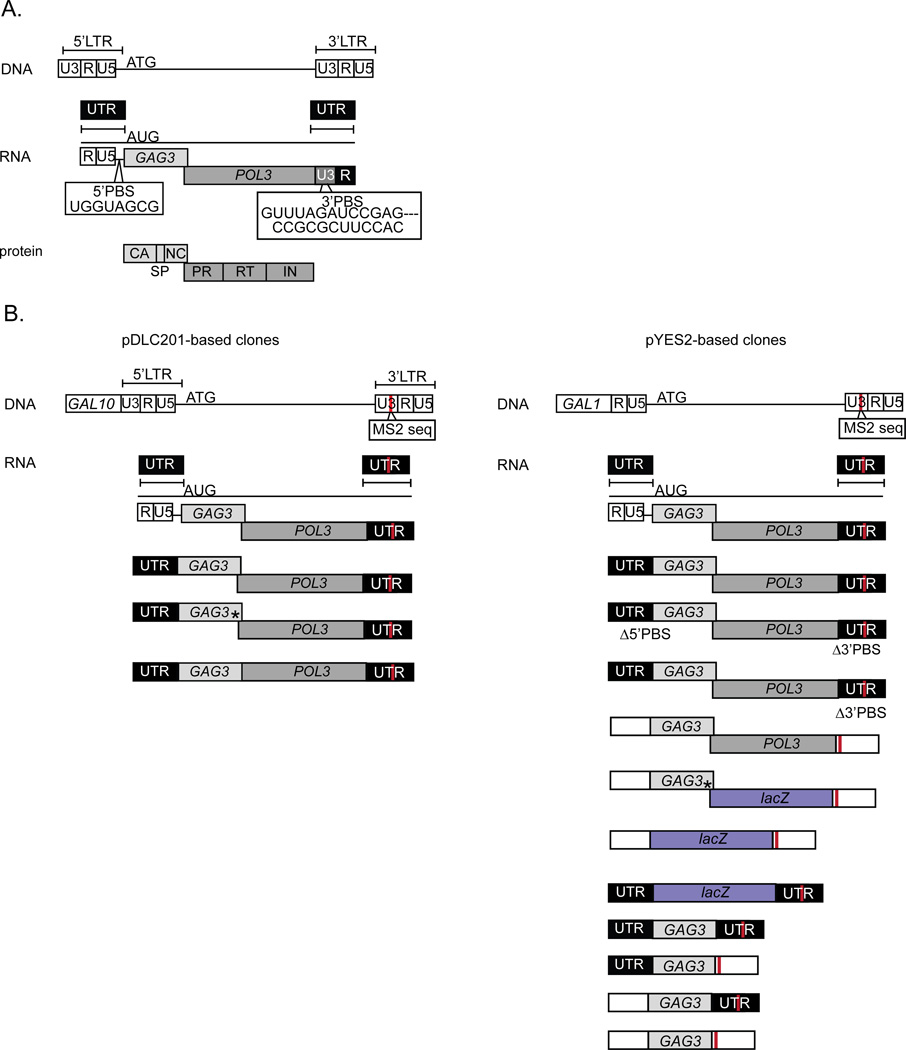
Plasmids used in this study are summarized in Table 1 and Ty3 variants are illustrated in Fig. 1. Ty3 expression vectors were based on either pDLC201 (Hansen et al., 1988) or pYES2.0 (Invitrogen Corp., Carlsbad, CA). The pDLC201 plasmid is a high-copy, URA3-marked plasmid in which the GAL1-10 UAS, oriented as it is in the GAL10 promoter, is fused to Ty3 at nt 123 of the LTR (i.e. between U3 and R) conferring galactose regulation of the native Ty3 transcript. Although micro-heterogeneity is observed for the native transcript, the majority of transcripts have 5’ and 3’ UTRs 193 and 227 nt in length, respectively (Hansen et al., 1988). The pSBS2183 plasmid (previously pTy3-MS2) is pDLC201 modified by insertion of two copies of the MS2-binding site downstream of POL3 to allow visualization of the Ty3 RNA (Beliakova-Bethell et al., 2006). Variants based on pSBS2183 express transcripts with the native Ty3 UTRs except for the presence of the MS2 sequence. The pYES2.0-based Ty3 expression plasmids are also high-copy and URA3-marked. Galactose-inducible Ty3 transcription initiates under regulation of the GAL1 promoter and UAS and terminates under control of the CYC1 terminator. In the pYES2.0 context, Ty3 transcripts have 5’ GAL1 (123–129 nts in length) and 3’ CYC1 (323–338 nts in length) UTRs. The expression plasmids and sequences of promoter, coding and terminator regions for each variant are provided in Supplemental Materials.
Table 1.
Plasmids
| Plasmid | Plasmid Contents | Genetic Markers (Base Plasmid) |
Ty3 Sequences (in Ty3 DNA, nt#) |
Reference |
|---|---|---|---|---|
| pYES2.0 | empty vector | URA3, 2μ | None | Invitrogen |
| pRS315 | empty vector | LEU2, ARS4/CEN6 | “ | (Sikorski & Hieter, 1989) |
| pRS314 | empty vector | TRP1, ARS4/CEN6 | ‘’ | (Sikorski & Hieter, 1989) |
| pDLC201* | Full-length, gal-inducible Ty3 element; no MS2 binding sequence | URA3, 2μ | 123–5351 | (Hansen et al., 1988) |
| pSBS2183 | Ty3-MS2 (UTR-GAG3-POL3-UTR-MS2) | URA3, 2μ (pDLC201) | “ | (Beliakova-Bethell et al., 2006) |
| pNB2241 | GAG3-POL3-MS2 | URA3, 2μ (pYES2) | 406–5057 | This work |
| pNB2355 | UTR-GAG3-STOP-POL3-UTR-MS2 | URA3, 2μ (pDLC201) | 123–5351 | “ |
| pNB2996 | GAG3-STOP-lacZ-MS2 | URA3, 2μ (pYES2) | 406–1288 | “ |
| pNB2995 | GAG3-FUS-POL3-MS2 | “ | 406–5057 | “ |
| pNB2354 | UTR-GAG3-FUS-POL3-UTR-MS2 | URA3, 2μ (pDLC201) | 123–5351 | “ |
| pNB3027 | UTR-GAG3-UTR-MS2 | URA3, 2μ (pYES2) | 223–1288, 5012–5351 | “ |
| pNB2242 | GAG3-MS2 | “ | 406-1288 | “ |
| pNB3026 | UTR-GAG3-MS2 | “ | 223–1288 | “ |
| pNB3007 | GAG3-UTR-MS2 | “ | 406–1288, 5012–5351 | “ |
| pNB2629 | MET25P -MS2-RFP-CYC1 TT | LEU2, ARS4/CEN6 (pRS315) | None | “ |
| pNB2259 | LacZ MS2 | URA3 2μ, (pYES2) | None | ‘’ |
| pNB2247 | 5’UTR-LacZ-UTR | URA3, 2μ (pDLC201) | 123–415, 5012–5351 | |
| pNB2788 | GAL-GAG3-CYC TT | TRP1, ARS4/CEN6 (pRS314) | 406–1288 | ‘’ |
| pKC3700 | Ty3-MS2 (UTR-GAG3-POL3-UTR-MS2) | URA3, 2μ (pYES2) | 223–5351 | “ |
| pKC3702 | UTRΔPBS-GAG3-POL3-UTRΔPBS-MS2) | URA3, 2μ (pYES2) | 223–5351, Δ343–350 Δ5169–5196 | “ |
| pKC3703 | UTR-GAG3-POL3-ΔPBSUTR-MS2) | URA3, 2μ (pYES2) | 223–5351, Δ5169–5196 | “ |
Sequences of plasmids pDLC201 and below are available in Supplemental Materials
Figure 1. Ty3 genome and expression variants.
A. Full-length Ty3 DNA (U3RU5-coding region-U3RU5) and RNA (RU5-coding region-U3R) sequences. First and second ORFs GAG3 and POL3 are translated into Gag3 and Gag3-Pol3 polyproteins and processed into the mature species indicated. The Ty3 gRNA 5’UTR is comprised of RU5 and 75 nt of the internal domain upstream of the initiator AUG. POL3 extends into the downstream LTR by 46 nts. For simplicity in the text, Ty3 “UTRs” refers to the 5’ UTR and the complete 3’ U3R (of which 46 nts is translated). B. Ty3-related expression constructs. The GAL1-10 UAS (GAL10 orientation) partially replaced U3 in the upstream LTR to allow galactose-regulated expression of Ty3-related transcripts (left-hand column). *Indicates stop codon. Right-hand column displays pYES2.0-based full length Ty3, GAG3 and LacZ reporter expression constructs. pYES2.0-based constructs containing a 5’ UTR derived from Ty3 have the RU5 sequence placed downstream of the GAL1 promoter. hUTRs were derived by transcription from pYES2.0 under control of the GAL1-10 UAS, the GAL1 promoter and 5’ UTR, and the CYC1 terminator. Empty white boxes denote hUTRs. Complete annotated plasmid sequences are presented in Supplemental Materials.
2.3 RNA and Dhh1 and Dcp2 reporter localization
MS2-tagged Ty3 RNAs (Ty3-MS2) were expressed in the presence of MS2 capsid protein fused to RFP (MS2-RFP) as described previously (Beliakova-Bethell et al., 2006). Cells were initially visualized using a Zeiss Axioplan 2 fluorescence microscope (Carl Zeiss Inc.). A subset of these cells was then quantitatively analyzed using confocal fluorescence microscopy. BY4741 cells transformed with Ty3-MS2 and MS2-RFP expression plasmids were grown at 24°C in synthetic raffinose lacking Leu and Ura to late logarithmic phase, diluted in the same medium and grown overnight to OD600= 0.2–0.3. Galactose was added to a final concentration of 2% in order to induce expression of Ty3 and cells were grown for 6 h at 24° C. At 30 min prior to live-cell imaging, cells were transferred to fresh medium of the same type, but lacking Met to induce MS2-RFP expression. Confocal laser scanning microscopy was performed on an inverted LSM510 laser scanning microscope (Carl Zeiss, Göttingen, Germany) using an oil immersion Plan-Apochromat 100×/1.4 NA objective. For the localization of RFP (Ty3 mRNA) light from a helium-neon laser at 543 nm was directed over a HFT KP 700/543 beam splitter and fluorescence was detected using an NFT 545 beam splitter in combination with a BP 565–615 nm band pass filter; for localization of GFP (PB proteins), light from a 488 nm argon laser was directed over a HFT488 beam splitter in combination with a BP 500–550 nm band pass filter. The pinhole was set to 1 Airy unit. Laser power and detector sensitivity were optimized to avoid pixel saturation. Images were collected with a frame size of 1024 × 1024 pixels, and processed for publication with Adobe Photoshop CS3 (Adobe Systems Incorporated, San Jose, CA). The experiments were repeated a minimum of two times with several independent transformants. The results were all similar. For quantitation and colocalization studies, single-plane confocal images from samples consisting of at least 150 cells from a single representative experiment were analyzed with Volocity Software 2.1 (PerkinElmer, Waltham, MA).
2.4 Ty3 RNA expression and nuclease protection assays
Ty3 RNA expression was quantified using a RT-qPCR assay. Expression of Ty3 RNA variants was induced as for microscopy, and 10 OD600 of cells was harvested. A Qiagen RNeasy kit was used to isolate total RNA according to the manufacturer’s protocol. RNA was quantified using a Nano-drop spectrophotometer (Nanodrop Technologies, Inc. Wilmington, DE), and RNA was used to generate cDNAs for RT-qPCR analysis. Briefly, 1.5 µg total RNA was used as a template for cDNA synthesis using an oligo(dT) primer (final concentration of 0.025 µg/µL) in a Superscript II Reverse Transcriptase reaction according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA). The cDNAs were diluted 1:10 and used as template for qRT-PCR amplification. Two individual transformants were analyzed, and each cDNA sample was tested in triplicate. The following primers were used to amplify the gene targets of interest: Ty3 GAG3, F5’-CGAACTTGATGCTGATGGAGAC-3’, R5’- GATCTTCTTGTCCTTACGGTATGG-3’; ACT1, F5’- ATTCTGAGGTTGCTGCTTTGG-3’, R5’- TGTCTTGGTCTACCGACGATAG-3’. The amplification of each cDNA sample was monitored using iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s protocol. The RT-qPCR cycles were performed using a RT-qPCR Cycler (Bio-Rad Laboratories, Hercules, CA) and the ratio of Ty3 GAG3 to ACT1 signal was calculated for each construct. GAG3 and ACT1 signals were compared to a standard curve to ensure that detection was within the linear range of the machine. Nuclease protection assays were performed with Benzonase (Sigma-Aldrich Corp., St. Louis, MO) or TurboNuclease (Accelagen, San Diego, CA) essentially as described (Clemens et al., 2011; Larsen et al., 2007).
2.5 Protein analysis
Yeast cells expressing wt or mutant derivative constructs were induced as for microscopy. Harvested cells were lysed under denaturing conditions to generate whole cell extracts as described previously (Clemens et al., 2011). Fifteen micrograms of total protein was subjected to SDS polyacrylamide gel electrophoresis and fractionated proteins were transferred to polyvinylidene fluoride membrane (Millipore Corp., Billerica, MA). Membranes were blocked with 2.5% non-fat milk in 1× PBST and incubated with primary rabbit polyclonal antibody against Ty3 CA (1:10,000), and primary mouse monoclonal antibody against Pgk1p (1:5,000) (Molecular Probes – Invitrogen, Carlsbad, CA). Protein bands were visualized with an ECL+ western blotting reagent kit (GE Healthcare, UK Ltd). A Fuji LAS-4000 imaging system and Multigauge V3.0 analysis software were used to quantify the chemiluminescent signal in each lane within the linear range of detection. Ty3 CA protein signal was normalized to the Pgk1p loading control signal for all samples, and mutant ratios were normalized to the wt Ty3 signal.
3. Results and Discussion
3.1 Ty3 UTRs and POL3 act independently to promote localization of cytoplasmic Ty3 RNA with PB protein foci
One of the most distinctive features of cells expressing Ty3 is the formation of one or two large foci containing Ty3 protein and RNA, and PB components (Beliakova-Bethell et al., 2006). The current study was undertaken to define the roles of Ty3 RNA non-coding and coding sequences in this process. To this end, a set of Ty3-derived RNAs (Fig. 1) tagged downstream of the Ty3 coding region with the bacteriophage MS2 capsid protein binding site were expressed under control of the GAL1-10 UAS in cells expressing MS2-RFP (Beliakova-Bethell et al., 2006). For simplicity, Ty3 RNA regions beginning at the 5’ end from RU5 through to the initiator ATG and the 3’ U3R region (containing a short portion of POL3) are referred to as UTRs. The contribution of Ty3 UTRs to gRNA function was assessed by substitution of Ty3 UTRs with hUTRs derived at the 5’ and 3’ ends from GAL1 and CYC1, respectively (Fig. 1). For the purpose of clarity, a construct name that does not contain a specific UTR designation represents a RNA variant that is flanked by hUTRs. The role of the POL3 coding sequence was tested by deleting POL3, substituting a heterologous ORF for POL3 and modulating the level of POL3 translation. Transformants were evaluated for Ty3 RNA and protein expression, and confocal microscopy was performed to determine RNA localization by observing the number of foci greater than 0.1 µm2 per MS2-RFP-positive cell and to determine overlap with Dhh1- and Dcp2-GFP reporter foci. Despite selection for Ty3 and MS2-RFP expression plasmids, only some cells are positive for both (Beliakova-Bethell et al., 2006) and (data not shown).
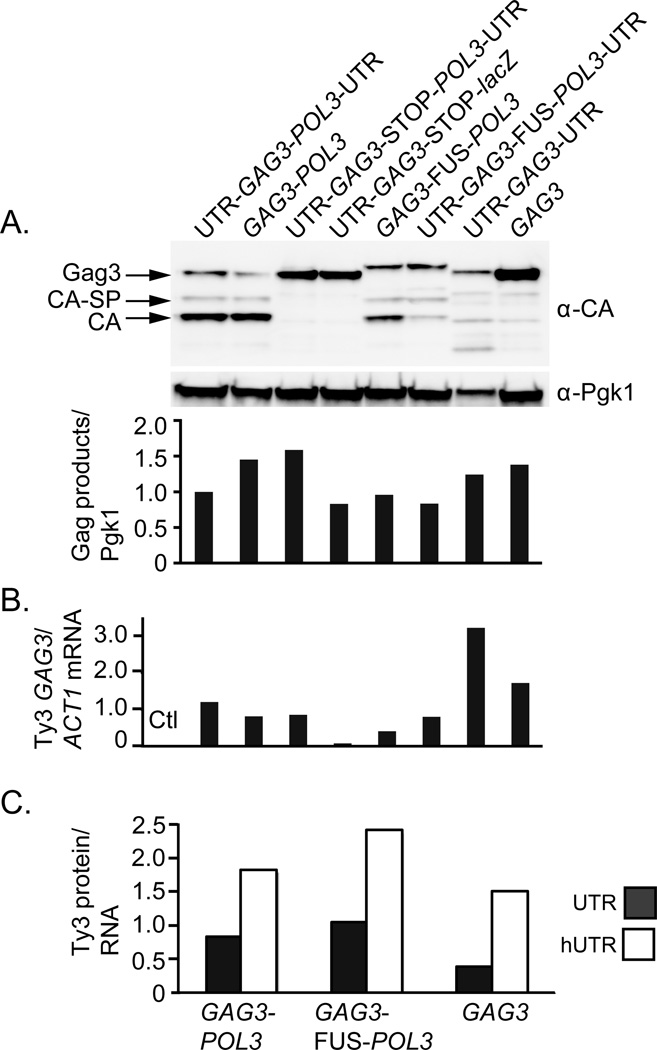
Since differing levels of expression might affect localization with PB factors and focus formation, Ty3 protein and RNA in cells expressing the Ty3 constructs were quantified. Immunoblot analysis showed that levels of Ty3 Gag3 and processing products did not differ among transformants by more than two-fold (Fig. 2A). In cells expressing wt Ty3, programmed frameshifting results in production of Gag3 and Gag3-Pol3 in a ratio of about 20:1 (Kirchner et al., 1992). Cells expressing a Ty3 in which GAG3 is fused to POL3 (GAG3-FUS-POL3) do not make Gag3 through run-off translation of GAG3. However, Gag3-Pol3 is processed into a slightly longer Gag3 product (Kirchner et al., 1992; Orlinsky & Sandmeyer, 1994), which was present in amounts comparable to levels of wt Gag3 (Fig. 2A). These results indicated that for the most part similar amounts of Gag3 protein were generated from the constructs tested for RNA localization. Ty3 wt and variant RNA levels were quantified by RT-qPCR, using primers that annealed within GAG3, which was present in all constructs. Ty3 RNA levels were normalized to the level of ACT1 RNA per cell (Fig. 2B). Most Ty3 RNA levels were within two-fold of the control. However, despite significant amounts of Gag3 produced, the normalized level of the RNA in which POL3 was substituted with lacZ (GAG3-STOP-lacZ) was quite low compared to the normalized level of control Ty3 RNA (0.08 versus 1.20). Although the stop codon at the end of the GAG3 ORF could have triggered NMD, a comparable decrease was not observed in constructs containing a stop codon downstream of the GAG3 programmed frameshift site (UTR-GAG3-POL3-UTR and GAG3-STOP-POL3). Interestingly, despite similar levels of protein, RNA levels of UTR-GAG3-UTR and GAG3 including and lacking Ty3 UTRs, were elevated by three- and two-fold, respectively, relative to control Ty3 RNA (Fig. 2A and B).
Figure 2. Expression of Ty3-related RNAs and proteins.
A. Ty3 Gag3 expression. Total Ty3 Gag3 and Gag3 processing products in WCEs were quantified using a LAS-4000 Fuji imaging system and Multigauge V3.0 analysis software. This level was normalized to the Pgk1 loading control, and these values were normalized to the wt Ty3 protein signal. Two transformants of each sample were quantified and averaged, and a representative blot is shown. α-CA, anti-capsid. α-Pgk1, anti-Pgk1. B. Ty3 RNA levels. Two independent transformants were analyzed by RT-qPCR using pairs of primers that annealed in GAG3 and in ACT1 RNA. RT-qPCR was performed in the linear concentration range for two independent transformants, each in triplicate. Triplicates were averaged and Ty3 GAG3 signals were normalized to those of ACT1. The average normalized GAG3 signal of the two transformants is presented. Ctl, uninduced control. C. RNAs containing Ty3 UTRs have lower Ty3 protein/RNA ratios. The ratio of Ty3 protein/RNA (determined as in A and B) was determined for cells expressing Ty3 RNA variants. UTRs, grey shaded boxes; hUTRs, white boxes.
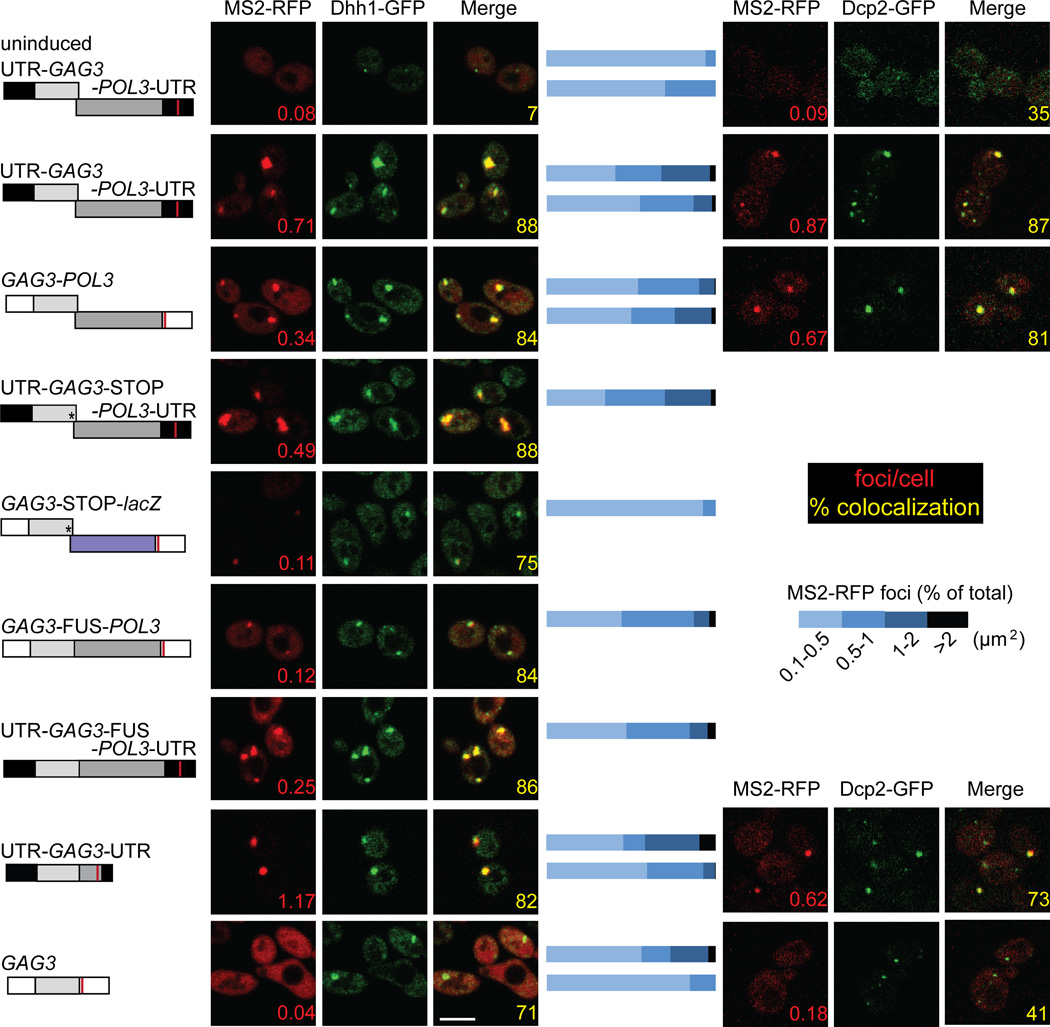
Cells expressing wt Ty3 RNA (UTR-GAG3-POL3-UTR) and Dhh1-GFP and Dcp2-GFP formed 0.71 and 0.87 Ty3 RNA foci per cell, respectively (Fig. 3). About half of the UTR-GAG3-POL3-UTR foci had an area of 0.5 µm2 or greater. Variations in the size of MS2-RFP foci are most likely influenced by many factors including Ty3 RNA and protein levels. In addition, strains expressing different PB reporters, Dhh1-GFP and Dcp2-GFP showed differences in size of foci and in foci per cell. These differences could reflect effects of the fluorescent reporter fusions on reporter function. Nevertheless, almost all MS2-RFP foci overlapped with Dhh1- and Dcp2-GFP reporter foci, indicating that the two PB reporters localized similarly (Fig. 3).
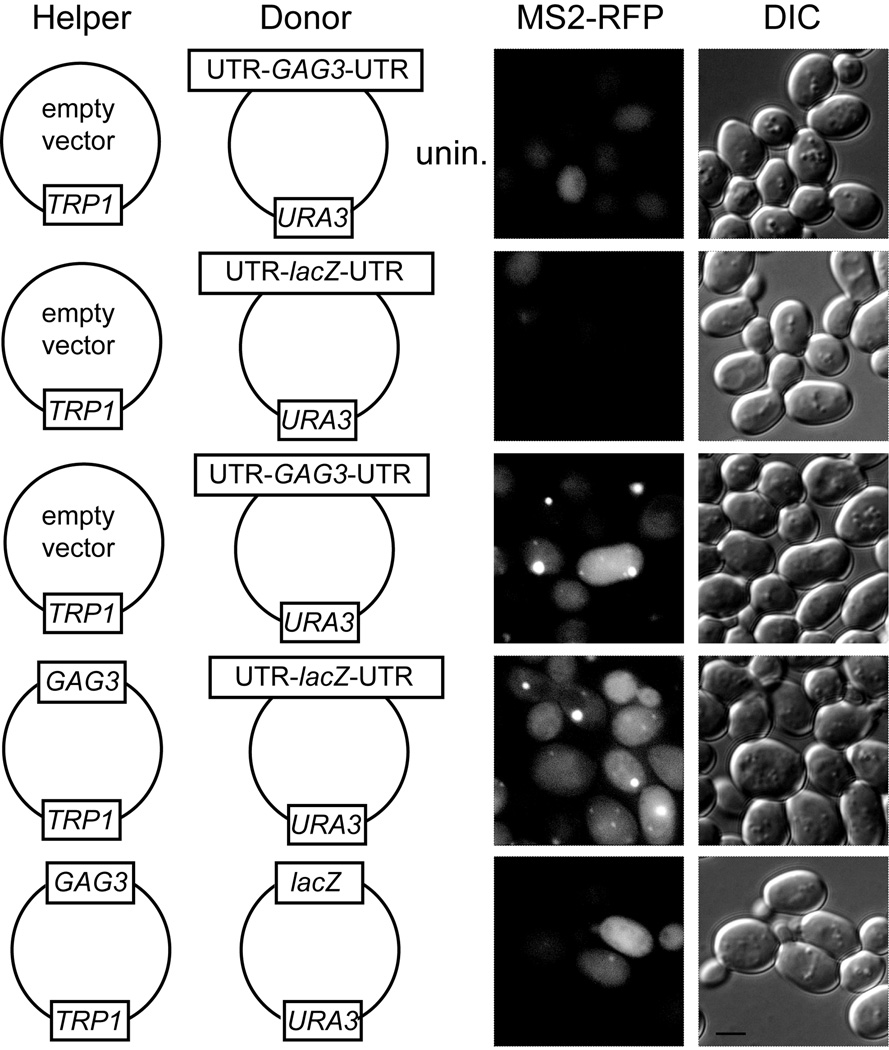
Figure 3. Ty3 sequences in UTRs and POL3 independently mediate localization to foci with PB reporters.
Confocal fluorescence microscopy localization of Ty3-related RNAs. BY4741 expressing the reporter protein Dhh1-GFP or Dcp2-GFP was transformed with Ty3-MS2 and MS2-RFP expression plasmids and induced as described in Methods. Images are representative of two independent transformants of each type. A minimum of 150 MS2-RFP-expressing cells were analyzed for each variant. The galactose-induced Ty3-MS2 RNA is indicated schematically to the left of the fluorescence image (for details see Fig. 1 legend). The top row of panels depict cells that were uninduced, while in the rest of the panels show induced cells. Inset numbers in red indicate number of RFP foci (greater than 0.1µm2)/MS2-RFP-expressing cells. Inset numbers in yellow indicate the percentage of MS2-RFP foci overlapping Dhh1-GFP or Dcp2-GFP reporter foci. The scale bar is 5 µm. Horizontal bars between images: relative size distribution of MS2-RFP foci measured as area of foci (in µm2). Upper bar of pair: Dhh1-GFP; lower bar: Dcp2-GFP. Scale (white bar) is 5 µm. Data processing was as described in Methods.
Ty3 UTRs are sufficient when supplied in cis to mediate retrotransposition of the marker gene HIS3 (Kirchner et al., 1992). Therefore, we first tested the contribution of Ty3 UTRs to formation of RNA foci by comparing localization of wt UTR-GAG3-POL3-UTR RNA to RNA in which the native Ty3 5’ and 3’ UTRs were substituted with hUTRs (GAG3-POL3)(Fig. 3). GAG3-POL3 RNA formed fewer foci with Dhh1-GFP (0.34 versus 0.71 foci per cell) and with Dcp2-GFP (0.67 versus 0.87 foci per cell) than did the native Ty3 RNA. However, GAG3-POL3 RNA foci that did form co-localized with Dhh1- and Dcp2-GFP reporter foci similarly to wt Ty3 RNA (84 and 81%, versus 88% and 87%, respectively), and were similar in size distribution to those of wt Ty3 RNA (Fig. 3). These results clearly indicate that Ty3 UTRs promote inclusion of Ty3 RNA in foci with PB proteins. However, the result that GAG3-POL3 lacking Ty3 UTRs localized to assembly foci was unanticipated.
Previous analysis of Gag3 mutants disrupted for T y3 RNA-binding demonstrated that Gag3 competent to bind RNA is required to form Ty3 RNA foci with PB proteins (Larsen et al., 2008). We therefore tested whether POL3 was dispensable for focus formation by examining localization of RNAs lacking POL3 with Ty3 UTRs (UTR-GAG3-UTR) and hUTRs (GAG3) compared to localization of RNAs containing POL3, wt Ty3 (UTR-GAG3-POL3-UTR) and GAG3-POL3 with hUTRs (GAG3-POL3). Cells expressing UTR-GAG3-UTR showed a higher level of focus formation than cells expressing wt Ty3 (1.17 versus 0.71), but a similar or slightly lower level of coincidence with the PB reporter proteins (82% versus 88%, Dhh1-GFP and 73% versus 87%, Dcp2-GFP). Cells expressing GAG3 with hUTRs showed diffuse fluorescence, and very few foci (0.04 and 0.18 foci per cell in the presence of the Dhh1- and Dcp2-GFP reporters, respectively). In addition, a lower percentage of these foci co-localized with PB reporters (71% and 41% for GAG3 compared to 84% and 81% for GAG3-POL3 with Dhh1- and Dcp2-GFP, respectively). Thus, in the full-length construct with hUTRs, POL3 played an essential role in the localization of the RNA to foci and foci formed by RNAs containing POL3 coincided to a greater extent with PB foci.
The POL3 requirement for localization of GAG3-POL3 RNA to foci with PB proteins prompted further exploration of the contribution of POL3 to localization of Ty3 transcripts with Ty3 UTRs. As described above, programmed frameshifts are a prominent feature of retroelement gRNAs. As ribosomes translating Gag3 terminate approximately 20 times more frequently than reading through the programmed frameshift separating the two reading frames (Farabaugh et al., 1993; Kirchner et al., 1992), RNA downstream of the programmed frameshift site is expected to have lower ribosomal occupancy than GAG3. We first tested whether read-through translation into POL3 and production of Gag3-Pol3 were important by introducing a stop codon downstream of the programmed frameshift between GAG3 and POL3 (UTR-GAG3-STOP-POL3-UTR). Compared to wt, cells expressing this construct showed fewer foci (0.49 versus 0.71 foci per cell). This result showed that Gag3-Pol3 protein is not essential for formation of Ty3 RNA foci with PB proteins. However, the lower number of foci per cell compared to those of wt UTR-GAG3-POL3-UTR suggested that RNA sequence of POL3 alone did not constitute the basis of GAG3-POL3 localization. The foci that formed with UTR-GAG3-STOP-POL3-UTR were somewhat larger than wt, and co-localized to a similar extent with the Dhh1- and Dcp2-GFP reporter foci. We hypothesized that either a low density of ribosomes in the POL3 reading frame or specific sequences could promote localization by providing access to PB proteins. The possibility that low ribosome occupancy was important to focus formation was tested indirectly by fusing GAG3 to POL3 (UTR-GAG-FUS-POL3-UTR). This RNA formed fewer foci than either UTR-GAG3-STOP-POL3-UTR or wt Ty3 (0.25 versus 0.49 or 0.71 foci per cell). However, foci that formed were similar in terms of size and co-localization with the Dhh1-GFP PB reporter (86% versus 88% or 88%). It was possible that the occurrence of programmed frameshifting might target Ty3 RNA for association with PB factors responsible for NMD. However, as discussed above, wt UTR-GAG3-POL3-UTR, UTR-GAG3-STOP-POL3-UTR and UTR-GAG3-FUS-POL3-UTR RNAs were present at comparable concentrations (Fig. 2B), suggesting that NMD was not a determining factor. Additional tests of GAG3-STOP-lacZ and GAG3-FUS-POL3 showed dramatic decreases when low-level translation of the downstream reading frame was disrupted (0.11 and 0.12 foci per cell with the Dhh1 reporter). However, the former result was confounded by a low level of RNA (Fig. 2B). Thus, in the context of the Ty3 UTRs and full-length RNA, low ribosome occupancy of POL3 could operate to allow focus formation in association with PB proteins. This is consistent with the ability of some of these proteins to associate with polysomes and down regulate translation [e.g. Lsm1-7, Pat1, and Dhh1 (Coller & Parker, 2004)].
The observations that inclusion of GAG3 and GAG3-POL3 RNAs in foci with PB proteins was significantly higher when coding sequences were flanked with Ty3 UTRs than when flanked with hUTRs indicates that Ty3 UTRs provide the dominant feature conferring inclusion of Ty3-related RNAs in foci with PB proteins. Positive-strand RNA viruses are characterized by relatively long and structured 5’ UTRs, and, retroviruses rely on both cap-dependent and cap-independent mechanisms as well as specialized helicases to promote translation initiation (Balvay et al., 2007; Bolinger & Boris-Lawrie, 2009; Bushell & Sarnow, 2002). Retroelements may generally have features that antagonize translation, thereby promoting association of factors such as PB proteins. With a length of 181–193 nts, the Ty3 5’ UTR is significantly longer than the average yeast 5’ UTR (50 nts) (Nagalakshmi et al., 2008) and is predicted to be structured (unpublished results). Our quantification of RNA and protein produced from the GAG3-POL3 and GAG3 templates with Ty3 UTRs showed lower protein to RNA ratios than analogous transcripts with hUTRs (Fig. 2C). This suggests that Ty3 UTRs could function to antagonize translation and in that way promote PB protein association. However, other factors also appear to influence translation and packaging. In the absence of native UTRs, inclusion of POL3 significantly enhanced the association of GAG3-containing RNA with PB proteins. Focus formation was diminished if the POL3 ORF was either untranslated or fused to GAG3 resulting in constitutive read-through translation. These data are consistent with roles for Gag3-Pol3 as well as Gag3 in focus formation. In addition, we hypothesize that frameshifting by attenuating the ribosomal occupancy of POL3 could also promote PB protein association with Ty3 RNA. Our data do not exclude the possibility that specific sequences in the POL3 region are important as binding sites for host factors involved in Ty3 RNA localization to foci.
3.2 Requirements for localization and packaging of GAG3-POL3 RNAs differ
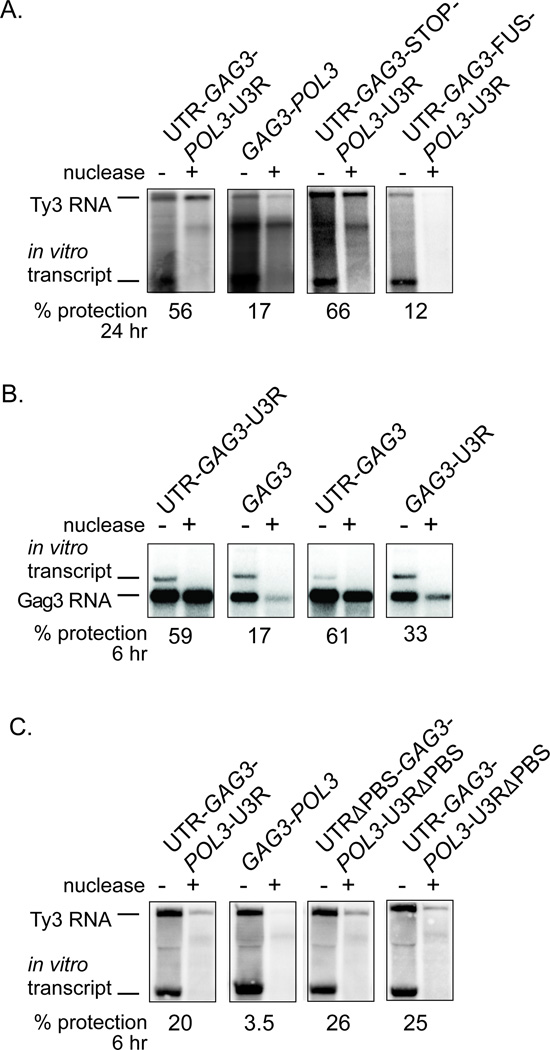
We next examined the relationship between Ty3 RNA localization to foci and packaging into VLPs. Packaging of retrotransposon gRNA into VLPs confers resistance to in vitro nuclease digestion (Lin & Levin, 1997) and protection increases with time of Ty3 induction (Clemens et al., 2011; Larsen et al., 2007) and (unpublished data). The packaging of Ty3 RNA with and without Ty3 UTRs was compared. After 24 h of Ty3 expression, 56% of wt Ty3 RNA (UTR-GAG3-POL3-UTR) was protected (Fig. 4A). GAG3-POL3 RNA with hUTRs, which localized in foci with Dhh1- and Dcp2-GFP reporters, was significantly less protected than the native Ty3 RNA (17% versus 56%) (Fig. 4A). This result indicated that Ty3 gRNA localization into foci with Gag3 and PB proteins is not sufficient to insure wt levels of packaging; rather, one or both of the Ty3 UTRs are required.
Figure 4. Packaging of variant Ty3 RNAs into VLPs.
Extracts of cells expressing Ty3 RNAs were prepared under non-denaturing conditions after 24 h (A) or 6 h (B and C) of galactose induction. An equal amount of in vitro transcribed, truncated Ty3 RNA (Ty3 runoff RNA) was added to each WCE to monitor nuclease digestion. Nuclease treatment was performed as described in Methods. Northern Blots were probed with a 32P-labeled DNA fragment specific to Ty3 RNA and quantified with Quantity One Software (Bio-Rad, Hercules, CA). Two or more independent experiments were performed for each Ty3 RNA variant, and a representative example is shown. % protection represents the amount of nuclease-treated RNA divided by the amount of untreated RNA. A. Localization of Ty3 RNA with PB proteins in foci does not insure packaging. B. The 5’ UTR of Ty3 is sufficient to direct packaging of short GAG3-containing RNAs. C. Deletion of the bipartite PBS sequences does not disrupt full-length Ty3 RNA packaging.
As indicated above, low-level translation of POL3 produced maximal focus formation. The negative effect of GAG3-POL3 read-through translation on focus formation [compare in Fig. 3: UTR-GAG3-POL3-UTR and UTR-GAG3-FUS-POL3-UTR (0.71 versus 0.25 foci/cell) and GAG3-POL3 and GAG3-FUS-POL3 (0.34 versus 0.12 foci/cell)] suggested that ribosomal occupancy of the downstream ORF antagonized PB protein and Gag3 association with the gRNA, but that this was of greater consequence in the absence of the UTRs. In order to explicitly assess the effect of read-through translation on packaging, UTR-GAG3-STOP-POL3-UTR RNA was compared to UTR-GAG3-FUS-POL3-UTR RNA for protection from nuclease digestion. These experiments showed that UTR-GAG3-STOP-POL3-UTR was modestly more efficiently packaged than wt Ty3 RNA (66% versus 56%). In contrast, the UTR-GAG3-FUS-POL3-UTR RNA showed much less protection than wt (12% versus 56%) (Fig. 4A). While reduced retrosome formation is associated with this reduced protection, this construct has elevated Gag3-Pol3 to Gag3 ratios (Fig. 2A), and consequently may be reduced for polyprotein processing. In the case of HIV-1, reduced processing is associated with unstable dimers and reduced RNA stability (L'Hernault et al., 2012). Together, these results are consistent with the model that RNAs that associate more efficiently into foci with PB proteins are more readily packaged into VLPs. However, requirements for processed Gag3 also likely contributed to reduced packaging of UTR-GAG3-FUS-POL3-UTR RNA (Fig. 4A).
The experiments described above implicated one or both of the UTRs in packaging. The role of UTRs was tested by examining extracts of cells expressing UTR-GAG3-UTR, GAG3 with 5’ and 3’ hUTRs, and GAG3 with only the Ty3 5’ UTR or 3’ UTR (Fig. 3) and one hUTR. Because differences in packaging might be most apparent in the early phase after induction, protection was measured after 6 h rather than 24 h of induction. UTR-GAG3-UTR RNA, which localized efficiently to PB protein-containing foci was packaged to a much greater extent than GAG3 with hUTRs (59% versus 17%) (Fig.4B). GAG3 RNA with the 5’ UTR a lone was protected to a similar extent as RNA with both UTRs (61% versus 59%). GAG3 RNA with only the Ty3 3’UTR was protected to a significantly lesser extent (33%), but still more than GAG3 RNA with hUTRs (17%) (Fig. 4B).
In summary, these results argue that while Ty3 RNAs can be delivered to PB foci through independent pathways mediated by UTRs and POL3, Ty3 UTRs provide the key signal for productive packaging into VLPs. In the case of the GAG3 RNA, both the Ty3 5’ and 3’ UTRs enhanced packaging, but the Ty3 5’ UTR had the greater effect. Independent of UTRs, translation of POL3 correlated inversely with packaging. Lastly, although GAG3 RNA with hUTRs represents a synthetic RNA that was less well packaged than UTR-GAG3-UTR, these results indicate that at least some packaging into Ty3 particles likely occurs in the absence of complete localization to macroscopic PB foci.
3.3 Importance of 5’-3’ bipartite primer binding sequences to Ty3 RNA packaging
Known retrovirus gRNA packaging sites contain DIS regions which contribute to packaging (Chen et al., 2009; Moore & Hu, 2009; Russell et al., 2004), consistent with the packaging of dimer species. In the case of Ty3, reverse transcription primer initiator tRNAMet has been shown to bind to a bipartite PBS in the 5’ and 3’ UTRs (Gabus et al., 1998). Furthermore, in vitro initiator tRNAMet primers bound to Ty3 gRNA have been proposed to anneal in anti-parallel orientation to each other at the tRNA 5’ ends and mediate mini-Ty3 dimerization (Gabus et al., 1998). Given a bipartite PBS, the ability of the 5’UTR alone to direct packaging of the GAG3 RNA suggested that primer-mediated dimerization is dispensable for packaging. Full-length Ty3 RNA variants were constructed with the 5’ and 3’ PBS or 3’ PBS alone deleted from the UTRs to test the role of the bipartite PBS in packaging (Fig. 4C). Surprisingly, deletion of both the 5’ and 3’ segments of the bi-partite PBS or the 3’ PBS segment alone resulted in similar increases in protection to the full-length Ty3 (26% and 25% versus 20%). Because we observed RNA protection in the absence of part or all of the bipartite PBS, our results suggest that if the tRNA primer bound to gRNA mediates dimerization of Ty3 gRNA in vivo, then, unlike what is observed for some retrovirus gRNAs, formation of the dimer interface is not a pre-requisite for Ty3 gRNA packaging. We also assayed these mutants for the ability to produce cDNA. As previously reported, disruption of the 5’ PBS resulted in an inability to generate cDNA. As predicted by the bipartite priming model, and as suggested by in vitro experiments, the mutant that retained the 5’ PBS but lacked the 3’ PBS also did not produce cDNA (unpublished results).
3.4 Gag3 can mediate packaging of Ty3 UTR-containing RNAs in trans
In this study we found that the Ty3 RNA variant GAG3 with Ty3 UTRs forms PB protein-associated foci and is protected against nuclease digestion, whereas GAG3 with hUTRs is neither localized nor protected, thereby implicating UTRs in packaging. UTRs were previously shown to be sufficient to mediate transposition of a genetic marker if complemented in trans by expression of Ty3 proteins from a helper element (Kirchner et al., 1992) and (unpublished data), suggesting that Gag3 function can promote packaging of RNA in trans. Because the experiments in the current study examined constructs in which Gag3 was expressed in cis, an important outstanding question was whether Gag3 could localize RNA to PB protein foci when expressed in trans. LacZ flanked by heterologous or Ty3 UTRs was expressed in the presence or absence of Ty3 Gag3. LacZ formed visible foci only when flanked by Ty3 UTRs and in the presence of Gag3 (Fig. 5). These foci were less intense than when Gag3 was expressed in cis. Nevertheless, this demonstrated that in the presence of trans-acting Gag3, Ty3 UTRs mediate localization consistent with the previously observed ability of Ty3 expression to mediate donor transposition in trans.
Figure 5. Gag3 packages Ty3 UTR-containing RNA in trans.
Yeast strain BY4741 containing trp1Δ was transformed with a low-copy vector plasmid carrying lacZ with Ty3-specific UTRs or hUTR; these were co-transformed with low-copy helper plasmid carrying GAG3 with hUTRs and low-copy plasmid carrying MS2-RFP reporter (see Table 1 and Methods). Samples were induced by addition of galactose and allowed to grow for 6 h (see Methods). The scale bar is 5 µm.
4. Summary and Conclusions
Retroelement VLP assembly results from a balance of competing translation and packaging processes. We proposed previously that PB localization facilitates Ty3 gRNA packaging into particles by sequestering the gRNA from the translation apparatus (Beliakova-Bethell et al., 2006). Based on previous investigations and the current study, we propose that as Gag3 accumulates, it interacts with a packaging site contained in the Ty3 UTRs, thereby antagonizing translation and allowing association with PB proteins, which further antagonize translation. In addition, we hypothesize that POL3, because of specific sequences or reduced ribosomal occupancy or both, interacts with host factors including PB proteins and independently promotes development of retrosome foci. Host protein interaction with Ty3 proteins and RNA allow for RNA exit from translation and for concentration of Gag3, Gag3-Pol3, and RNA, thereby driving assembly.
Supplementary Material
Highlights.
Ty3 UTRs or GAG3-POL3 mediate RNA localization to Ty3 assembly foci.
The Ty3 5’ UTR is sufficient for packaging of Ty3 GAG3 or GAG3-POL3 RNA into VLPs.
The 5’-3’- bipartite PBS is not essential for packaging of Ty3 gRNA.
Acknowledgements
This research was supported by funds from National Institutes of Health grants R01 GM33281 to SBS. NBB was supported on National Institutes of Health Training Grants 5 T32 AI 07319. We thank M. Oakes (UCI) for assistance with fluorescence microscopy. This work was made possible in part, through access to the Optical Biology Core facility of the Developmental Biology Center, a Shared Resource supported in part by the Cancer Center Support Grant (CA-62203) and Center for Complex Biological Systems Support Grant (GM-076516) the at the University of California, Irvine.
Abbreviations
- aa
amino acids
- ABCE1
ATP binding cassette sub-family E member 1
- CA
capsid
- CBC
cap binding complex
- cDNA
copy DNA
- Crm1
chromosome region maintenance protein 1
- Dcp2
decapping protein 2
- Dhh1/hDDX6
dead box helicase homolog/human DEAD (Asp-Glu-Ala-Asp) box polypeptide 6
- DIS
dimerization initiation sequence
- Edc3
enhancer of decapping protein 3
- FRET
Förster (fluorescence) resonance energy transfer
- GFP
green fluorescent protein
- gRNA
genomic RNA
- hUTRs
heterologous untranslated regions
- HIV-1
human immunodeficiency virus
- hnRNP A2
heterogeneous nuclear ribonucleoprotein A2
- IN
integrase
- Lsm1-7
Like-sm proteins 1-7
- LTR
long terminal repeat
- MHR
major homology region
- miRNA
micro RNA
- MLV
murine leukemia virus
- Mov10
murine leukemia virus 10 protein
- NC
nucleocapsid
- NMD
nonsense-mediated decay
- NMR
nuclear magnetic resonance
- NTD
amino terminal domain
- Pat1
protein associated with topoisomerase 1
- PB
(RNA) processing body
- PBS
primer binding site
- PBST
phosphate buffered saline tween solution
- PR
protease
- Psi
packaging signal
- RFP
red fluorescent protein
- RISC
RNA induced silencing complex
- RNP
Ribonucleoprotein
- RSV
Rous sarcoma virus
- RT
reverse transcriptase
- SDS
sodium dodecyl sulfate
- SG
stress granule
- UAS
upstream
- UTR
untranslated region
- VLP
virus-like particle
- WCE
whole-cell extract
- Xrn1
exo-ribonuclease 1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clement JF, Song R, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J Cell Sci. 2010;123:369–383. doi: 10.1242/jcs.055897. [DOI] [PubMed] [Google Scholar]
- Aizer A, Brody Y, Ler LW, Sonenberg N, Singer RH, Shav-Tal Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol Biol Cell. 2008;19:4154–4166. doi: 10.1091/mbc.E08-05-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D, BDSJD Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. 2005 [Google Scholar]
- Anderson EC, Lever AM. Human immunodeficiency virus type 1 Gag polyprotein modulates its own translation. J Virol. 2006;80:10478–10486. doi: 10.1128/JVI.02596-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH. Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology. 2012;9:53. doi: 10.1186/1742-4690-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F, FBRKR E. coli, plasmids, and bacteriophages. Current Protocols in Molecular Biology. 2002 [Google Scholar]
- Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat Rev Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basyuk E, Boulon S, Skou Pedersen F, Bertrand E, Vestergaard Rasmussen S. The packaging signal of MLV is an integrated module that mediates intracellular transport of genomic RNAs. J Mol Biol. 2005;354:330–339. doi: 10.1016/j.jmb.2005.09.071. [DOI] [PubMed] [Google Scholar]
- Basyuk E, Galli T, Mougel M, Blanchard JM, Sitbon M, Bertrand E. Retroviral genomic RNAs are transported to the plasma membrane by endosomal vesicles. Dev Cell. 2003;5:161–174. doi: 10.1016/s1534-5807(03)00188-6. [DOI] [PubMed] [Google Scholar]
- Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beliakova-Bethell N, Beckham C, Giddings THJ, Winey M, Parker R, Sandmeyer S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA. 2006;12:94–101. doi: 10.1261/rna.2264806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Styles CA, Fink GR. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol Cell Biol. 1986;6:3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Boris-Lawrie K. Mechanisms employed by retroviruses to exploit host factors for translational control of a complicated proteome. Retrovirology. 2009;6:8. doi: 10.1186/1742-4690-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger C, Yilmaz A, Hartman TR, Kovacic MB, Fernandez S, Ye J, Forget M, Green PL, Boris-Lawrie K. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 2007;35:2629–2642. doi: 10.1093/nar/gkm124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol. 2008;183:441–455. doi: 10.1083/jcb.200807043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Nissan T, Parker R. Analyzing P-bodies and stress granules in Saccharomyces cerevisiae. Methods Enzymol. 2010;470:619–640. doi: 10.1016/S0076-6879(10)70025-2. [DOI] [PubMed] [Google Scholar]
- Burdick R, Smith JL, Chaipan C, Friew Y, Chen J, Venkatachari NJ, Delviks-Frankenberry KA, Hu WS, Pathak VK. P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J Virol. 2010;84:10241–10253. doi: 10.1128/JVI.00585-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Sarnow P. Hijacking the translation apparatus by RNA viruses. J Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, Konig R, Bandyopadhyay S, Ideker T, Goff SP, Krogan NJ, Frankel AD, Young JA, Chanda SK. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000437. e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. Translation is not required to generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J Virol. 2000;74:11531–11537. doi: 10.1128/jvi.74.24.11531-11537.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butsch M, Boris-Lawrie K. Destiny of unspliced retroviral RNA: ribosome and/or virion? J Virol. 2002;76:3089–3094. doi: 10.1128/JVI.76.7.3089-3094.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chable-Bessia C, Meziane O, Latreille D, Triboulet R, Zamborlini A, Wagschal A, Jacquet JM, Reynes J, Levy Y, Saib A, Bennasser Y, Benkirane M. Suppression of HIV-1 replication by microRNA effectors. Retrovirology. 2009;6:26. doi: 10.1186/1742-4690-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol Cell Biol. 2010;30:382–398. doi: 10.1128/MCB.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U S A. 2009;106:13535–13540. doi: 10.1073/pnas.0906822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MP, Chen J, Nikolaitchik OA, Hu WS. Molecular determinants of HIV-1 intersubtype recombination potential. Virology. 2007;363:437–446. doi: 10.1016/j.virol.2007.01.034. [DOI] [PubMed] [Google Scholar]
- Cimarelli A, Darlix JL. Assembling the human immunodeficiency virus type 1. Cell Mol Life Sci. 2002;59:1166–1184. doi: 10.1007/s00018-002-8495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens K, Larsen L, Zhang M, Kuznetsov Y, Bilanchone V, Randall A, Harned A, Dasilva R, Nagashima K, McPherson A, Baldi P, Sandmeyer S. The Ty3 Gag3 Spacer Controls Intracellular Condensation and Uncoating. J Virol. 2011;85:3055–3066. doi: 10.1128/JVI.01055-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Ficheux D, Darlix JL. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem. 2000;275:19210–19217. doi: 10.1074/jbc.M001371200. [DOI] [PubMed] [Google Scholar]
- Cristofari G, Gabus C, Ficheux D, Bona M, Le Grice SF, Darlix JL. Characterization of active reverse transcriptase and nucleoprotein complexes of the yeast retrotransposon Ty3 in vitro. J Biol Chem. 1999;274:36643–36648. doi: 10.1074/jbc.274.51.36643. [DOI] [PubMed] [Google Scholar]
- D'Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- D'Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- Darlix JL, Lapadat-Tapolsky M, de Rocquigny H, Roques BP. First glimpses at structure-function relationships of the nucleocapsid protein of retroviruses. J Mol Biol. 1995;254:523–537. doi: 10.1006/jmbi.1995.0635. [DOI] [PubMed] [Google Scholar]
- Dooher JE, Schneider BL, Reed JC, Lingappa JR. Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production. Traffic. 2007;8:195–211. doi: 10.1111/j.1600-0854.2006.00524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman N, Lever A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J Virol. 2000;74:11413–11417. doi: 10.1128/jvi.74.23.11413-11417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 5' to 3' mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol. 2010;84:5052–5066. doi: 10.1128/JVI.02477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F, Salmon K, Shen MW, Aeling KA, Ito E, Irwin B, Tran UP, Hatfield GW, Da Silva NA, Sandmeyer S. A vector set for systematic metabolic engineering in Saccharomyces cerevisiae. Yeast. 2011;28:123–136. doi: 10.1002/yea.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh PJ. Post-transcriptional regulation of transposition by Ty retrotransposons of Saccharomyces cerevisiae. J Biol Chem. 1995;270:10361–10364. doi: 10.1074/jbc.270.18.10361. [DOI] [PubMed] [Google Scholar]
- Farabaugh PJ, Zhao H, Vimaladithan A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YX, Moore SP, Garfinkel DJ, Rein A. The genomic RNA in Ty1 virus-like particles is dimeric. J Virol. 2000;74:10819–10821. doi: 10.1128/jvi.74.22.10819-10821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JA, An W, King SR, Telesnitsky A. Nonrandom dimerization of murine leukemia virus genomic RNAs. J Virol. 2004;78:12129–12139. doi: 10.1128/JVI.78.22.12129-12139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Heyman T, Bystrom AS, Wilhelm M, Wilhelm FX. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol Cell Biol. 1998;18:799–806. doi: 10.1128/mcb.18.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtak V, Mulky A, Rawlings SA, Kozhaya L, Lee K, Kewalramani VN, Unutmaz D. Perturbation of the P-body component Mov10 inhibits HIV-1 infectivity. PLoS One. 2010;5:e9081. doi: 10.1371/journal.pone.0009081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabus C, Ficheux D, Rau M, Keith G, Sandmeyer S, Darlix JL. The yeast Ty3 retrotransposon contains a 5'-3' bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998;17:4873–4880. doi: 10.1093/emboj/17.16.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JWJ, Gorelick RJ, Rein A, Weeks KM. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc Natl Acad Sci U S A. 2010;107:19248–19253. doi: 10.1073/pnas.1006897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin SD, Allen JF, Lever AM. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J Virol. 2001;75:12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, Devine SE. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics. 2003;164:867–879. doi: 10.1093/genetics/164.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorov B, Decimo D, Smagulova F, Pechoux C, Mougel M, Muriaux D, Darlix JL. Intracellular HIV-1 Gag localization is impaired by mutations in the nucleocapsid zinc fingers. Retrovirology. 2007;4:54. doi: 10.1186/1742-4690-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LJ, Chalker DL, Orlinsky KJ, Sandmeyer SB. Ty3 GAG3 and POL3 genes encode the components of intracellular particles. J Virol. 1992;66:1414–1424. doi: 10.1128/jvi.66.3.1414-1424.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen LJ, Chalker DL, Sandmeyer SB. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol Cell Biol. 1988;8:5245–5256. doi: 10.1128/mcb.8.12.5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng X, Kharytonchyk S, Garcia EL, Lu K, Divakaruni SS, LaCotti C, Edme K, Telesnitsky A, Summers MF. Identification of a minimal region of the HIV-1 5'-leader required for RNA dimerization, NC binding, and packaging. J Mol Biol. 2012;417:224–239. doi: 10.1016/j.jmb.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housset V, De Rocquigny H, Roques BP, Darlix JL. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J Virol. 1993;67:2537–2545. doi: 10.1128/jvi.67.5.2537-2545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65–74. doi: 10.1083/jcb.200707010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Irwin B, Aye M, Baldi P, Beliakova-Bethell N, Cheng H, Dou Y, Liou W, Sandmeyer S. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 2005;15:641–654. doi: 10.1101/gr.3739005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SF, Telesnitsky A. Retroviral RNA dimerization and packaging: the what, how, when, where, and why. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001007. e1001007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye JF, Lever AM. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J Virol. 1999;73:3023–3031. doi: 10.1128/jvi.73.4.3023-3031.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- Keeney JB, Chapman KB, Lauermann V, Voytas DF, Astrom SU, von Pawel-Rammingen U, Bystrom A, Boeke JD. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol Cell Biol. 1995;15:217–226. doi: 10.1128/mcb.15.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J, Sandmeyer S. Proteolytic processing of Ty3 proteins is required for transposition. J Virol. 1993;67:19–28. doi: 10.1128/jvi.67.1.19-28.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner J, Sandmeyer SB, Forrest DB. Transposition of a Ty3 GAG3-POL3 fusion mutant is limited by availability of capsid protein. J Virol. 1992;66:6081–6092. doi: 10.1128/jvi.66.10.6081-6092.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein KC, Reed JC, Lingappa JR. Intracellular destinies: degradation, targeting, assembly, and endocytosis of HIV Gag. AIDS Rev. 2007;9:150–161. [PubMed] [Google Scholar]
- Kutluay SB, Bieniasz PD. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001200. e1001200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov YG, Zhang M, Menees TM, McPherson A, Sandmeyer S. Investigation by atomic force microscopy of the structure of Ty3 retrotransposon particles. J Virol. 2005;79:8032–8045. doi: 10.1128/JVI.79.13.8032-8045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L'Hernault A, Weiss EU, Greatorex JS, Lever AM. HIV-2 genome dimerization is required for the correct processing of Gag: a second-site reversion in matrix can restore both processes in dimerization-impaired mutant viruses. J Virol. 2012;86:5867–5876. doi: 10.1128/JVI.00124-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen LS, Beliakova-Bethell N, Bilanchone V, Zhang M, Lamsa A, Dasilva R, Hatfield GW, Nagashima K, Sandmeyer S. Ty3 nucleocapsid controls localization of particle assembly. J Virol. 2008;82:2501–2514. doi: 10.1128/JVI.01814-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen LS, Zhang M, Beliakova-Bethell N, Bilanchone V, Lamsa A, Nagashima K, Najdi R, Kosaka K, Kovacevic V, Cheng J, Baldi P, Hatfield GW, Sandmeyer S. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J Virol. 2007;81:6957–6972. doi: 10.1128/JVI.02207-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler JF, Jr, Merkulov GV, Boeke JD. A nucleocapsid functionality contained within the amino terminus of the Ty1 protease that is distinct and separable from proteolytic activity. J Virol. 2002;76:346–354. doi: 10.1128/JVI.76.1.346-354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque K, Halvorsen M, Abrahamyan L, Chatel-Chaix L, Poupon V, Gordon H, DesGroseillers L, Gatignol A, Mouland AJ. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic. 2006;7:1177–1193. doi: 10.1111/j.1600-0854.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- Levin JG, Grimley PM, Ramseur JM, Berezesky IK. Deficiency of 60 to 70S RNA in murine leukemia virus particles assembled in cells treated with actinomycin D. J Virol. 1974;14:152–161. doi: 10.1128/jvi.14.1.152-161.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JG, Rosenak MJ. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: evidence for persistence of viral messenger RNA. Proc Natl Acad Sci U S A. 1976;73:1154–1158. doi: 10.1073/pnas.73.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Levin HL. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes Dev. 1997;11:270–285. doi: 10.1101/gad.11.2.270. [DOI] [PubMed] [Google Scholar]
- Liu C, Zhang X, Huang F, Yang B, Li J, Liu B, Luo H, Zhang P, Zhang H. APOBEC3G Inhibits MicroRNA-mediated Repression of Translation by Interfering with the Interaction between Argonaute-2 and MOV10. J Biol Chem. 2012;287:29373–29383. doi: 10.1074/jbc.M112.354001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5'-leader RNA that regulate genome packaging. Science. 2011;334:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malagon F, Jensen TH. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol Cell Biol. 2008;28:6022–6032. doi: 10.1128/MCB.00684-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc Natl Acad Sci U S A. 2009;106:15657–15662. doi: 10.1073/pnas.0908305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Fu W, Nikolaitchik O, Chen J, Ptak RG, Hu WS. Dimer initiation signal of human immunodeficiency virus type 1: its role in partner selection during RNA copackaging and its effects on recombination. J Virol. 2007;81:4002–4011. doi: 10.1128/JVI.02589-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MD, Hu WS. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 2009;11:91–102. [PMC free article] [PubMed] [Google Scholar]
- Moser JJ, Fritzler MJ. Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int J Biochem Cell Biol. 2010;42:828–843. doi: 10.1016/j.biocel.2009.11.018. [DOI] [PubMed] [Google Scholar]
- Mouland AJ, Xu H, Cui H, Krueger W, Munro TP, Prasol M, Mercier J, Rekosh D, Smith R, Barbarese E, Cohen EA, Carson JH. RNA trafficking signals in human immunodeficiency virus type 1. Mol Cell Biol. 2001;21:2133–2143. doi: 10.1128/MCB.21.6.2133-2143.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathans R, Chu CY, Serquina AK, Lu CC, Cao H, Rana TM. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol Cell. 2009;34:696–709. doi: 10.1016/j.molcel.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni N, Nikolaitchik OA, Dilley KA, Chen J, Galli A, Fu W, Prasad VV, Ptak RG, Pathak VK, Hu WS. Mechanisms of human immunodeficiency virus type 2 RNA packaging: efficient trans packaging and selection of RNA copackaging partners. J Virol. 2011;85:7603–7612. doi: 10.1128/JVI.00562-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaitchik O, Rhodes TD, Ott D, Hu WS. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J Virol. 2006;80:4691–4697. doi: 10.1128/JVI.80.10.4691-4697.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark-McMahon MH, Beliakova-Bethell NS, Darlix JL, Le Grice SF, Sandmeyer SB. Ty3 integrase is required for initiation of reverse transcription. J Virol. 2002;76:2804–2816. doi: 10.1128/JVI.76.6.2804-2816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics. 2008;178:197–214. doi: 10.1534/genetics.107.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlinsky KJ, Gu J, Hoyt M, Sandmeyer S, Menees TM. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J Virol. 1996;70:3440–3448. doi: 10.1128/jvi.70.6.3440-3448.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlinsky KJ, Sandmeyer SB. The Cys-His motif of Ty3 NC can be contributed by Gag3 or Gag3-Pol3 polyproteins. J Virol. 1994;68:4152–4166. doi: 10.1128/jvi.68.7.4152-4166.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott DE. Cellular proteins detected in HIV-1. Rev Med Virol. 2008;18:159–175. doi: 10.1002/rmv.570. [DOI] [PubMed] [Google Scholar]
- 94.Parent LJ. New insights into the nuclear localization of retroviral Gag proteins. Nucleus. 2011;2:92–97. doi: 10.4161/nucl.2.2.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman M, Resh MD. Identification of an intracellular trafficking and assembly pathway for HIV-1 gag. Traffic. 2006;7:731–745. doi: 10.1111/j.1398-9219.2006.00428.x. [DOI] [PubMed] [Google Scholar]
- Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6:741–755. doi: 10.1111/j.1600-0854.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- Reed JC, Molter B, Geary CD, McNevin J, McElrath J, Giri S, Klein KC, Lingappa JR. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J Cell Biol. 2012;198:439–456. doi: 10.1083/jcb.201111012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RS, Liang C, Wainberg MA. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology. 2004;1:23. doi: 10.1186/1742-4690-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer SB, Clemens KA. Function of a retrotransposon nucleocapsid protein. RNA Biol. 2010;7:642–654. doi: 10.4161/rna.7.6.14117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S, Aye M, Menees TM. Ty3: A position-specific gypsy-like element in Saccharomeyes cerevisiae. Mobile DNA II ASM press. 2002:663–682. [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher KD, Parker R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS One. 2010;5:e10006. doi: 10.1371/journal.pone.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voytas DF, Boeke JD. Ty1 and Ty5 of Saccharomyces cerevisiae. 2002:631–662. [Google Scholar]
- Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, Zheng YH. Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J Biol Chem. 2010;285:14346–14355. doi: 10.1074/jbc.M110.109314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Lloyd RE. Regulation of stress granules in virus systems. Trends Microbiol. 2012;20:175–183. doi: 10.1016/j.tim.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Boeke JD. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol Cell Biol. 1990;10:2695–2702. doi: 10.1128/mcb.10.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- Yu SF, Lujan P, Jackson DL, Emerman M, Linial ML. The DEAD-box RNA helicase DDX6 is required for efficient encapsidation of a retroviral genome. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002303. e1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, Nicolas A, Thermes C, Morillon A. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.