Abstract
A variety of mechanotransduction pathways mediate the response of fibroblasts or chondrocytes to ultrasound stimulation. In addition, regulatory pathways that co-ordinate stimulus-specific cellular responses are likely to exist. In this study, analysis was confined to the hypothesis that ultrasound stimulation (US) influences the chromatin structure, and that these changes may reflect a regulatory pathway that connects nuclear architecture, chromatin structure and gene expression. Murine fibroblasts seeded on tissue culture plates were stimulated with US (5.0 MHz (14 kPa), 51-s per application) and the thermal denaturation profiles of nuclei isolated from fibroblasts were assessed by dynamic scanning calorimetry (DSC). When compared to the thermal profiles obtained from the nuclei of non-stimulated cells, the nuclei obtained from stimulated cells showed a change in peak profiles and peak areas, which is indicative of chromatin remodeling. Independently, US was also observed to impact the histone (H1):chromatin association as measured indirectly by DAPI staining. Based on our work, it appears plausible that US can produce a remodeling of chromatin, thus triggering signal cascade and other intracellular mechanisms.
Keywords: Ultrasound, Stimulation, Chromatin, Gene expression, Transition peaks
1. Introduction
In the past two decades, tissue engineering strategies seeking to engineer functional tissues in the laboratory have taken advantage of cell adhesion and proliferation on a wide range of substrates, under the influence of a variety of soluble factors such as growth factors and hormones, and under conditions of mechanical stimulation (Carver and Heath, 1999; Bonassar et al., 2001; Sah et al., 1989; Parvizi et al., 1999; Zhang et al., 2003; Nishikori et al., 2002; Lee et al., 2006). In theory, as applied to tissue engineering research, the cells are exposed to a variety of stimuli to achieve proliferation and differentiation. The impacts of substrate morphology, surface chemistry and external stress on the cellular morphology and actin organization have also been evaluated (Angele et al., 2003; Bonassar et al., 2001; Bougault et al., 2008; Buchmann et al., 1995; Dalby et al., 2004; Ingber, 2008, 2006; Chen and Grinnell, 1995; Fioravanti et al., 2007; Geiger et al., 2001; Knight et al., 2006). Substrate induced changes in cellular morphology have been correlated to the remodeling of in situ chromatin as judged by the thermal denaturation profiles obtained by differential scanning calorimetry (DSC) (Nicolini et al., 1983; Vergarini et al., 2004). However, the ability of a stimulus that is external to the cells to modulate chromatin structure is less well understood.
The research in this study focuses on the determination of cell function under external stimulation, specifically stimulation via ultrasound (US). We employ intermittent applications of low-intensity-diffuse-ultrasound (LIDUS) signals ranging from 1.5 MHz to 8.0 MHz and at varying application frequencies to stimulate bovine chondrocytes and murine fibroblasts (unpublished data and (Hasanova et al., 2011)). Our data provides strong evidence that the US stimulation at 5.0 MHz (14 kPa) modulated the proliferative capacity, biosynthetic activity and the gene expression of integrins of articular chondrocytes maintained in 3D matrices. Moreoever, upon exposure to US increases in the levels of gene expression of Sox-5 and Sox-9, transcription factors known to impact collagen-II expression were observed (Noriega et al., 2007; Noriega, 2009; Hasanova et al., 2011).
Researchers theorize that nuclear architecture and chromatin organization play vital roles in the control of gene expression. The extent of large-scale chromatin organization will likely impact the extent of DNA transcription and modulate accessibility to transcription factors (Ingber et al., 1987). Considering the cellular response obtained in response to US exposure, we hypothesize that US is able to temporarily modify chromatin organization in stimulated cells. Therefore, this study assesses the changes in chromatin structure upon US stimulation via differential scanning calorimetry (DSC). We selected fibroblasts as a model system since (1) reference thermal denaturation profiles of fibroblasts have been reported (Vergarini et al., 2004) and are available for valuable comparison; (2) fibroblasts have also been reported to respond to US exposure (unpublished data, (Zhou et al., 2004)); and (3) association of chondrocytes and fibroblasts is very close, and differentiation of either cell type from the same precursor is known to occur.
2. Materials and methods
2.1. Cell culture
L929 murine fibroblasts were purchased from ATCC (Manassas, VA) and expanded according to ATCC protocols and recommendations. Briefly, frozen cells were thawed and seeded on a 75-cm2 T flask until 80% confluence was attained; cells were expanded and cells from passage 2 or 3 were used in our experiments. Fibroblasts were grown in DMEM medium in the presence of 10% fetal calf serum (GIBCO), 1% non-essential amino acids (GIBCO), 2 mM L-glutamine and 1.0% penicillin–streptomycin (Sigma Chemical Company, St. Louis, MO). All cells were grown at 37 °C under a 5% CO2-humidified atmosphere.
2.2. Cell seeding and US stimulation
Fibroblasts were seeded on 6-well TCP plates at a seeding density of 1 × 104 cells/cm2 and maintained in a CO2 incubator (37 °C under a 5% CO2-humidified atmosphere) until 80% confluence was attained (~48-h), and then subjected to US exposure. The cells were exposed to US for 1 day (regimen-US-1D) or 3 days (regimen-US-3D). US application was as follows: 5.0 MHz, 14 kPa, 51-s per application, 3-applications/day at an interval of 4 h. US was applied as detailed elsewhere (Noriega et al., 2007). The cells seeded on TCP plates that were not subjected to US stimulation served as controls. Both controls and test plates were handled similarly.
2.3. Nuclei isolation
Upon completion of US exposure, the fibroblasts were collected from the TCP plates (4 × 107 cells total) by trypsinization as detailed elsewhere (Noriega et al., 2007; Hasanova et al., 2011). One TCP plate (i.e. 6 wells) constituted one test condition. The wells were individually exposed to US, and the cells released, pooled and processed to make cell pellets. Equivolume aliquots (1.5 ml containing ~2 × 107 cells) were centrifuged at 200 × g for 10 min at 4 °C; each cell pellet was drained and resuspended in 0.5 ml of nuclei buffer with sucrose (0.3 M sucrose, 50 mM Tris–HCI, pH 6.8, 25 mM KCl, 4 mM MgCl2, 1 mM CaC12, 1 mM phenylmethyl-sulfonyl fluoride (PMSF in ethanol), 0.01% 2-mercaptoethanol) and the samples (~0.6 ml) were transferred into Eppendorf tubes. The cells were lysed and membrane fractions were removed by the addition of 0.05 ml 10% Triton X-100 (Sigma) followed by the addition of 0.55 ml of 80% glycerol in nuclei buffer without sucrose (50 mM Tris–HCI, pH 6.8, 25 mM KCl, 4 mM MgCl2, 1 mM CaC12, 1 mM phenylmethylsulfonyl fluoride (PMSF in ethanol), 0.01% 2-mercaptoethanol). The nuclei suspension was vortexed in an Eppendorf removed and the nuclei were transferred to aluminum pans to run DSC scans. The nuclei analyzed immediately after isolation were denoted as (A) whereas the nuclei analyzed after rest (4 °C for 40 min) were denoted as (B). The experiments were repeated 3–4 times.
2.4. Differential scanning calorimetry (DSC)
All calorimetry experiments were run on a Q100 (TA Instruments, New Castle, DE) thermal analyzer. An empty hermetic Al pan was used as a reference. The DSC was calibrated for temperature using water (melting point, 0.0 °C) and indium (melting point, 156.6 °C). Heat flow was calibrated using indium (ΔHm, 28.45 J/g). Typically, 5–15 mg of nuclei were sealed in the hermetic aluminum pan and placed in the sample compartment, equilibrated at −10 °C and heated to a final temperature of 120 °C. A heating rate of 10 °C/min was used and the sample compartment was flushed with dry nitrogen flowing at 25 mL/min.
2.5. Staining of nuclei
A small fraction (~0.5 mg) of isolated nuclei were resuspended in 200 μl of 4% paraformaldehyde in PBS and spread over a silane coated glass slide (EMS, Hatfield, PA), rinsed with PBS and incubated with 1 ml of perm solution (0.25% Triton-100X; 0.025% CHAPS in PBS) for 20 min. Next, they were counterstained with 0.05 mg/ml of DAPI (Invitrogen-D1306, excitation/emission maxima: 358/461 nm) before mounting on coverslips. The nuclei were visualized by confocal microscopy (Olympus FV500 Inverted, Olympus IX 81) at 100× magnification and at 2048 × 2048 resolution.
2.6. Quantitative real-time PCR
Upon completion of US exposure, the cell seeded plates were kept in the CO2 incubator for 2 h. The media was then removed and the cells were washed with ice-cold HBSS and incubated with 200 μl/well of Trizol reagent. Finally, the cell homogenate was collected and the RNA was isolated by using a QIAGEN RNeasy mini kit (Invitrogen, CA). The mRNA level was quantified by using quantitative real-time PCR (qRT-PCR). The qRT-PCR analysis was carried out using a QuantiFast Probe RT-PCR kit (Invitrogen, CA), where 50 ng of total RNA was added per 10 μl reaction vial with RT mix, RT-PCR master mix, sequence-specific primers and Taqman probes. The sequences for all target gene primers and probes were purchased commercially from Applied Biosystems (Carlsbad, CA). GAPDH was used as the internal control. The qRT-PCR assays were carried out in triplicate on Eppendorf’s mastercycler Realplex real time PCR system. The cycling conditions were 10 min for cDNA formation by reverse transcriptase enzyme at 50 °C and 5 min for polymerase activation at 95 °C, followed by 40 cycles at 95 °C for 30 s, at 55 °C for 30 s and 72 °C for 1 min. The threshold was set above the non-template control background and within the linear phase of target gene amplification to calculate the cycle number at which the transcript was detected.
2.7. Quantification analysis
Fluorescent intensities of DAPI stained nuclei were quantified using ImageJ™ software (www.NIH.gov). Data represented the mean and standard deviation values of 60–100 stained nuclei from 5 independent estimations.
2.8. Statistical analyses
Unless otherwise indicated, all experiments were run at least 3 times. One way analysis of variance followed by Tukey’s test was used to establish significance at an alpha level of 0.05.
2.8.1. Results and discussion
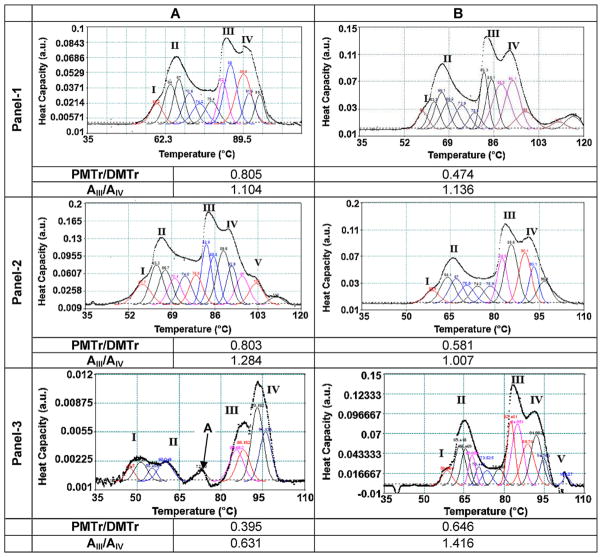
The effect of ultrasound stimulation on the in situ dynamics of chromatin folding/unfolding in fibroblasts subjected to US was assessed by DSC. Representative DSC profiles are shown in Fig. 1 Panels 1, 2, and 3 correspond to the DSC profiles obtained from the nuclei isolated from control cells, cells stimulated via regimen-US-1D and regimen-US-3D, respectively. As observed in Fig. 1, all of the DSC profiles obtained showed four major transitions. An additional transition at 103 °C, transition V, was also detected in some DSC profiles.
Fig. 1.
Differential scanning calorimetric scans. Cells were exposed to US (5.0 MHz, 14 kPa, 51-s/application, 3-applications/day) and upon completion of the US exposure, the nuclei were isolated from the cells and subjected to DSC. Panel-1, panel-2 and panel-3 correspond to control (no-US), US-1-day and US-3-days, respectively. “A” denotes nuclei that were analyzed immediately with minimal time lag and “B” denotes nuclei that were assayed upon incubation at 4 °C for 45-min. The experiments were repeated at least 3 times. A representative scan is shown; the deconvoluted peaks are shown as colored lines. The peak areas were computed and the ratios calculated and shown. PMTr/DMTr: ratio of area under protein melting transition peaks to area under DNA melting transition peaks. AIII/AIV: ratio of area under DSC transition peak III to DSC transition peak IV. (For interpretation of references to color in this figure legend, the reader is referred to the web version of this article.)
Previous research has enabled the identification and assignment of attributes to these transitions, which are summarized in Table 1. In our study, transitions I and II were noted at 58 °C and 66 °C, respectively. Similar to values reported elsewhere, transition III was located at 84 °C, although Touchette et al. (Touchette and Cole, 1992) reported two transitions at 76 °C and 89 °C, respectively. In this study, transition IV was noted at 91 °C, or about 6 °C lower than the values reported by Vergarini et al. (2004) and about 2–4 °C higher than the values reported elsewhere (Touchette and Cole, 1992, Lepock et al., 2001; Giartosio et al., 1992). In select DSC profiles, an additional thermal transition V was also noted at 103 °C. In previous studies, transition V (107 °C) has been attributed to unstacking of bases in topologically constrained DNA (Almagor and Cole, 1989), and to interactions of core particles within condensed domains (Russo et al., 1995). When treated with high salt concentrations, the endotherm at 107 °C was observed to convert into a transition at 90 °C. Another key observation in the DSC profile obtained from the nuclei subjected to regimen-US-3D that was analyzed immediately was the presence of a peak at 72.9 °C (peak A). This peak was not noticeable in control cells or in scans obtained for incubations for 40 min.
Table 1.
Peak nomenclatures and thermal transitions.
| Transition | (T °C)* | Assignment | Reference |
|---|---|---|---|
| I (0) | 58 (57) | Melting of nuclear matrix, lamins and intermediate filaments | Lepock( 2001) and Balbi(1989) |
| II (I) | 66 (64) | Conformation change of histone complement and denaturation nuclear-RNA associated proteins | Cavazza(1991), Balbi (1989) |
| III (II) | 84 (84) | Melting of nucleosome organized into 10nm filaments | |
| IV (III) | 91 (97) | Organization of the nucleosome into higher order structures (~30 nm fibers), unstacking of base pairs in unconstrained DNA, compaction of DNA | Cavazza(1991), Balbi (1989) |
| V (IV) | 103 (107) | Unstacking of bases in topographically constrained DNA, interaction of the core particle with condensed chromatin structures, reorganization of histone(H1) chromatin complex | Almagor and Cole (1989) |
Black Roman Numerals refer to the nomenclature adopted in this paper and thermal transition temperatures noted in DSC scans presented in Fig. 1. Number in red parentheses refer to the nomenclature adopted by Vergarini et al. (2004) and thermal transition temperatures noted by Vergarini et al. (2004).
As a next step, the peaks were deconvoluted and the areas under the peaks were computed and compared between different study groups. The values (arbitrary units, a.u.) of areas under the peaks and their ratios were considered representative of the amount of material being degraded under each transition. As listed in Table 1, peaks I and II were relatable to the protein component of the DNA-binding proteins and thus were grouped together (Protein Melting transitions, PMTr). As shown in Fig. 1, peaks III to V were attributable to DNA/chromatin melting and thus were grouped together (DNA Melting transitions, DMTr).
In the control cells, the PMTr/DMTr ratio decreased from 0.805 to 0.474 for cells that were analyzed with minimum time delay compared to cells that were analyzed after an incubation of 40 min. A similar trend in the PMTr/DMTr ratio was noted for US stimulated cells via regimen-US-1D, where the ratio decreased from 0.803 to 0.581. However, for the cells stimulated via regimen-US-3D, the PMTr/DMTr ratio increased from 0.395 to 0.646. Considering that the nuclei were isolated at the same time and incubated for identical time periods, these comparisons were only possible between samples at the time points indicated.
In both control and samples treated via regimen-US-1D, the peak area under transition III was higher when compared to the peak area under transition IV. This observation is in agreement with results reported elsewhere (Vergarini et al., 2004). Further, this feature was similar in the samples analyzed after 40 min of incubation. However, in the cells treated via regimen-US-3D and analyzed immediately, the area under transition peak III was lower when compared to the peak area under transition IV. In order to make a comparative analysis, the ratio of the area under peak III to the area under peak IV (AIII/AIV) was calculated. Fig. 1 illustrates that despite the differences in absolute area values, the AIII/AIV ratio showed a trend, i.e. an AIII/AIV ratio between 1.104 and 1.136 was noted for the control cells immediately analyzed and analyzed after a time delay, respectively. An AIII/AIV ratio of 1.284 was noted for the cells stimulated via regimen-US-1D and analyzed immediately whereas a ratio of 1.007 was noted for the cells stimulated via regimen-US-1D and analyzed after the time delay. The cells stimulated under regimen-US-3D showed an AIII/AIV ratio of 0.631 and 1.416 analyzed immediately or after 40-min of incubation, respectively. For the control cells and cells stimulated via regimen-US-1D, as per the peak assignment adopted in Table 1, the area corresponding to the unfolding of nucleosomes organized in 10 nm filaments (i.e. peak III) was higher than the area corresponding to the unfolding of nucleosomes organized into higher order structures (peak IV) under all conditions studied. For the cells stimulated via regimen-US-3D, the area corresponding to the unfolding of nucleosomes organized into higher order structures (i.e. peak IV) was higher for the cells analyzed immediately after US application. Further, when assayed after 40 min of incubation, the ratio is similar to that obtained with the control cells and cells stimulated with regimen-US-1D. This last observation indicates that cells stimulated via regimen-US-3D possess more chromatin organized in higher order structures than control cells.
Elsewhere it has been shown that the accessibility of DAPI for DNA increases when histone (H1) is loosely associated with chromatin, thus increasing the availability of DAPI binding sites (Rundquist and Lindner, 2006; Loborg and Rundquist, 2000). Thus, the fluorescence intensity of DAPI (fluorescent DNA stain) was used to correlate the change in chromatin conformation upon US stimulation. Therefore, the nuclei from the control cells and the stimulated cells were stained with DAPI and the fluorescent images were processed with ImageJ™ software (Fig. 2). A decrease in DAPI intensity was noted in the cells that were subjected to ultrasound stimulation via regimen A-US-1D when compared to the control cells. DAPI/H1 staining was not performed since the transition peak V was not noted in DSC scans obtained from cells stimulated via regimen-US-3D. The finding that a decrease in intensity of the nuclei obtained from the cells subjected to US differed statistically different (p < 0.01, n = 60) when compared to the nuclei obtained from non-stimulated cells, suggests that H1 is more firmly associated with the chromatin of cells subjected to regimen-US-1D. These results strongly support and correlate with the appearance of peak V in the DSC profiles of the nuclei obtained from US stimulated cells under regimen-US-1D.
Fig. 2.
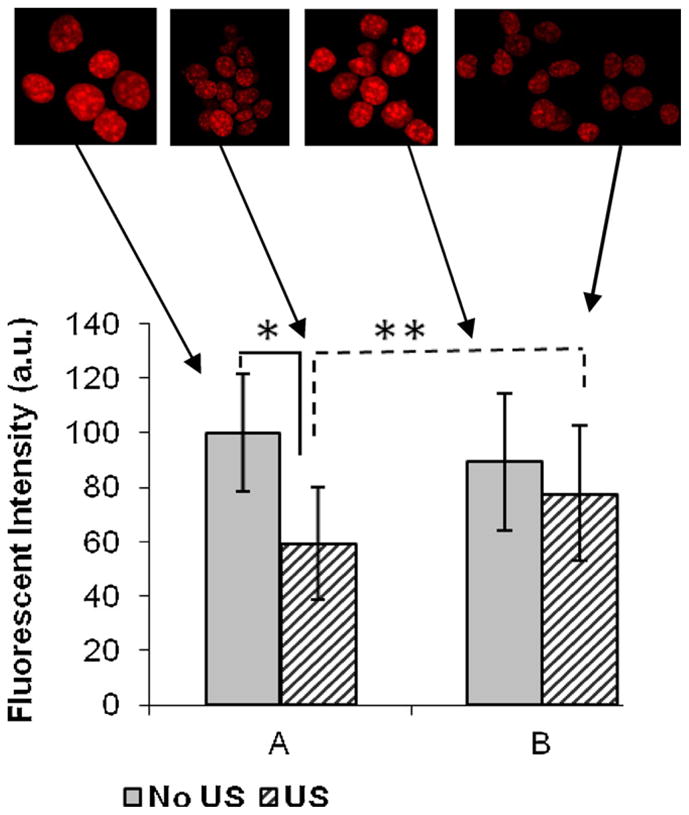
Quantitative evaluation of the relative fluorescent intensities of DAPI stained nuclei. The fluorescent intensity DAPI stained nuclei were imaged using ImageJ™. “A” denotes nuclei that were analyzed immediately with minimal time lag and “B” denotes nuclei that were assayed upon incubation at 4 °C for 45-min. The decrease in intensity in US nuclei (*) is statistically significant (p < 0.01, n = 60) compared to control (non-stimulated) cells. The increase in DAPI intensity after 40 min at 4 °C is statistically different from the initial intensity (**p < 0.001, n = 100).
Changes in cell shape induced by variations in substrate morphology are reflected in changes in the nuclear architecture (Vergarini et al., 2004). Based on our analyses, we noted that the nuclear architecture was also impacted by US as determined by the DSC profiles of the nuclei taken immediately and after an incubation of 40 min, respectively, upon US application. A distinct change in shape in the thermal denaturation profiles of the nuclei obtained from cells exposed to US was observed as compared to the nuclei obtained from the control cells. Further, there were distinct differences in the DSC profiles of the nuclei between cells that were stimulated via regimen-US-1D or regimen-US-3D and analyzed immediately.
We noted several insights. First, the AIII/AIV ratio reversed between these regimen-US-1D and regimen-US-3D. The data in this study alludes to large scale chromatin organization in cells subjected to US via regimen-US-3D. Second, the presence of a transient transition peak at 72.9 °C in the DSC scans of cells obtained after stimulation via regimen-US-3D was also noted. In a previous study (Touchette and Cole, 1992), two different thermal transitions at 76 °C and 89 °C were noted when cells were isolated in presence of NaCl, and when an NaCl-free buffer was employed in the nuclei-isolation protocols, two transition peaks at 76 °C and 89 °C fused to result in a singular peak under 85 °C. Interestingly, the nuclei isolation protocol only used KCl, MgCl2 and CaC12, yet still showed the appearance of a peak that could be assigned to the presence of Na+ ion. This introduces the possibility that, however transiently, ultrasound alters the balance of ions into the nucleus by allowing more Na+ ions from the cytoplasm upon the application of US stimulation. As we have noted, an influx of ions may lead to a greater interaction of ions with the chromatin structure and change the peak appearances. However, these observed changes were more pronounced for the nuclei that were extracted from cells subjected to US stimulation via regimen-US-3D when compared to regimen-US-1D.
Analysis of the confocal images upon DAPI staining and the DSC profiles of the nuclei suggests that histone-1 (H1) is closely associated to the chromatin of cells that have been subject to ultrasound via regimen-US-1D. This observation, along with the presence of a transition peak at 107 °C–an indicator of H1-DNA association in DSC scans of nuclei analyzed after US exposure via regimen-US-1D – points to the possibility that US at low intensities can have a transient effect on chromatin organization. At the same time, it raises the issues of whether this temporal reorganization is enough to trigger different signaling pathways that induce up-regulation of chondrogenic markers and transcription factors. Interestingly, our published data provides evidence that a US stimulation regimen similar to that used in this study induced a marked increase in the expression of chondrogenic markers, including type-II collagen, aggrecan and transcription factor, Sox-9 (Hasanova et al., 2011). We have also shown that US stimulation of chondrocytes induced phosphorylation of focal adhesion kinase (FAK), Src, p130Crk-associated substrate (p130Cas), Crk-II and extracellular-regulated kinase (ERK), which implicates these intracellular signaling molecules in a US-mediated signaling pathway (Whitney et al., 2012). To correlate the application of US and expression of early transcription factors that are reported to be rapidly and transiently induced by mechanical stimulus, the level of expression of the early response genes c-jun, c-fos and c-myc were assayed via qRT-PCR as shown in Fig. 3. An elevated transient expression of early response genes was noted and the levels of gene expression were observed to return to control levels when assayed 6 h after US exposure (data not shown). In summary, it appears plausible that US can produce a transient remodeling of chromatin, and thus trigger a signal cascade and other intracellular mechanisms.
Fig. 3.
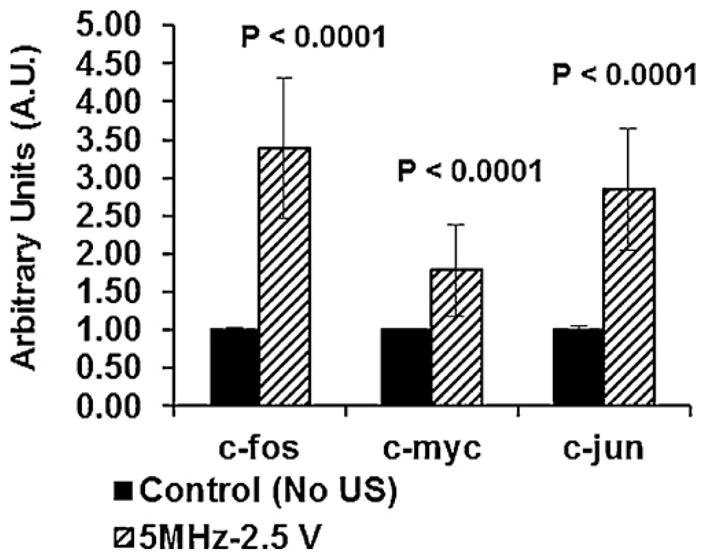
Effect of US stimulation (5.0 MHz, 14 kPa, 51-s/application, 3-applications/day) on the level of expression of early response genes. Cells were stimulated with US and mRNA was isolated at 2 h after the last US exposure. The levels of expression of c-jun, c-fos and c-myc genes were determined by qRT-PCR. Non-stimulated cells served as controls. One way analysis of variance followed by Tukey’s test was used to establish significance at an alpha level of 0.05.
While this study uses US stimulation at 5.0 MHz (14 kPa), our future efforts will focus on the evaluation of different frequencies of US and chromatin remodeling. We suggest that more work is needed to fully evaluate the role of the elevated expression of early response genes at varying US parameters. These combined studies will be crucial in helping to better explain the mechanisms of US action on cells. At issue is whether the change in the transient organization of chromatin, as noted in ratio of AIII/AIV, can possibly facilitate the transport of nuclear or transcriptional factors. Our future studies will also evaluate a heat shock factor (HSF-1) mechanism activated under ultrasound stimulation is a possible mechanism for cells to adapt to the stimulus.
References
- Almagor M, Cole D. Changes in chromatin structure during the aging of cell cultures as revealed by differential scanning calorimetry. Biochemistry. 1989;28:5688–93. doi: 10.1021/bi00439a052. [DOI] [PubMed] [Google Scholar]
- Angele P, Yoo JU, Smith C, Mansour J, Jepsen KJ, Nerlich M, et al. Cyclic hydrostatic pressure enhances the chondrogenic phenotype of human mesenchymal progenitor cells differentiated in vitro. Journal of Orthopaedic Research. 2003;21:451–7. doi: 10.1016/S0736-0266(02)00230-9. [DOI] [PubMed] [Google Scholar]
- Bonassar LJ, Grodzinsky AJ, Frank EH, Davila SG, Bhaktav NR, Trippel SB. The effect of dynamic compression on the response of articular cartilage to insulin-like growth factor I. Journal of Orthopaedic Research. 2001;19:11–7. doi: 10.1016/S0736-0266(00)00004-8. [DOI] [PubMed] [Google Scholar]
- Bougault C, Paumier A, Aubert-Foucher E, Mallein-Gerin F. Molecular analysis of chondrocytes cultured in agarose in response to dynamic compression. BMC Biotechnology. 2008:8. doi: 10.1186/1472-6750-8-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann MD, Gluzband YD, Grodzinsky AJ, Hunzinker EB. Mechanical compression modulates matrix biosynthesis in chondrocyte/agarose culture. Journal of Cell Science. 1995;108:1497–508. doi: 10.1242/jcs.108.4.1497. [DOI] [PubMed] [Google Scholar]
- Carver SE, Heath CA. Semi-continuous perfusion system for delivering intemittent physiological pressure to regenarating cartilage. Tissue Engineering. 1999;5:1–11. doi: 10.1089/ten.1999.5.1. [DOI] [PubMed] [Google Scholar]
- Chen BM, Grinnell AD. Integrins and modualtion of transmitter realize from motor nerve. Science. 1995;269:1578–80. doi: 10.1126/science.7667637. [DOI] [PubMed] [Google Scholar]
- Dalby MJ, Riehle MO, Sutherland DS, Agheli H, Curtis ASG. Fibroblast response to a controlled nanoenvironment produced by colloidal lithography. Journal of Biomedical Materials Research. 2004;69A:314–22. doi: 10.1002/jbm.a.20138. [DOI] [PubMed] [Google Scholar]
- Fioravanti A, Moretti E, Scapigliati G, Cervone R, Galeazi M, Collodel G. Morphological, immunocytochemical and biochemical studies in human osteroarthritic chondrocytes exposed to IL-1β; and cyclical hydrostatic pressure. Clinical and Experimental Rheumatology. 2007:25. [PubMed] [Google Scholar]
- Geiger B, Bershadsky A, Pankov R, Yamada KM. Transmembrane extracellular matrix-cytoskelton crosstalk. Nature Reviews Molecular Cell Biology. 2001;2:793–805. doi: 10.1038/35099066. [DOI] [PubMed] [Google Scholar]
- Giartosio A, Wang C, D‘Alessio S, Ferraro A, Altieri F, Eufemi M, et al. Differential scanning calorimetry of chicken erythrocyte nuclei. European Journal of Biochemistry. 1992;208:17–22. doi: 10.1111/j.1432-1033.1992.tb17154.x. [DOI] [PubMed] [Google Scholar]
- Hasanova GI, Noriega SE, Mamedov TG, Thakurta SG, Turner JA, Subramanian A. The effect of ultrasound stimulation on the gene and protein expression of chondrocytes seeded in chitosan scaffolds. Journal of Tissue Engineering and Regenerative Medicine. 2011 doi: 10.1002/term.384. [DOI] [PubMed] [Google Scholar]
- Ingber DE. Tensegrity-based mechanosensing from macro to micro. Progress in Biophysiscs and Molecular Biology. 2008;97:163–79. doi: 10.1016/j.pbiomolbio.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB Journal. 2006;20:811–27. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- Ingber DE, Madri JA, Folkman J. Endothelial growth factors and extracellular matrix regulate DNA synthesis through modulation of cell and nuclear expansion. In Vitro Cellular and Developmental Biology. 1987;23:387–94. doi: 10.1007/BF02620997. [DOI] [PubMed] [Google Scholar]
- Knight MM, Toyoda T, Lee DA, Bader DL. Mechanical compression and hydrostatic pressure induce reversible changes in actin cytoskeletal organisation in chondrocytes in agarose. Journal of Biomechanics. 2006;39:1547–51. doi: 10.1016/j.jbiomech.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Choi BH, Min B-H, Son YS, Park SR. Low-intensity ultrasound stimulation enhances chondrogenic differentiation in alginate culture of mesenchymal stem cells. Artificial Organs. 2006;30:707. doi: 10.1111/j.1525-1594.2006.00288.x. [DOI] [PubMed] [Google Scholar]
- Lepock JR, Frey JE, Heynen ML, Senisterra GA, Warters RL. The nuclear matrix is a thermolabile cellular structure. Cell Stress & Chaperones. 2001;6:136–47. doi: 10.1379/1466-1268(2001)006<0136:tnmiat>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loborg H, Rundquist I. Affinity of linker histones for chromatin in situ analyzed using DAPI as a cytochemical probe. Cytometry. 2000;40:1–9. [PubMed] [Google Scholar]
- Nicolini C, Trefiletti V, Cavazza B, Cuniberti C, Patrone E, Carlo P, et al. Quaternary and quinternary structures of native chromatin DNA in liver nuclei: differential scanning calorimetry. Science. 1983;219:176–8. doi: 10.1126/science.6849127. [DOI] [PubMed] [Google Scholar]
- Nishikori T, Ochi M, Uchio Y, Maniwa S, Kataoka H, Kawasaki K, et al. Effects of low-intensity pulsed ultrasound on proliferation and chondroitin sulfate synthesis of cultured chondrocytes embedded in Atelocollagen gel. Journal of Biomedical Materials Research. 2002;59:201–6. doi: 10.1002/jbm.1226. [DOI] [PubMed] [Google Scholar]
- Noriega SE. PhD dissertation. Chemical and Biomolecular Engineering. Lincoln: Universtiy of Nebraska Lincoln; 2009. Role of scaffold topography and stimulation via ultrasound on the biosynthetic activity of chondrocytes seeded in 3D matrices; p. 328. [Google Scholar]
- Noriega SE, Mammedov T, Turner JA, Subramanian A. Intermittent applications of continuous ultrasound on the viability, proliferation, morphology and matrix production of chondrocytes in 3D matrices. Tissue Engineering. 2007;13:611–8. doi: 10.1089/ten.2006.0130. [DOI] [PubMed] [Google Scholar]
- Parvizi J, Wu C, Lewallwen DG, Greenleaf JF, Bolander ME. Low intensity ultrasound stimulates proteoglycan synthesis in rat chondrocytes by increasing aggrecan gene expression. Journal of Orthopaedic Research. 1999;17:487–94. doi: 10.1002/jor.1100170405. [DOI] [PubMed] [Google Scholar]
- Rundquist I, Lindner H. Analyses of linker histone–chromatin interactions in situ. Biochemistry and Cell Biology. 2006;84:427–36. doi: 10.1139/o06-071. [DOI] [PubMed] [Google Scholar]
- Russo I, Barboro P, Alberti I, Parodi S, Balbi C. Role of H1 in chromatin folding. A thermodynamic study of chromatin reconstitution by differential scanning calorimetry. Biochemistry. 1995;34:301–11. doi: 10.1021/bi00001a037. [DOI] [PubMed] [Google Scholar]
- Sah RL, Kim YJ, Doong JH, Grodzinsky AJ, Plaas AH, Sandy JD. Biosynthetic response of cartilage explants to dynamic compression. Journal of Orthopaedic Research. 1989;7:619–36. doi: 10.1002/jor.1100070502. [DOI] [PubMed] [Google Scholar]
- Touchette NA, Cole D. Effect of salt concentration and H1 histone removal on the differential scanning calorimetry of nuclei. Biochemistry. 1992;31:1842–9. doi: 10.1021/bi00121a037. [DOI] [PubMed] [Google Scholar]
- Vergarini L, Grattarola M, Nicolini C. Modifications of chromatin structure and gene expression following induced alterations of cellular shape. The International Journal of Biochemistry & Cell Biology. 2004;36:1447–61. doi: 10.1016/j.biocel.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Whitney NP, Lamb A, Subramanian A. Integrin-mediated mechanotransduction pathway of low-intensity continuous ultrasound in human chondrocytes. Ultrasound in Medicine and Biology. 2012 doi: 10.1016/j.ultrasmedbio.2012.06.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Huckle J, Francomano CA, Spencer RGS. The effects of pulsed low-intensity ultrasound on chondrocyte viability, proliferation, gene expression and matrix production. Ultrasound in Medicine and Biology. 2003;29:1645–51. doi: 10.1016/j.ultrasmedbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Zhou S, Schmelz A, Seufferlein T, Li Y, Zhao J, Bachem MG. Molecular mechanisms of low intensity pulsed ultrasound in human skin fibroblasts. The Journal of Biological Chemistry. 2004;279:54563–54469. doi: 10.1074/jbc.M404786200. [DOI] [PubMed] [Google Scholar]