Abstract
Vaccines against emerging pathogens such as the 2009 H1N1 pandemic virus can benefit from current technologies such as rapid genomic sequencing to construct the most biologically relevant vaccine. A novel platform (Ad5 [E1-, E2b-]) has been utilized to induce immune responses to various antigenic targets. We employed this vector platform to express hemagglutinin (HA) and neuraminidase (NA) genes from 2009 H1N1 pandemic viruses. Inserts were consensuses sequences designed from viral isolate sequences and the vaccine was rapidly constructed and produced. Vaccination induced H1N1 immune responses in mice, which afforded protection from lethal virus challenge. In ferrets, vaccination protected from disease development and significantly reduced viral titers in nasal washes. H1N1 cell mediated immunity as well as antibody induction correlated with the prevention of disease symptoms and reduction of virus replication. The Ad5 [E1-, E2b-] should be evaluated for the rapid development of effective vaccines against infectious diseases.
Keywords: Viral shedding; Influenza; H1N1 vaccine; Adenovirus vector; Ad5 [E1-, E2b-]; Hemagglutinin; Neuraminidase; Cell mediated immunity; Pandemic; Horizontal transmission
1. Article
Emerging infectious diseases, pandemics, and bioterrorism have the potential for catastrophic impact. Healthcare workers have faced chronic challenges in producing countermeasures against such threats, as exemplified during the recent H1N1 pandemic. The new H1N1 virus was identified in Mexico in April 2009 but sufficient vaccine for widespread immunization was not available until late fall that year, at which time the virus had already spread globally. In order to react to such threats we must have the capability to produce medical countermeasures rapidly in the presence of emerging pathogens, whether known or unknown, novel or reemerging, natural or intentional.
The 2009 H1N1 pandemic virus represents the greatest incidence of human infection with a swine originated influenza to date with laboratory-confirmed cases in over 200 countries and more than 15,000 deaths [1]. The advancement of efficient PCR and sequencing techniques has allowed agencies such as the National Institutes of Heath (NIH), Centers for Disease Control (CDC) and the World Health Organization (WHO) to rapidly identify and characterize newly emerging pathogens. Isolation and sequence analysis of the 2009 H1N1 viruses determined that the novel pathogen was the product of multiple-rounds of re-assortment of three influenza virus stains which included portions of avian, swine and human influenza viruses [2]. Genetic information on such pathogens is made public to researchers though sequence repositories such as GenBank and the NCBI Influenza Virus Sequence Database. Although sequence data of emerging pathogens is now being made available, this information is not often employed in current vaccine development.
During the H1N1 pandemic in 2009 the first line treatment options were antivirals including Oseltamivir (Tamiflu™) and Andzanamivir and included archaic preventative methods such as school-closures, limitation of travel and quarantine [3,4]. Sequence data from patients infected with pandemic 2009 H1N1 virus reveled that the virus developed resistance to Oseltamivir. The first Oseltamivir-resistant strain was identified four months after the first appearance of the virus and by May 2010 there were over fifty resistant 2009 H1N1 strains [5,6]. The developed drug resistance and the capability to recombine with other influenza strains demonstrates the potential of the pathogen to rapidly mutate which left available treatments for infected individuals ineffective [5,6].
Vaccines remain the most effective counter measure to protect immunologic naïve populations from emerging pathogens. The vaccine development response to 2009 H1N1 was slow and plagued with technical problems. Traditional influenza vaccines are grown in chicken-egg-based culture and in response to the 2009 influenza pandemic, this timely manufacturing process was further hindered by suboptimal-yields of H1N1 vaccine [7]. By the time 2009 H1N1 vaccine was available in October of 2009 the virus had already spread globally. Even once the vaccine was available there were persistent shortages followed by an over abundance due to the lag in response time [8]. Utilizing the limited number of eggs available to generate vaccines against pandemic viruses such as the 2009 H1N1 also caused shortages of seasonal flu vaccine supply by limiting manufacturing capabilities [9]. All of these issues outline the importance of developing new technology strategies that can respond quickly and produce adequate amounts of effective countermeasures against emerging pathogens.
We have developed a vaccine platform that can be used to rapidly produce vaccines against multiple agents at high numbers of doses in a minimal time frame. The platform is a novel recombinant adenovirus serotype 5 (Ad5) delivery platform with unique deletions in the early 1 (E1), early 2 (E2b) and early 3 (E3) regions (Ad5 [E1-, E2b-]) [10–12]. Antigenic gene targets are cloned into the vector, which once administered in vivo infect antigen-presenting cells that express the inserted antigen gene and induce immune responses to the pathogenic target. The platform induces both antibodies and cell-mediated immunity (CMI) [13–17]. We have utilized this platform to produce vaccines against viral antigens such as HIV-1 as well as immunotherapeutics against antigenic cancer targets such as carcinoembryonic antigen and human epidermal growth factor receptor 2 (HER2/neu) [13–17]. The Ad5 [E1-, E2b-]-CEA immunotherapeutic is now undergoing clinical evaluation for the treatment of CEA expressing malignancies including colon and breast cancers.
In the study described herein, we constructed two Ad5 [E1-, E2b-] vectors designed to express the 2009 H1N1 hemagglutinin (HA) and neuraminidase (NA) proteins. The HA and NA genes which were inserted in the Ad5 [E1-, E2b-] vector were created by generating consensus sequences off of real-time sequence data from the then circulating 2009 H1N1 sequences originating in North and South America. This sequence data was provided and made public by the NCBI Influenza Virus Sequence Database. The H1N1 specific immune responses, protection from H1N1 challenge and inhibition of virus shedding was induced by vaccinations with Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA in murine and ferret models.
2. Results
2.1. Induction of immune responses against Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA
The Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA virus particles were constructed as previously described [10–12]. It was approximately 6 weeks from gene insert design to production of purified vaccines. Confirmation of HA and NA gene expression was confirmed by Western blot analysis prior to immunization (data not shown). A study was performed in mice to determine the immunologic effect of immunizations with increasing doses of Ad5 [E1-, E2b-]-HA, Ad5 [E1-, E2b-]-NA or both combined. BALB/c mice (n = 5/group) were immunized three times at a weekly interval with 108, 109 or 1010 VP of Ad5 [E1-, E2b-]-HA, Ad5 [E1-, E2b-]-NA or Ad5 [E1-, E2b-]-HA + NA or buffer alone (controls). Fourteen days after the final immunization, splenocytes from the mice were assessed for HA and NA specific IFN-γ and IL-2 secretion by ELISpot analysis, which was performed as previously described [13–16]. Splenocytes were stimulated with HA or NA peptides (NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH). The HA 94-peptide array spans the hemagglutinin protein of the A/New Caledonia/20/99 strain of H1N1 (GenPept ABF21272). The NA 78-peptide array spans the NA of the A/New Caledonia/20/99 strain of H1N1 (GenPept CAD57252).
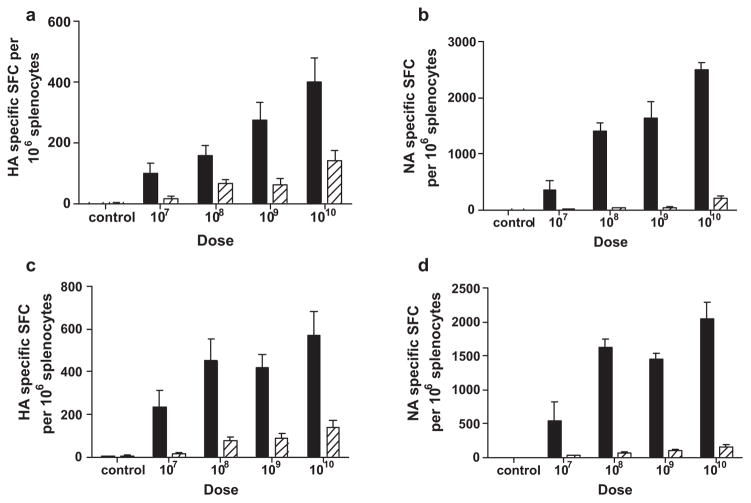
A CMI response was induced to HA and NA in mice which received Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA, respectively. A dose response effect to the vaccine was observed and the greatest induction of CMI correlated to the highest dose of 1010 VP of Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA (Fig. 1). Mice immunized with both constructs simultaneously had H1N1 specific CMI responses to both HA and NA. Based on these results, the dose of 1010 VP was chosen for further evaluation. To assess the induction of CD4 T cell responses, splenocytes from mice immunized three times with 1010 VP of Ad5 [E1-, E2b-]-HA or Ad5 [E1-, E2b-]-HA + NA were isolated and CD4+ cells were assessed for IL-2 production after exposure to HA or NA peptide pools using flow cytometry. CD4+ T cells from mice immunized with Ad5 [E1-, E2b-]-HA or Ad5 [E1-, E2b-]-HA/NA exhibited 1.62 ± 0.47% or 0.87 ± 0.44% reactivity, respectively, following exposure to an HA peptide pool whereas unstimulated control splenocytes exhibited 0.5% reactivity. CD4 T cells from mice immunized with Ad5 [E1-, E2b-]-HA or Ad5 [E1-, E2b-]-HA/NA exhibited 0.65 ± 0.5% or 2.45 ± 1% reactivity, respectively, upon exposure to an NA peptide pool whereas unstimulated control splenocytes exhibited 0.5% reactivity. These results indicate that specific CD4+ T cell responses were induced during immunizations with Ad5 [E1-, E2b-]-HA or Ad5 [E1-, E2b-]HA/NA.
Fig. 1.
CMI dose response titration of Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA. Ad5 naïve BALB/c mice (n = 5/group) were immunized three times every two weeks using a dose of 108, 109 or 1010 VP Ad5 [E1-, E2b-]-HA (a), Ad5 [E1-, E2b-]-NA (b) or Ad5 [E1-, E2b-]-HA + NA (c and d) or buffer alone (controls). Fourteen days after the final immunization, splenocytes from the mice were assessed by ELISpot analysis for IFN-γ (black bars) or IL-2 (striped bars) secretion following antigen peptide stimulation. The greatest induction of CMI was achieved using 1010 VP of the vector. For positive controls, splenocytes were pulsed with concanavalin A in all ELISpot assays (data not shown). The error bars depict the standard error of the mean (SEM).
2.2. Immunized mice were protected from lethal H1N1 challenge
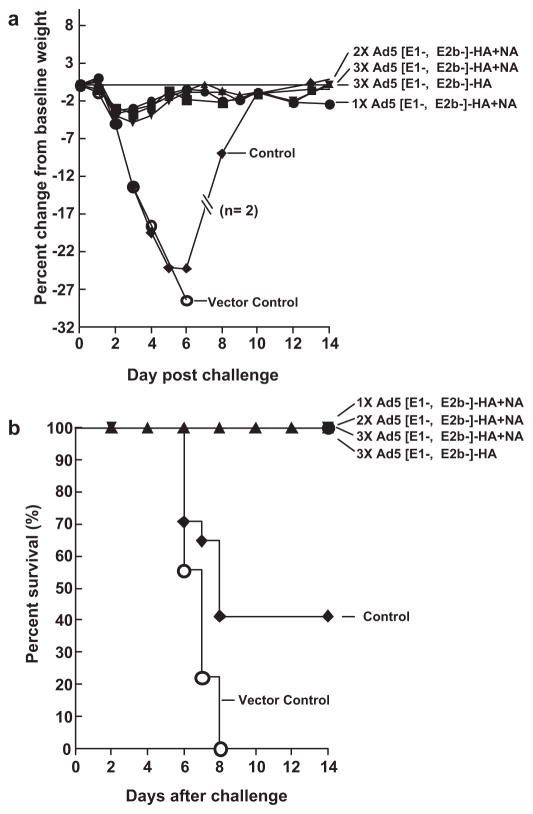
To determine the efficacy of immunization with Ad5 [E1-, E2b-]HA and Ad5 [E1-, E2b-]-NA to protect from challenge with virulent H1N1 virus, groups of mice (n = 15) were immunized subcutaneously one, two or three times at two week intervals with 1010 VP of Ad5 [E1-, E2b-]-HA + NA (2 × 1010 VP total). To determine the effect of Ad5 [E1-, E2b-]-HA immunization alone, a fourth group of mice were immunized three times with 1010 VP of the vaccine. A vector control group consisted of mice (n = 15) immunized twice at a two-week interval with 1010 VP of Ad5-null, an Ad5 vector with no inserted transgene. This construct was used to control against any non-specific responses induced by the Ad5 viral backbone. Non-immunized mice were used as additional experimental controls (n = 10). Forty-six days after the last vaccination, mice were inoculated intranasally with 105 TCID50/animal of Influenza H1N1 A/CA/07/2009 (equivalent to 100 LD50). Mice were observed daily for clinical signs of disease including weight loss, core temperature fluctuation or death over 21 days following viral infection.
Mice immunized 1, 2 or 3 times with Ad5 [E1-, E2b-]-HA + NA or Ad5 [E1-, E2b-]-HA alone exhibited nonsignificant, transient weight loss (Fig. 2A) and no detectable clinical signs of disease such change in temperature (data not shown). One hundred percent of immunized mice survived H1N1 challenge and their survival curves were significantly different as assessed by the log rank (Mantel–Cox) test when compared with vector control (p < 0.01; log rank test) and experimental control mice (p < 0.01; log rank test). All animals in the vector control and non-immunized control groups became moribund and experienced significant weight loss by day three post-H1N1 challenge. One hundred percent of mice in the vector control group died by day 8 post-H1N1 infection and 59% of control mice died 8–10 days post-challenge (Fig. 2B). The variation in survival between the two control groups was within the normal range. The induction of H1N1 specific antibody was assessed using a hemagglutination inhibition assay (HAI) as previously described [18]. Mice immunized with Ad5 [E1-, E2b-]-HA + NA or Ad5 [E1-, E2b-]-HA developed HA neutralizing antibody responses following vaccination which were further boosted after H1N1 challenge (Table 1).
Fig. 2.
Immune protection from lethal H1N1 challenge in mice. Mice (n = 15) were immunized with Ad5 [E1-, E2b-]-HA + NA (2 × 1010 VP total) once (●), twice (■) or three times (▲) at a two week interval. A fourth group of mice were immunized three times at a two week interval with 1010 VP of Ad5 [E1-, E2b-]-HA alone (▼). Mice immunized with Ad5-null, an Ad5 vector with no transgene insert, were used as a vector control group (○) and non-immunized mice were used as experimental controls (◆). Forty-six days after the last vaccination, mice were inoculated intranasally with 105 PFU/animal of Influenza H1N1 A/CA/07/2009 and weight loss (a) and survival (b) were recorded and evaluated. p < 0.01; log rank test between vaccinated and vector control mice survival and p < 0.01; log rank test, between vaccinated and experimental control mice survival by Mantel–Cox analysis.
Table 1.
Viral load in the lung of mice after challenge with H1N1 virus and H1N1 virus specific humoral response pre- and post challenge.
| Group | Viral titer in lung (log 10 TCID50 /g)a
|
HAI
|
||
|---|---|---|---|---|
| Day +3 | Day +6 | Day −3 | Day +21 | |
| 1X-Ad5 [E1-, E2b-]-HA+NA | <LDb | <LD | 1:640 | 1:2560 |
| 2X-Ad5 [E1-, E2b-]-HA+NA | <LD | <LD | 1:1280 | >1:5120 |
| 3X-Ad5 [E1-, E2b-]-HA+NA | <LD | <LD | 1:1280 | >1:5120 |
| 3X-Ad5 [E1-, E2b-]-HA | <LD | <LD | 1:896 | 1:1706 |
| Vector Control | 5.3;5.3;5.5 | 4.5; 5.5; 3.6 | <1:40 | NA |
| Experimental Control | 6.3; 5.7; 4.7 | 4.5; 5.0; 3.5 | <1:40 | 1:1280 |
Challenge virus was Influenza H1N1 A/CA/07/2009.
LD, limit of detection.
Three randomly pre-selected animals from each group were terminated on day 3 and day 6 post challenge and the H1N1 viral titers in the lungs of mice were determined as previously described [18]. Immunized mice had no detectable H1N1 virus in their lungs but mice in both control groups had high-levels of H1N1 virus present (Table 1). Histological evaluation of the lungs of H1N1 challenged mice 6 days post challenge revealed extensive hemorrhagic pulmonary inflammation in both control groups (Fig. 3). The lungs from control mice displayed dense granulocytic and lymphocytic cell infiltrates in the interstitum, around vessels and airways with focally denuded lamina propria due to epithelial necrosis and desquamation. In contrast, the lungs of all vaccinated mice were normal without the presence of histopathological changes associated with viral infection (Fig. 3).
Fig. 3.
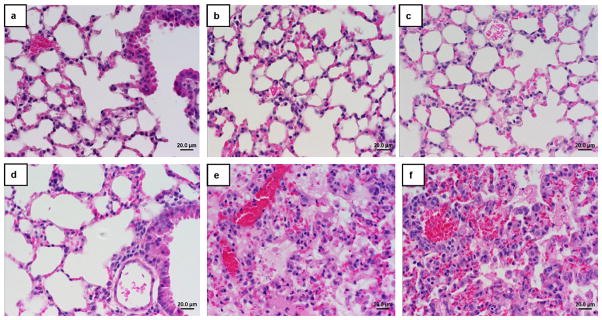
Photomicrographs of the lung sections of H1N1 challenged mice. Lung sections from mice at 6 days post challenge were hematoxylin and eosin stained. Lungs from mice immunized one (a), two (b) or three times (c) with Ad5 [E1-, E2b-]-HA + NA (2 × 1010 VP total), three times with Ad5 [E1-, E2b-]-HA alone (d) exhibited no signs of H1N1 infection and the airway epithelium remained intact (original magnification 40×). Lung samples from vector control mice (e) and non-immunized mice (f) had dense granulocytic and lymphocytic cell infiltrates.
2.3. Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA immunization protected ferrets from H1N1 challenge
Ferrets are widely used as an animal model to predict vaccine outcome or antiviral efficacy in humans [18,19]. To investigate the effectiveness of immunization with Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA to protect from infection with H1N1 virus in ferrets, groups of three to four month old ferrets were immunized three times at two-week intervals with 1010 VP of Ad5 [E1-, E2b-]-HA + NA (2 × 1010 VP total) (n = 6) or 1010 VP of Ad5 [E1-, E2b-]-HA (n = 5). An experimental control group consisted of ferrets (n = 4), which were injected three times at two-week intervals with normal saline. Vaccine-induced HAI titers were determined 27 days after the final immunization. Vaccinated ferrets had robust levels of H1N1 specific antibodies present with HAI titers averaging 1:160–1:640 in Ad5 [E1-, E2b-]-HA + NA immunized ferrets and 1:320–1:1280 in Ad5 [E1-, E2b-]-HA immunized ferrets (Table 3). Thirty days after the final vaccination, all ferrets were inoculated intranasally with 106 TCID50/animal of Influenza H1N1 A/CA/07/2009.
Table 3.
Hemagglutinin inhibition (HAI) titers in ferrets pre-challenge and viral load in nasal washes of ferrets 4 days post challenge with H1N1.
| Group | HAI
|
Viral load in nasal discharge (TCID50 /ml)a | |
|---|---|---|---|
| GMT | Range | ||
| Ad5 [E1-, E2b-]-HA+NA | 452.5 | 1:160–1:640 | <LDb |
| Ad5 [E1-, E2b-]-HA | 640 | 1:320–1:1280 | <LD |
| Saline | <1:40 | NA | 5.7;5.0;4.5;4.7 |
Challenge virus was Influenza H1N1 A/CA/07/2009.
LD, limit of detection.
Ferrets were observed for disease development for 21 days following challenge. The animals in the control group experienced significant weight loss, averaging −6.61% change from baseline at day seven. Ferrets immunized with Ad5 [E1-, E2b-]-HA + NA experienced no weight loss following challenge and continually gained weight during the course of the study. Ferrets in the control group weighted significantly less than Ad5 [E1-, E2b-]-HA + NA vaccinated mice on study day 5 (p = 0.017), day 7 (p < 0.01), day 10 (p = 0.015) and day 13 (p = 0.013). Ferrets immunized with Ad5 [E1-, E2b-]-HA alone experienced only mild and transient weight loss. Ferrets in the control group weighed significantly less than Ad5 [E1-, E2b-]-NA on study day 7 (p = 0.041). All ferrets in the control group presented clinical signs of disease including nasal discharge, ocular discharge and sneezing beginning on day 3 post challenge (Table 2). The vaccinated ferrets immunized with Ad5 [E1-, E2b-]-HA + NA or Ad5 [E1-, E2b-]-HA remained healthy with no clinical signs of H1N1 infection with the exception of one ferret from each group that experienced short-lived, mild nasal discharge. The H1N1 virus challenge was not lethal in any of the groups of ferrets.
Table 2.
Clinical symptoms of disease in ferrets following H1N1 challenge. Number of ferrets with clinical symptomsa/total number of ferrets.
| Day post challenge | Saline | Ad5 [E1-, E2b-]-HA + NA | Ad5 [E1-, E2b-]-HA |
|---|---|---|---|
| 0 | 0/4 | 0/6 | 0/5 |
| 1 | 0/4 | 0/6 | 0/5 |
| 2 | 4/4 | 0/6 | 1/5 |
| 3 | 2/4 | 0/6 | 1/5 |
| 4 | 4/4 | 1/6 | 1/5 |
| 5 | 4/4 | 0/6 | 0/5 |
| 6 | 4/4 | 0/6 | 0/5 |
| 7 | 4/4 | 0/6 | 0/5 |
| 8 | 4/4 | 0/6 | 0/5 |
| 9 | 2/4 | 0/6 | 0/5 |
| 10 | 1/4 | 0/6 | 0/5 |
| 11 | 1/4 | 0/6 | 0/5 |
| 12 | 0/4 | 0/6 | 0/5 |
Clinical symptoms of disease include nasal discharge, fatigue, ocular discharge and/or sneezing.
Four days following challenge, nasal washes were performed on the ferrets and the H1N1 vial load was determined as previously described [18]. Nasal washes from vaccinated ferrets contained no detectable infectious H1N1 virus as opposed to control ferrets, which had high titers of H1N1 virus present in their nasal washes (Table 2). Vaccination with Ad5 [E1-, E2b-]-HA + NA or Ad5 [E1-, E2b-]-HA alone may prevent viral shedding in this animal model of human infection.
3. Discussion
The studies reported herein demonstrate the potential utility of the Ad5 [E1-, E2b-] platform for vaccine development to protect against emerging pathogenic threats. The Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA vaccines were designed and constructed rapidly from the then currently circulating 2009 H1N1 viral isolates which may have provided for the most biologically relevant vaccine to the pandemic virus. The vaccine was produced and constructed in less than six-weeks after insert sequence identification. This rapid production process could serve as a great advantage over traditional, egg-based manufacturing processes to quickly produce vaccines to emerging pathogens.
T cell immunity directed against influenza viral antigens have been reported to provide protection in animal models as well as in humans [20,24–27]. CMI responses play a major role in the control of viral infection and viral clearance through either direct lysis of infected cells and/or secretion of antiviral cytokines and chemokines [20–23]. To develop complete immunologic memory against influenza, especially heterosubtypic immunity, it has been reported that the vaccination must induce both B-cell and CD4 immunity [28]. Traditional subunit influenza vaccines only induce neutralizing antibodies targeting external viral coat proteins. Vaccination with Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA induced robust CMI and humoral immune responses to H1N1, which translated into significant protection from disease development and death following H1N1 challenge in mice. Vaccinated ferrets also displayed significant protection from H1N1 challenge. They had minimal or no clinical symptoms of H1N1 infection and nasal washes revealed a complete blockade of the virus production in the upper respiratory tract. Reducing the clinical symptoms of influenza such as coughing and sneezing as well as potentially blocking H1N1 virus shedding can greatly reduce horizontal transmission, an important aspect in containing pandemic infectious diseases.
Although the protective effects of immunization against H1N1 was observed following immunization with Ad5 [E1-, E2b-]-HA alone, the most effective vaccination strategy in both the mouse and ferret models was immunizing against both the HA and NA targets. Immunity to NA reduced clinical symptoms of disease in both mice and ferrets. This finding is in agreement with other reports that vaccination against both targets results in a broader protective immunity against influenza infection than vaccination against HA alone [29,30].
The Ad5 [E1-, E2b-] is a platform technology that can be utilized to produce vaccines against numerous pathogens that pose serious threats such as pandemic viruses but also potential bioterrorism agents. Antigenic gene targets to NIH Category A bioterrorism agents such as the filoviruses Ebola and Marburg or the arenaviruse Lassa have been identifeid and could be inserted into the viral backbone to produce a vaccine such as we have done here [31–33]. The antigen insert could also be re-designed and a vaccine rapidly produced if the pathogen is modified intentially or through natural adaptive processes.
The advancement of molecular technologies, such as rapid sequencing techniques and recombinant viral vectors, should be employed in the development of vaccines against novel and emerging pathogens. Historically, the response rate to pathogen outbreaks or pandemic pathogen emergence has been slow and vaccine development has lagged, resulting in major negative public heath and economic cost worldwide [34]. New technologies that can be employed in the rapid development of effective vaccines against pathogenic threats, such as the Ad5 [E1-, E2b-] platform, should be explored and exploited. The Ad5 [E1-, E2b-] antigen delivery platform and the manufacturing human E.C7 cell line has been successfully used to manufacture human grade vaccine material which has been approved by the FDA for clinical use and is now being administered to humans [35]. In future studies, we will continue to develop this platform for a H1N1 vaccine as well as insert antigenic targets from other influenza strains to induce broad influenza immunity.
4. Methods
4.1. Influenza virus
Influenza A/CA/07/2009 (H1N1) virus was obtained from the U.S. Centers for Disease Control and Prevention. The viral stock was grown in 10-day-old chicken eggs, harvested, and frozen at −80 °C in 1 ml portions that contain approximately 1 × 107 TCID50/ml.
4.2. Gene synthesis and Ad5 vectors construction
Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA was constructed and produced as previously described [13–16] using 1743 bp HA and 1421 bp NA consensus gene sequences generated from HA and NA viral coat proteins of pandemic 2009 H1N1 strains from North and South America.
4.3. Animal studies
Six to eight-week-old female BALB/c mice (Charles River, Wilmington, MA) were housed in animal facilities at the Infectious Disease Research Institute (IDRI) (Seattle, Washington) or at the University of Texas, Medical Branch (Galveston, TX). Three to four month old ferrets (Triple F Farms, Sayre, PA) were housed at the University of Texas, Medical Branch. All procedures were conducted according to Institutional Animal Care and Usage Committee (IACUC) approved protocols. Animals were given food and water ad libitum.
4.4. Western blot analysis
Western blot analysis to determine HA and NA expression by the Ad5 [E1-, E2b-]-HA and Ad5 [E1-, E2b-]-NA vectors was performed as previously described [13]. The probing antibody used was mouse anti-HA or anti-NA antibody (1:250) (provided by NIH Biodefense and Emerging Infections (BEI) Research Resources Repository, NIAID, NIH). Western blotting analysis system (GE Healthcare, Piscataway, NJ) was run according to the manufacturer’s specifications.
4.5. Hemagglutination inhibition (HAI) testing
A HAI assay was performed as previously described [18]. HI antibodies to the circulating strains of influenza A were measured by a standard method [36]. The HI titers of ferret serum samples are reported as the reciprocal of the highest dilution at which hemagglutination was inhibited.
4.6. Enzyme-linked immunospot (ELISpot) assay for IFN-γ and IL-2 secreting cells
A previously described ELISpot assay [13–16] was used to detect H1N1 HA and NA specific IFN-γ and IL-2–producing T cells from mouse splenocytes stimulated with HA or NA peptides. Cells stimulated with concanavalin A (ConA) at a concentration of 0.0625 μg/well served as positive controls. Colored spot-forming cells (SFC) were counted using an Immunospot ELISpot plate reader (Cellular Technology, Shaker Heights, OH) and responses were considered to be positive if, 1) 50 SFC were detected/106 cells after subtraction of the negative control and, 2) SFC were ≥2-fold than those in the negative control wells.
4.7. Flow cytometry
To assess CD4 T cell responses, CD4+ cells in splenocytes were assayed for IL-2 production employing flow cytometry methods previously described [37,38]. Splenocytes from immunized and control mice (n = 5/group) were harvested and incubated 5 h with 0.5 μg/ml H1N1-HA peptide, in the presence of GolgiStop, a protein transport inhibitor. Cells were then fixed, permeabilized, stained for IL-2, and analyzed by flow cytometry.
4.8. Median tissue culture infectious dose (TCID50) assay
Serial ten-fold dilutions of the virus stock or of a 10% tissue homogenate was prepared in MEM without serum. MDCK cells were grown to confluency in 96-well tissue culture plates, washed twice with 100 μl of DPBS, followed by inoculation of 100 μl of each virus dilution of virus into four replicate wells, or, as negative control, DPBS. Plates were incubated for 90 min at 37 °C, 5% CO2, after which an additional 100 μl of MEM was added to each well. Plates were incubated for 4 days at 37 °C, 5% CO2. HA assay [36] was performed by removing 50 μL of supernatant from each well and transferring it to a 96-well plate, followed by addition of 50 μL per well of a 0.5% solution of whole chicken erythrocytes (Charles River) suspended in DPBS w/Ca+ and Mg2+. Erythrocytes were allowed to settle and hemagglutination was documented for each replicate. Virus concentration of stocks for infection was determined as TCID50 per ml. For organ titrations, infectious virus titers were expressed as median tissue culture infectious (TCID50) dose per gram (g) of tissue [36]
4.9. Statistical analysis
Statistically significant differences in the mean immune responses between groups of animals were determined by Student’s t-test with a p-value of ≤0.05 being considered significant (GraphPad Prism® Software, Inc.). Survival curve analysis was performed using the log rank (Mantel–Cox) test (GraphPad Prism® Software, Inc.).
Acknowledgments
The authors want to thank Ms. Carol Jones for her contribution and management of the study and Ms. Stephanie Balcaitis for excellent technical assistance. This study was funded in part by NIH-NIAID grant 2R44AI1071733 awarded to FRJ and Etubics Corporation.
Footnotes
Competing interests: FRJ, ESG, YX, SB are employees of Etubics Corporation, JPB is a consultant to the Company. All of the UTMB authors do not have any financial interest in Etubics Corporation and are employees of UTMB.
References
- 1.Taubenberger JK, Morens DM. Influenza: the once and future pandemic. Public Health Rep. 2010;3:16–26. [PMC free article] [PubMed] [Google Scholar]
- 2.Lu J, Liu D, Lui W, Shi T, Tong Y, Cao W. Genetic stability and linkage analysis of the 2009 influenza A(H1N1) virus based on sequence homology. Arch Virol. 2009;154:1883–90. doi: 10.1007/s00705-009-0526-2. [DOI] [PubMed] [Google Scholar]
- 3.Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) viruses circulating in humans. Science. 2009;325:197–201. doi: 10.1126/science.1176225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uyeki TM. 2009 H1N1 virus transmission and outbreaks. N Engl J Med. 2010;362:2221–3. doi: 10.1056/NEJMe1004468. [DOI] [PubMed] [Google Scholar]
- 5.Janies DA, Voronkin IO, Studer J, Hardman J, Alexandrov BB, Treseder TW, et al. Selection for resistance to oseltamivir in seasonal and pandemic H1N1 influenza and widespread co-circulation of the lineages. Int J Health Geogr. 2010;9:13. doi: 10.1186/1476-072X-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.http://www.cdc.gov/flu/weekly/fluactivity.htm.
- 7.http://www.npr.org/blogs/health/2009/07/manufacturing_problems_with_sw.html.
- 8.Fitzpatrick M. Excess H1N1 vaccine could be sold, kept in reserve. The Vancouver Sun. 2010 Jan10 [Google Scholar]
- 9.http://www.medscape.com/viewarticle/702229.
- 10.Amalfitano A, Begy CR, Chamberlain JS. Improved adenovirus packaging cell lines to support the growth of replication-defective gene-delivery vectors. Proc Natl Acad Sci USA. 1996;93:3352–6. doi: 10.1073/pnas.93.8.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodges BL, Serra D, Hu H, Begy CA, Chamberlain JS, Amalfitano A. Multiply deleted [E1, polymerase-, and pTP-] adenovirus vector persists despite deletion of the preterminal protein. J Gene Med. 2000;2:250–9. doi: 10.1002/1521-2254(200007/08)2:4<250::AID-JGM113>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Amalfitano A, Hauser MA, Hu H, Serra D, Begy CR, Chamberlain JS. Production and characterization of improved adenovirus vectors with the E1, E2b, and E3 genes deleted. J Virol. 1998;72:926–33. doi: 10.1128/jvi.72.2.926-933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabitzsch ES, Xu Y, Yoshida LH, Balint J, Gayle RB, Amalfitano A, et al. A preliminary and comparative evaluation of a novel Ad5 [E1-, E2b-] recombinant based vaccine used to induce cell mediated immune responses. Immunol Lett. 2009;122:44–51. doi: 10.1016/j.imlet.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gabitzsch ES, Xu Y, Balint JP, Jr, Hartman ZC, Lyerly HK, Jones FR. Anti-tumor immunotherapy despite immunity to adenovirus using a novel adenoviral vector Ad5 [E1-, E2b-]-CEA. Cancer Immunol Immunother. 2010;59:1131–5. doi: 10.1007/s00262-010-0847-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabitzsch ES, Xu Y, Yoshida LH, Balint J, Amalfitano A, Jones FR. Novel adenovirus type 5 vaccine platform induces cellular immunity against HIV-1 Gag, Pol, Nef despite the presence of Ad5 immunity. Vaccine. 2009;27:6394–8. doi: 10.1016/j.vaccine.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gabitzsch ES, Xu Y, Balcaitis S, Balint JP, Jones FJ. An Ad5 [E1-, E2b-]-HER2/neu vector induces immune responses and inhibits HER2/neu expressing tumor progression in Ad5 immune mice. Cancer Gene Ther. 2011 Jan;(14) doi: 10.1038/cgt.2010.82. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osada T, Yang XY, Hartman ZC, Glass O, Hodges BL, Niedzwiecki D, et al. Optimization of vaccine responses with an E1, E2b and E3-deleted Ad5 vector circumvents pre-existing anti-vector immunity. Cancer Gene Ther. 2009;16:673–82. doi: 10.1038/cgt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun NE, Linde NS, Zacks MA, Barr IG, Hurt AC, Smith JN, et al. Injectable peramivir mitigates disease and promotes survival in ferrets and mice infected with the highly virulent influenza virus, A/Vietnam/1203/04 (H5N1) Virology. 2008;374:198–209. doi: 10.1016/j.virol.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Laan JW, Herberts C, Lambkin-Williams R, Boyers A, Mann AJ, Oxford J. Animal models in influenza vaccine testing. Expert Rev Vaccines. 2008;7:783–93. doi: 10.1586/14760584.7.6.783. [DOI] [PubMed] [Google Scholar]
- 20.Yap KL, Ada GL, McKenzie IF. Transfer of specific cytotoxic T lymphocytes protects mice inoculated with influenza virus. Nature. 1978;273:238–9. doi: 10.1038/273238a0. [DOI] [PubMed] [Google Scholar]
- 21.Bender BS, Rowe CA, Taylor SF, Wyatt LS, Moss B, Small PA., Jr Oral immunization with a replication-deficient recombinant vaccinia virus protects mice against influenza. J Virol. 1996;70:6418–24. doi: 10.1128/jvi.70.9.6418-6424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 23.Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, et al. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J Exp Med. 1999;189:991–8. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epstein SL, Kong WP, Misplon JA, Lo CY, Tumpey TM, Xu L, et al. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine. 2005;23:5404–10. doi: 10.1016/j.vaccine.2005.04.047. [DOI] [PubMed] [Google Scholar]
- 25.Lalvani A, Brookes R, Hambleton S, Britton WJ, Hill AV, McMichael AJ. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–65. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambhara S, Woods S, Arpino R, Kurichh A, Tamane A, Underdown B, et al. Heterotypic protection against influenza by immunostimulating complexes is associated with the induction of cross-reactive cytotoxic T lymphocytes. J Infect Dis. 1998;177:1266–74. doi: 10.1086/515285. [DOI] [PubMed] [Google Scholar]
- 27.Sambhara S, Kurichh A, Miranda R, Tumpey T, Rowe T, Renshaw M, et al. Heterosubtypic immunity against human influenza A viruses, including recently emerged avian H5 and H9 viruses, induced by FLU-ISCOM vaccine in mice requires both cytotoxic T-lymphocyte and macrophage function. Cell Immunol. 2001;211:143–53. doi: 10.1006/cimm.2001.1835. [DOI] [PubMed] [Google Scholar]
- 28.Epstein SL, Lo CY, Misplon JA, Lawson CM, Hendrickson BA, Max EE, et al. Mechanisms of heterosubtypic immunity to lethal influenza A virus infection in fully immunocompetent, T cell-depleted, beta2-microglobulin-defiecnt, and J chain deficient-mice. J Immunol. 1997;158:1222–30. [PubMed] [Google Scholar]
- 29.Fiers W, Neirynck S, Deroo T, Saelens X, Jou WM. Soluble recombinant influenza vaccines. Philos Trans R Soc Lond B Biol Sci. 2001;356:1961–3. doi: 10.1098/rstb.2001.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F, Yang W, Fang F, Chang H, Yu P, Chen Z. Essential sequence of influenza B virus hemagglutinin DNA to provide protection against lethal homologous viral infection. DNA Cell Bio. 2008;27:377–85. doi: 10.1089/dna.2007.0706. [DOI] [PubMed] [Google Scholar]
- 31.Ledgerwood JE, Costner P, Desai N, Holman L, Enama ME, Yamshchikov G, et al. A replication defective recombinant Ad5 vaccine expressing Ebola virus GP is safe and immunogenic in healthy adults. Vaccine. 2010;29:304–13. doi: 10.1016/j.vaccine.2010.10.037. [DOI] [PubMed] [Google Scholar]
- 32.Geisbert TW, Baily M, Geisbert JB, Asiedu C, Roederer M, Grazia-Pau M. Vector choice determines immunogenicity and potency of genetic vaccines against Angola Marburg virus in nonhuman primates. J Virol. 2010;84:10386–94. doi: 10.1128/JVI.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Branco LM, Grove JN, Geske FJ, Boisen ML, Muncy IJ, Magliato SA, et al. Lassa virus-like particles displaying all major immunological determinants as a vaccine candidate for Lassa hemorrhagic fever. Virol J. 2010;7:279. doi: 10.1186/1743-422X-7-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKibbin W, Sidorenko D. Global macroeconmic consequences of pandemic influenza. Presentations at ACHERH Forum; 2006; 2006. [Google Scholar]
- 35.http://www.clinicaltrials.gov/ct2/results?term=Etubics.
- 36.WHO. WHO Manual on Animal Influenza Diagnosis and Surveillance. 2002. [Google Scholar]
- 37.Hobeika AC, Clay TM, Mosca PJ, Lyerly HK, Morse MA. Quantitating therapeutically relevant T-cell responses to cancer vaccines. Crit Rev Immunol. 2001;21:287–97. [PubMed] [Google Scholar]
- 38.Hobeika AC, Morse MA, Osada T, Ghanayem M, Niedzwiecki D, Barrier R, et al. Enumerating antigen-specific T-cell responses in peripheral blood: a comparison of peptide MHC Tetramer, ELISpot, and intracellular cytokine analysis. J Immunother. 2005;28:63–72. doi: 10.1097/00002371-200501000-00008. [DOI] [PubMed] [Google Scholar]