Abstract
Imaging in vivo dynamics of cellular behavior throughout a developmental sequence can be a powerful technique for understanding the mechanics of tissue patterning. During animal development, key cell proliferation and patterning events occur very quickly. For instance, in Caenorhabditis elegans all cell divisions required for the larval body plan are completed within six hours after fertilization, with seven mitotic cycles1; the sixteen or more mitoses of Drosophila embryogenesis occur in less than 24 hr2. In contrast, cell divisions during plant development are slow, typically on the order of a day 3,4,5 . This imposes a unique challenge and a need for long-term live imaging for documenting dynamic behaviors of cell division and differentiation events during plant organogenesis. Arabidopsis epidermis is an excellent model system for investigating signaling, cell fate, and development in plants. In the cotyledon, this tissue consists of air- and water-resistant pavement cells interspersed with evenly distributed stomata, valves that open and close to control gas exchange and water loss. Proper spacing of these stomata is critical to their function, and their development follows a sequence of asymmetric division and cell differentiation steps to produce the organized epidermis (Fig. 1).
This protocol allows observation of cells and proteins in the epidermis over several days of development. This time frame enables precise documentation of stem-cell divisions and differentiation of epidermal cells, including stomata and epidermal pavement cells. Fluorescent proteins can be fused to proteins of interest to assess their dynamics during cell division and differentiation processes. This technique allows us to understand the localization of a novel protein, POLAR6, during the proliferation stage of stomatal-lineage cells in the Arabidopsis cotyledon epidermis, where it is expressed in cells preceding asymmetric division events and moves to a characteristic area of the cell cortex shortly before division occurs. Images can be registered and streamlined video easily produced using public domain software to visualize dynamic protein localization and cell types as they change over time.
Keywords: Plant Biology, Issue 70, Molecular Biology, Developmental Biology, Cellular Biology, Botany, plant, live imaging, epidermis, stomata, confocal, time lapse, Arabidopsis, cotyledon
Protocol
1. Seed Sterilization
Prepare seed sterilization solution: 33% household bleach, 0.1% Triton X-100.
Place seeds carrying desired fluorescent reporter construct(s) and genotype(s) in 1.7 ml tube and apply 1 ml of sterilization solution. Incubate on nutator for 15 min.
In a sterile hood, use a pipettor to remove sterilization solution from tube, leaving seeds behind. Rinse with 1 ml sterile water. Repeat four times.
Incubate at 4 °C for two days or more.
2. Preparation of Chamber Slide Media
Mix 20 ml of 0.5% solution of Bacto Agar (not pure agarose) in water in a 200 ml flask, and microwave until dissolved. Be cautious when boiling the solution.
Cool the solution to around 60 °C.
Collect 1 ml of agar solution with a pipettor and slowly apply to the cover glass inside the chamber slide (Lab-Tek II Chambered #1.5 German Coverglass System). Solution which is too hot or applied too quickly may melt glue or crack glass.
Immediately add a second ml of agar solution. Agar media should now completely cover the bottom of the slide chamber. This minimal media is sufficient to support development in a plant cotyledon, which contains stored nutrients, for four to five days by maintaining moisture and allowing gas diffusion.
Cool slide on benchtop with chamber covered. If flask is covered tightly, solution may be saved and reheated for future use.
3. Seed Dissection
Remove tube of sterile seed from 4 °C storage.
Place a paper tissue on the stage of a dissecting microscope and moisten with water. Adjust tissue to create a smooth surface. Tissue is used to stabilize seeds for dissection.
Pipet 20-30 seeds from the tube to the tissue, being careful to place them in the dissecting scope's field of view.
Using sharp forceps (Roboz, #5 biology tip), carefully remove the seed coat from the seedling inside. Both outer and inner integuments must be removed.
When imaging the cotyledon, it is beneficial to remove the hypocotyl and radicle. Cotyledons contain sufficient nutrients to develop for several days under this protocol. If intact, the hypocotyl will straighten and lengthen dramatically, moving the cotyledon out of the time lapse field of view. Use a scalpel to slice the cotyledons free of the hypocotyl.
Hold dissected cotyledons immersed in water, in a microtube or similar.
Repeat 3.4-3.6 until the desired number of cotyledons is reached. 15-20 successful dissections are recommended before mounting.
4. Mounting Cotyledons in the Chamber Slide
Using a small (18 mm2) cover slip, cut through the agar media and lift the entire layer up from the chamber slide's glass base.
Pipet dissected cotyledons with a minimum of water from holding tube into the chamber slide, under the lifted agar layer.
Gently lower the agar onto the cotyledons. If excess water is present, soak it away with a tissue, being careful not to disturb the cotyledons. Specimens should no longer move easily when mount is complete.
Place cover on chamber slide and move slide to microscope stage.
5. Time Lapse Imaging
Set up inverted confocal microscope to image the desired fluorophore(s). LSM700 parameters used: for GFP, excitation at 488 nm and collection with a band pass filter at 440-530 nm; for RFP, excitation at 555 nm and collection with a band pass filter at 570-610 nm.
Inspect mounted specimens for damage. Program the locations of intact, properly mounted cotyledons into microscope software. In Zen 2009: Focus on a cotyledon with 20x objective and move objective to center of chamber slide. Under Acquisition Mode, change Zoom to 0.5 and Frame Size to 48x48 pixels. Select Positions checkbox and Scan overview image... button under Positions, then increase Horizontal and Vertical tile numbers to 30-35. When scan is complete, click the Positions button under the Dimensions tab and place crosshairs at each cotyledon to observe.
Set up z series encompassing the cotyledon epidermis of all samples.In Zen 2009: Click the Z-Stack checkbox. Select each position under Position List and click the Move to button. Focus on the uppermost plane of the cotyledon epidermis and click the Set First button under Z-Stack. Focus to the lowest position at which epidermis is usefully visible and click the Set Last button. Click the button next to Optimal, which shows the best z-slice thickness, to set. Check each cotyledon to confirm that these settings encompass all samples; the z parameters cannot be set for individual positions in Zen 2009.
Set up time series at desired resolution and length. Intervals of 15-30 min for three days are effective for protein dynamics in epidermal cell division. This interval can be changed as necessary. In Zen 2009: Click the Time Series checkbox. Under Time Series, set Cycles for the number of images desired using the slider or typing in the text field. Set Interval to 30 and use the pull-down menu to select min.
Begin xyzt multi-position scan. Note that the number of positions must be limited by the scan specifications; if total scan time is greater than the desired interval, the time points will not be correct. A maximum of six simultaneous positions are possible when imaging GFP and RFP at 59 z-slices in a 30-min interval using our system. In Zen 2009: Change Frame Size to desired resolution and click Start Experiment.
6. Video Editing
From the confocal data file, make a maximum intensity z-projection of each time/position combination and export as image files in a lossless format such as TIFF. File names should be in one series per position (e.g., POLAR-GFP_pos1_0001, POLAR-GFP_pos1_0002, etc., not POLAR-GFP_0001_pos1) for software to combine them into a movie. In Zen 2009: Use Copy:Subset under the Processing tab to split positions into separate files, then Maximum Intensity Projection. Finally, export as TIFF (File:Export, Full resolution image window - series).
Use FIJI7,8 to align successive images by running the bUnwarpJ macro9(Plugins:Registration:bUnwarpJ). Change Registration Mode to Mono. All Advanced Options can use default values. For long time lapse sequences, download and install the macro "Affine + Consistent Elastic 2D Image Registration," which will apply bUnwarpJ to a series of images automatically, and open your data using File:Import:Image Sequence to use it. Uncheck MOPS landmarks extraction and run.
Use FIJI/ImageJ or Quicktime Pro to open the sequence of aligned images and save in the desired video format (AVI is a good choice).
Representative Results
A set of informative time points collected with this method is shown in Figure 3. Cell membranes are labeled with RFP (pm-rb) and GFP is fused to POLAR protein under its native promoter (POLAR::POLAR-GFP)6 At the 30-min time scale, we see cell divisions along with the changes in protein localization preceding them. Asymmetric cell divisions in the stomatal lineage form stem-cell-like stomatal precursors called meristemoids, which retain the ability to undergo further asymmetric divisions, and their sister cells, called stomatal lineage ground cells (SLGCs). SLGCs do not assume a stomatal precursor fate; they frequently transdifferentiate into pavement cells, but may resume asymmetric division capability to produce another meristemoid spaced away from the first as the leaf expands. All of these fates are reflected in the expression and localization of POLAR::POLAR-GFP (Fig. 3, Video 1).
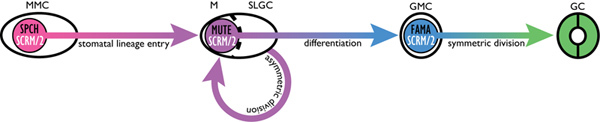 Figure 1. Schematic of stomatal development. Protodermal cells enter the stomatal lineage as meristemoid mother cells (MMC) in a step controlled by the basic helix-loop-helix transcription factors SPEECHLESS (SPCH), SCREAM (SCRM), and SCRM2. MMCs divide asymmetrically to produce a meristemoid (M) and stomatal lineage ground cell (SLGC); this step may occur multiple times and provides one mechanism by which stomata are spaced apart. The cell-state transition of the meristemoid to the guard mother cell (GMC) identity is directed specifically by the bHLH MUTE as well as SCRM/2. A final symmetric division of the GMC and its differentiation into two mature guard cells (GC) requires the bHLH FAMA with SCRM/2 10. Click here to view larger figure.
Figure 1. Schematic of stomatal development. Protodermal cells enter the stomatal lineage as meristemoid mother cells (MMC) in a step controlled by the basic helix-loop-helix transcription factors SPEECHLESS (SPCH), SCREAM (SCRM), and SCRM2. MMCs divide asymmetrically to produce a meristemoid (M) and stomatal lineage ground cell (SLGC); this step may occur multiple times and provides one mechanism by which stomata are spaced apart. The cell-state transition of the meristemoid to the guard mother cell (GMC) identity is directed specifically by the bHLH MUTE as well as SCRM/2. A final symmetric division of the GMC and its differentiation into two mature guard cells (GC) requires the bHLH FAMA with SCRM/2 10. Click here to view larger figure.
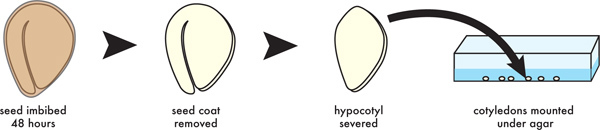 Figure 2. Diagram of seed dissection and mounting procedure. Seeds are sterilized and held at 4 °C, the seed coats and hypocotyls removed, and the cotyledons mounted under agar media in a chamber slide for imaging on an inverted confocal microscope.
Figure 2. Diagram of seed dissection and mounting procedure. Seeds are sterilized and held at 4 °C, the seed coats and hypocotyls removed, and the cotyledons mounted under agar media in a chamber slide for imaging on an inverted confocal microscope.
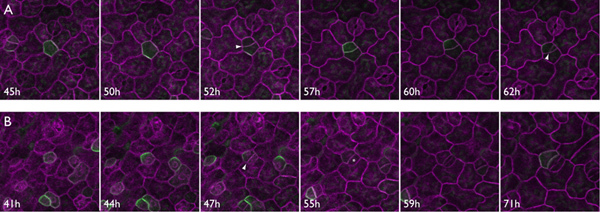 Figure 3. Time-lapse imaging demonstrates POLAR::POLAR-GFP distal localization preceding asymmetric cell division. Series A: POLAR-GFP initially appears evenly distributed, then approximately two hours before asymmetric divisions (arrowheads) POLAR-GFP segregates away from the site of the incipient meristemoid. Series B: Following the initial asymmetric division (arrowhead), POLAR-GFP disappears in both cells, implying rapid transition to guard mother cell (GMC) state by the meristemoid. The GMC (asterisk) divides symmetrically and differentiates to form stomatal guard cells, while the sister cell regains POLAR-GFP expression, presumably presaging a later asymmetric division.
Figure 3. Time-lapse imaging demonstrates POLAR::POLAR-GFP distal localization preceding asymmetric cell division. Series A: POLAR-GFP initially appears evenly distributed, then approximately two hours before asymmetric divisions (arrowheads) POLAR-GFP segregates away from the site of the incipient meristemoid. Series B: Following the initial asymmetric division (arrowhead), POLAR-GFP disappears in both cells, implying rapid transition to guard mother cell (GMC) state by the meristemoid. The GMC (asterisk) divides symmetrically and differentiates to form stomatal guard cells, while the sister cell regains POLAR-GFP expression, presumably presaging a later asymmetric division.
Video 1. Streamlined, registered video of POLAR::POLAR-GFP localization during amplification and spacing of one stomatal precursor cell. Initially, POLAR-GFP appears uniformly in the cell; by 39 hr after germination, it polarizes strongly, just before a division (40.0h) placing a smaller meristemoid at the opposite end of the cell. The larger cell at right also regains stomatal lineage identity, and by 45 hr POLAR-GFP localization moves adjacent to the stomatal precursor. The division at 47 hr produces a new meristemoid (right) oriented away from the existing meristemoid (left) from the first. The end result is two stomata separated by one cell. (Images missing from the video were not collected and do not represent significant changes.) Click here to view movie.
Discussion
This time-lapse confocal technique allows longitudinal studies of fluorescently tagged protein expression and localization in individual cells of the Arabidopsis cotyledon epidermis, which in the case of POLAR and other dynamically changing proteins is critical to a proper understanding of their function. Previously, sustained time lapse imaging has been used to examine Arabidopsis root fungal infection11 and meristem growth5,12, but adding the cotyledon epidermis expands the versatility of this technique and allows its use for additional proteins, particularly assisting the expanding field of stomatal development.
The protocol's main limitation is that it is currently restricted to cotyledons, which contain the nutrients they need for early development. Further, cotyledon development is not exactly identical to that undergone in air because the gel medium restricts gas exchange. Scanning laser exposure and room lights are adequate for photomorphogenesis, inducing cotyledon expansion and chloroplast maturation, but stomata may occasionally develop in adjacent pairs due to low CO2 concentration. This tendency can be reduced by using only a thin layer of media and minimal mounting water as directed in this protocol.
Fluorophore selection is also important for success with this technique. Both GFP and RFP work well, and allow double labeling to visualize the cell periphery or assess protein co-localization. Custom filters greatly reduce autofluorescence and allow sensitive detection of fluorescent protein tags even when chlorophyll is present. However, even filtered CFP autofluorescence in germinating cotyledons is strong enough to preclude visibility of almost all signal using the LSM700. For instruments with a spectral unmixing capability, a wider selection of fluorophores may be feasible.
Disclosures
No conflicts of interest declared.
Acknowledgments
We thank Amanda Rychel for assistance in developing the time lapse protocol and Lynn Pillitteri for constructing POLAR::POLAR-eGFP. We are also grateful to ABRC for providing the pm-rb construct. This protocol was developed through a support from the PRESTO award from Japan Science Technology and Agency. Research on POLAR was also supported by the University of Washington Royalty Research Fund (RRF-4098) and the National Science Foundation (MCB-0855659). KMP is an NSF Graduate Research Fellow (DGE-0718124), and KUT is an HHMI-GBMF investigator.
References
- Sulston JE, Schierenberg E, White JG, Thomson JN. The Embryonic Cell Lineage of the Nematode Caenorhabditis elegans. Developmental Biology. 1983;100:64. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Foe V, Odell G, Edgar BA. Chapter 3 Mitosis and Morphogenesis: Point and Counterpoint. In: Bate M, Martinez Arias A, editors. Development of Drosophila melanogaster. Plainview, NY: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Vincent CA, Carpenter R, Coen ES. Cell Lineage Patterns and Homeotic Gene Activity During Antirrhinum Flower Development. Current Biology. 1995;5:1449. doi: 10.1016/s0960-9822(95)00282-x. [DOI] [PubMed] [Google Scholar]
- Beemster GTS, De Vusser K, De Tavernier E, De Bock K, Inzé D. Variation in Growth Rate between Arabidopsis Ecotypes Is Correlated with Cell Division and A-Type Cyclin-Dependent Kinase Activity. Plant Physiology. 2002;129(2):854. doi: 10.1104/pp.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandjean O, Vernoux T, Laufs P, Belcram K, Mizukami Y, Traas J. In Vivo Analysis of Cell Division, Cell Growth, and Differentiation at the Shoot Apical Meristem in Arabidopsis. Plant Cell. 2004;16(1):74. doi: 10.1105/tpc.017962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillitteri LJ, Peterson KM, Horst RJ, Torii KU. Molecular Profiling of Stomatal Meristemoids Reveals New Component of Asymmetric Cell Division and Commonalities among Stem Cell Populations in Arabidopsis. Plant Cell. 2011;23(9):3260. doi: 10.1105/tpc.111.088583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiji Is Just ImageJ. Fiji; 2008. Available from: http://fiji.sc/wiki/index.php/Fiji. [Google Scholar]
- Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11(7):36. [Google Scholar]
- Arganda-Carreras I, Sorzano Sánchez, Marabini CO, Carazo R, de Solorzano JMOrtiz-, C , Kybic J. Computer Vision Approaches to Medical Image Analysis, ser. Lecture Notes in Computer Science. Vol. 4241. Heidelberg, Berlin: Springer; 2006. Consistent and Elastic Registration of Histological Sections Using Vector-Spline Regularization; pp. 85–95. [Google Scholar]
- Peterson KM, Rychel AL, Torii KU. Out of the Mouths of Plants: The Molecular Basis of the Evolution and Diversity of Stomatal Development. Plant Cell. 2010;22(2):296. doi: 10.1105/tpc.109.072777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czymmek KJ, Fogg M, Powell DH, Sweigard J, Park S-Y, Kang S. In vivo Time-lapse Documentation Using Confocal and Multi-Photon Microscopy Reveals the Mechanisms of Invasion into the Arabidopsis Root Vascular System by Fusarium oxysporum. Fungal Genetics and Biology. 2007;44(10):1011. doi: 10.1016/j.fgb.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Campilho A, Garcia B, Toorn HV, Wijk HV, Campilho A, Scheres B. Time-Lapse Analysis of Stem-cell Divisions in the Arabidopsis thaliana Root Meristem. Plant Journal. 2006;48:619. doi: 10.1111/j.1365-313X.2006.02892.x. [DOI] [PubMed] [Google Scholar]


