Abstract
Rapid detection and differentiation of methicillin-resistant Staphylococcus aureus (MRSA) is critical for the early diagnosis of difficult-to-treat nosocomial and community acquired clinical infections and improved epidemiological surveillance. We developed a microfluidics chip coupled with surface enhanced Raman scattering (SERS) spectroscopy (532 nm) “lab-on-a-chip” system to rapidly detect and differentiate methicillin-sensitive S. aureus (MSSA) and MRSA using clinical isolates from China and the United States. A total of 21 MSSA isolates and 37 MRSA isolates recovered from infected humans were first analyzed by using polymerase chain reaction (PCR) and multilocus sequence typing (MLST). The mecA gene, which refers resistant to methicillin, was detected in all the MRSA isolates and different allelic profiles were identified assigning isolates as either previously identified or novel clones. A total of 17,400 SERS spectra of the 58 S. aureus isolates were collected within 3.5 hours using this optofluidic platform. Intra- and inter-laboratory spectral reproducibility yielded a differentiation index value of 3.43 to 4.06 and demonstrated the feasibility of using this optofluidic system at different laboratories for bacterial identification. A global SERS-based dendrogram model for MRSA and MSSA identification and differentiation to the strain level was established and cross-validated (Simpson index of diversity of 0.989) and had an average recognition rate of 95% for S. aureus isolates associated with a recent outbreak in China. SERS typing correlated well with MLST indicating that it has high sensitivity and selectivity and would be suitable for determining the origin and possible spread of MRSA. A SERS-based partial least-squares regression model could quantify the actual concentration of a specific MRSA isolate in a bacterial mixture at levels from 5 to 100% (regression coefficient, > 0.98; residual prediction deviation, >10.05). This optofluidic platform has advantages over traditional genotyping for ultrafast, automated and reliable detection and epidemiological surveillance of bacterial infections.
Keywords: MRSA, microfluidics, Raman spectroscopy, chemometrics
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a leading pathogen associated with serious hospital and community-acquired disease 1. Early diagnosis of MRSA infection in humans is critical to determine a suitable treatment to reduce morbidity and mortality 2. MRSA can be detected using chromogenic and selective media 3, and various DNA-based genotyping techniques, including commercially available BD GeneOhm and Xpert MRSA assays 4, polymerase chain reaction (PCR) 5, pulsed-field gel electrophoresis (PFGE) 6 and multilocus sequence typing (MLST) 7. However, all of these techniques are time consuming, laborious, require highly trained personnel, dedicated facilities and sometimes provide false-negative or false-positive results 8. Biosensor and bioelectronics techniques offer an alternative to these more traditional detection methods for rapid detection of MRSA 9.
Confocal micro-Raman spectroscopy provides the basis for epidemiological typing method for various pathogenic bacteria, such as Pseudomonas aeruginosa 10, Mycobacterium spp. 11, Campylobacter spp. 12, and S. aureus 13. Conducting SERS to detect bacteria at the single cell level without cultivation shows great potential 14. However, the major disadvantage of SERS to date has been the requirement for metal nanocolloids to serve as the SERS active substrate which, for bacterial detection, often results in poor spectral reproducibility due to variations in the properties of different batches of nanocolloid and phenomenon that bacterial detection is dependent upon nanocolloid concentration and this relationship needs to be empirically established for different analytes 15. Signal enhancement effects can vary within the same batch of nanocolloid because of the nonhomogeneity and unpredictable aggregation properties of the nanoparticles within the colloidal suspension 16. Thus, a high number of spectra for each biological sample are required to confirm the spectral features and subsequently establish and validate reliable chemometric models for prediction of analyte concentration in samples of unknown composition 17.
Optofluidic platforms with SERS detection have been constructed for detection of chemical and biological samples in a well-controlled microenvironment 18. For example, Walter et al. 19 designed a SERS microfluidic system to rapidly discriminate between nine strains of Escherichia coli. This optofluidic system showed substantial advantages compared to traditional SERS detection under a static condition. Spectral measurement taken under controlled flow conditions reduces problems with differing scattering geometry of the nanoparticles, instable mixing times, and localized heating 20. In addition, a microfluidic system with this design could reliably handle small sample volumes and a high sample throughput 21. By creating a segmented flow format using mineral oil as the segregation medium 22, memory effects were reduced 23 as were peak tailing 24 and baseline drift 25.
Here, we report an automated PDMS-based optofluidic platform combining microfluidics and SERS to rapidly detect and investigate the epidemiology of MRSA isolated from infected humans. SERS typing results correlated well with reference genotyping methods, including PCR and MLST. To the best of our knowledge, this is the first study conducting an optofluidic based “lab-on-a-chip” detection system to perform epidemiological monitoring of contagious bacterial infections.
EXPERIMENTAL SECTION
The detailed information about chemicals and reagents, bacterial sample collection and preparation, DNA extraction, detection of mecA by PCR, and MLST was in Supplementary Information section.
Microfluidic chip fabrication
The fabrication process is shown in Fig. S1 and the detailed processing procedures were in Supplementary Information section.
Lab-on-a-chip device for SERS measurements
The microfluidic system with T-junction geometry was designed and fabricated to produce droplets using standard soft lithography technique 26. A microfluidic syringe pump system (NE-1002X, New Era Pump Systems Inc., Farmingdale, NY) was used to inject the reagents and control flow at ultra-low flow rates of less than 0.01 μl/s. The programmed linear gradients were flow-rates of 0.008 μl/s for the mineral oil, 0.006 μl/s for the silver nanocolloid solution, 0.001 μl/s for 0.5 molar sodium chloride solution (aggregation agent), and 0.01 μl/s for the bacterial suspension. The principle of silver nanocolloid preparation is based on the reduction of silver cations and the synthesis procedure was mainly followed by previous method 27. Multiple batches of fresh silver nanocolloids were prepared in each laboratory to validate the SERS-based pattern recognition. Under these conditions, the bacterial suspension formed a series of droplets in the carrier oil in the microchannel. These droplets were then mixed with silver nanocolloid solution as well as the aggregation agent. Chaotic advection inside the droplets enhanced mixing within the designed winding channels 28.
Confocal micro-Raman spectroscopic instrumentation
Two confocal micro-Raman spectroscopic systems were set up at two different laboratories, one in China (Renishaw inVia) and one in the United States (WITec alpha300) as described in detail in our recent publication 12. Both systems have a charge-coupled device (CCD) array detector. A 532-nm double Nd:YAG laser (10 mW, 0.5 s integration time) was introduced through a 100× objective into the microchannel and focused on the sample droplet (Fig. 1) for spectral collection over a simultaneous wavenumber shift range of 3700 to 200 cm−1 in an extended mode. During the ongoing segmented flow of the droplets, the individual Raman spectra of mineral oil and bacterial samples within the mineral oil were collected accordingly. Spectral processing and subtraction could be employed to remove the spectral interference from mineral oil, as described previously 21–22.
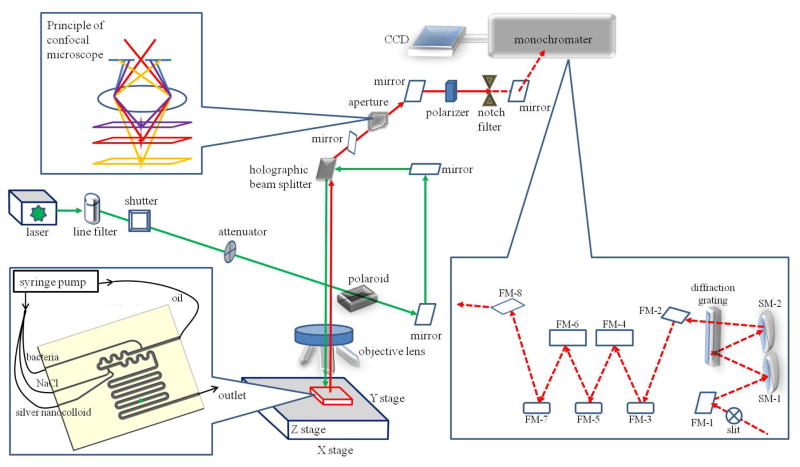
Figure 1.
Scheme of the optofluidic platform including a confocal micro-Raman spectroscopic system coupled with microfluidic “lab-on-a-chip” system. The microfluidic chip was connected with syringe pumps and placed in the measurement stage, where the droplets inside of the microchannel were illuminated with the incident laser (green color) and the scattered light (red color) was separated by the notch filter to remove Rayleigh scattering light and only allow Raman signal (red color dash line) to pass through. The Raman signal was dispersed on a diffraction grating and collected using a charge-coupled device (CCD) detector. The principle of the confocal technique was demonstrated that only the scattering light derived from the focal point could pass through the aperture.
The detailed information about spectral processing, discriminatory power calculation and chemometric analysis was in Supplementary Information section.
RESULTS
Optofluidic system
We fabricated the microfluidic chip using PDMS (Fig. S1) and detailed processing steps can be found in Experimental Section. A detailed scheme of the optofluidic platform including a confocal micro-Raman spectroscopic system coupled with microfluidic “lab-on-a-chip” system is shown in Fig. 1 and the configuration of the real platform is shown in Fig. S2a. Mineral oil was used as a carrier fluid in the microchannel. Droplets of bacteria suspension were generated using a T-junction structure, as shown in Fig. S2b. Depending on the fluid properties and flow rates, droplets can form due to three distinct mechanisms: squeezing, dripping and jetting. In our case, the capillary number Ca, which measures the ratio of viscous force and surface tension force, is estimated to be 10−3. It has been reported32 that the transition from squeezing to dripping at a T-junction happens at Ca = 10−2. Therefore, in our case, the droplets were generated via the squeezing mechanism, meaning that surface tension forces dominated shear stresses and the size of the droplets was controlled by the ratio of the flow rates of the two fluids only. Based on the equations proposed by Garstecki et al. 29, the droplet length in our case is estimated to be 0.9 mm, which is very close to the experimentally measured value of 1.1 mm. Once the bacteria suspension droplets were formed, they were transported downstream to mix with another droplet containing silver nanocolloid and NaCl using the dosing technique developed by Henkel et al. 30. The merged droplets were mixed thoroughly when they were pushed through a winding microchannel within a short time period. The fast mixing can be credited to the microfluidic baker’s transformation that are best characterized by a series of stretches, folds, and reorientations that decrease the striation length.
Raman spectral features of MRSA and MSSA
The optofluidic-based SERS spectra of S. aureus were collected along with the segmented flow in the microchannel and the representative processed SERS spectral features of MRSA and MSSA are shown in Fig. S3. The band assignments are summarized in Table S3 and could be referred to previous publications 12, 31. MRSA and MSSA had similar SERS spectral features but some variations of band assignments were observed between MRSA and MSSA. For example, there were several band shifts between MRSA and MSSA, including a shift of the α-helix of amide I band from 1659 cm−1 for MRSA to 1667 cm−1 for MSSA, a shift of the C-C stretch of carbohydrate band from 911 cm−1 for MRSA to 923 cm−1 for MSSA and a shift of the C-H bending of tyrosine band from 1176 cm−1 for MRSA to 1210 cm−1 for MSSA (Fig. S3). These band shifts may be due to a different interaction mode between biological samples and the silver nanocolloids 12, 14b, 14e. In addition, MRSA and MSSA had unique bands. For nucleic acids, MRSA had unique bands at 1373 cm−1 (ring breathing modes of the DNA/RNA bases) and 1093 cm−1 (DNA backbone-phosphate backbone) while MSSA had a unique band at 1084 cm−1 (phosphodiester groups in nucleic acid). For proteins, MRSA had unique bands at 935 cm−1 (α-helix of protein) and 819 cm−1 (protein band) while MSSA had a unique protein band at 1270 cm−1 (amide III). Additionally, MSSA had a unique lipid band at 966 cm−1.
The spectral reproducibility (Dy1y2 value) was calculated using the wavenumbers between 1800 cm−1 to 400 cm−1 and the Dy1y2 values were between 3.43 and 4.06, indicating a better spectral reproducibility than our previous work where confocal micro-Raman spectroscopy was conducted to record the spectral features of Campylobacter 12. Preliminary experiment was conducted using the commercially available Klarite substrate (Renishaw Diagnostics) for the SERS detection of MRSA. We found that the spectral reproducibility (Dy1y2 values between 11.63 and 16.29) was worse than the current optofluidic SERS system, indicating that the current engineering design significantly alleviates the problems of inhomogeneity for SERS.
The correlation between optofluidic-based SERS typing and PCR typing for S. aureus antibiotic resistance
The result of the PCR amplification of the mecA gene in 38 S. aureus isolates from China and 20 S. aureus isolates from the United States is summarized in Table S3 (37 MRSA isolates and 21 MSSA isolates). Amplification of the mecA gene for selective S. aureus isolates is shown in Fig. S4.
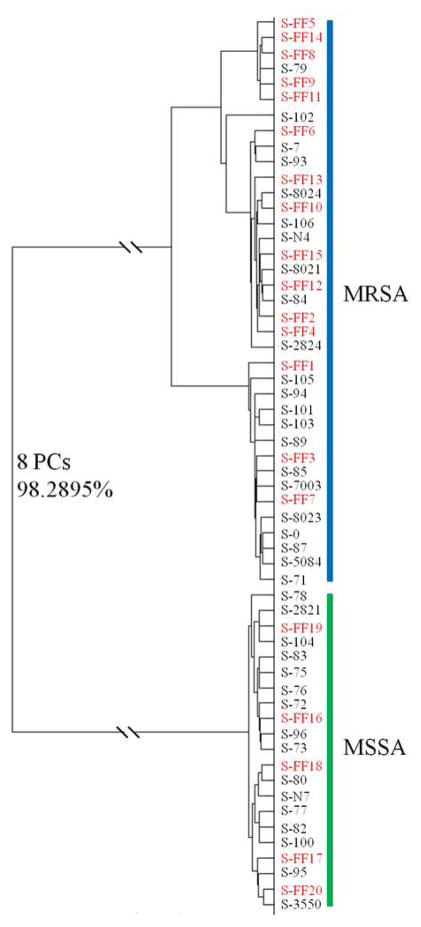
A global supervised DFA dendrogram model on the basis of SERS spectral features of intact S. aureus cells was established using the 38 S. aureus isolates from China and cross-validated using the 20 S. aureus isolates from the United States (Fig. 2). The spectral feature of each bacterial isolate in the supervised DFA dendrogram model has an average of 300 spectra collected from three independent experiments. Therefore, this model contained a total of 17,400 Raman spectra. The interclass distance was calculated based on Mahalanobis distance measurements between groups in the supervised DFA dendrogram model and the values were higher than 8.50. Clusters with interclass distance values higher than 3 are believed to be significantly different from each other 32. This result further validated the discrimination between MRSA and MSSA using optofluidic-based SERS typing.
Figure 2.
Composite dendrogram representing the global chemometric model to identify MRSA and MSSA from China and the United States. Entries in black represent the training data using MRSA and MSSA samples from China, and those in red denote the validation data using MRSA and MSSA samples from the United States. The model was established and validated as described in the text.
This DFA dendrogram model achieved a classification sensitivity of 96.5% and a specificity of 92.6% using Wards cluster algorithm at the cut-off value established at 99% similarity. The ROC curves were generated to further evaluate the group segregation (Fig. S5). Each point on the ROC curve represents a sensitivity/specificity pair corresponding to a particular decision threshold. The AUC under the ROC curve is 0.989, which indicated that the optofluidic SERS typing can well distinguish between two diagnostic groups (i.e., MRSA and MSSA).
The numerical index of discrimination of the optofluidic-based SERS typing was determined to be 0.989 by calculating the Simpson’s index of diversity. A value higher than 0.9 indicates a good discrimination typing method for bacterial samples 12. Bayesian probability analysis was then employed to compare the top 15 significant features with PCs determined by using the DFA model. We found good agreement in the results of the two methods. The stability of the DFA model was determined using three different Monte Carlo estimations, one using the 15 most significant features, a second with the 15 least significant features, and one in which all features were selected and compared. The highest stability of the DFA model was derived from the use of the 15 most significant features (0.65 ± 0.12). A model with a slightly lower stability (0.58 ± 0.08) was achieved when all features were selected, indicating the contribution of non-significant features and possibly interference and noise. The lowest stability model was obtained when the 15 least significant features (0.02 ± 0.01) were used.
Prediction from a recent nosocomial S. aureus outbreak
An outbreak associated with multiple S. aureus isolates recently occurred in second hospital affiliated to Tianjin Medical University in summer 2012. Twenty S. aureus isolates were obtained from both patients and staff members in this hospital which may or may not have comprised different strains. Both PCR typing and optofluidic-based SERS typing were conducted and the spectra were incorporated into our cross-validated global dendrogram model (Fig. 2). The recognition results for each isolate are summarized in Table S4. The average recognition rate was 95% for these twenty isolates including 12 MRSA isolates encoding the mecA gene and 8 MSSA isolates without the mecA gene.
The correlation between optofluidic-based SERS typing and MLST for S. aureus epidemiology
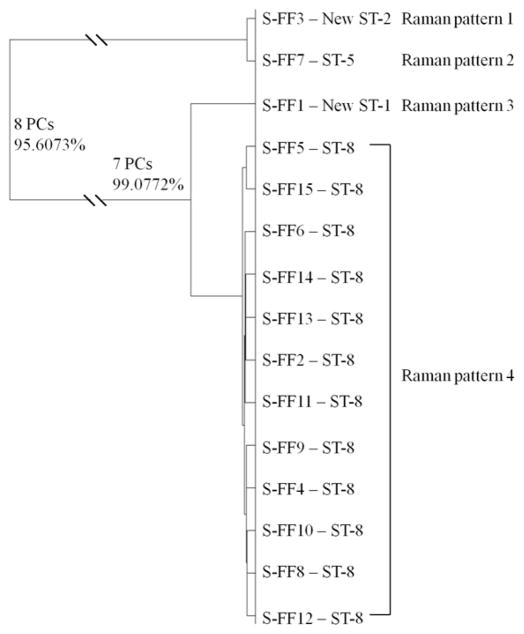
MLST was conducted to determine the epidemiology of S. aureus isolates from the United States. The allelic fragments of S. aureus isolates for MLST are shown in Fig. S6 and the properties of the sequence types of S. aureus isolates from the United States are summarized in Table S2. The unsupervised HCA based dendrogram model is shown in Fig. 3, indicating a good concordance between optofluidic-based SERS typing and MLST. The optofluidic SERS spectral features of four different patterns of MRSA are shown in Fig. S7.
Figure 3.
Hierarchical cluster analysis of MRSA clinical isolates from the United States. Full details of the numbers of PCs determined at each level and the associated percentages of explained variance are shown. This dendrogram shows the reproducibility and concordance of optofluidic-based Raman typing and MLST.
Identification of MRSA in a bacterial mixture sample
Two clinical S. aureus isolates (i.e., MRSA S-93 and MSSA S-N7) were mixed in a ratio of 5% to 100% MRSA isolate in the mixture. The corresponding SERS spectra of these 11 different bacterial cell mixtures were recorded using the optofluidic platform and a PLSR model was established and a leave-one-out cross-validation conducted (Table S5). This supervised chemometric model included a total of 3300 spectra (11 MRSA-MSSA mixtures, 300 spectra for each mixture) with a high R and RPD values for both calibration and validation models, low RMSE values for both calibration and validation models and 6 of latent variables (Table S5). These parameters indicated that the PLSR model could accurately determine and predict the actual concentration of MRSA in a mixture of MRSA and MSSA strains.
DISCUSSION
Microfluidic chip design
Microfluidic systems have obvious advantages over traditional analytical methods by providing a miniaturized device in the form of a “lab-on-a-chip” system that integrates sample preparation, analyte separation, biochemical reaction and detection together 21, providing a well-controlled microenvironment. Recently, microfluidic systems have been employed to couple nucleic acid extraction and amplification for rapid diagnosis of MRSA from clinical specimens 33.
Most microfluidic platforms for detection are manufactured with glass or a glass-PDMS-glass three-layer composite 34. PDMS is a soft and biocompatible material and is well known for its interfacial adhesion property to assist in bonding 20, 35 that is easy to fabricate and is extremely precise 26. Besides, the bonding process, which uses oxygen plasma treatment, is strong and reliable and has no leaking problems 36.
In the present study, we employed a droplet-based segmented flow system for the optofluidic platform. Adjacent biological samples are separated in the continuous flow system by a mineral oil droplet. Deposition of nanoparticles inside the microchannel, which may adversely affect detection due to the conjugation of analytes to the optical window, creating a “memory effect” can be reduced using segmented flow 22, 37. The method of Strehle et al.23 using crystal violet as the analyte was used to check the “memory effect” of our microfluidic system. The flow was abruptly ceased at 200 s and the corresponding Raman spectrum collected for a sample of silver nanocolloid containing only water droplets and no obvious spectral features were detected (Fig. S8), indicating that the flow was appropriately segmented and the “memory effect” was minimal.
SERS implementation into microfluidic system
On-line detection techniques, including chemiluminescence and fluorescence require a tagging step 38. The width of the fluorescence bands are broad and band overlapping may occur if several analytes are simultaneously determined 18f. Raman spectroscopy provides an attractive alternative. In the present study, we used silver nanocolloid for signal enhancement from microbes flowing in the microchannel and optimized this system. Because individual single nanoparticles could only provide a faint Raman scattering signal enhancement 39, aggregation by the addition of salt, such as nitrate and chloride, is required 20 and incorporating silver and gold nanoparticles can amplify the weak Raman scattering signal significantly, theoretically 1014 times, placing detection of single molecule within the scope of this analytical method 12, 14a, 14e. Compared to gold nanocolloid, silver nanocolloid is less stable and the size distribution is more difficult to control, resulting in lower homogeneity 15b from various aggregation behaviors but this can be compensated for in a flow-through system by efficient mixing. However, the advantage of silver nanocolloid is that enhancement effect (factor) is 1000 times higher than that of gold nanocolloid 39.
Here a higher intensity of green laser (532 nm, 10 mW, 0.5 s integration time) was introduced into the microchannel for SERS spectra collection than that employed by Walter et al.19 for SERS spectral collection (532 nm laser, 5 mW, incident 1 s integration time) for E. coli strains in a microfluidic system. Compared to fluorescence, photodecomposition is reduced for Raman spectroscopy because of rapid quenching of excited states for the analytes 18f making rapid spectral collection possible. In addition, the fluidic condition can evenly disperse the energy from the incident laser light, which is the major factor requiring employment of a higher laser power compared to traditional Raman and SERS for this optofluidic system. Further, flow behavior was controlled, preventing sample burning.
Use of a confocal technique allowed removal of interfering scattering signals from PDMS which is a Raman active material. Each of our confocal micro-Raman spectroscopic systems (Renishaw system in China and WITec system in the United States) has an adjustable confocal slit coupled with the aperture located in the back image plane of the microscope objective lens (Fig. 1) to collect only the green laser (532 nm)-induced spectral signal from a definite focal volume (see red layer in Fig. 1), blocking diffracted light from outside the focal plane (see purple and yellow layers in Fig. 1). This confocal mode can effectively remove the background stray light derived from the out-of-focus regions of the PDMS 20, 35.
Minor variations in SERS spectral features between MRSA and MSSA were observed (Fig. S3, Table S3). Spectral profiles of MRSA and MSSA differed in the mass range of m/z 500–3500 Da and cluster analysis could be constructed for visual segregation 40 for matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. A MALDI-TOF method for phenol-soluble modulin α1 and α2 peptides as a biomarker for MRSA and vancomycin-intermediate S. aureus isolates has also been developed 41.
Timeline advantage
MRSA is the most frequently identified antibiotic-resistant pathogenic bacteria in the world and the infection rates have increased over the past decades. The prevalence of MRSA varies by country 42. The highest proportion of MRSA infection is in large hospitals and the transmission between patients is mainly responsible for the emerging deadly multidrug-resistant S. aureus strains 43. The European Food Safety Authority has recently (October 5, 2012) published a scientific report recommending continuous monitoring and reporting program for MRSA in food-producing animals, food and feed. Improved analytical methodology for identification and typing is required for this zoonotic agent because MRSA colonization in production animals and the dissemination of this zoonosis into the food production chain is still not clearly understood 44. Therefore, further study and surveillance of epidemiology and dissemination for MRSA, especially the development of new techniques for rapid detection of MRSA in clinical and now environmental specimens is critical to decrease infection rates acquired from hospital and community and from agricultural products, is necessary to improve environmental health.
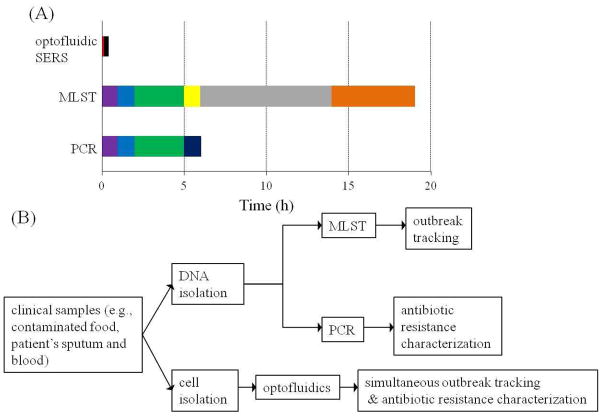
One of the major incentives for conducting this research work was to shorten the diagnostic time. Traditional genotyping methods (e.g., PCR, PFGE and MLST) have a long turnaround time (2–3 days) and a low sample throughput for bacterial detection starting from a pure culture (Fig. 4a). Therefore, these methods are not suitable for routine uses in hospital and community surveillance. While, PCR is useful for rapid identification (~ 6 h) of MRSA or MSSA isolates from a clinical specimen including DNA extraction (1 h), PCR setup (1 h), PCR amplification (3 h) and gel electrophoresis (1 h), it does not permit determination of the source of an outbreak. Recently, recombinase polymerase amplification (RPA) has been widely conducted to rapidly detect bacteria; however, RPA cannot address the bacterial epidemiological surveillance issues. To address this concern, scientists have turned to MLST or PFGE 8. The time frame for MLST (or PGFE) is long, requiring DNA extraction (1 h), PCR setup (1 h), PCR amplification (3 h), PCR cleanup (1 h), sequencing (8 h), and alignment and sequence typing (5 h). An important caveat with these techniques is that they require an additional step to distinguish between MRSA and MSSA isolates (e.g., S-FF2 and S-FF18 in Table S2), that have identical sequence types.
Figure 4.
Optofluidic SERS biosensor shows advantages compared to traditional molecular typing. Panel (A): Time table to compare the detection speed of one S. aureus isolate starting from a pure culture using different methods (PCR, MLST and optofluidic-SERS). Purple color denotes DNA extraction, blue color denotes PCR setup, green color denotes PCR amplification, dark blue color denotes gel electrophoresis, yellow color denotes PCR clean up, grey color denotes DNA sequencing, orange color denotes alignment and sequence typing, red color denotes spectral collection and black color denotes data analysis. S. aureus was grown in LB broth at 37°C overnight before performing any detection method. Panel (B): Flow chart of comparing the working efficiency between optofluidic SERS typing and genotyping techniques to study bacterial epidemiology and antibiotic-resistant profiles.
The optofluidic platform described here provides a much more rapid analysis. On the basis of 25–30 mins for the generation time of MRSA 1a, cultivation for 200 mins starting from single bacterial cell is enough (100 cells per droplet) for detection using SERS spectra. It will be important to optimize the relationship between bacterial cell density in each droplet and the SERS signal intensity and further studies with this system are needed. A decrease in cultivation time for less bacterial cell numbers necessary for accurate identification and classification based on spectral features in this optofluidic platform is proposed.
A total of 300 SERS spectra were recorded for each bacterial isolate to establish and validate the relevant chemometric model. Considering 0.5 s as the integration time for collecting each spectrum, a total of 3.3 min was needed for spectral recording, including approximate 1 min for spectral collection of mineral oil used for sample segregation which also provides an optical baseline to show that the flow and optical behavior within the optofluidic device remained stable.
Simultaneous determination of S. aureus antibiotic-resistance and epidemiological dissemination
PFGE, spa typing and MLST are the major tools to study the epidemiology and dissemination of S. aureus in the food production chain, community, and hospitals 45. Even though most research agencies still used PFGE as the gold standard for S. aureus epidemiological studies, the application of MLST has increased significantly during recent years 46. In addition, the detection of the mecA gene for MRSA has to be conducted individually using PCR or other related techniques, such as loop-mediated isothermal amplification. Our study indicated that optofluidic-based SERS typing can not only detect and differentiate MRSA from MSSA, but can also be used to successfully investigate the epidemiology of MRSA isolates (e.g., distinguish between isolates that were part of the outbreak and isolates that were not) in a fast turnaround time (Fig. 4b). Further, many clinical specimens may contain a mixture of Staphylococci and this may presents challenges for reliable and direct detection of MRSA from clinical specimens 47. Our optofluidic-based SERS PLSR model could accurately determine the concentration of an MRSA strain in a mixture containing MSSA (Table S5), a finding with significant clinical application.
Finally, this method is robust. Experimental protocols were conducted in different laboratories by using two different confocal micro-Raman spectroscopic systems coupled with microfluidic platform for recording the SERS spectral features of S. aureus clinical isolates from China and the United States. We created a global dendrogram model from the data from these two laboratories and used this to monitor a recent S. aureus-associated infection outbreak (Table S4) denoting the possibility of employing this optofluidic technique as a reliable routine tool for rapid detection and epidemiological surveillance of MRSA in the world.
Supplementary Material
Acknowledgments
We thank Dr. Ferric C. Fang (University of Washington), Dr. Yanjin He (Tianjin Medical University), and Dr. Jinhai Huang (Tianjin University) for providing MRSA and MSSA clinical isolates.
This work was supported from funds awarded to J.X. by DOD (DARPA N66001-11-1-4127), funds awarded to M.E.K. by NIH (R56 AI088518-01A1), funds awarded to B.A.R. by USDA-NIFA (2011-68003-20096), funds awarded to S.W. by National Basic Program (973) of China (2012CB720803) and funds awarded to D.R.S. by NIH (T32GM083864).
Footnotes
Additional information as noted in text.
References
- 1.(a) Cooper BS, Medley GF, Stone SP, Kibbler CC, Cookson BD, Roberts JA, Duckworth G, Lai R, Ebrahim S. Methicillin-resistant Staphylococcus aureus in hospitals and the community: stealth dynamics and control catastrophes. Proc Natl Acad Sci U S A. 2004;101(27):10223–8. doi: 10.1073/pnas.0401324101. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327(5964):469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bootsma MC, Diekmann O, Bonten MJ. Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing. Proc Natl Acad Sci U S A. 2006;103(14):5620–5. doi: 10.1073/pnas.0510077103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson JF, Riebe KM, Hall GS, Wilson D, Whittier S, Palavecino E, Ledeboer NA. Spectra MRSA, a new chromogenic agar medium to screen for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2010;48(1):215–9. doi: 10.1128/JCM.01555-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley PG, Grabsch EA, Howden BP, Gao W, Grayson ML. Comparison of the Xpert methicillin-resistant Staphylococcus aureus (MRSA) assay, BD GeneOhm MRSA assay, and culture for detection of nasal and cutaneous groin colonization by MRSA. J Clin Microbiol. 2009;47(11):3769–72. doi: 10.1128/JCM.00303-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tokue Y, Shoji S, Satoh K, Watanabe A, Motomiya M. Comparison of a polymerase chain reaction assay and a conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36(1):6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanc DS, Struelens MJ, Deplano A, De Ryck R, Hauser PM, Petignat C, Francioli P. Epidemiological validation of pulsed-field gel electrophoresis patterns for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2001;39(10):3442–5. doi: 10.1128/JCM.39.10.3442-3445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–15. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. Escmid; Esgem, Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infec. 2007;13:1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 9.Ceylan Koydemir H, Kulah H, Ozgen C, Alp A, Hascelik G. MEMS biosensors for detection of methicillin resistant Staphylococcus aureus. Biosens Bioelectron. 2011;29(1):1–12. doi: 10.1016/j.bios.2011.07.071. [DOI] [PubMed] [Google Scholar]
- 10.Willemse-Erix DF, Jachtenberg JW, Schut TB, van Leeuwen W, van Belkum A, Puppels G, Maquelin K. Towards Raman-based epidemiological typing of Pseudomonas aeruginosa. J Biophotonics. 2010;3(8–9):506–11. doi: 10.1002/jbio.201000026. [DOI] [PubMed] [Google Scholar]
- 11.Buijtels PC, Willemse-Erix HF, Petit PL, Endtz HP, Puppels GJ, Verbrugh HA, van Belkum A, van Soolingen D, Maquelin K. Rapid identification of mycobacteria by Raman spectroscopy. J Clin Microbiol. 2008;46(3):961–5. doi: 10.1128/JCM.01763-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu XN, Huang Q, Miller WG, Aston DE, Xu J, Xue F, Zhang HW, Rasco BA, Wang S, Konkel ME. Comprehensive Detection and Discrimination of Campylobacter Species by Use of Confocal Micro-Raman Spectroscopy and Multilocus Sequence Typing. Journal of Clinical Microbiology. 2012;50(9):2932–2946. doi: 10.1128/JCM.01144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Willemse-Erix HFM, Jachtenberg J, Barutci H, Puppels GJ, van Belkum A, Vos MC, Maquelin K. Proof of Principle for Successful Characterization of Methicillin-Resistant Coagulase-Negative Staphylococci Isolated from Skin by Use of Raman Spectroscopy and Pulsed-Field Gel Electrophoresis. Journal of Clinical Microbiology. 2010;48(3):736–740. doi: 10.1128/JCM.01153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wulf MWH, Willemse-Erix D, Verduin CM, Puppels G, van Belkum A, Maquelin K. The use of Raman spectroscopy in the epidemiology of methicillin-resistant Staphylococcus aureus of human- and animal-related clonal lineages. Clin Microbiol Infec. 2012;18(2):147–152. doi: 10.1111/j.1469-0691.2011.03517.x. [DOI] [PubMed] [Google Scholar]; (c) Willemse-Erix DF, Scholtes-Timmerman MJ, Jachtenberg JW, van Leeuwen WB, Horst-Kreft D, Bakker Schut TC, Deurenberg RH, Puppels GJ, van Belkum A, Vos MC, Maquelin K. Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J Clin Microbiol. 2009;47(3):652–9. doi: 10.1128/JCM.01900-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Jarvis RM, Goodacre R. Discrimination of bacteria using surface-enhanced Raman spectroscopy. Anal Chem. 2004;76(1):40–47. doi: 10.1021/ac034689c. [DOI] [PubMed] [Google Scholar]; (b) Ellis DI, Brewster VL, Dunn WB, Allwood JW, Golovanov AP, Goodacre R. Fingerprinting food: current technologies for the detection of food adulteration and contamination. Chem Soc Rev. 2012;41(17):5706–27. doi: 10.1039/c2cs35138b. [DOI] [PubMed] [Google Scholar]; (c) Alvarez-Puebla RA, Liz-Marzan LM. SERS-based diagnosis and biodetection. Small. 2010;6(5):604–10. doi: 10.1002/smll.200901820. [DOI] [PubMed] [Google Scholar]; (d) Liu TY, Tsai KT, Wang HH, Chen Y, Chen YH, Chao YC, Chang HH, Lin CH, Wang JK, Wang YL. Functionalized arrays of Raman-enhancing nanoparticles for capture and culture-free analysis of bacteria in human blood. Nat Commun. 2011;2:538. doi: 10.1038/ncomms1546. [DOI] [PubMed] [Google Scholar]; (e) Kneipp J, Kneipp H, Kneipp K. SERS - a single-molecule and nanoscale tool for bioanalytics. Chemical Society Reviews. 2008;37(5):1052–1060. doi: 10.1039/b708459p. [DOI] [PubMed] [Google Scholar]
- 15.(a) Liu Y, Chen YR, Nou X, Chao K. Potential of surface-enhanced Raman spectroscopy for the rapid identification of Escherichia coli and Listeria monocytogenes cultures on silver colloidal nanoparticles. Appl Spectrosc. 2007;61(8):824–31. doi: 10.1366/000370207781540060. [DOI] [PubMed] [Google Scholar]; (b) Zeiri L, Efrima S. Surface-enhanced Raman spectroscopy of bacteria: the effect of excitation wavelength and chemical modification of the colloidal milieu. J Raman Spectrosc. 2005;36(6–7):667–675. [Google Scholar]
- 16.(a) Kahraman M, Yazici MM, Sahin F, Bayrak OF, Culha M. Reproducible surface-enhanced Raman scattering spectra of bacteria on aggregated silver nanoparticles. Appl Spectrosc. 2007;61(5):479–85. doi: 10.1366/000370207780807731. [DOI] [PubMed] [Google Scholar]; (b) Levene C, Correa E, Blanch EW, Goodacre R. Enhancing Surface Enhanced Raman Scattering (SERS) Detection of Propranolol with Multiobjective Evolutionary Optimization. Anal Chem. 2012;84(18):7899–905. doi: 10.1021/ac301647a. [DOI] [PubMed] [Google Scholar]
- 17.Lu XN, Al-Qadiri HM, Lin MS, Rasco BA. Application of Mid-infrared and Raman Spectroscopy to the Study of Bacteria. Food Bioprocess Tech. 2011;4(6):919–935. [Google Scholar]
- 18.(a) Quang LX, Lim C, Seong GH, Choo J, Do KJ, Yoo SK. A portable surface-enhanced Raman scattering sensor integrated with a lab-on-a-chip for field analysis. Lab Chip. 2008;8(12):2214–9. doi: 10.1039/b808835g. [DOI] [PubMed] [Google Scholar]; (b) Tong L, Righini M, Gonzalez MU, Quidant R, Kall M. Optical aggregation of metal nanoparticles in a microfluidic channel for surface-enhanced Raman scattering analysis. Lab Chip. 2009;9(2):193–5. doi: 10.1039/b813204f. [DOI] [PubMed] [Google Scholar]; (c) Ashok PC, Singh GP, Rendall HA, Krauss TF, Dholakia K. Waveguide confined Raman spectroscopy for microfluidic interrogation. Lab Chip. 2011;11(7):1262–70. doi: 10.1039/c0lc00462f. [DOI] [PubMed] [Google Scholar]; (d) Chon H, Lim C, Ha SM, Ahn Y, Lee EK, Chang SI, Seong GH, Choo J. On-Chip Immunoassay Using Surface-Enhanced Raman Scattering of Hollow Gold Nanospheres. Anal Chem. 2010;82(12):5290–5295. doi: 10.1021/ac100736t. [DOI] [PubMed] [Google Scholar]; (e) Dochow S, Krafft C, Neugebauer U, Bocklitz T, Henkel T, Mayer G, Albert J, Popp J. Tumour cell identification by means of Raman spectroscopy in combination with optical traps and microfluidic environments. Lab on a Chip. 2011;11(8):1484–1490. doi: 10.1039/c0lc00612b. [DOI] [PubMed] [Google Scholar]; (f) Fan X, White IM. Optofluidic Microsystems for Chemical and Biological Analysis. Nat Photonics. 2011;5(10):591–597. doi: 10.1038/nphoton.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Hwang H, Kim SH, Yang SM. Microfluidic fabrication of SERS-active microspheres for molecular detection. Lab Chip. 2011;11(1):87–92. doi: 10.1039/c0lc00125b. [DOI] [PubMed] [Google Scholar]; (h) Xu BB, Ma ZC, Wang L, Zhang R, Niu LG, Yang Z, Zhang YL, Zheng WH, Zhao B, Xu Y, Chen QD, Xia H, Sun HB. Localized flexible integration of high-efficiency surface enhanced Raman scattering (SERS) monitors into microfluidic channels. Lab on a Chip. 2011;11(19):3347–3351. doi: 10.1039/c1lc20397e. [DOI] [PubMed] [Google Scholar]
- 19.Walter A, Marz A, Schumacher W, Rosch P, Popp J. Towards a fast, high specific and reliable discrimination of bacteria on strain level by means of SERS in a microfluidic device. Lab Chip. 2011;11(6):1013–21. doi: 10.1039/c0lc00536c. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Choo J. Recent advances in surface-enhanced Raman scattering detection technology for microfluidic chips. Electrophoresis. 2008;29(9):1815–28. doi: 10.1002/elps.200700554. [DOI] [PubMed] [Google Scholar]
- 21.Marz A, Henkel T, Cialla D, Schmitt M, Popp J. Droplet formation via flow-through microdevices in Raman and surface enhanced Raman spectroscopy--concepts and applications. Lab Chip. 2011;11(21):3584–92. doi: 10.1039/c1lc20638a. [DOI] [PubMed] [Google Scholar]
- 22.Marz A, Bocklitz T, Popp J. Online-calibration for reliable and robust lab-on-a-chip surface enhanced Raman spectroscopy measurement in a liquid/liquid segmented flow. Anal Chem. 2011;83(21):8337–40. doi: 10.1021/ac2015799. [DOI] [PubMed] [Google Scholar]
- 23.Strehle KR, Cialla D, Rosch P, Henkel T, Kohler M, Popp J. A reproducible surface-enhanced raman spectroscopy approach. Online SERS measurements in a segmented microfluidic system. Anal Chem. 2007;79(4):1542–7. doi: 10.1021/ac0615246. [DOI] [PubMed] [Google Scholar]
- 24.Lim C, Hong J, Chung BG, deMello AJ, Choo J. Optofluidic platforms based on surface-enhanced Raman scattering. Analyst. 2010;135(5):837–844. doi: 10.1039/b919584j. [DOI] [PubMed] [Google Scholar]
- 25.Cecchini MP, Hong J, Lim C, Choo J, Albrecht T, Demello AJ, Edel JB. Ultrafast surface enhanced resonance Raman scattering detection in droplet-based microfluidic systems. Anal Chem. 2011;83(8):3076–81. doi: 10.1021/ac103329b. [DOI] [PubMed] [Google Scholar]
- 26.(a) Xia H, Whitesides GM. Soft Lithography. Angewandte Chemie International Edition. 1998;37(5):550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]; (b) Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nat Protoc. 2010;5(3):491–502. doi: 10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- 27.Lee PC, Meisel D. Adsorption and Surface-Enhanced Raman of Dyes on Silver and Gold Sols. J Phys Chem-Us. 1982;86(17):3391–3395. [Google Scholar]
- 28.Song H, Chen DL, Ismagilov RF. Reactions in droplets in microfluidic channels. Angew Chem Int Ed Engl. 2006;45(44):7336–56. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garstecki P, Fuerstman MJ, Stone HA, Whitesides GM. Formation of droplets and bubbles in a microfluidic T-junction-scaling and mechanism of break-up. Lab Chip. 2006;6(3):437–46. doi: 10.1039/b510841a. [DOI] [PubMed] [Google Scholar]
- 30.Henkel T, Bermig T, Kielpinski M, Grodrian A, Metze J, Köhler J. Chip modules for generation and manipulation of fluid segments for micro serial flow processes. Chemical Engineering Journal. 2004;101(1):439–445. [Google Scholar]
- 31.(a) De Gelder J, De Gussem K, Vandenabeele P, Moens L. Reference database of Raman spectra of biological molecules. J Raman Spectrosc. 2007;38(9):1133–1147. [Google Scholar]; (b) Movasaghi Z, Rehman S, Rehman DIU. Raman spectroscopy of biological tissues. Applied Spectroscopy Reviews. 2007;42(5):493–541. [Google Scholar]
- 32.Lu X, Rasco BA, Kang DH, Jabal JM, Aston DE, Konkel ME. Infrared and Raman spectroscopic studies of the antimicrobial effects of garlic concentrates and diallyl constituents on foodborne pathogens. Anal Chem. 2011;83(11):4137–46. doi: 10.1021/ac2001498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(a) Wang CH, Lien KY, Wu JJ, Lee GB. A magnetic bead-based assay for the rapid detection of methicillin-resistant Staphylococcus aureus by using a microfluidic system with integrated loop-mediated isothermal amplification. Lab Chip. 2011;11(8):1521–31. doi: 10.1039/c0lc00430h. [DOI] [PubMed] [Google Scholar]; (b) Hwang KY, Kwon SH, Jung SO, Namkoong K, Jung WJ, Kim JH, Suh KY, Huh N. Solid Phase DNA Extraction with a Flexible Bead-Packed Microfluidic Device to Detect Methicillin-Resistant Staphylococcus aureus in Nasal Swabs. Anal Chem. 2012;84(18):7912–8. doi: 10.1021/ac3016533. [DOI] [PubMed] [Google Scholar]; (c) Sabat AJ, Chlebowicz MA, Grundmann H, Arends JP, Kampinga G, Meessen NE, Friedrich AW, van Dijl JM. Microfluidic-chip-based multiple-locus variable-number tandem-repeat fingerprinting with new primer sets for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2012;50(7):2255–62. doi: 10.1128/JCM.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duffy DC, McDonald JC, Schueller OJ, Whitesides GM. Rapid Prototyping of Microfluidic Systems in Poly(dimethylsiloxane) Anal Chem. 1998;70(23):4974–84. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 35.Park T, Lee S, Seong GH, Choo J, Lee EK, Kim YS, Ji WH, Hwang SY, Gweon DG. Highly sensitive signal detection of duplex dye-labelled DNA oligonucleotides in a PDMS microfluidic chip: confocal surface-enhanced Raman spectroscopic study. Lab Chip. 2005;5(4):437–42. doi: 10.1039/b414457k. [DOI] [PubMed] [Google Scholar]
- 36.Li P, Xue W, Xu J. The fabrication of PDMS interconnecting interface assisted by tubing fixation. Lab Chip Chips and Tips. 2011 [Google Scholar]
- 37.Cristobal G, Arbouet L, Sarrazin F, Talaga D, Bruneel JL, Joanicot M, Servant L. On-line laser Raman spectroscopic probing of droplets engineered in microfluidic devices. Lab Chip. 2006;6(9):1140–6. doi: 10.1039/b602702d. [DOI] [PubMed] [Google Scholar]
- 38.Monat C, Domachuk P, Eggleton B. Integrated optofluidics: A new river of light. Nature Photonics. 2007;1(2):106–114. [Google Scholar]
- 39.Tantra R, Brown RJC, Milton MJT. Strategy to improve the reproducibility of colloidal SERS. J Raman Spectrosc. 2007;38(11):1469–1479. [Google Scholar]
- 40.Du Z, Yang R, Guo Z, Song Y, Wang J. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 2002;74(21):5487–91. doi: 10.1021/ac020109k. [DOI] [PubMed] [Google Scholar]
- 41.Lu JJ, Tsai FJ, Ho CM, Liu YC, Chen CJ. Peptide biomarker discovery for identification of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus strains by MALDI-TOF. Anal Chem. 2012;84(13):5685–92. doi: 10.1021/ac300855z. [DOI] [PubMed] [Google Scholar]
- 42.Grundmann H, Aires-de-Sousa M, Boyce J, Tiemersma E. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet. 2006;368(9538):874–85. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 43.Ke W, Huang SS, Hudson LO, Elkins KR, Nguyen CC, Spratt BG, Murphy CR, Avery TR, Lipsitch M. Patient sharing and population genetic structure of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2012;109(17):6763–8. doi: 10.1073/pnas.1113578109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.European, Technical specifications on the harmonised monitoring and reporting of antimicrobial resistance in methicillin-resistant Staphylococcus aureus in food-producing animals and food. EFSA Journal. 2012;10:2897. [Google Scholar]
- 45.Faria NA, Carrico JA, Oliveira DC, Ramirez M, de Lencastre H. Analysis of typing methods for epidemiological surveillance of both methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Clin Microbiol. 2008;46(1):136–44. doi: 10.1128/JCM.01684-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goering RV, Shawar RM, Scangarella NE, O’Hara FP, Amrine-Madsen H, West JM, Dalessandro M, Becker JA, Walsh SL, Miller LA, van Horn SF, Thomas ES, Twynholm ME. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus isolates from global clinical trials. J Clin Microbiol. 2008;46(9):2842–7. doi: 10.1128/JCM.00521-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ, van Belkum A, Ouellette M, Roy PH, Bergeron MG. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J Clin Microbiol. 2004;42(5):1875–84. doi: 10.1128/JCM.42.5.1875-1884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.