Abstract
The most potent tumorigen identified among the polycyclic aromatic hydrocarbons (PAH) is the non-planar fjord region dibenzo[a,l]pyrene (DB[a,l]P). It is metabolically activated in vivo through the widely-studied diol epoxide (DE) pathway to form covalent adducts with DNA bases, predominantly guanine and adenine. The (+)-11S,12R,13R,14S DE enantiomer forms adducts via its C14-position with the exocyclic amino group of guanine. Here, we present the first NMR solution structure of a DB[a,l]P-derived adduct, the 14R (+)-trans-anti-DB[a,l]P–N2-dG (DB[a,l]P-dG) lesion in double-stranded DNA. In contrast to the stereochemically identical benzo[a]pyrene-derived N2-dG adduct (B[a]P-dG) in which the B[a]P rings reside in the B-DNA minor groove on the 3’-side of the modifed deoxyguanosine, in the DB[a,l]P-derived adduct the DB[a,l]P rings intercalate into the duplex on the 3’-side of the modified base from the sterically crowded minor groove. Watson-Crick base pairing of the modified guanine with the partner cytosine is broken, but these bases retain some stacking with the bulky DB[a,l]P ring system. This new theme in PAH DE - DNA adduct conformation differs from: (1) the classical intercalation motif where Watson-Crick base-pairing is intact at the lesion site, and (2) the base-displaced intercalation motif in which the damaged base and its partner are extruded from the helix . The structural considerations that lead to the intercalated conformation of the DB[a,l]P-dG lesion in contrast to the minor groove alignment of the B[a]P-dG adduct, and the implications of the DB[a,l]P-dG conformational motif for the recognition of such DNA lesions by the human nucleotide excision repair apparatus, are discussed.
The most potent tumorigen identified among the polycyclic aromatic hydrocarbons (PAHs) is the non-planar fjord region compound dibenzo[a,l]pyrene (DB[a,l]P).1-4 It is present in cigarette smoke, fossil fuel combustion products such as smoke and coal particulates, and in soil and sediment samples.3,5 Fjord PAH such as DB[a,l]P are non-planar in order to avoid a steric clash between adjacent hydrogen atoms in the crowded fjord region (Figure 1).6 On the other hand, the aromatic ring system is planar in the case of the more extensively studied bay region PAH benzo[a]pyrene (B[a]P).7 Interestingly, in rodent model systems, DB[a,l]P is up to 100 times more tumorigenic than B[a]P.1,3,8,9 Upon metabolic activation through the widely-studied diol epoxide (DE) pathway3,10,11, the predominant product is the (–)-anti-14R,13S,12S,11R-DB[a,l]P diol epoxide (anti-DB[a,l]PDE), while the (+)-syn stereoisomer is formed in lesser amounts.12-14 The DEs derived from DB[a,l]P are highly tumorigenic8,13,15,16 and mutagenic3,17,18. DB[a,l]PDE-induced mutations targeted to both adenine and guanine have been observed in a Big Blue mouse cell CII transgene study.19 DNA adducts derived from the DB[a,l]PDEs have been observed in mouse lung20, and in oral21 and other rat tissues22. Both guanine and adenine adducts are produced by the DEs, and the proportion varies with the system, the identity of the DEs and their concentration, and other variables.3,11,22-27 The treatment of mouse skin with DB[a,l]P yields stable anti-DB[a,l]PDE-derived adenine and guanine adducts in DNA in comparable proportions.27 Upon treatment of human cells that were either proficient in nucleotide excision repair (NER+) or -deficient (NER–) with racemic anti-DB[a,l]PDE, both adenine and guanine adducts were generated within the cellular DNA, and two of the major guanine adducts with unknown stereochemistry were resistant to removal in the NER+ cells.26 The levels of anti-DB[a,l]PDE-dG and related adducts have been correlated with the levels of mutations and tumor initiation in rodents.28,29
Figure 1.
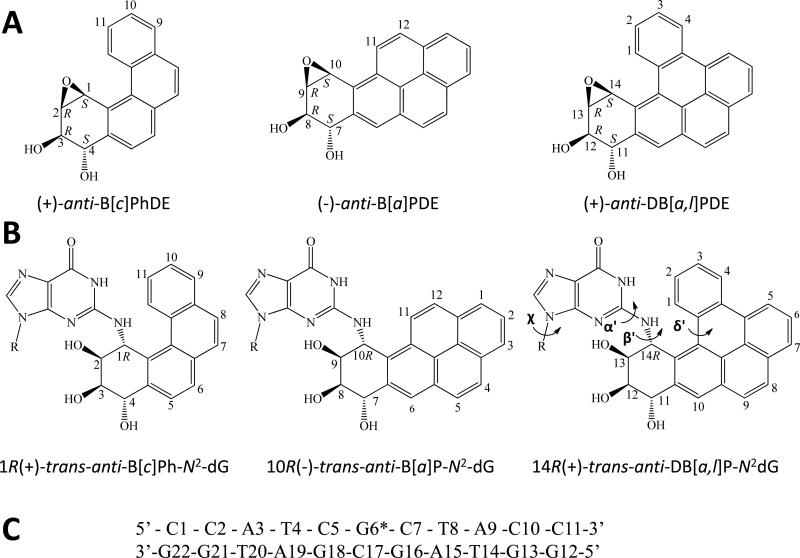
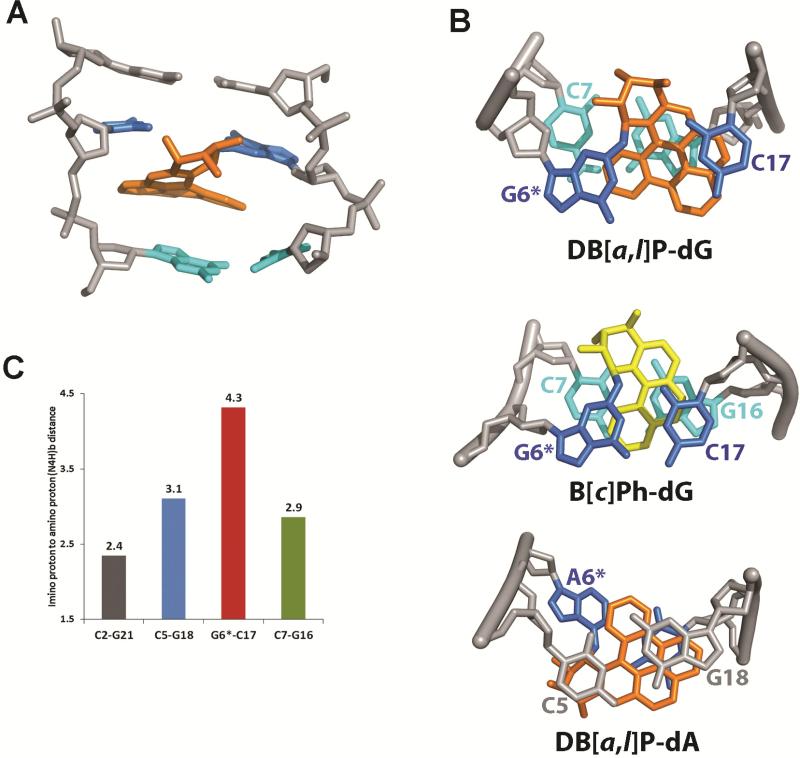
(A) Structures of the S,R,R,S anti-PAH diol epoxides. These react with guanine in DNA by trans-addition to the 1 position of (+)-anti-B[c]PhDE, the 10 position of (–)-anti-B[a]PDE, and the 14 position of (+)-anti-DB[a,l]PDE to form the stereochemically defined adducts with the same absolute configurations defined in (B). The sequence of the modified 11-mer duplex is shown in (C), where G6* denotes the site of the modified guanine. The torsion angles α' (N1-C2-N2-C14), β' (C2-N2-C14-C13), and δ'(C15-C17-C20-C1) are indicated for the DB[a,l]P-dG adduct.
It is often assumed that DNA adducts that are most resistant to DNA repair are also the most persistent ones in vivo, and are therefore particularly important in the etiology of cancer. We have thus been interested in studying the relationships between the conformational features of PAH diol epoxide-derived DNA lesions, reviewed in30-33, and their resistance to repair by the nucleotide excision repair (NER) mechanism. The relationships between NER and the structural features of the adducts derived from the fjord region PAH diol epoxides are of particular interest34 because of their high tumorigenic activities3,35, unique fjord aromatic ring topology, and the non-planar and flexible aromatic ring in the fjord region6,36.
Previously, an interesting difference in adduct conformations, elucidated by NMR methods, was found in identical sequence contexts between structurally different but stereochemically identical lesions with R absolute configuration at the linkage site; the adducts were all derived from the trans-addition of the S,R,R,S anti-diol epoxides with identical stereochemistry (Figure 1). It was previously found that the 10R (–)-trans-anti-B[a]P–N2-dG (B[a]P-dG) adduct is positioned in the minor groove oriented towards the 3′-end of the modified strand37, while the stereochemically analogous 1R (+)-trans-anti-B[c]Ph–N2-dG (B[c]Ph-dG) adduct is intercalated from the sterically crowded minor groove on the 3′-side of the modified deoxyguanosine residue with Watson-Crick pairing maintained at the lesion site38. This striking difference in adduct conformations was attributed to the smaller physical size (three aromatic rings), curved topology and non-planarity of the fjord B[c]Ph-dG adduct, in contrast to the planar, rigid bay region B[a]P-dG adduct with four aromatic rings. Modeling showed that intercalation by the B[a]P ring system produced a less well stacked structure than for the B[c]Ph case38. Since the 14R (+)-trans-anti-DB[a,l]P–N2-dG (DB[a,l]P-dG) adduct is still larger with five aromatic rings (Figure 1B), our initial hypothesis was that this bulky aromatic ring system would also adopt a minor groove rather than an intercalated conformation. However, in this work we demonstrate that the DB[a,l]P-dG adduct adopts an intercalated conformation with the aromatic ring system inserted from the minor groove on the 3’-side of the damaged guanine but with a novel conformational theme: the Watson-Crick base pair at the lesion site is broken, while pairing is intact in the stereochemically related B[c]Ph-dG 38 and B[a]P-dG adducts37. In both the B[c]Ph and DB[a,l]P cases, the damaged base and its partner base are stacked over the PAH ring system; however, stacking is surprisingly weaker for the DB[a,l]P case, because of the ruptured base pair. Thus, the relatively large bulk and the particular arrangement of the aromatic rings in the DB[a,l]P residue produces yet another conformational theme30 for the family of PAH-derived lesions.
Methods
Synthesis of anti-DB[a,l]PDE modified oligonucleotides
The single-stranded, site-specifically modified 11-mers, 5’-CCATCG*CTACC with a DB[a,l]P-dG residue at the central G* were generated employing automated DNA synthesis methods, utilizing the diastereomerically pure DB[a,l]P-dG phosphoramidite as described previously39,40. The purified DB[a,l]P-modified oligonucleotides were dissolved in 10 mM sodium phosphate buffer solution, pH 6.8, 100 mM NaCl, and were annealed with the fully complementary strand by heating the solutions at 70 °C for 10 min, followed by cooling to room-temperature overnight.
Wang et al.39 observed an unusual migration of the DB[a,l]PDE residue in the same sequence (Figure 1C) to neighboring guanine residues on the complementary strand. This migration also involved a stereochemical inversion of the initial 14R trans adduct to a 14S cis adduct, i.e., an inversion of the orientation of the guanine residue at the C14 position that occurs on a time scale of several days39. In order to minimize the effects of this migration, the modified DNA duplexes were annealed immediately before the NMR experiments were conducted. The duplexes thus prepared for the NMR experiments were stored at –20 °C and were used within less than one week after the annealing process. The samples that were studied by NMR here showed contributions of the migrated lesion which is most likely the migrated cis-anti adduct at G16, and is present in less than 10% range concentrations. It is therefore likely that these migrated adducts account for some of the observed but unidentified NOEs in the 2-D NMR spectra.
NMR measurements
The 2D NOESY spectrum was recorded in-house using a Bruker Avance 500 MHz NMR spectrometer with mixing times of 100 and 200 ms at 28 °C and 300 ms at 15 °C in D2O buffer, and also in 90% H2O-10% D2O buffer solution at a mixing time of 150 ms at 1 °C. All 2D NOESY data sets were acquired with phase sensitive pulse sequences using the States-TPPI method. The relaxation delay was set to 1.5 sec in D2O (1.2 sec in H2O) with 512 FIDs in the indirect dimension, and each FID was recorded with 104 transient scans with 2048 complex data points and spectral width of 10.5 ppm in both dimensions for the spectra in D2O, and 21 ppm for the corresponding spectra in H2O. The data sets were processed with the standard Bruker processing software package (TOPSPIN 1.3), and peak assignments were done using the SPARKY program41. The Total Correlation SpectroscopY (TOCSY) method with mixing times of 60 ms in D2O buffer solution at 28 °C and 80 ms at 15 °C were used for the assignment of the critical cytosine H5 H6 and DB[a,l]P aromatic ring protons in the adducted duplex. Data acquisition for the 2D TOCSY spectra were the same as for the 2D NOESY spectra. The NMR buffer solution had the following composition: 10 mM sodium phosphate, 100 mM NaCl, pH 6.8, and 25 nM DSS (2,2-dimethyl-2-silapentane-5-sulfonate, sodium salt) as the zero ppm reference marker.
Molecular dynamics computations and distance refinement
We built the initial model for the DB[a,l]P-dG adduct based on the B-DNA high resolution NMR solution structure of the fjord region analogue with the same absolute configurations at the linkage site and the three hydroxyl groups in the benzylic ring (Figure 1B) of the 1R (+)-trans-anti-B[c]Ph–N2-dG adduct38 (PDB ID42: 1HX4). Molecular modeling was employed to extract the modified nucleoside from the 11-mer duplex which was capped with hydrogen atoms at the O3’ and O5’ positions. Then we added two aromatic rings to the B[c]Ph residue to create the DB[a,l]P adduct initial model. This model for the DB[a,l]P-dG adduct on the nucleoside level was then subjected to geometry optimization utilizing Gaussian 03 43 at the B3LYP/6-31G* level. The B[c]Ph in the NMR solution structure was then replaced by the geometry optimized DB[a,l]P and minor close contacts were alleviated with molecular modeling. This provided a 3’-side classical intercalation conformation in the starting model as was indicated by the NMR data (see Results).
The protocol we utilized to obtain the NMR distance-refined structure entailed first 3 ns of unrestrained MD simulation based on the initial model, followed by 1 ns of distance-restrained MD simulation (Table 1) with weights in the range of 20-40 kcal/mol·Å. The following distance restraint ranges were utilized: strong 2.5-3.3 Å, medium 3.3-4.5 Å, weak 4.5-6.5 Å. Additionally, the G6* imino to C17 amino proton distances were restrained to greater than 4 Å in accordance with the NMR data that demonstrates the absence of an NOE between the G6* imino and the C17 amino protons. These computations were carried out using the AMBER 9 simulation package44, the Cornell et al. force field45 with the parm99.dat parameter set46. The partial charges of the modified nucleotide were computed utilizing quantum mechanical Hartree Fock calculations with the 6-31G* basis set employing the Gaussian 03 package43. The charges were then fitted to each atomic center with the RESP algorithm45, 47. Other force field parameters for the DB[a,l]P-dG adduct were assigned to be consistent with the rest of the AMBER force field. Tables S1 and S2 (Supporting Information) give the force field parameters for the DB[a,l]P-dG adduct. The details of the MD simulation protocol are given in Supporting Information. Five structures that best represented the NMR data were extracted from the restrained MD simulation and used for analyses.
Table 1.
Observed intermolecular NOEs in the DB[a,l]P-dGdC 11-mer duplex, and achieved distances in the restrained MD simulations.
| DB[a,l]P proton | Chemical Shift | NOE | Intensity | Achieved distancesa |
|---|---|---|---|---|
| H1 | 7.81 ppm | C7 (H1′) | w | 5.6(0.5) |
| G6 (H1′) | m | 4.4(0.2) | ||
| H2 | 6.38 ppm | C7 (H1′) | w | 6.0(0.3) |
| C7 (H6) | w | 5.1(0.1) | ||
| G6 (H1′) | s | 3.0(0.2) | ||
| H3 | 6.77 ppm | G6(H2″) | w | 6.3(0.3) |
| H7 | 7.35 ppm | G16 (H8) | m | 3.9(0.1) |
| G16 (H1′) | w | 5.4(0.1) | ||
| G16 (H2′) | w | 5.2(0.2) | ||
| H8 | 7.10 ppm | G16 (H8) | m | 4.0(0.1) |
| G16 (H1′) | m | 4.1(0.1) | ||
| G16 (H2′) | w | 4.5(0.2) | ||
| G16 (H2″) | s | 2.9(0.1) | ||
| H9 | 7.25 ppm | G16 (H8) | w | 5.3(0.1) |
| G16 (H1′) | m | 3.8(0.1) | ||
| G16 (H2′) | w | 4.0(0.1) | ||
| G16 (H2″) | w | 5.5(0.1) | ||
| G16 (H3′) | w | 5.1(0.2) | ||
| H10 | 7.94 ppm | G16 (H1′) | m | 4.0(0.1) |
| H14 | 6.48 ppm | C7 (H1′) | w | 6.0(0.5) |
Ensemble average for the selected five structures, followed by the standard deviation.
Structural Analyses
The PTRAJ module of AMBER 9 was employed for structural analyses. Stacking interactions were evaluated by computing the van der Waals interaction energies between the DB[a,l]P or B[c]Ph aromatic rings and bases of adjacent base pairs (G6*:C17 and C7:G16) with the program ANAL of AMBER 9.44 DNA duplex groove dimensions and helicoidal parameters were analyzed using MD Toolchest48,49. The INSIGHTII program (Accelrys Software, Inc.) was employed for visualizing and model building. The figures and movies were prepared with Pymol50. Computations were carried out on our own cluster of Dell workstations, the multi-purpose high performance computing resources of New York University (NYU-ITS), and the National Science Foundation's Extreme Science and Engineering Discovery Environment (XSEDE) computational resources.
Results
NMR characterization of the DB[a,l]P-dG•dC duplex
A combination of NMR and computational methods were used to generate a model of the structure of the duplex 11-mer containing the DB[a,l]P-dG adduct embedded in the oligonucleotide 5’-CCATCG*CTACC at position G6* (Figure 1C), annealed with its unmodified complementary strand.
Exchangeable proton spectra
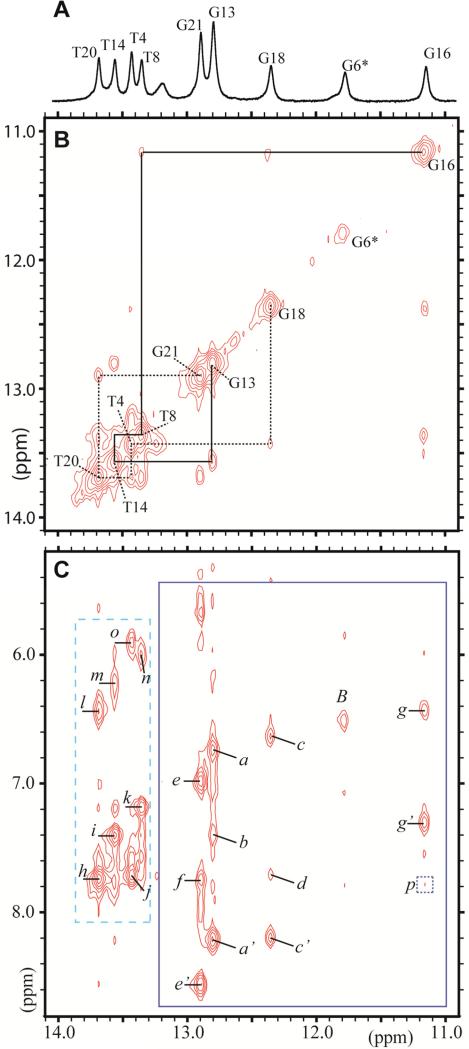
The 1D exchangeable proton spectrum (10.9–14.1 ppm) of the DB[a,l]P-dG•dC 11-mer duplex in H2O buffer, pH 6.8 at 5 °C, is shown in the top part of Figure 2A. The imino proton resonances were assigned by analyzing the imino-imino proton and imino-other proton NOE cross-peaks in the NOESY (150 ms mixing time) spectrum at 1 °C (Figure 2B and 2C) by standard procedures as described earlier51,52. Nine well resolved imino protons are observable including two up-field shifted imino protons at 11.83 and 11.16 ppm, identified as the imino protons of the modified guanine G6* and guanine G16, respectively. The imino protons of the two guanines G13 and G21 were clearly distinguishable, while those of the terminal G22 and G12 could not be identified due to fraying at the two ends of the duplex. The imino proton of G18, on the 5’-side of the adduct G6*, is characterized by a chemical shift of 12.4 ppm, while the G16 imino proton resonance on the 3’-side of G6* is upfield-shifted to 11.16 ppm. These observations suggest that the polycyclic aromatic residue is positioned on the 3’-side of G6* as described previously for the B[c]Ph-dG adduct 38. The imino-imino proton sequential assignments (Figure 2B) are shown for the central nine base pairs starting from G21 to G13. The sequential assignments can be traced without interruption up to the lesion site, the G6*:C17 base pair, as well as beyond this base pair. There is no detectable NOE corresponding to an interaction between the G6* imino proton and G16 (NH1), or G18 (NH1).
Figure 2.
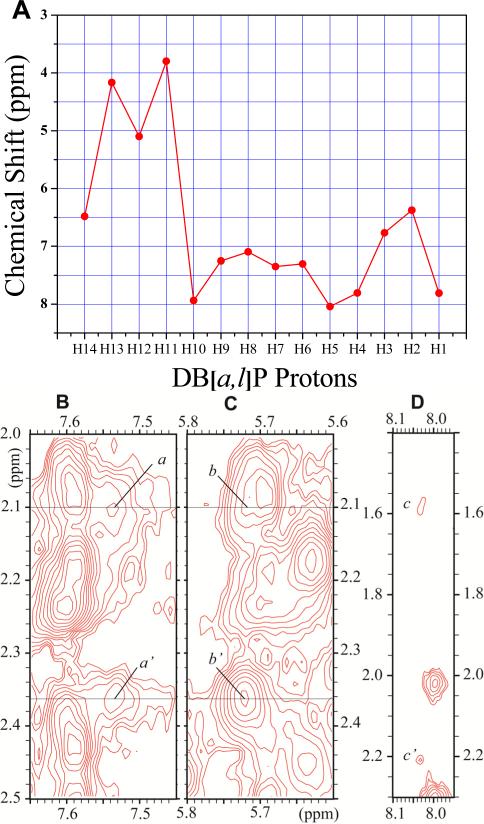
The 1-D and 2-D NMR spectra of the DB[a,l]P-dG adduct in the 11mer duplex in H2O buffer (pH 6.8) was recorded using a 500 MHz spectrometer. (A) The 1-D spectrum (10.9–14.1 ppm) at 5°C indicates the imino proton assignments. (B) Portion of a 2D NOESY (150 ms mixing time) contour plot at 1°C, in H2O buffer solution showing the neighboring imino-imino NOE connectivities that can be followed from G18 to G21 on the 5’-side of G6*:C17 (dotted line), and from G16 to G13 on the 3’-side of G6*:C17 ( solid line). The connectivities between G* and G16, G18 are missing. (C) Portion of a 2D NOESY (same conditions as B) showing the NOE connectivities between imino (11.0–13.8 ppm) and amino/base (5.5–8.8 ppm) protons. The rectangle with the dashed borders shows the characteristic Watson-Crick hydrogen bonded thymine imino to adenine H2 proton NOEs. The rectangle with solid borders shows the guanine imino proton - cytosine amino proton connectivities.
The cross peaks labeled a to o identifing DNA NOEs are assigned as follows, where ‘b’ and ‘nb’ define hydrogen-bonded and non-hydrogen-bonded amino protons, respectively): : a, C10(N4H)nb – G13(N1H); a’, C10(N4H)b – G13(N1H); b, A9(H2) – G13(N1H); c, C5(N4H)nb – G18(N1H); c’, C5(N4H)b – G18(N1H); d, A19(H2) – G18(N1H); e, C2(N4H)nb – G21(N1H); e’, C2(N4H)b – G21(N1H); f, A3(H2) – G21(N1H); g, C7(N4H)nb – G16(N1H); g’, C7(N4H)b – G16(N1H); h, A3(H2) – T20 (N3H); i, A9(H2) – T14 (N3H); j, A19(H2) – T4 (N3H); k, A15(H2) – T8 (N3H); l, A3(N6H) – T20 (N3H); m, A9(N6H) – T14 (N3H); n, A15(N6H) – T8 (N3H); o, A19(N6H) – T4 (N3H); p (square with dotted borders),G16(N1H) – DB[a,l]P(H1).
A portion of the NOESY contour plot corresponding to the NOE imino (11.0–13.8 ppm) to amino or base H2 (5.5–8.8 ppm) proton region, (150 ms mixing time) in H2O buffer at 1 °C, is shown in Figure 2C. The Watson-Crick base pairing of G6*:C17, the site of the lesion, is disrupted since there are no NOE cross-peaks between the G6* imino and the C17(NH2) amino protons (Figure 2C). By contrast, the NOE cross-peaks between the two flanking base pairs, the G16 imino and C7 amino protons (g,g’), as well as NOEs between the G18 imino and C5 amino protons (c,c’), are consistent with intact Watson-Crick hydrogen bonding patterns. However, these NOE cross-peaks are somewhat weaker in intensity than those characterizing the C2:G21 (e,e’) and the C10:G13 NOEs (a,a’), indicating that the Watson-Crick hydrogen bonding at the base pairs adjacent to the modification site are weakened. The amino protons (7.3 and 6.43 ppm) of C7, the partner base of G16, are upfield shifted relative to the amino protons of C2, C5, and C10 (Figure 2, and Table S3). On the other hand, the resonances of the amino protons of C5 are not upfield shifted. Taken together, the signicant upfield shifts of the C7 amino protons, and the large upfield shift of the G16 imino proton, are attributed to ring current shielding effects that arise from the intercalation of the aromatic DB[a,l]P ring system between the G6*:C17 and C7:G16 base pairs, as observed in the case of the B[c]Ph-dG adduct 38. The imino proton of G6* is also upfield shifted, although to a lesser extent than the imino proton of G16. While this proton could be more solvent-exposed than the adjacent base pair imino protons because NOEs between the G6* imino proton and its partner base C17 amino protons are missing, the linewidths of the G18, G6*, and G18 imino protons are similar (Figure 2A). This suggests that there is no rapid solvent exchange of the G6* imino proton with water protons and that its upfield shift is thus most likely due to ring current shielding effects arising from the stacking interactions between the DB[a,l]P moiety and the base pairs G6*:C17 and C7:G16.
Non-exchangeable proton spectra
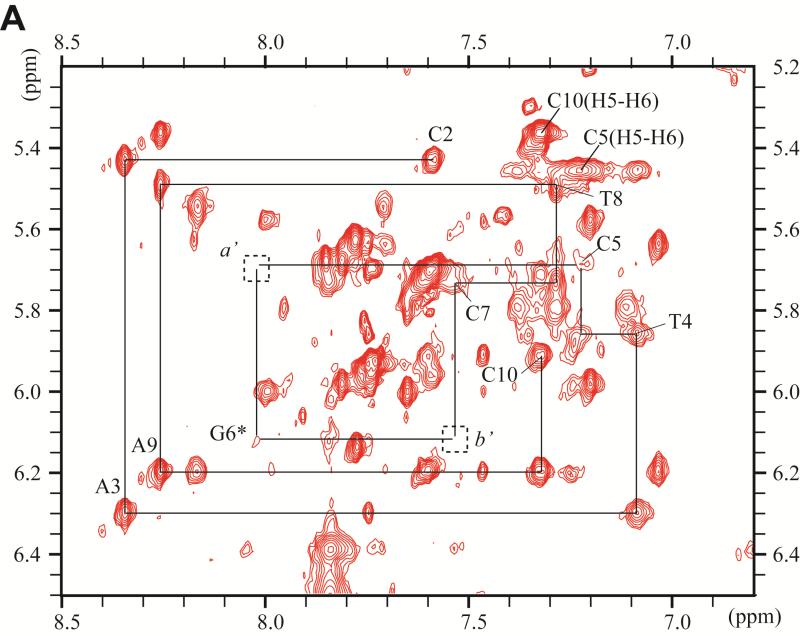
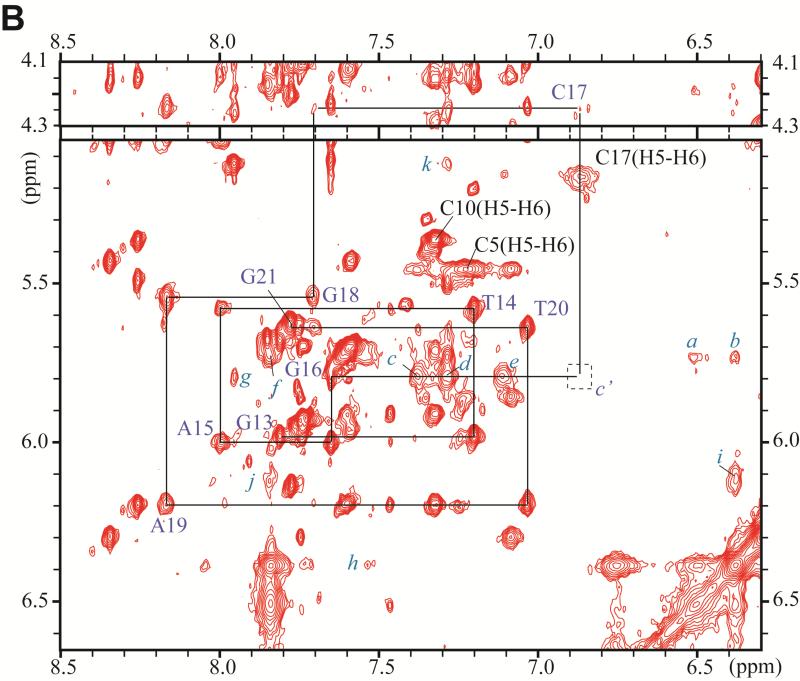
The non-exchangeable carcinogen protons have been assigned from the analysis of through-bond TOCSY and through space NOESY connectivities using standard procedures 52. Examples of these spectra are shown in Figures S1 and S2 of the Supporting Information. Figure 3 depicts an expanded NOESY spectrum (200 ms mixing time, 500MHz spectrometer) in D2O buffer solution at 28 °C. The NOE connectivities between base protons (6.3–8.5 ppm) and sugar H1’ proton (4.1–4.3 and 5.1–6.6 ppm) regions, are shown in this figure.
Figure 3.
Expanded contour plot of a NOESY spectrum (200ms mixing time) of the DB[a,l]P-dG lesion in the 11-mer duplex in D2O aqueous buffer solutions using a 500MHz spectrometer equipped with a cryoprobe at 28°C; in these 2-D plots, the horizontal axis defines the purine H8, and pyrimidine H5 or H6 chemical shifts, while the ordinate defines the deoxyribose H1’ proton chemical shifts. (A) Sequential assignment based on the connectivities between base protons and their own and 5′-flanking nucleotide H1′ deoxyribose protons for the C2-A3-T4-C5-G6*-C7-T8-A9-C10 segment. The NOEs can be followed from C2 to C5, and from C7 to C10. Connectivities are not observable between G6* and any of its flanking bases in this 2-D plot, but see Figure 4D for connectivities between G6* and C5. The squares labeled a′ and b′ identify the positions of the missing NOEs between C5 and G6*, and G6* and C7. (B) Sequential assignments for the unmodified strand in the segment G13-T14-A15-G16-C17-G18-A19-T20. The upper panel is cut lower than the bottom panel. Identification of NOE cross-peaks labeled a to k: between the DB[a,l]P moiety and DNA protons: a, C7(H1’) – DBP(H14); b, C7(H1’) – DBP(H2); c, G16(H1’) – DBP(H7); d, G16(H1’) – DBP(H9); e, G16(H1’) – DBP(H8); f, C7(H1’) – DBP(H1); g, G16(H1’) – DBP(H10); h, C7(H6) – DBP(H2); i, G6(H1’) – DBP(H2); j, G6(H1’) – DBP(H1); k, G6(H3’) – DBP(H9).
The square labeled c' identifies the position of the missing NOE beyweem G16(H1’) and C17(H6).
The sequential assignment of base protons and their own and 5’-flanking sugar H1’ protons can be traced on the modified strand within the C2-A3-T4-C5 and the C7-T8-A9-C10 segments (Figure 3A). The connectivities can also be traced within the G21-T20-A19-G18 and G16-A15-T14-G13 segments in the unmodified strand (Figure 3B). The NOE's between C5-G6*, G6*-C7, and G16-C17 are missing (boxes a’, b’ in Figure 3A and c’ in Figure 3B, respectively,). The NOEs between T4-C5 and C17-G18 are weak, while the C7-T8 NOE is comparable in intensity to the other, analogous NOEs on its 3′-side. These observations confirm that the normal B-DNA conformation is maintained in these sections of the 11-mer duplex, although NOEs at the T4-C5 and C17-G18 steps are weaker than normal. The two terminal base pairs of the oligonucleotide duplex are destabilized by end-fraying effects.
The bulky DB[a,l]P residue causes significant perturbations within the central (...C5-G6*-C7...)•(...G18-C17-G16...) region of the duplex with the loss of Watson-Crick hydrogen bonding at G6*:C17 that is evident from the H2O NOESY spectrum (Figure 2C). While the C5(H1′)-G6*(H8) NOE on the modified strand is not observable (Figure 3A, box a’), the C5(H2′ and H2″)-G6*(H8) NOEs are discernable, but very weak (Figure 4D, c and c’). Theses NOEs, as well as the absence of the G16(H1′)-C17(H6) NOE (Figure 3B, c’), and the very weak C17(H1′)−G18(H8) NOE (Figure 3B), provide important distance restraints for defining the structural alignments in the central region of the duplex (see below). The weak intranucleotide NOE's between base protons and their own H1′ protons at C17, G18, C5, C7, and G6* (Figure 3) are attributed to a broadening of these resonances, suggesting that these bases are more mobile than those in the adjacent, more stable regions of the duplex. The C17(H5-H6) and the C5(H5-H6) intranucleotide NOEs are also significantly broadened relative to the C10(H5-H6) NOE that is distant from the site of the lesion. These broadening effects are consistent with enhanced mobilities due to the disrupted hydrogen bonding at G6*:C17 that contributes to the low intensity of the C17(H6-H1′) and other NOEs in the central region of the duplex (Figure 3). Similarly, broadening effects diminish the intensities of the C7(H1′-H2′,H2″) (Figure 4C, b and b’) and C7(H6-H2′,H2″) NOEs (Figure 4B, a and a’).
Figure 4.
(A) Plot of the chemical shifts of the DB[a,l]P aromatic ring protons. (B) - (D): Expanded contour plots of the NOESY spectrum (200ms mixing time) of the DB[a,l]P-dG lesion in the 11mer duplex in D2O aqueous buffer solution using a 500MHz spectrometer with a cryoprobe at 28 °C. (B) NOEs between the C7 base H6 proton and its own 2′, and 2″ sugar protons: a, C7(H6) – C7(H2′); a’, C7(H6) – C7(H2″). (C) NOEs between C7-H1′ and its own 2′ and 2″ sugar protons: b, C7(H1′) – C7(H2′); b’: C7(H1′) C7(H2″). (D) NOEs between the G6* base proton H8 and flanking base C5 sugar protons: c, G6*(H8) – C5(H2′); c’, G6*(H8) – C5(H2″).
The C17-H1’ proton is dramatically upfield shifted to 4.25 ppm (Table S3 and Figure 3B) from values of ~ 5.3 – 6.2 exhibited by H1’ protons of other C residues in the same duplex positioned in the minor grove (Table S3). In addition, several other upfield chemical shifts are noted (a) the H2’ and H2” sugar protons of C17 are upfield shifted by 0.35 – 0.65 ppm, and (b) the H6 proton of C17 also exhibits a significant upfield shift of 0.3 – 0.8 ppm relative to the same proton in other C residues (Table S3). Such proton chemical shifts are consistent with position-dependent ring current contributions from the DB[a,l]P aromatic ring system. In contrast to these upfield shifts, G6*(H1′) is downfield shifted by 0.55 ppm relative to the unmodified case in the same duplex (Table S3), and is attributed to the unstacking of the G6* base and its greater exposure to solvent. Compared to their corresponding values in the unmodified duplex (Table S4), the two protons, H8 of G6* and H6 of C7, were also downfield shifted around ~ 0.2 - 0.3 ppm, consistent with a decrease in the normal stacking interactions.
Carcinogen NMR assignments
The chemical shifts of the aliphatic and aromatic DB[a,l]P ring protons are listed in Table 1 and are graphically represented in Figure 4A. The non-exchangeable carcinogen protons have been assigned from analysis of through-bond TOCSY (Figure S1) and through-space NOESY connectivities. There are strong TOCSY cross-peaks between most well resolved benzylic and aliphatic protons, except for the H6 (7.31 ppm) and H7 (7.35 ppm) protons which overlap with one another.
Intermolecular NOEs
A total of 20 intermolecular NOEs have been identified and assigned between the non exchangeable DB[a,l]P protons and non-exchangeable nucleic acid protons which involve both the modified and unmodified strands. While there may be additional intermolecular NOEs, we did not consider them if they were not shown as well separated NOE cross-peaks because of significant overlap and/or peak broadening. These intermolecular NOEs were classified as strong, medium and weak (Table 1).
The DB[a,l]P protons are separated into three groups. One group includes DB[a,l]P residue protons H1, H2, H3 and H14 (Figure 1) that exhibit NOEs with G6* and C7 in the modified strand. The second group comprises protons H7 to H10 that exhibit NOE connectivities only to G16 protons in the unmodified strand. The third group includes protons that do not exhibit any NOEs under the conditions of our experiments. No intermolecular NOEs were observed between the non-exchangeable DB[a,l]P protons and the exchangeable DNA protons, including the imino protons. We note that most of the intermolecular NOEs are observed between the DB[a,l]P residue protons and DNA nucleotide moieties on the 3’-side of G6*.
An unidentified NOE cross-peak between G16 (NH1) and G18 (NH1) in Figure 2 results, most likely, from the presence of a small fraction of the migrated 14S (+)-cis-anti-DB[a,l]P–N2-dG adduct at G16 and/or G1839, as described in the Methods section. We infer that this cross-peak stems from this migration because it was not possible to bring this interproton distance within NOE range in our modeling computations that were based on all the reliable NOEs (see below). Some other unidentified cross-peaks may also stem from this migrated adduct and were not further investigated since the appearance of such NOE connectivities increased markedly with increasing storage time of the samples (weeks to months) after the initial annealing of the two strands to form duplexes in solution (data not shown).
NMR distance-restrained structure: Intercalation of the DB[a,l]P ring system on the 3’-side of the modified guanine with disrupted base pairing at the lesion site
We utilized MD simulations with distance restraints to obtain structural models consistent with the NOEs and the NMR-derived data indicating ruptured Watson-Crick pairing at the lesion site, as shown in Table 1. The NMR data indicate that the DB[a,l]P aromatic rings are intercalated on the 3’-side of the modified guanine; in this respect it is conformationally similar to an NMR solution structure of the B[c]Ph-dG adduct (Figure 1B) with the same stereochemical properties38. Accordingly, we created an initial model of the DB[a,l]P-dG adduct based on the B-DNA NMR solution structure of this B[c]Ph adduct, and carried out restrained MD simulations in explicit solvent and with neutralizing sodium ions, as detailed in the Methods section. We then selected five structures from the ensemble derived from the 500 – 1000 ps interval of the restrained MD simulation that best represent the NMR data. Table 1 shows the mean achieved distances with standard deviations for these five structures. We note that all achieved inter-proton distances, when their standard deviations are included, are very near the assigned distance bounds. The ensemble of the five structures is shown in Figure S3 and one selected structure is shown in Figures 5A and 6. All the analyses described below are the average values for these five structures.
Figure 5.
(A) A DB[a,l]P-dG structure in the 11-mer duplex (Figures 1B,C), selected from the five structures extracted from the restrained MD simulation that best represent the NMR data; the view is from the minor groove of the central trimer. The DB[a,l]P moiety is in orange, the modified guanine and its partner cytosine are in blue, the 3’-side base pair C7:G16 are in cyan, and the rest of the DNA is in light gray. The hydrogen atoms in the DNA duplexes are not displayed for clarity.
(B) Comparison of the stacking patterns of DB[a,l]P-dG, B[c]Ph-dG38 and DB[a,l]P-dA68 adducts in double-stranded oligonucleotides; the views are looking down the helix axis along the 5′→ 3′ direction. In the guanine adducts the DB[a,l]P and B[c]Ph rings are intercalated between G6*:C17 and C7:G16, while for the adenine adduct intercalation is between C5:G18 and G6*:C17. The color code is the same as Figure 5A, except that the B[c]Ph moiety is in yellow. The hydrogen atoms in the DNA duplexes are not displayed for clarity except for the hydrogens of the C7 amino group in DB[a,l]P-dG. A stereoview of the DB[a,l]P-dG adduct is shown in Figure S4 of the Supporting Information. (C) Comparisons of guanine imino to cytosine amino distances (Å) in the NMR distance-refined model (mean values of five structures) for the designated base pairs; the NMR data indicate weakened or absent Watson-Crick pairing at C5:G18, G6*:C17 and C7:G16, while C2:G21 which is more distant from the site of modification has a normal base pair value.
Figure 6.
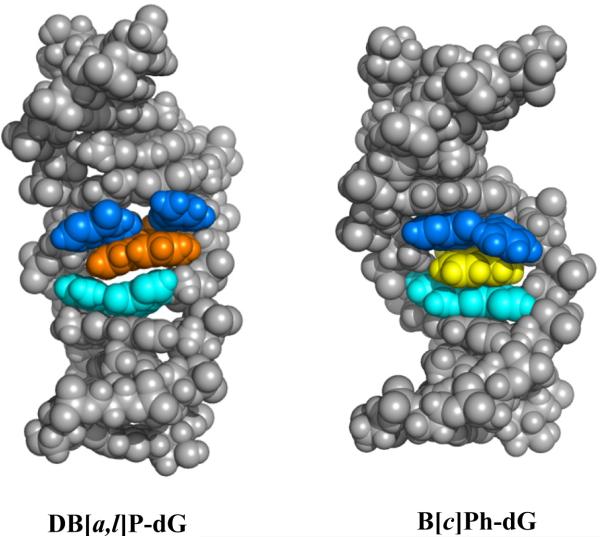
Comparison of DB[a,l]P-dG and B[c]Ph-dG modified duplexes rendered in CPK. The color code is as in Figure 5B.
As shown in Figure 5A, the intercalation of the DB[a,l]P rings on the 3’-side of the G6*, between the G6*:C17 and C7:G16 base pairs, is from the minor groove side. The modified base pair is fully disrupted, as indicated by the absence of an NOE connectivity between the G6* imino and the amino protons of the C17 partner base (Figure 2C). However, the adjacent bases within the lesion site remain stacked with the DB[a,l]P aromatic ring system. The C17 base stacks mostly with the 8-9 ring (Figure 1) of the aromatic DB[a,l]P residue, while G6* stacks with the 2-3-4 ring. However, the C17 is quite mobile as shown in Figure S3, which is consistent with our inability to establish any NOEs between the DB[a,l]P and C17 protons. On the 3’-side of the DB[a,l]P ring system, the G16 stacks well with the DB[a,l]P aromatic rings at DB[a,l]P H7, H8,H9 and H10 (Table 1), and C7 stacks modestly, but with one amino proton stacked well with the DB[a,l]P 2-3 ring edge (Figure 5B; a stereoview of the DB[a,l]-dG adduct is shown in Figure S4). The observed upfield shifts of G6*:C17 and C7:G16 protons are a reflection of these stacking interactions. All other base pairs are maintained, although the base pairs adjacent to the lesion on its 3′- and 5′-sides are perturbed; this is consistent with the NMR data, which shows somewhat weaker imino to amino NOEs in the case of the C5:G18 and C7:G16 base pairs (Figure 2C, NOEs c,c’ and g,g’, respectively) as compared to normal Watson-Crick base pairs (Figure 2C, NOEs a,a’ and e,e’). In our models these distances are longer than normal for B-DNA as shown in Figure 5C. Other structural parameters also manifest disturbances as described below.
With the DB[a,l]P moiety attached to the minor groove exocyclic amino group of G6* (Figure 1B), the benzylic ring of the adduct is positioned in the minor groove, and its hydroxyl groups protrude into the solvent. The covalent linkage bond torsion angles α' and β' (Figure 1B), and the glycosidic torsion angle χ in our model have average values of 154±14°, 110±10°, and 328±9°, respectively. The glycosidic torsion is in the anti domain, as observed by NMR. The α' and β' values are within the low energy regions computed previously for benzo[a]pyrene- and benzo[c]phenanthrene diol epoxide-derived nucleoside adducts with R stereochemistry at the linkage site.53,54 The fjord region deviates from planarity with the torsion angle δ' (C15-C17-C20-C1) (Figure 1) having an average value of 28±4°. This twist optimizes stacking between the DB[a,l]P aromatic rings and the C7:G16 base pair; its positive value is characteristic of R stereoisomeric intercalated fjord region adducts derived from B[c]P and DB[a,l]P38,55,56.
We analyzed the helicoidal and base pairing parameters in our model to assess further the distortions to the B-DNA structure produced by the minor groove intercalation of the bulky DB[a,l]P ring system. This stacked position of the bulky DB[a,l]P moiety with its five aromatic rings has its distal 5-6-7 and 8-9 rings in close steric contact with the backbone, and this steric hindrance drives the rupture of the G6*:C17 base pair (Figure 5). The rupturing is manifested in the distances between the heavy atoms that would have comprised the hydrogen bond (all greater than 4 Å, while the normal values are ~ 3 Å,57), the G6* imino to C17 amino distances ( 4.3 and 5.9 Å, normal values of ~ 2.4 and 3.9 Å, Figure 5C) and base pair stretch (2.8 Å, normal value ~ 0 Å, Figure S5A). The ruptured base pairing also causes extensive groove opening on both major and minor groove sides (Figure S5E and S5F). These disturbances are propagated to the adjacent residues involving the C5:G18 and C7:G16 base pairs. The ruptured base pair accommodates the strain of the intercalated DB[a,l]P residue without the normal untwisting at that residue58, but there is severe untwisting at the C5:G18 and C7:G16 residues (Figure S5D). Rise is enlarged between the G6*:C17 and C7:G16 residues (Figure S5C) to form the intercalation pocket as usual58. Although not hydrogen bonded, the damaged base G6* and partner C17, being stacked into the helix over the DB[a,l]P aromatic ring system, are positioned in buckled and propeller-twisted orientations. The buckle is 29±10° at G6*:C17 but adopts a value of –38±7° at the C7:G16 pair; this creates a wedge-shaped intercalation pocket which, together with the fjord region twist, optimizes stacking of the DB[a,l]P ring system with adjacent base pairs. The NMR data indicate weakened Watson-Crick pairing for the C5:G18 and C7:G16 base pairs which is manifested by their high buckle compared to the unmodified DNA (Figure S5B). Elongated C5:G18 and C7:G16 imino to amino distances are seen in our structures (Figure 5C) for these two base pairs consistent with the experimentally observed weakened NOE connectivities.
Discussion
The DB[a,l]P ring system shares structural features with both the planar, rigid bay region B[a]P and the twisted fjord B[c]Ph aromatic ring systems (Figure 1). It has one additional ring, as compared to B[a]P, which creates the fjord region. The smaller B[c]Ph has the same fjord topology as DB[a,l]P, but has two fewer aromatic rings (Figure 1). All three PAH adducts, DB[a,l]P-dG, B[a]P-dG and B[c]Ph-dG, form stereochemically identical adducts with the amino groups of guanine. While the B[c]Ph-dG adduct is intercalated into the helix from the minor groove on the 3’-side of the modified guanine and retains Watson-Crick base pairing at G6*:C17, the B[a]P-dG adduct is positioned in the minor groove on the 3’-side of the damaged guanine. In this work we wished to elucidate the impact on adduct conformation of the two additional distal aromatic rings in the DB[a,l]P residue, relative to the three aromatic rings in the B[c]Ph-dG case38. We sought to distinguish between two hypotheses: the DB[a,l]P-dG adduct would assume (1) a minor groove conformation like the B[a]P-dG adduct37, or (2) a 3’-side intercalated structure like the fjord B[c]Ph-dG adduct38. We reasoned that the additional two aromatic rings in the DB[a,l]P-dG, relative to the B[c]Ph-dG adduct might hinder intercalation from the narrow minor groove, and produce instead a minor groove alignment like the B[a]P-dG adduct.
The NMR distance-restrained MD simulation clearly demonstrates that the DB[a,l]P-dG adduct assumes a 3’ intercalative conformation like the B[c]Ph-dG adduct. However, the G6*:C17 Watson-Crick base pair at the lesion site is disrupted. This effect is attributed to the greater number and organization of aromatic rings in the DB[a,l]P-dG adduct; the two additional distal rings cause steric hindrance that disfavors the simultaneous intercalation and the maintenance of intact G6*:C17 hydrogen bonding, as in the structure of the B[c]Ph-dG adduct. The NMR evidence shows that the DB[a,l]P ring system is intercalated on the 3’-side of G6*. This is revealed by (1) the up-field shifts of the imino protons of G6* and G16, which arise from ring current shielding effects due to the stacking interactions between the DB[a,l]P moiety and the adjacent bases, and (2) the NOEs that are observed between the DB[a,l]P and C7:G16 base pair protons on the 3’-side of G6* (Table 1). Disruption of Watson-Crick pairing at the lesion site G6*:C17 is indicated by the absence of NOEs between the G6* imino proton to its Watson-Crick partner C17 amino protons. The NMR data also shows that G6* adopts the anti glycosidic bond conformation, based on the weak NOE between the base G6* (H8) and the H1’ sugar proton, and that all other Watson-Crick base pairs, except the terminal ones, are intact. However, structural perturbations are in evidence at the base pairs flanking the damaged base pairs on both sides, as shown by weakened imino to amino NOEs in the case of the C5:G18 and C7:G16 base pairs which are buckled and propeller twisted (Figures 5 and S5). In addition, the duplex structure within the central trimer is significantly distorted with enlarged minor and major grooves and untwisting of base pairs (Figure S5). While the G6*:C17 base pair is ruptured, these two bases remain stacked with the DB[a,l]P ring system, consistent with the observed upfield shifts of the G6* imino and the C17 sugar and base protons.
The type of structure adopted by the DB[a,l]P-dG adduct is a novel motif, distinct from the classical intercalation family which is characterized by intact Watson-Crick base pairing even at G6*:C17 in the NMR structures of the 1R (+) and 1S (–)-trans-anti-B[c]Ph–N2-dG, 1R (+) and 1S (–)-trans-anti-B[c]Ph–N6 dA adducts38,56,59 and fjord PAH 1R (+)-trans-anti-benzo[g]chrysene–N6-dA adduct60. However, the G6*:C17 Watson-Crick pairing is fully disrupted in the present DB[a,l]P-dG adduct because the distal 5-6-7 and 8-9 rings are in steric close contact with the DNA backbone of the partner strand. This steric hinderance is alleviated by the rupture of the G6*:C17 hydrogen bonds, but without displacement of the bases into a groove. This new conformational theme is therefore also very distinct from the well established base displaced intercalation structures, in which the bulky intercalated PAH aromatic ring system displaces the modified guanine and partner cytosine residues from the helix30, 61-63. It is also distinct from the recently detailed aristolactam II-dA lesion in which the aromatic ring system, together with the linked adenine residue, is inserted into the helix, but the partner T is displaced64.
To gain further insights into the structural impact of the additional two rings in the DB[a,l]P-dG adduct, compared to the smaller B[c]Ph-dG adduct, we analyzed the characteristics of the nine simulated structures of the B[c]Ph-dG adduct (PDB ID 1HX4) in the same sequence context38. These are compared with our selected five representative NMR solution structures of the DB[a,l]P adduct (see Methods) as shown in Figures 5B and 6. With the Watson-Crick base pair preserved in the B[c]Ph-dG case, the grooves are much less enlarged, and untwisting is less pronounced and spread over three base pair steps, while in the case of the DB[a,l]P-dG adduct, untwisting at the lesion site is not needed due to the ruptured base pair (Figure S5). The buckle follows similar trends in the B[c]Ph-dG as in the case of the DB[a,l]P-dG adduct, creating a similar wedge-shaped intercalation pocket. Computed Van der Waals stacking interactions (see Methods) between the B[c]Ph aromatic rings and bases within the intercalation pocket are –21 kcal/mol, which is greater by ~ 2.0 kcal/mol than in the DB[a,l]P-dG adduct case, even though the latter has two more aromatic rings. The reason for this surprisingly lower stacking energy for the DB[a,l]P-dG adduct stems from the ruptured Watson-Crick base pairing which shifts the G6* and C17 into positions that are less favorable for overlap interactions (Figure 5B).
A further comparison can be made with the bay region B[a]P-dG adduct (Figure 1) which is characterized by a B-DNA minor groove conformation with the aromatic ring system oriented towards the 3’-end of the modified strand. The question that arises is why the rigid and planar bay B[a]P-dG region adduct is located in the minor groove, while the B[c]Ph-dG and DB[a,l]P-dG adducts assume intercalated conformations with insertion of the aromatic ring systems into the double-helix from the minor groove side. We speculate that intercalation from the narrow minor groove is facilitated by the flexibility of the fjord region 9,10,11,12 (B[c]Ph) and the 1,2,3,4 (DB[a,l]P aromatic rings, while the rigidity of the bay region B[a]P residue inhibits the intercalative insertion from the minor groove. Furthermore, prior modeling studies suggested that an intercalated B[a]P-dG adduct does not stack as well in the intercalation pocket as the B[c]Ph-dG adduct38. The advantages associated with the minor groove conformation is that Watson-Crick base pairing is maintained, the helix is not stretched and not extensively unwound, but the disadvantage is that one face of the aromatic ring system is exposed to the aqueous solvent. On the other hand, the intercalated structures require DNA stretching and unwinding58, but have the advantage that the aromatic rings are shielded from the solvent and are stabilized by van der Waals interactions with the neighboring bases pairs within the intercalation pocket. In the DB[a,l]P-dG structure, Watson-Crick pairing is disrupted at the site of the lesion, but the carcinogen-base stacking interactions and shielding of the aromatic rings from solvent, nevertheless, favor the intercalated conformation.
Conclusions and biological relevance
A combination of NMR methods and computational approaches was used to determine the conformational properties of the DB[a,l]P-dG adduct with 14R absolute configuration at the covalent N2-dG linkage site. The bulky polycyclic aromatic DB[a,l]P ring system is intercalated on the 3’-side of the modified guanine from the sterically crowded minor groove side, causing ruptured Watson-Crick base pairing at the lesion site and distortions to the local B-DNA structure, while retaining van der Waals stacking interactions of the DB[a,l]P ring system with adjacent DNA base pairs. This represents a novel conformational theme differing from the previously established classical intercalation and base displaced intercalation motifs. The NMR data provides a foundation for structural comparisons with other PAH diol epoxide-derived lesions and their biological response characteristics such as nucleotide excision repair. While the DB[a,l]P-dG adduct is an excellent NER substrate, the stereochemically identical 14R (+)-trans-anti-DB[a,l]P–N6-dA (DB[a,l]P-dA) adduct is resistant to repair in HeLa cell extracts65. Although both adducts are intercalated, the DB[a,l]P-dA lesion stabilizes the modified DNA duplex66, while the DB[a,l]P-dG adduct is strongly destabilizing67. The structural origin of this difference is essentially due to the intercalation from the minor groove on the 3’-side of the modified guanine in the case of the DB[a,l]P-dG adduct, in contrast with the major groove 5’-side intercalation of the DB[a,l]P-dA adduct; in the DB[a,l]P-dA case Watson-Crick pairing at the lesion site is maintained and there is excellent stacking between the DB[a,l]P rings and the intercalation pocket (Figure 5B)55,68. Future work is needed to further elucidate the intriguing structure-function relationships in NER recognition mechanisms that are attributed to the impact of DNA adduct topology and stereochemistry on the recognition of the lesions.
Supplementary Material
Acknowledgement
The dibenzo[a,l]pyrene diol epoxide used in some of the experiments was obtained from Dr. S. Amin at the NCI Chemical Carcinogen Storage Facility at Pennsylvania State University (Hershey, PA). The major fraction of the site-specifically modified oligonucleotides used in this work was supplied by Dr. Haruhiko Yagi and Dr. Albrecht Seidel which is gratefully acknowledged. This work used computational resources of the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number MCB060037. Components of this work were conducted in the Shared Instrumentation Facility at NYU that was constructed with support from the Research Facilities Improvement Grant C06 RR-16572 from the National Center for Research Resources, NIH. Furthermore, NMR resources were also used at the New York Structural Biology Center (NYSBC) where N.E.G. is an affiliated Principal Investigator. The NYSBC is a STAR site supported by the New York State Office of Science, Technology and Academic Research and its NMR resources are supported by NIH P41 GM66354. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Funding: The research reported in this publication was supported by the National Cancer Institute (NCI) of the National Institutes of Health under award numbers CA-099194 (N.E.G.) and CA-28038 (S.B). Computational infrastructure and systems management was partially supported by NIH Grant CA-75449 to S.B. This study was supported in part by NCI Contract NO2-CB-81013-74 (S.Amin).
Abbreviations
- PAH
polycyclic aromatic hydrocarbon
- DE
diol epoxides
- DB[a,l]P
dibenzo[a,l]pyrene
- B[c]Ph
benzo[c]phenanthrene
- B[a]P
benzo[a]pyrene
- DB[a,l]P-dG
14R (+)-trans-anti-DB[a,l]P–N2-dG
- B[a]P-dG
10R (–)-trans-anti-B[a]P–N2 dG adduct
- B[c]Ph-dG
1R (+)-trans-anti-B[c]Ph–N2-dG
- DB[a,l]P-dA
14R (+)-trans-anti-DB[a,l]P–N6-dA
- (−)-anti-DB[a,l]PDE
(–)-anti-14R,13S,12S,11R-DB[a,l]P diol epoxide
- (+)-anti-DB[a,l]PDE
(+)-anti-14S,13R,12R,11S-DB[a,l]P diol epoxide
- MD
molecular dynamics
- NER
nucleotide excision repair
- RMSD
root mean square deviation
- NOESY
Nuclear Overhauser effect spectroscopy
- COSY
correlated spectroscopy
- TOCSY
total COSY
- DSS
2,2-dimethyl-2-silapentane-5-sulfonate, sodium salt
Footnotes
Supporting Information
Experimental NMR TOCSY and NOSEY spectra and nucleic acid chemical shifts of the DB[a,l]P-dG modified and unmodified duplexes are provided. Details of MD simulation protocol and force field parameters, superposition of five selected structures, stereoview of the DB[a,l]P-dG intercalation pocket and helical parameters and groove widths analyses are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
NMR structures of the DB[a,l]P-dG adduct have been deposited as Protein Data Bank entry 2lzk.
References
- 1.Cavalieri EL, Higginbotham S, RamaKrishna NV, Devanesan PD, Todorovic R, Rogan EG, Salmasi S. Comparative dose-response tumorigenicity studies of dibenzo[alpha,l]pyrene versus 7,12 dimethylbenz[alpha]anthracene, benzo[alpha]pyrene and two dibenzo[alpha,l]pyrene dihydrodiols in mouse skin and rat mammary gland. Carcinogenesis. 1991;12:1939–1944. doi: 10.1093/carcin/12.10.1939. [DOI] [PubMed] [Google Scholar]
- 2.Cavalieri EL, Higginbotham S, Rogan EG. Dibenzo[a,I]pyrene: The most potent carcinogenic aromatic hydrocarbon. Polycycl. Aromat. Comp. 1994;6:177–183. [Google Scholar]
- 3.Luch A. On the impact of the molecule structure in chemical carcinogenesis. Experientia, Suppl. 2009;99:151–179. doi: 10.1007/978-3-7643-8336-7_6. [DOI] [PubMed] [Google Scholar]
- 4.Luch A. Nature and nurture – Lessons from chemical carcinogenesis. Nat. Rev. Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 5.Bostrom CE, Gerde P, Hanberg A, Jernstrom B, Johansson C, Kyrklund T, Rannug A, Tornqvist M, Victorin K, Westerholm R. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ. Health Perspect. 2002;110(Suppl 3):451–488. doi: 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz AK, Carrell HL, Glusker JP. Dibenzo[a,l]pyrene (dibenzo[def,p]chrysene): fjord-region distortions. Carcinogenesis. 1998;19:1641–1648. doi: 10.1093/carcin/19.9.1641. [DOI] [PubMed] [Google Scholar]
- 7.Karle IL, Yagi H, Sayer JM, Jerina DM. Crystal and molecular structure of a benzo[a]pyrene 7,8-diol 9,10-epoxide N2-deoxyguanosine adduct: absolute configuration and conformation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:1433–1438. doi: 10.1073/pnas.0307305101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amin S, Desai D, Dai W, Harvey RG, Hecht SS. Tumorigenicity in newborn mice of fjord region and other sterically hindered diol epoxides of benzo[g]chrysene, dibenzo[a,l]pyrene (dibenzo[def,p]chrysene), 4H-cyclopenta[def]chrysene and fluoranthene. Carcinogenesis. 1995;16:2813–2817. doi: 10.1093/carcin/16.11.2813. [DOI] [PubMed] [Google Scholar]
- 9.Hecht SS, el-Bayoumy K, Rivenson A, Amin S. Potent mammary carcinogenicity in female CD rats of a fjord region diol-epoxide of benzo[c]phenanthrene compared to a bay region diol-epoxide of benzo[a]pyrene. Cancer Res. 1994;54:21–24. [PubMed] [Google Scholar]
- 10.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 11.Szeliga J, Dipple A. DNA adduct formation by polycyclic aromatic hydrocarbon dihydrodiol epoxides. Chem. Res. Toxicol. 1998;11:1–11. doi: 10.1021/tx970142f. [DOI] [PubMed] [Google Scholar]
- 12.Gill HS, Kole PL, Wiley JC, Li KM, Higginbotham S, Rogan EG, Cavalieri EL. Synthesis and tumor-initiating activity in mouse skin of dibenzo[a,l]pyrene syn- and anti-fjord region diolepoxides. Carcinogenesis. 1994;15:2455–2460. doi: 10.1093/carcin/15.11.2455. [DOI] [PubMed] [Google Scholar]
- 13.Ralston SL, Lau HH, Seidel A, Luch A, Platt KL, Baird WM. The potent carcinogen dibenzo[a,l]pyrene is metabolically activated to fjord-region 11,12-diol 13,14-epoxides in human mammary carcinoma MCF-7 cell cultures. Cancer Res. 1994;54:887–890. [PubMed] [Google Scholar]
- 14.Luch A, Seidel A, Glatt H, Platt KL. Metabolic activation of the (+)-S,S- and (-)-R,R-enantiomers of trans-11,12-dihydroxy-11,12-dihydrodibenzo[a,l]pyrene: stereoselectivity, DNA adduct formation, and mutagenicity in Chinese hamster V79 cells. Chem. Res. Toxicol. 1997;10:1161–1170. doi: 10.1021/tx970005i. [DOI] [PubMed] [Google Scholar]
- 15.Amin S, Krzeminski J, Rivenson A, Kurtzke C, Hecht SS, el-Bayoumy K. Mammary carcinogenicity in female CD rats of fjord region diol epoxides of benzo[c]phenanthrene, benzo[g]chrysene and dibenzo[a,l]pyrene. Carcinogenesis. 1995;16:1971–1974. doi: 10.1093/carcin/16.8.1971. [DOI] [PubMed] [Google Scholar]
- 16.Zhang SM, Chen KM, Aliaga C, Sun YW, Lin JM, Sharma AK, Amin S, El-Bayoumy K. Identification and quantification of DNA adducts in the oral tissues of mice treated with the environmental carcinogen dibenzo[a,l]pyrene by HPLC-MS/MS. Chem. Res. Toxicol. 2011;24:1297–1303. doi: 10.1021/tx200188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahadevan B, Dashwood WM, Luch A, Pecaj A, Doehmer J, Seidel A, Pereira C, Baird WM. Mutations induced by (-)-anti-11R,12S-dihydrodiol 13S,14R-epoxide of dibenzo[a,l]pyrene in the coding region of the hypoxanthine phosphoribosyltransferase (Hprt) gene in Chinese hamster V79 cells. Environ. Mol. Mutagen. 2003;41:131–139. doi: 10.1002/em.10136. [DOI] [PubMed] [Google Scholar]
- 18.Guttenplan JB, Kosinska W, Zhao ZL, Chen KM, Aliaga C, Deltondo J, Cooper T, Sun YW, Zhang SM, Jiang K, Bruggeman R, Sharma AK, Amin S, Ahn K, El-Bayoumy K. Mutagenesis and carcinogenesis induced by dibenzo[a,l]pyrene in the mouse oral cavity: a potential new model for oral cancer. Int. J. Cancer. 2011;130:2783–2790. doi: 10.1002/ijc.26344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon JH, Besaratinia A, Feng Z, Tang MS, Amin S, Luch A, Pfeifer GP. DNA damage, repair, and mutation induction by (+)-Syn and (-)-anti-dibenzo[a,l]pyrene-11,12-diol-13,14-epoxides in mouse cells. Cancer Res. 2004;64:7321–7328. doi: 10.1158/0008-5472.CAN-04-1094. [DOI] [PubMed] [Google Scholar]
- 20.Mahadevan B, Luch A, Bravo CF, Atkin J, Steppan LB, Pereira C, Kerkvliet NI, Baird WM. Dibenzo[a,l]pyrene induced DNA adduct formation in lung tissue in vivo. Cancer Lett. 2005;227:25–32. doi: 10.1016/j.canlet.2004.11.056. [DOI] [PubMed] [Google Scholar]
- 21.Zhang SM, Chen KM, Aliaga C, Sun YW, Lin JM, Sharma AK, Amin S, El-Bayoumy K. Identification and quantification of DNA adducts in the oral tissues of mice treated with the environmental carcinogen dibenzo[a,l]pyrene by HPLC-MS/MS. Chem. Res. Toxicol. 2011;24:1297–1303. doi: 10.1021/tx200188j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arif JM, Smith WA, Gupta RC. DNA adduct formation and persistence in rat tissues following exposure to the mammary carcinogen dibenzo[a,l]pyrene. Carcinogenesis. 1999;20:1147–1150. doi: 10.1093/carcin/20.6.1147. [DOI] [PubMed] [Google Scholar]
- 23.Devanesan P, Ariese F, Jankowiak R, Small GJ, Rogan EG, Cavalieri EL. A novel method for the isolation and identification of stable DNA adducts formed by dibenzo[a,l]pyrene and dibenzo[a,l]pyrene 11,12-dihydrodiol 13,14-epoxides in vitro. Chem. Res. Toxicol. 1999;12:796–801. doi: 10.1021/tx980203p. [DOI] [PubMed] [Google Scholar]
- 24.Dreij K, Seidel A, Jernstrom B. Differential removal of DNA adducts derived from anti-diol epoxides of dibenzo[a,l]pyrene and benzo[alpha]pyrene in human cells. Chem. Res. Toxicol. 2005;18:655–664. doi: 10.1021/tx0497090. [DOI] [PubMed] [Google Scholar]
- 25.Ralston SL, Seidel A, Luch A, Platt KL, Baird WM. Stereoselective activation of dibenzo[a,l]pyrene to (-)-anti (11R,12S,13S,14R)- and (+)-syn(11S,12R,13S,14R)-11,12-diol-13,14-epoxides which bind extensively to deoxyadenosine residues of DNA in the human mammary carcinoma cell line MCF-7. Carcinogenesis. 1995;16:2899–2907. doi: 10.1093/carcin/16.12.2899. [DOI] [PubMed] [Google Scholar]
- 26.Spencer WA, Singh J, Orren DK. Formation and differential repair of covalent DNA adducts generated by treatment of human cells with (+/-)-anti-dibenzo[a,l]pyrene-11,12-diol-13,14-epoxide. Chem. Res. Toxicol. 2009;22:81–89. doi: 10.1021/tx8001675. [DOI] [PubMed] [Google Scholar]
- 27.Todorovic R, Devanesan P, Rogan E, Cavalieri E. Identification and quantification of stable DNA adducts formed from dibenzo[a,l]pyrene or its metabolites in vitro and in mouse skin and rat mammary gland. Chem. Res. Toxicol. 2005;18:984–990. doi: 10.1021/tx049681s. [DOI] [PubMed] [Google Scholar]
- 28.Nesnow S, Davis C, Nelson G, Ross JA, Allison J, Adams L, King LC. Comparison of the morphological transforming activities of dibenzo[a,l]pyrene and benzo[a]pyrene in C3H10T1/2CL8 cells and characterization of the dibenzo[a,l]pyrene-DNA adducts. Carcinogenesis. 1997;18:1973–1978. doi: 10.1093/carcin/18.10.1973. [DOI] [PubMed] [Google Scholar]
- 29.Prahalad AK, Ross JA, Nelson GB, Roop BC, King LC, Nesnow S, Mass MJ. Dibenzo[a,l]pyrene-induced DNA adduction, tumorigenicity, and Ki-ras oncogene mutations in strain A/J mouse lung. Carcinogenesis. 1997;18:1955–1963. doi: 10.1093/carcin/18.10.1955. [DOI] [PubMed] [Google Scholar]
- 30.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem. Res. Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 31.Cho BP. Dynamic conformational heterogeneities of carcinogen-DNA adducts and their mutagenic relevance. J. Environ. Sci. Health, Part C: Environ. Carcinog. Ecotoxicol. Rev. 2004;22:57–90. doi: 10.1081/LESC-200038217. [DOI] [PubMed] [Google Scholar]
- 32.Lukin M, de Los Santos C. NMR structures of damaged DNA. Chem. Rev. 2006;106:607–686. doi: 10.1021/cr0404646. [DOI] [PubMed] [Google Scholar]
- 33.Stone MP, Huang H, Brown KL, Shanmugam G. Chemistry and structural biology of DNA damage and biological consequences. Chem. Biodivers. 2011;8:1571–1615. doi: 10.1002/cbdv.201100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Y, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Broyde S, Patel DJ, Geacintov NE. Recognition and removal of bulky DNA lesiosn by the nucleotide excision repair system. In: Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Wiley-VCH; Weinheim, Germany: 2010. pp. 261–298. [Google Scholar]
- 35.Luch A, Friesel H, Seidel A, Platt KL. Tumor-initiating activity of the (+)-(S,S)- and (-)-(R,R)-enantiomers of trans-11,12-dihydroxy-11,12-dihydrodibenzo[a,l]pyrene in mouse skin. Cancer Lett. 1999;136:119–128. doi: 10.1016/s0304-3835(98)00265-1. [DOI] [PubMed] [Google Scholar]
- 36.Hirshfeld FL. The structure of overcrowded aromatic compounds. Part VII. Out-of-plane deformation in benzo[c] phenanthrene and 1,12-dimethyl-benzo[c]phenanthrene. J. Chem. Soc. 1963:2126–2135. [Google Scholar]
- 37.de los Santos C, Cosman M, Hingerty BE, Ibanez V, Margulis LA, Geacintov NE, Broyde S, Patel DJ. Influence of benzo[a]pyrene diol epoxide chirality on solution conformations of DNA covalent adducts: the (-)-trans-anti-[BP]G.C adduct structure and comparison with the (+)-trans-anti-[BP]G.C enantiomer. Biochemistry. 1992;31:5245–5252. doi: 10.1021/bi00138a002. [DOI] [PubMed] [Google Scholar]
- 38.Lin CH, Huang X, Kolbanovskii A, Hingerty BE, Amin S, Broyde S, Geacintov NE, Patel DJ. Molecular topology of polycyclic aromatic carcinogens determines DNA adduct conformation: a link to tumorigenic activity. J. Mol. Biol. 2001;306:1059–1080. doi: 10.1006/jmbi.2001.4425. [DOI] [PubMed] [Google Scholar]
- 39.Wang B, Sayer JM, Yagi H, Frank H, Seidel A, Jerina DM. Facile interstrand migration of the hydrocarbon moiety of a dibenzo[a,l]pyrene 11,12-diol 13,14-epoxide adduct at N(2) of deoxyguanosine in a duplex oligonucleotide. J. Am. Chem. Soc. 2006;128:10079–10084. doi: 10.1021/ja0608038. [DOI] [PubMed] [Google Scholar]
- 40.Yagi H, Frank H, Seidel A, Jerina DM. Revised assignment of absolute configuration of the cis- and trans-N6-deoxyadenosine adducts at C14 of (+/-)-11beta,12alpha-dihydroxy-13alpha,14alpha-epoxy-11,12,13,14-tetrahydrodibenz o[a,l]pyrene by stereoselective synthesis. Chem. Res. Toxicol. 2008;21:2379–2392. doi: 10.1021/tx800268f. [DOI] [PubMed] [Google Scholar]
- 41.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: 2006. [Google Scholar]
- 42.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JJA, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03. Gaussian, Inc.; Wallingford CT: 2003. [Google Scholar]
- 44.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. University of California; San Francisco: 2006. [Google Scholar]
- 45.Cieplak P, Cornell WD, Bayly C, Kollman PA. Application of the Multimolecule and Multiconformational Resp Methodology to Biopolymers - Charge Derivation for DNA, Rna, and Proteins. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 46.Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 47.Bayly CI, Cieplak P, Cornell WD, Kollman PA. A Well-Behaved Electrostatic Potential Based Method Using Charge Restraints for Deriving Atomic Charges - the Resp Model. J. Phys. Chem. 1993;97:10269–10280. [Google Scholar]
- 48.Ravishanker G, Swaminathan S, Beveridge DL, Lavery R, Sklenar H. Conformational and helicoidal analysis of 30 PS of molecular dynamics on the d(CGCGAATTCGCG) double helix: “curves”, dials and windows. J. Biomol. Struct. Dyn. 1989;6:669–699. doi: 10.1080/07391102.1989.10507729. [DOI] [PubMed] [Google Scholar]
- 49.Ravishanker G, Wang W, Beveridge DL. MD Toolchest. Wesleyan University; Middletown, CT: [Google Scholar]
- 50.The PyMOL Molecular Graphics System . Version 1.3 ed. Schrödinger; LLC: [Google Scholar]
- 51.Patel DJ, Shapiro L, Hare D. DNA and RNA: NMR studies of conformations and dynamics in solution. Q. Rev. Biophys. 1987;20:35–112. doi: 10.1017/s0033583500004224. [DOI] [PubMed] [Google Scholar]
- 52.Reid BR. Sequence-specific assignments and their use in NMR studies of DNA structure. Q. Rev. Biophys. 1987;20:1–34. doi: 10.1017/s0033583500004212. [DOI] [PubMed] [Google Scholar]
- 53.Xie XM, Geacintov NE, Broyde S. Stereochemical origin of opposite orientations in DNA adducts derived from enantiomeric anti-benzo[a]pyrene diol epoxides with different tumorigenic potentials. Biochemistry. 1999;38:2956–2968. doi: 10.1021/bi9825605. [DOI] [PubMed] [Google Scholar]
- 54.Wu M, Yan SF, Tan J, Patel DJ, Geacintov NE, Broyde S. Conformational searches elucidate effects of stereochemistry on structures of deoxyadenosine covalently bound to tumorigenic metabolites of benzo[C] phenanthrene. Front. Biosci. 2004;9:2807–2818. doi: 10.2741/1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai Y, Geacintov NE, Broyde S. Nucleotide excision repair efficiencies of bulky carcinogen - DNA adducts are governed by a balance between stabilizing and destabilizing interactions. Biochemistry. 2012;51:1486–1499. doi: 10.1021/bi201794x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cosman M, Fiala R, Hingerty BE, Laryea A, Lee H, Harvey RG, Amin S, Geacintov NE, Broyde S, Patel D. Solution conformation of the (+)-trans-anti-[BPh]dA adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrene to the 5'-side of the adduct site without disruption of the modified base pair. Biochemistry. 1993;32:12488–12497. doi: 10.1021/bi00097a029. [DOI] [PubMed] [Google Scholar]
- 57.Saenger W. Principles of Nucleic Acid Structure. Springer Verlag; New York: 1984. [Google Scholar]
- 58.Neidle S, Pearl LH, Herzyk P, Berman HM. A molecular model for proflavine-DNA intercalation. Nucleic Acids Res. 1988;16:8999–9016. doi: 10.1093/nar/16.18.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cosman M, Laryea A, Fiala R, Hingerty BE, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (-)-trans-anti-benzo[c]phenanthrene-dA ([BPh]dA) adduct opposite dT in a DNA duplex: intercalation of the covalently attached benzo[c]phenanthrenyl ring to the 3'-side of the adduct site and comparison with the (+)-trans-anti-[BPh]dA opposite dT stereoisomer. Biochemistry. 1995;34:1295–1307. doi: 10.1021/bi00004a024. [DOI] [PubMed] [Google Scholar]
- 60.Suri AK, Mao B, Amin S, Geacintov NE, Patel DJ. Solution conformation of the (+)-trans-anti-benzo[g]chrysene dA adduct opposite dT in a DNA duplex. J. Mol. Biol. 1999;292:289–307. doi: 10.1006/jmbi.1999.2974. [DOI] [PubMed] [Google Scholar]
- 61.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 62.Cosman M, Hingerty BE, Luneva N, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (-)-cis-anti-benzo[a]pyrenyl-dG adduct opposite dC in a DNA duplex: intercalation of the covalently attached BP ring into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1996;35:9850–9863. doi: 10.1021/bi9605346. [DOI] [PubMed] [Google Scholar]
- 63.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 64.Lukin M, Zaliznyak T, Johnson F, de los Santos C. Structure and stability of DNA containing an aristolactam II-dA lesion: implications for the NER recognition of bulky adducts. Nucleic Acids Res. 2012;40:2759–2770. doi: 10.1093/nar/gkr1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kropachev K, Kolbanovskiy M, Rodriguez FA, Cai YQ, Ding S, Zhang L, Amin S, Broyde S, Geacintov NE. Dibenzo a,l pyrene Diol Epoxide-adenine but Not -Guanine Adducts Are Resistant to Nucleotide Excision Repair in Human Cell Extracts. Chem. Res. Toxicol. 2010;23:282–282. [Google Scholar]
- 66.Ruan Q, Kolbanovskiy A, Zhuang P, Chen J, Krzeminski J, Amin S, Geacintov NE. Synthesis and characterization of site-specific and stereoisomeric fjord dibenzo[a,l]pyrene diol epoxide-N(6)-adenine adducts: unusual thermal stabilization of modified DNA duplexes. Chem. Res. Toxicol. 2002;15:249–261. doi: 10.1021/tx010157k. [DOI] [PubMed] [Google Scholar]
- 67.Tang Y. Optical spectroscopic and NMR Studies of Covalent Polycyclic Aromatic Hydrocarbon–DNA adducts: Influence ofBase Sequence Context and Carcinogen Topology. New York Universtiy; 2008. [Google Scholar]
- 68.Cai Y, Ding S, Geacintov NE, Broyde S. Intercalative conformations of the 14R (+)- and 14S (-)-trans-anti-DB[a,l]P-N-dA adducts: molecular modeling and MD simulations. Chem. Res. Toxicol. 2011;24:522–531. doi: 10.1021/tx1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.