Summary
During embryonic cell cycles, B-cyclin-CDKs function as the core component of an autonomous oscillator. Current models for the cell-cycle oscillator in non-embryonic cells are slightly more complex, incorporating multiple G1, S-phase, and mitotic cyclin-CDK complexes. However, periodic events persist in yeast cells lacking all S-phase and mitotic B-cyclin genes, challenging the assertion that cyclin-CDK complexes are essential for oscillations. These and other results led to the proposal that a network of sequentially activated transcription factors functions as an underlying cell-cycle oscillator. Here we examine the individual contributions of a transcription-factor network and cyclin-CDKs to the maintenance of cell-cycle oscillations. Our findings suggest that while cyclin-CDKs are not required for oscillations, they do contribute to oscillation robustness. A model emerges in which cyclin expression (thereby, CDK activity) is entrained to an autonomous transcriptional oscillator. CDKs then modulate oscillator function and serve as effectors of the oscillator.
Introduction
In order to divide, cells must properly execute the sequence of duplication and segregation events making up the cell cycle. Repeated cycles of cell division generate the exponential growth in cell number essential for early embryogenesis in multi-cellular organisms. These rapid cycles of cell division are dependent on oscillations in cyclin-CDK activity (reviewed in Murray, 2004). In embryonic cells, cyclin is constitutively synthesized from stores of maternal mRNA, allowing cyclin-CDK activity to build throughout interphase. When cyclin-CDK activity accumulates to critical levels, it triggers the events of mitosis and the degradation of cyclin protein. Thus, cyclin-CDK activity forms the self-limiting biochemical oscillator responsible for embryonic cell-cycle oscillations and acts as an effector of that oscillator.
The discovery that CDKs are essential for cell-cycle progression in yeast (Hartwell et al., 1974; Nurse et al., 1976) suggested oscillations in cyclin-CDK activity constitute a universal cell-cycle oscillator in eukaryotes. However, this widely-accepted model does not account for fundamental differences between the early embryonic cell cycle and other eukaryotic cell cycles. Embryonic cleavage divisions consist of rapid cycles of replication and division; whereas other eukaryotic cell cycles are considerably longer and highly regulated in order to coordinate cell growth and extra-cellular signals with cell division. Additionally, in early embryonic cells, cyclin is synthesized at a constant rate from a pool of maternal mRNA, but in yeast and other eukaryotic systems, cyclin synthesis is regulated transcriptionally (reviewed in Fung and Poon, 2005; Wittenberg and Reed, 2005). Thus, in non-embryonic cells, cyclin oscillations are not autonomous; they rely on transcriptional inputs. Although much is understood about the transcriptional regulation of cyclins, the role of transcription in cell-cycle oscillations remains unclear.
Previous studies have suggested CDK activities are not essential for oscillations associated with the cell cycle in the budding yeast, Saccharomyces cerevisiae (Haase and Reed, 1999; Orlando et al., 2008). In the absence of B-cyclin homologues required for S phase and mitosis (CLB1-6), G1 events repeat at cell-cycle intervals, even though DNA replication, mitosis and cytokinesis fail to occur (Haase and Reed, 1999). Recently, these cells have been shown to maintain oscillations in the cell-cycle-regulated transcriptional program, demonstrating that S-phase and mitotic cyclins are not required to maintain periodicity in the transcriptional program (Orlando et al., 2008). It was proposed that a transcription factor (TF) network, composed of sequentially activated TFs, with the intrinsic ability to oscillate, is important for maintaining periodic transcription in the absence of B-cyclins. Further, this TF-network oscillator is coupled to cyclin-CDK activity during the normal cell cycle, and together, they are hypothesized to produce cell-cycle oscillations (Orlando et al., 2008).
The TF-network model parsimoniously explains both oscillations and maintenance of the cell-cycle-regulated transcriptional program in the absence of S-phase and mitotic cyclins. However, it remains possible that B-cyclin-independent oscillations are driven by a transcription-extrinsic oscillator that regulates one or more periodically active transcription factors. Here, we investigate the mechanism of cell-cycle oscillations in budding yeast. We examine the roles of both CDK activities and TFs in the regulation of B-cyclin-independent oscillations. Our data support a model in which a TF-network comprises the primary cell-cycle oscillator, while CDKs are important for promoting robust oscillations and serve as effectors of the oscillator by triggering essential cell-cycle events.
Results
Periodic expression of G1 cyclins is not required for B-cyclin-independent oscillations
In yeast cells lacking the six B-type cyclins essential for S phase and mitosis, oscillations in both bud formation and in the cell-cycle regulated transcriptional program have been shown to occur with a period very similar to that of wild-type cells (Haase and Reed, 1999; Orlando et al., 2008). While these cells lack the cyclin-CDK activities necessary for S phase or mitosis, they still periodically express G1 cyclins.
To test whether periodic expression of G1 cyclins is required for oscillations in cells lacking B-cyclin-CDK activities, we examined B-cyclin-independent budding cycles in cells with constitutive G1 cyclin by expressing a non-degradable allele of CLN2 from the heterologous GAL1 promoter. This allele, CLN24t3s, lacks seven CDK phosphorylation sites (Lanker et al., 1996). Phosphorylation of these sites by G1-cyclin-Cdk1, targets Cln2 protein for ubiquitination by the SCF ubiquitin ligase (Lanker et al., 1996). Thus, expression of this hyper-stable allele produces constant G1-cyclin-CDK levels throughout the cell cycle (Figure 1C and 1D) (Lanker et al., 1996).
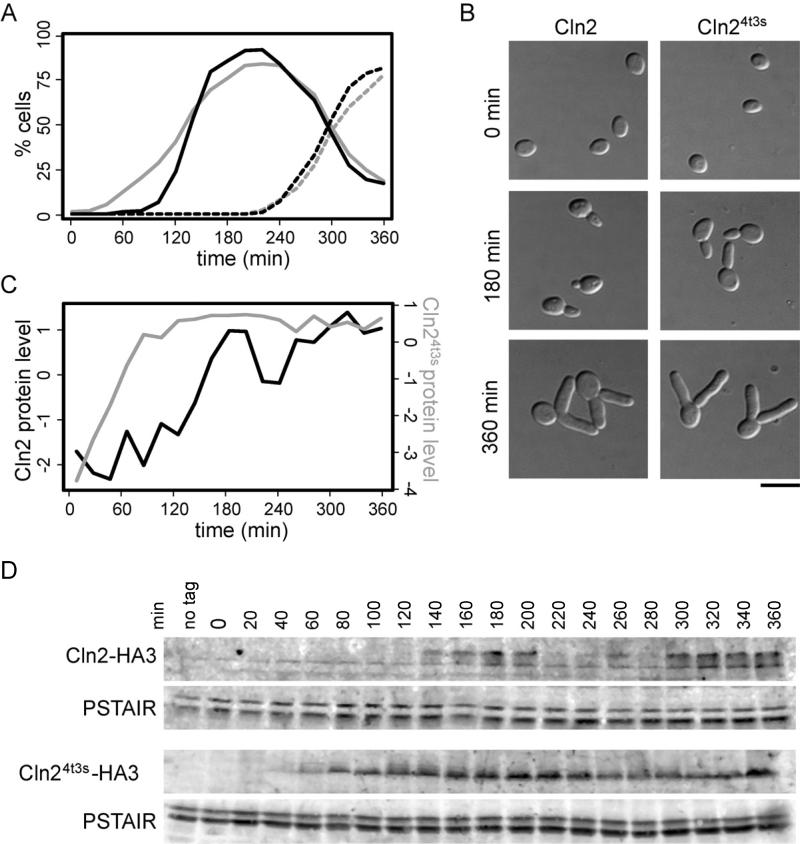
Figure 1. G1 cyclin oscillations are not required for B-cyclin-independent oscillations.
Cells were synchronized in G1 and released in the presence of galactose to induce expression of SIC1Δ3P and CLN24t3s from the GAL1 promoter. Cells expressing CLN24t3s, grey; cells not expressing CLN24t3s, black. (A) Bud formation was measured over time and averaged among three experiments at each time point; percent cells with one bud, solid line, percent cells with two or more buds, broken line. Representative images are shown (B); bar 5μm. Protein levels of Cln24t3s-HA3 or Cln2-HA3 were measured (D) in cells expressing CLN24t3s (bottom panel) or control cells (top panel) and normalized to the anti-PSTAIR loading control. (C) Normalized protein levels are expressed as log2-fold change relative to the mean protein level for each experiment.
Bud formation and Cln2 expression were monitored in cells synchronized by elutriation and expressing both the hyper-stable B-cyclin-specific inhibitor, Sic1Δ3P (Verma et al., 1997) and Cln24t3s or only Sic1Δ3P (Figure 1). Both strains continued to oscillate, as evidenced by the formation of multiple buds (Haase and Reed, 1999) (Figure 1A and 1B). Cln2-HA3 exhibits multiple phosphorylation forms (Lanker et al., 1996) and protein levels oscillate. Cln24t3s-HA3, however, runs as a single band and protein levels stay uniformly high (Figure 1C and 1D). Together, these data indicate that G1-cyclin-Cdk1 oscillations are not required for B-cyclin-independent oscillations.
Transcriptional oscillations in the absence of all Cdk1 activity
Although periodic expression of G1 cyclins is not required for B-cyclin-independent budding cycles, it remains possible that G1-cyclin-Cdk1 activity plays a critical role in the regulation of transcriptional oscillations. In yeast, several studies have shown that approximately 20 percent of the genome is expressed during a discrete portion of the cell cycle (Orlando et al., 2008; Pramila et al., 2006; Spellman et al., 1998). This cell-cycle-regulated transcriptional program continues to oscillate in cells lacking B-cyclins (Orlando et al., 2008).
To test whether G1 cyclins play a role in transcriptional oscillations, we measured global transcript dynamics in cells lacking all Cdk1 activity by means of a temperature-sensitive allele of the yeast Cdk1 gene, cdc28-4 (Hartwell et al., 1973). At the restrictive temperature, cdc28-4 cells arrest in the G1 phase of the cell cycle as unbudded cells with 1C DNA content (Hartwell et al., 1973) and do not appear to undergo any periodic events, including budding. To measure mRNA levels over time, cdc28-4 cells were synchronized in early G1 by centrifugal elutriation and shifted to the restrictive temperature, 37°C. Aliquots were harvested at 20-minute intervals. To confirm loss of CDK activity, bud formation was monitored for cells incubated at 37°C (Figure S1A-C). Total mRNA was isolated from samples collected at each time-point and hybridized to Affymetrix Yeast 2.0 oligonucleotide arrays. Mean transcript levels from two independent replicate experiments were highly reproducible, with an r2 of 0.995 (Figure S1D).
To determine whether cell-cycle-regulated transcripts continue to oscillate independent of all Cdk1 activities, we first identified an oscillatory period for cdc28-4 cells. In previous studies, the period of wild-type cells and B-cyclin mutant cells was established by tracking bud emergence as a landmark cell-cycle event (Orlando et al., 2007; Orlando et al., 2008). However, cdc28-4 cells do not undergo budding cycles, and lack other measurable landmark events. Thus, we used the mRNA dynamics of genes known to be periodically transcribed in wild-type cells (Orlando et al., 2008) to infer the period of transcriptional oscillations. We reasoned if some subset of the cell-cycle-regulated transcriptional program continues to be periodically expressed in the absence of Cdk1 activity, then their mRNA dynamics should be similar in cdc28-4 cells and wild-type cells within a single oscillatory period.
To determine the cdc28-4 period in an unbiased fashion, we systematically aligned the entire set of transcript profiles from cdc28-4 cells with respect to profiles of periodic genes from wild-type cells grown at 37°C, and then maximized global correlation between the data sets in order to infer the oscillatory period using an Markov chain Monte Carlo (MCMC) algorithm to sample parameters from a mixture model (see Supplementary Methods sections B-E for a detailed description). Along with estimating the average cdc28-4 oscillatory period, this algorithm also clusters genes into distinct sets of oscillating and non-oscillating genes.
For each cdc28-4 replicate, the algorithm identified a cluster corresponding to a set of genes that maintains periodic transcription in the absence of all Cdk1 activities. 838 genes out of 1276 genes that are cell-cycle regulated in wild-type cells continued to be periodic in both cdc28-4 replicates (the intersection of the two clusters; Table S1). The global pattern of transcript dynamics for these 838 genes is very similar in cdc28-4 cells and wild-type cells (Figure 2A-2B). Example transcript dynamics of individual genes are shown in Figure 2C-2H. Given that these experiments are performed on populations of cells, the second peak of mRNA levels appears reduced compared to the first peak since the population loses synchrony over time (Figure 2A-2H) (Orlando et al., 2007). This effect appears more pronounced in cdc28-4 cells as the cycle time is greater than in wild-type cells. Thus we conclude that approximately 66% of cell-cycle-regulated transcripts continue to be periodically expressed in the absence of Cdk1 activity.
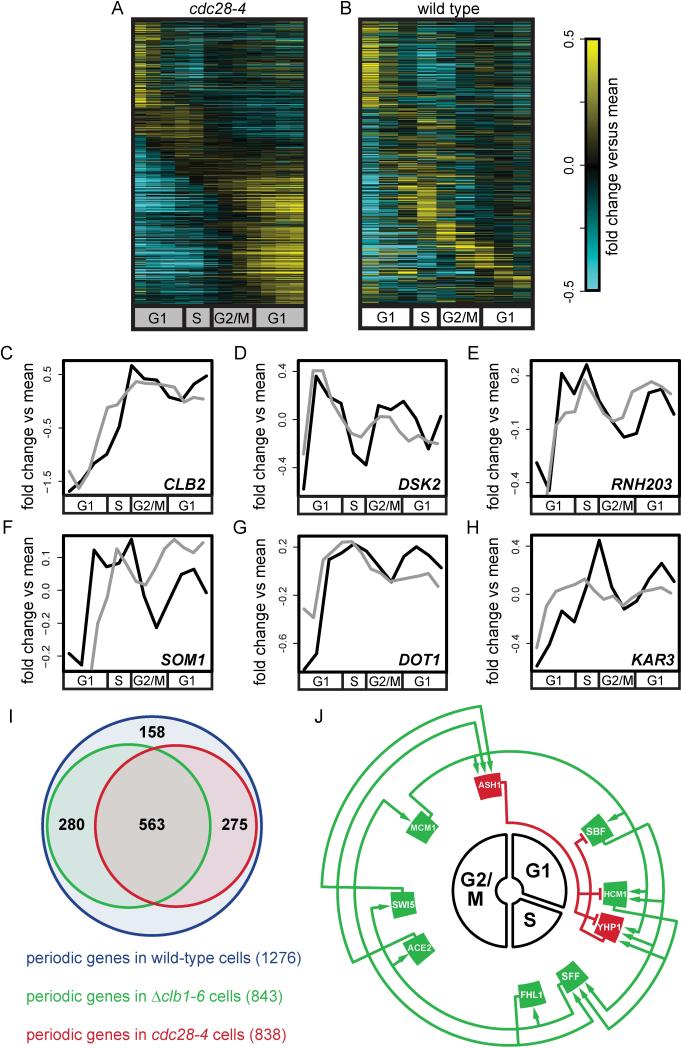
Figure 2. Periodic transcript dynamics in the absence of Cdk1 activity.
Transcript dynamics were measured in a synchronous population of cdc28-4 cells at the restrictive temperature, and periodic transcripts were identified by comparison to transcript dynamics in wild-type cells grown under the same conditions. Genes that are periodically expressed in wild-type cells and cdc28-4 cells are shown in (A) cdc28-4, (B) wild-type cells. Genes are aligned to a cell-cycle time line and expressed as log2-fold change relative to mean expression in the interval shown. Each row represents transcript levels for a single gene in (A, B). The grey bar designates cell cycle phases through which cells do not progress. (C-H) Line graphs showing the expression of single genes expressed as log2-fold change relative to the mean in wild-type cells (black), and cdc28-4 cells (grey). (I) Venn diagram showing overlap of periodically expressed genes in wild-type cells (blue), Δclb1-6 cells (red) and cdc28-4 cells (green). (J) Network graph of TFs remaining periodic in cdc28-4 cells. All nodes were placed on a cell-cycle time-line based on time of peak expression; activators, green; repressors, red. Edges were drawn based on evidence for TF-promoter interactions in the literature. Also see Figure S1, Tables S1 and S2.
The bulk of transcripts in the cell-cycle-regulated transcriptional program continue to accumulate on schedule in cells lacking S-phase and mitotic B-cyclin genes (Orlando et al., 2008). In order to compare gene sets maintaining periodicity in B-cyclin mutant cells to cdc28-4 cells, we used the algorithm outlined above to identify 843 periodic genes from global transcript dynamics previously collected in B-cyclin mutant cells (Orlando et al., 2008) (Table S1). Our analysis found that 563 genes maintain periodicity in both the B-cyclin mutant cells and cdc28-4 cells (Figure 2I). These results indicate that although the periodic expression of many genes is dependent on cyclin-CDK activity, a substantial fraction of the cell-cycle-regulated transcriptional program is independent of all Cdk1 activities.
An oscillatory TF network in the absence of Cdk1 activity
It has been proposed that a TF-network of cascading transcription factors controls the cell-cycle-regulated transcriptional program independent of S-phase and mitotic cyclin-CDK activities (Orlando et al., 2008). It has also been proposed that this TF network has intrinsic oscillatory capability and may serve as an underlying cell-cycle oscillator (Orlando et al., 2008). To determine if a similar transcription network model could explain transcript periodicity in cells lacking all Cdk1 activity we constructed a graphical network model (Figure 2J).
The nodes in the network graph, depicted in Figure 2J, correspond to TFs periodically expressed in cdc28-4 cells (Figure 2A). Two additional nodes, SBF and MCM1, were also included as both MCM1 and the DNA binding component of SBF, SWI4 show clear periodic expression by visual inspection (Figure S1F and S1G). All nodes were placed on a cell-cycle timeline at the point where their transcript levels peak and connected by edges based on evidence for a physical interaction between a TF and the promoter of a gene encoding a downstream TF (Table S2, Supplementary Methods, section F).
The TF network shown in Figure 2J may behave as a transmission oscillator, transmitting waves of activity around a loop (Sevim et al., 2010). To determine whether the architecture of this network has the capacity to produce oscillations, we converted the static network to a synchronously updating Boolean model (Table 1). In this framework, expression of TFs is binary, and when active, each TF affects its targets through logical functions that are applied synchronously (reviewed in Shmulevich and Aitchison, 2009). To simplify the network, SFF (Swi-five factor) and FHL1, as well as ACE2 and SWI5, were combined into single nodes (Figure S2A). When endowed with Boolean logic functions (Table 1A), the model enters an oscillatory attractor, progressing cyclically through three distinct states (Table 1B; Attractor #1). These oscillations are qualitatively similar to the oscillations we observe in cdc28-4 cells (Figure S2B and S2C).
Table 1.
Boolean model of TF-network oscillations. Logic Rules (A), attractors (B) and robustness to starting state (C) of a synchronously updating Boolean network constructed from the network in (Figure 2J; also see Figure S2).
| A | ||||
|---|---|---|---|---|
| TF | A | B | C | D |
| SBF | MCM1 ^ ¬ YHP1 | MCM1 ^ ¬ YHP1 | MCM1 ^ ¬ YHP1 | MCM1 ^ ¬ YHP1 |
| HCM1 | (SBF ⋁ MCM1) ^ ¬ ASH1 | (SBF ⋁ MCM1) ^ ¬ ASH1 | (SBF ⋁ MCM1) ^ ¬ ASH1 | (SBF ⋁ MCM1) ^ ¬ ASH1 |
| YHP1 | (MCM1 ⋁ SBF ⋁ HCM1) ^ ¬ ASH1 | (MCM1 ⋁ SBF ⋁ HCM1) ^ ¬ ASH1 | (MCM1 ⋁ SBF ⋁ HCM1) ^ ¬ ASH1 | (MCM1 ⋁ SBF ⋁ HCM1) ^ ¬ ASH1 |
| SFF/FHL1 | HCM1 ⋁ SBF | HCM1 ^ SBF | HCM1 ⋁ SBF | HCM1 ^ SBF |
| ACE2/SWI5 | SFF/FHL1 | SFF/FHL1 | SFF/FHL1 | SFF/FHL1 |
| MCM1 | SFF/FHL1 | SFF/FHL1 | SFF/FHL1 | SFF/FHL1 |
| ASH1 | ACE2/SWI5 ⋁ MCM1 | ACE2/SWI5 ⋁ MCM1 | ACE2/SWI5 ^ MCM1 | ACE2/SWI5 ^ MCM1 |
| B | |||||||
|---|---|---|---|---|---|---|---|
| Attractor #1 | SBF | YHP1 | HCM1 | SFF/FHL1 | ACE2/SWI5 | MCM1 | ASH1 |
| State 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 |
| State 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| State 3 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| Attractor #2 | SBF | YHP1 | HCM1 | SFF/FHL1 | ACE2/SWI5 | MCM1 | ASH1 |
|---|---|---|---|---|---|---|---|
| State 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Attractor #3 | SBF | YHP1 | HCM1 | SFF/FHL1 | ACE2/SWI5 | MCM1 | ASH1 |
|---|---|---|---|---|---|---|---|
| State 1 | 1 | 0 | 0 | 1 | 1 | 1 | 1 |
| C | |||
|---|---|---|---|
| Activation Logic | Attractor #1 | Attractor #2 | Attractor #3 |
| A | 87.50% | 7.81% | 4.69% |
| B | 28.12% | 71.88% | 0.00% |
| C | 89.84% | 7.81% | 2.34% |
| D | 35.94% | 64.06% | 0.00% |
To examine the robustness of network oscillations, we explored the behavior of the model when initialized from all possible starting states. Of the 256 possible starting states, the model enters the oscillatory attractor 89.50% of the time (Table 1C; Attractor #1). From the remaining states, the network enters one of two non-oscillating attractors (Table 1B; Attractors #2 and #3). We then tested whether oscillations in this model are sensitive to choices in Boolean logic. We found that the network enters the oscillatory attractor under several logic sets (Table 1A and 1C), indicating that network oscillations are robust to changes in logic and starting state. The robust capacity of this network to oscillate suggests that this TF network could plausibly function as an autonomous oscillator in the absence of Cdk1 activities.
Does a TF-network regulate oscillations in vivo?
Although we have demonstrated that a TF-network has the capacity to maintain oscillations in silico, it is not clear that a TF-network is involved in cyclin-independent oscillations. It remains possible that a transcription-extrinsic biochemical oscillator maintains periodicity in the absence of B-cyclin-CDK activity and entrains the global transcriptional program by controlling the activity of one or more TFs. We reasoned if a TF-network controls the timing of oscillations, altering expression levels of individual TFs would change the oscillatory period or cause more extreme oscillatory defects.
We manipulated the proposed TF-network oscillator by perturbing the expression of TFs in the network oscillator model (SWI6, YHP1, and HCM1), as well as TFs that are known to affect the expression of TFs in the model (MBP1, YOX1, SFG1) (Table S2). We used perturbations that were likely to be dominant to avoid any potential redundancies caused by network complexity. Specifically, we deleted TFs that serve as repressors or co-repressors (YOX1, YHP1 (Pramila et al., 2002), SFG1 (White et al., 2009), and MBP1 (de Bruin et al., 2006)), and over-expressed TFs that positively regulate transcription from the GAL1 promoter (SWI6 (Taba et al., 1991), and HCM1 (Pramila et al., 2006)).
We tested the effect of these network perturbations in cells undergoing B-cyclin-independent oscillations. Cells were arrested in G1 by alpha factor treatment and then released in the presence of galactose to induce expression of the hyper-stable B-cyclin-specific CDK inhibitor, Sic1Δ3P. Bud formation was monitored for 1.5 to 2 cycles and compared between perturbed and control cells (Figure 3A - 3G).
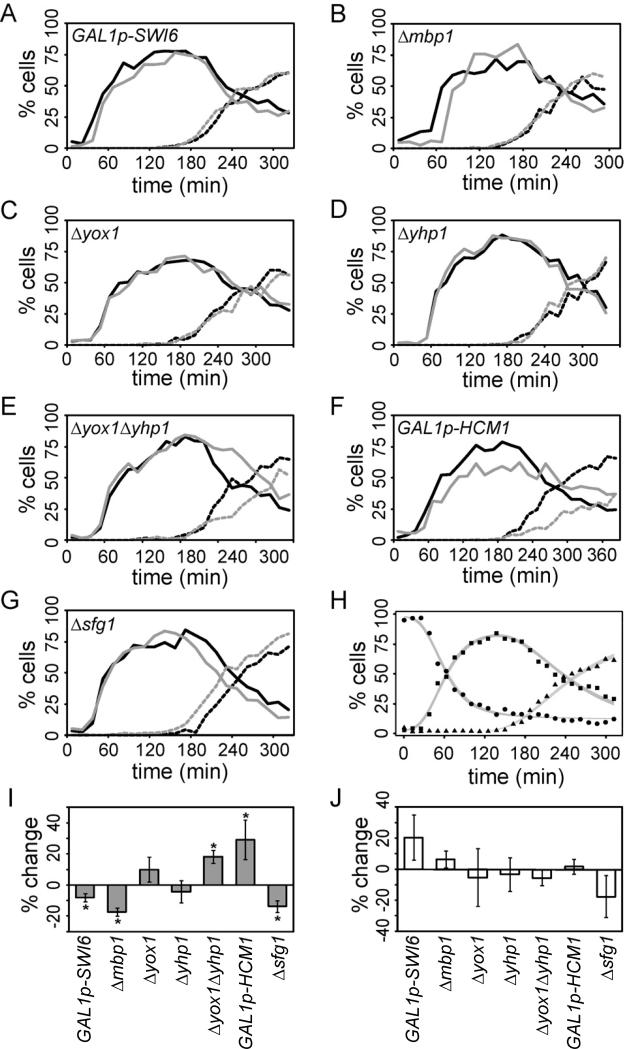
Figure 3. Perturbing TF expression alters the period of B-cyclin independent oscillations.
Cells with altered TF expression and control cells were synchronized in G1 by mating pheromone and released into the cycle in the presence (3J) or absence (3A-3I) of B-cyclin-CDK activity. Oscillations in bud formation were monitored. Representative budding curves for cells lacking B-cyclin-CDK activities are shown for GAL1p-SWI6 cells (A), Δmbp1 cells (B), Δyox1 cells (C), Δyhp1 cells (D), Δyox1; Δyhp1 cells (E), GAL1p-HCM1 cells (F) and Δsfg1 cells (G). Control cells, black; perturbed cells, grey; cells with 1 bud, solid line; cells with 2 buds, broken line. (H) A representative CLOCCS fit for cells lacking B-cyclin-CDK activity; cells with 0 buds, circles; cells with 1 bud, squares; cells with 2 buds, triangles. The width of the colored band reflects the degree of posterior uncertainty in the fit data. Learned period lengths for three experiments were averaged and are shown as % change from control for cells lacking B-cyclin activities (I) and cycling cells (J). Bars indicate standard deviation and asterisks (*) indicate periods significantly different than control cells (paired t-test; p<.05). Also see Figure S3, Table S3.
To quantify the changes in period, we used the CLOCCS (characterizing loss of cell cycle synchrony) algorithm (Orlando et al., 2007; Orlando et al., 2008) to learn the oscillatory period of the population of cells in each experiment. The algorithm uses a MCMC approach to fit each budding curve (Figure 3H) and outputs several parameters, including period length (Supplementary Methods section H). Learned periods were compared between perturbed and control cells (Figure 3I, Table S3A).
While some perturbations (Δyox1 or Δyhp1) did not significantly affect the period of cyclin-independent oscillations, most of the perturbations we tested did cause considerable changes to the period. Interestingly, we found that some perturbations shortened the period (GAL1p-SWI6, Δmbp1, Δsfg1), while others lengthened the period (Δyox1Δyhp1 and GAL1p-HCM1) (Figure 3I). Although perturbations that lengthen the period do not cause distinct changes in morphology (Figure S3), it is still possible that these perturbations are affecting some fundamental process such as nutrient uptake or bud formation. To control for these secondary effects, we also tested the effect of the network perturbations on normally cycling cells (Figure 3J). Some perturbations caused small changes in cell-cycle period length; however, neither Δyox1Δyhp1 nor GAL1p-HCM1 caused notable changes in period (Table S3B). These data demonstrate that these perturbations affect the timing of cyclin-independent oscillations, rather than causing a delay by indirectly affecting the health of the cells.
Together, the changes in period we observe when the expression of TFs is manipulated point to a role for regulators of periodic transcription in controlling the timing of B-cyclin-independent oscillations. Thus, these data support a model in which a TF-network promotes oscillations in the absence of B-cyclin-CDK activity.
How do CDKs affect oscillations?
By eliminating all Cdk1 activities, we revealed the capability of cells to maintain oscillations in the absence of all CDK activity. Although we present evidence that TFs are important for the timing of B-cyclin-independent oscillations, period length is not significantly affected by TF perturbations when all CDK activities are present (Figure 3). These findings suggest CDKs have a role in maintaining period length, although they are not required for oscillations.
CDKs are known to interact with and phosphorylate several TFs thought to control the cell-cycle-regulated transcriptional program (Table S4). In addition, CDK regulation of TFs is known to modulate transcription through both positive and negative feedback (Figure 4A and 4B). Thus, CDK-mediated phosphorylation likely regulates the activity of the TF-network oscillator during the normal cell cycle by modulating the expression level of periodic genes.
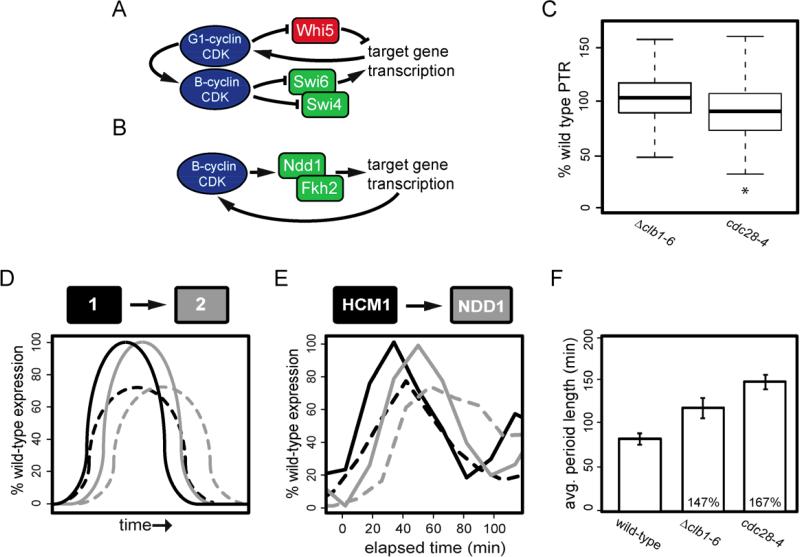
Figure 4. CDKs affect oscillations though feedback on the network oscillator.
(A and B) Examples of CDK regulation of transcription through positive (B) and both positive and negative feedback (A); kinase activities, blue; transcriptional activators, green; transcriptional repressors, red. (C) Box-and-whisker plots of PTRs for genes periodic in both Δclb1-6 and cdc28-4 cells calculated as the percent of wild-type control PTR (grown at 30° C for and 37° C for cdc28-4 cells). Whiskers extend to 1.5 times the interquartile range; asterisk (*) indicates significantly different PTRs between Δclb1-6 or cdc28-4 cells with respect to wild-type cells. (D) Model of amplitude loss; solid line, wild-type expression; broken line, expression in the absence of CDK activities. (E) Expression of HCM1, black, and NDD1, grey, normalized to maximum expression in wild-type cells; wild-type cells, solid line; Δclb1-6 cells, broken line. (F) Average period length in wild-type cells (including similar periods observed at both 30° and 37° C), B-cyclin mutant cells, and cdc28-4 cells. Values for wild-type cells were determined by CLOCCS (Orlando et al., 2007). For Δclb1-6 cells and cdc28-4 cells, average period length is determined through MCMC mixture modeling. Error bars represent standard deviation between the replicates, percent increase over appropriate wild-type periods are indicated for Δclb1-6 and cdc28-4 cells. Also see Table S4.
To determine the effects of Cdk1 activity on the amplitude of transcriptional oscillations, we compared peak-to-trough ratios (PTRs) of the 563 genes that are periodically expressed in wild-type cells, B-cyclin mutant cells (Δclb1-6) and cdc28-4 cells. PTRs were calculated as the mean ratio of maximum transcript level to minimum transcript level between replicate experiments (Table S1, and Supplementary Methods, section I).
To compare PTRs across experiments, PTRs from B-cyclin mutant cells and cdc28-4 cells were compared to the appropriate wild-type control (grown at 30° or 37°C, Figure 4C). By examining global trends in PTRs across conditions, we observed that PTRs in cdc28-4 cells are lower than in B-cyclin mutant or wild-type cells (Figure 4C). These data indicate that CDK activities impact the transcriptional oscillator by regulating the amplitude of transcription. Although the PTRs in Δclb1-6 cells are, in bulk, not significantly different from wild-type cells (Figure 4C), the PTRs for several network TFs are substantially reduced (Figure 4E, Table S1).
Role of CDKs in regulating network oscillations
If transcriptional oscillations are driven by a TF network, reduced amplitudes are likely to affect oscillatory dynamics of the network. Consider a simplified two-node network in which TF1 activates the transcription of the TF2 gene (Figure 4D). The rate of TF2 synthesis likely depends on the concentration of its activator, TF1, at any given time. If the amplitude of TF1's transcription over time is decreased, it is likely that the level of TF2 transcript accumulation will also be reduced and its expression will be delayed. Indeed, this behavior is observed in mRNA dynamics from cells lacking CDK activities. For example, Hcm1 normally promotes the transcriptional activation of the NDD1 gene that encodes a downstream TF in the network. In B-cyclin mutant cells, we observe a reduction in the amplitude of both HCM1 and NDD1 as compared to wild-type mRNA levels as well as a temporal delay in the expression of these two genes (Figure 4E).
Extending this behavior to the full oscillatory TF network, one would predict progression through the network in cells lacking Cdk1 activity would be slower than in wild-type cells, and thus, the period may be extended. Comparing the measured cell-cycle period of wild-type cells (Orlando et al., 2008) to learned oscillatory period lengths for B-cyclin-mutant and cdc28-4 cells (Supplementary methods, section C), we find that period length does, in fact, increase in cells that lack Cdk1 activities (Figure 4F). These observations support a role for CDKs in the regulation of amplitude and period of transcriptional oscillations.
Cyclins as effectors of the TF-network oscillator
In embryonic systems, CDK functions as a core component of the oscillator as well as the effector of the oscillator. Our findings demonstrate that in yeast cells, autonomous oscillations in the cell-cycle-regulated transcriptional program can occur independent of all Cdk1 activities. However, by conventional measures, the cell cycle is arrested in these cells, indicating that periodic gene expression is not sufficient to trigger hallmark cell-cycle events. These findings suggest that cyclin-CDKs are also required as the effectors of the TF-network oscillator. Unlike embryonic systems, cyclin expression in yeast is dependent on transcription, and CDK activities are therefore entrained, at least in part, to the cell-cycle-regulated transcriptional program. Thus, a role for cyclin-CDKs as effectors of the TF network would provide a mechanism for the entrainment of cell-cycle events to transcriptional oscillations.
If cyclins are indeed the effectors of TF-network-driven oscillations, then we expect cell-cycle events specific to each cyclin will be activated periodically as distinct cyclins are reintroduced to cells lacking cyclin-CDK activities. Furthermore, the timing of those events should be coordinated with transcript oscillations.
In order to test this hypothesis, we chose to start with cells lacking all B-type cyclin genes which have been shown to undergo autonomous transcriptional oscillations in the absence of cell-cycle progression (Orlando et al., 2008). We then “added back” the CLB6 gene by using a strain in which all other B-cyclins have been deleted (Δclb1,2,3,4,5 GAL1-CLB1; Δclb1-5) (Haase et al., 2001). The CLB6 gene is periodically transcribed in wild-type cells (Orlando et al., 2008), and the Clb6 protein is highly unstable (Jackson et al., 2006). Thus, when CLB6 is periodically transcribed in Δclb1-5 cells, the Clb6 protein rises and falls with each cycle of CLB6 expression (Figure S4).
Clb6-Cdk1 complexes can trigger entry into S phase, but not entry into mitosis (Kuhne and Linder, 1993; Schwob and Nasmyth, 1993). Therefore, if Clb6 is acting as an effector of transcriptional oscillations, one would expect that DNA replication will be initiated in these cells coincident with the rise of CLB6 transcript levels. Since Clb6 is an unstable protein, DNA replication origins should be allowed to re-license when Clb6 levels drop, as there are no other B-cyclins present to block licensing (Dahmann et al., 1995). Finally, when Clb6 levels rise in the second cycle, a new round of DNA replication should be initiated in these mitotically arrested cells.
To examine the effect oscillations of Clb6 have on DNA replication, a population of Δclb1-5 cells was synchronized in G1 by centrifugal elutriation. Samples were taken every 20 minutes for more than two oscillation cycles, and analyzed for bud formation, DNA content and CLB6 and CLN2 transcript levels by northern blot (Figure 5).
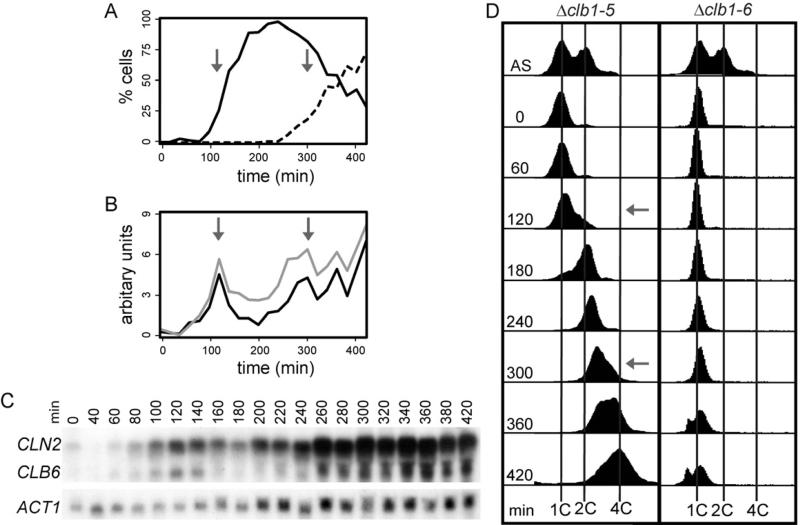
Figure 5. Periodic Expression of Clb6 drives cyclic activation of DNA replication.
Δclb1-5; GAL1pr-CLB1 cells were synchronized in G1 and released in presence of dextrose. Cells were monitored for bud emergence (A; solid line, one bud, broken line 2 buds), expression of CLN2 (grey) and CLB6 (black) by northern blot (B and C) and DNA content (D). (B) mRNA levels are plotted as the level of cyclin mRNA divided by the level of ACT1 at that time point. In all panels grey arrows signify the first (120 min) and second (300 min) peak of CLB6 expression. (D) DNA content is compared to Δclb1-6; GAL1pr-CLB1 cells. Accumulation of cells with less than 1C DNA content reflects the lysis of Δclb1-6 cells at late time points. Also see Figure S4.
As expected, oscillations in bud formation and both CLN2 and CLB6 transcript accumulation (Figure 5A-5C) were observed. However, unlike cells lacking all B-cyclins, Δclb1-5 cells enter S phase as CLB6 transcript levels rise, and then re-enter a new S phase when CLB6 transcript levels rise on the next cycle (Figure 5D). To ensure that increases in DNA content are due to chromosomal DNA replication, these experiments were done in cells lacking mitochondrial DNA (rho0). Strikingly, the budding cycles, DNA replication cycles, and transcription oscillations are all coordinated such that they occur with a relative timing similar to that observed in wild-type cells. These findings are consistent with a model in which the timing of cell-cycle events is determined by the kinetics of transcript oscillations driven by a TF network, and that cyclins, being entrained to this network, activate CDKs and function as the effectors triggering cell-cycle events (Figure 6).
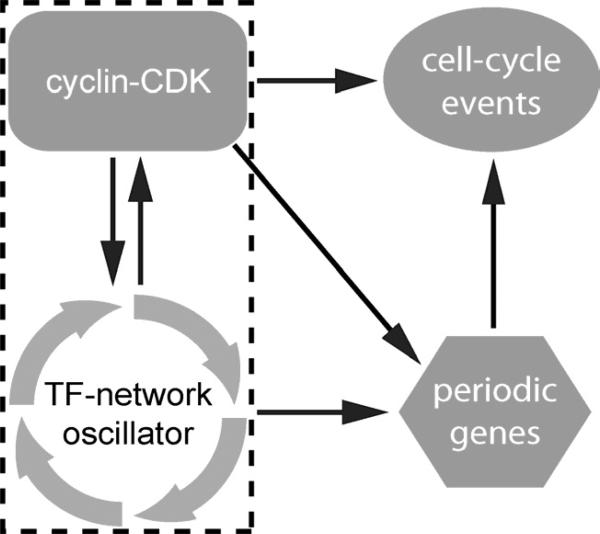
Figure 6. Model integrating CDK-independent oscillations, CDKs and cell-cycle events.
Box indicates elements required for robust oscillations.
Discussion
What defines the fundamental cell-cycle oscillator?
In early embryonic cells, self-sustaining oscillations in cyclin-CDK activity have been shown to drive periodic events during the cell cycle. CDKs are important for cell-cycle progression in non-embryonic cells and yeast, and the embryonic CDK-based oscillator model has been extrapolated to these systems. As cyclin mRNAs are synthesized periodically outside of early embryogenesis, new models must account for transcriptional control. Prevailing models have suggested that periodic transcription is controlled by CDKs, so most oscillator models for yeast and non-embryonic cells still place CDKs at the core of the oscillating mechanism.
CDK-based oscillator models have been challenged by studies indicating that cyclic activation of budding and the cell-cycle-regulated transcriptional program are largely independent of S-phase and mitotic cyclins (Haase and Reed, 1999; Orlando et al., 2008). It was proposed that periodic events in cyclin-mutant cells are driven by an oscillator based on a network of serially activated transcription factors. However, B-cyclin-independent oscillations could also be explained by some transcription-extrinsic biochemical oscillator that both maintains oscillations and triggers the activity of one or more TFs regulating the periodic transcriptional program.
One obvious candidate for a transcription-extrinsic oscillator is G1-cyclin-CDK activity. G1 cyclins are present in B-cyclin mutant cells and they continue to periodically activate Cdk1 (Haase and Reed, 1999). In addition, G1 cyclins trigger both their own expression and their own proteolysis, suggesting that G1-cyclins could form an autonomous oscillator (Cross and Tinkelenberg, 1991; Dirick and Nasmyth, 1991; Lanker et al., 1996). To address the possibility that autonomous oscillations in G1-cyclin-CDK activity drive B-cyclin-independent oscillations, oscillations in G1-cyclin-CDK activity were eliminated by constitutive expression of a hyper-stable allele of CLN2 in cells lacking B-cyclin-CDK activity (Figure 1). Under these conditions, cyclic activation of budding continues with similar kinetics to cells periodically expressing G1 cyclins. This finding argues that oscillations of G1 cyclins are not required to drive budding cycles, and is consistent with the assertion that G1 cyclin-CDKs are not acting as an autonomous oscillator in this system. Nonetheless, it remained to be determined whether the presence of G1 cyclins plays an essential role in oscillations. Using a global transcriptional profiling approach, we found a substantial fraction of transcripts from genes that are normally transcribed periodically in wild-type cells, continue to oscillate in cells that lack all Cdk1 activities, indicating that transcript oscillations do not require Cdk1 (Figure 2).
Mathematical models support the idea that a TF network could maintain oscillations in cells lacking all Cdk1 activities (Table 1). These models, however, do not rule out the possibility that an unknown transcription-extrinsic oscillator entrains the cell-cycle regulated transcriptional program. In order to directly test the TF-network oscillator model, we perturbed the expression of several different regulators of the transcriptional program. Although no single perturbation abolished B-cyclin independent oscillations, many perturbations caused changes in the period of these oscillations. These findings suggest a mechanism inherent to transcription itself, such as a TF-network oscillator, rather than a transcription-extrinsic oscillator, comprises the B-cyclin-independent oscillator. In addition, the observation that no single TF perturbation abolished oscillations, suggests there may be considerable functional redundancy in the TF-network oscillator. Interestingly, knockdown of important circadian clock genes in mammalian cells do not abolish oscillations, producing only modest changes to oscillation period length. The period changes observed when clock genes were knocked down were attributed to compensation mechanisms within the circadian oscillator network (Baggs et al., 2009). Taken together, these findings suggest robustness to perturbation may be a general property of network oscillators.
What is the role of CDK activity in transcriptional oscillations?
Although our data indicate oscillations associated with the cell cycle do not require cyclin-CDK activity, they also suggest that CDKs do have important roles in regulating these oscillations. Although perturbing the expression of TFs causes changes in the period of B-cyclin-independent oscillations, oscillations in the presence of all CDK activity appear more robust to these perturbations (Figures 3I and 3J). Additionally, in the absence of CDK activity, we see a global reduction in the amplitude of transcript oscillations (Figure 4C), as well as an increase in period length (Figure 4F). CDKs are known to regulate the activity of many periodic TFs (Figure 4A, 4B, Table S4), suggesting that while cyclin synthesis is entrained to the TF network, CDKs feed back on the network and modulate its activity.
Aside from periodic transcription, cdc28-4 cells exhibit no overt periodic cell-cycle behaviors, suggesting that periodic gene expression alone is not sufficient for triggering cell-cycle events. Thus, the activation of cell-cycle events may require the combined action of periodic gene expression and CDK activity, and/or periodic CDK activity. Consistent with this idea, cells expressing only G1 cyclins maintain periodic gene expression and undergo repeated rounds of budding, an event that requires G1-cyclin-CDK activity (Haase and Reed, 1999; Orlando et al., 2008). Furthermore, we have shown cells continue to re-bud even when G1 cyclins are expressed constitutively (Figure 1), indicating that some periodic activity beyond cyclic expression of G1 cyclins is important for initiating cycles of budding. Finally, when the expression of Clb6, an unstable S-phase cyclin, is entrained to transcriptional oscillations, cells undergo successive rounds of DNA replication that are coordinated with budding and transcription cycles (Figure 5). In a previous study, repeated rounds of spindle pole body duplication have also been observed under similar conditions (Haase et al., 2001). These findings indicate the timing of cell-cycle events is, at least in part, controlled by the entrainment of cyclin expression to the network oscillator.
An integrated cell-cycle oscillator model
Together, data presented in this study support a model for the regulation of cell-division cycles (Figure 6). In this model, a TF-network generates cell-cycle oscillations and controls the periodic transcription of a large collection of genes, including cyclins. Distinct cyclins activate Cdk1 during the cell-cycle intervals in which they are expressed, and these activated complexes have two functions. First, cyclin-CDKs feed back on the network oscillator by modulating TF activity. Although oscillations persist in the absence of CDK activities, CDK-mediated feedback appears to be required for robust oscillations (Figure 6, grey box). Second, CDKs serve as effectors, coupling the oscillator to the activation of cell-cycle events.
It was recently proposed that CDK oscillations coordinate several distinct autonomous oscillations through phase locking (Lu and Cross, 2010). Our model for cell-cycle oscillations does not rule out the possibility that the periodic behaviors we observe are influenced by other oscillators such as those controlling metabolic processes (Chen et al., 2007; Klevecz et al., 2004; Novak and Mitchison, 1987), nor does it rule out the possibility that other oscillations are entrained to CDK activity. Nevertheless, unlike the oscillations observed by Lu and Cross, the periodic behaviors (budding and transcript dynamics) revealed when Cdk1 activities are diminished are remarkably similar, both qualitatively and quantitatively to those observed in normally dividing cells. This similarity is particularly striking given that the cells lacking Cdk1 activities are fully cell-cycle arrested by all conventional measures and continue to grow in size (Goranov et al., 2009). Although CDK complexes can form autonomous oscillators in embryonic systems, in which cyclin is constitutively synthesized from pools of maternal mRNAs, autonomous oscillations cannot occur in yeast cells, as cyclin synthesis is dependent on transcriptional activation of cyclin genes. Accordingly, we propose a model in which nonembryonic cells integrate both transcription and CDK activities to produce robust oscillations in the cell-division cycle.
Experimental Procedures
Yeast strains, growth and synchronization
All strains used in this study are derivatives of BF264-15DU (MATa; ade1; his2; leu2-3,112; trp1-1; ura3Δns) (Richardson et al., 1992). All strains were made through standard yeast genetic procedures. Relevant genotypes are included in Table S5. Yeast cultures were grown in YEP medium (1% yeast extract, 2% peptone, 0.012% adenine, 0.006% uracil supplemented with 2% sugar (dextrose, sucrose or galactose). For GAL1-SIC1Δ3P experiments, cells were grown in YEP containing 2% sucrose and 0.1% dextrose prior to centrifugal elutriation or grown in YEP containing 2% sucrose and arrested in 20-50ng/mL alpha factor pheromone. The resulting population was resuspended in YEP medium containing 2% galactose. Identical protocols were carried out for control experiments in cycling cells. For experiments carried out at the cdc28-4 restrictive temperature, cells were grown at 25°C in YEP medium containing 2% dextrose prior to synchronization by centrifugal elutriation. The resulting population was resuspended in YEP media containing 2% dextrose pre-warmed to 37°C. Samples were taken every 20 minutes for 5 or more hours. For CLB1 shut-off experiments, B-cyclin mutant cells were grown in YEP medium containing 2% galactose prior to centrifugal elutriation. The resulting population was resuspended in YEP medium containing 2% dextrose and 1M sorbitol.
Microscopy
All samples analyzed by microscopy were fixed in 2% paraformaldehyde. Buds were scored for 200 or more cells per sample. Cells were imaged using a Zeiss Axio Imager widefield fluorescence microscope, a 100x objective and standard filter sets. Images were acquired with a Hamamatsu Orca ER monochrome cooled-CCD camera with IEEE and captured using Metamorph 7.1 (Universal Imaging).
Protein Isolation and Immunoblotting
Cell lysates were subjected to SDS-PAGE and immunoblotting using the following antibodies: mouse anti-HA (Roche Diagnostics, Indianapolis, IN), mouse anti-PSTAIR, (Abcam, Inc. Cambridge, MA), and IRDye 800 conjugated goat anti-mouse (Li-Cor Biosciences, Lincoln, NE). Membranes were analyzed with a Li-Cor Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). Signal was quantified using Image J 1.41o (National Institutes of Health, USA) and normalized to anti-PSTAIR.
RNA isolation and analysis
In all cases, total RNA was extracted from yeast as described previously (Haase and Reed, 1999). For microarray analysis, RNA was purified with the RNAeasy MinElute Cleanup Kit (Qiagen, Valencia, CA). cDNA synthesis and fluorescent labeling was done with either the GeneChip One cycle Labeling (Affymetrix, Santa Clara, CA) or the Ambion MessageAmp Premier kit (Ambion Biosystems, Foster City, CA) and hybridized to Yeast 2.0 Expression arrays (Affymetrix, Santa Clara, CA). Labeling, hybridization and image collection was performed at the Duke Institute for Genome Sciences and Policy DNA Microarray Core Facility (www.genome.duke.edu/cores/microarray). CEL files from the oligonucleotide arrays were normalized and summarized alongside previously published arrays (Supplementary Methods section A) (Orlando et al., 2008). Northern blot analysis was carried out as previously described (Haase and Reed, 1999) using probes complimentary to the CLB6, CLN2 and ACT1 transcripts.
Computational and statistical analyses
To compare transcript dynamics between wild-type cells and mutant cells, the datasets were systematically aligned using parameters for period length and offset described previously (Supplementary Methods section B) (Orlando et al., 2007). A Markov chain Monte Carlo (MCMC) mixture modeling algorithm (Supplementary Methods section C) was devised and evaluated in order to compare these alignments. From this algorithm, we computed a posterior ratio to distinguish between periodic and non-periodic models of gene expression (Supplementary Methods section D).
Construction of TF networks and the synchronously updating Boolean model, as well as calculation of PTRs were performed as described previously with some minor modifications (Supplementary Methods sections F, G, I) (Orlando et al., 2008).
The CLOCCS model was used to learn period lengths of cycling cells (Orlando et al., 2007), and cells lacking B-cyclin activity (Orlando et al., 2008) as previously described with some minor modifications (Supplementary Methods section H).
Supplementary Material
Highlights.
Transcriptional cell-cycle oscillations persist in the absence of all CDK activity.
Perturbing periodic transcription factors affects oscillation period.
CDKs modulate oscillator function by phosphorylating transcription factors.
CDKs serve as effectors of the oscillator by triggering cell-cycle events.
Acknowledgements
We thank J. Harer, D. Lew, M. Clyde, and members of the Haase lab for helpful discussions. We also thank D. Lew, S. Bristow, M. Chee, A. Leman and C. Lin for critical reading of the manuscript. This work was supported by the National Institutes of Health (NIH P50-GM081883-01 to S.B.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers.
Datasets generated in this study are available in NCBI's Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih .gov/gds) using accession number GSE32974.
References
- Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Odstrcil EA, Tu BP, McKnight SL. Restriction of DNA replication to the reductive phase of the metabolic cycle protects genome integrity. Science. 2007;316:1916–1919. doi: 10.1126/science.1140958. [DOI] [PubMed] [Google Scholar]
- Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth KA. S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a prereplicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- de Bruin RA, Kalashnikova TI, Chahwan C, McDonald WH, Wohlschlegel J, Yates J, 3rd, Russell P, Wittenberg C. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23:483–496. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–757. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- Fung TK, Poon RY. A roller coaster ride with the mitotic cyclins. Semin Cell Dev Biol. 2005;16:335–342. doi: 10.1016/j.semcdb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Goranov AI, Cook M, Ricicova M, Ben-Ari G, Gonzalez C, Hansen C, Tyers M, Amon A. The rate of cell growth is governed by cell cycle stage. Genes Dev. 2009;23:1408–1422. doi: 10.1101/gad.1777309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Reed SI. Evidence that a free-running oscillator drives G1 events in the budding yeast cell cycle. Nature. 1999;401:394–397. doi: 10.1038/43927. [DOI] [PubMed] [Google Scholar]
- Haase SB, Winey M, Reed SI. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nat Cell Biol. 2001;3:38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LP, Reed SI, Haase SB. Distinct mechanisms control the stability of the related S-phase cyclins Clb5 and Clb6. Mol Cell Biol. 2006;26:2456–2466. doi: 10.1128/MCB.26.6.2456-2466.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci U S A. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhne C, Linder P. A new pair of B-type cyclins from Saccharomyces cerevisiae that function early in the cell cycle. EMBO J. 1993;12:3437–3447. doi: 10.1002/j.1460-2075.1993.tb06018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanker S, Valdivieso MH, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- Lu Y, Cross FR. Periodic cyclin-Cdk activity entrains an autonomous Cdc14 release oscillator. Cell. 2010;141:268–279. doi: 10.1016/j.cell.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. Recycling the cell cycle: cyclins revisited. Cell. 2004;116:221–234. doi: 10.1016/s0092-8674(03)01080-8. [DOI] [PubMed] [Google Scholar]
- Novak B, Mitchison JM. Periodic cell cycle changes in the rate of CO2 production in the fission yeast Schizosaccharomyces pombe persist after a block to protein synthesis. J Cell Sci. 1987;87(Pt 2):323–325. doi: 10.1242/jcs.87.2.323. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Iversen ES, Hartemink AJ, Haase SB. A probabilistic model for cell cycle distributions in synchrony experiments. Cell Cycle. 2007;6:478–488. doi: 10.4161/cc.6.4.3859. [DOI] [PubMed] [Google Scholar]
- Orlando DA, Lin CY, Bernard A, Wang JY, Socolar JE, Iversen ES, Hartemink AJ, Haase SB. Global control of cell-cycle transcription by coupled CDK and network oscillators. Nature. 2008;453:944–947. doi: 10.1038/nature06955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Miles S, GuhaThakurta D, Jemiolo D, Breeden LL. Conserved homeodomain proteins interact with MADS box protein Mcm1 to restrict ECB-dependent transcription to the M/G1 phase of the cell cycle. Genes Dev. 2002;16:3034–3045. doi: 10.1101/gad.1034302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H, Lew DJ, Henze M, Sugimoto K, Reed SI. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes and Development. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- Sevim V, Gong X, Socolar JE. Reliability of transcriptional cycles and the yeast cell-cycle oscillator. PLoS Comput Biol. 2010;6:e1000842. doi: 10.1371/journal.pcbi.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmulevich I, Aitchison JD. Deterministic and stochastic models of genetic regulatory networks. Methods Enzymol. 2009;467:335–356. doi: 10.1016/S0076-6879(09)67013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taba MR, Muroff I, Lydall D, Tebb G, Nasmyth K. Changes in a SWI4,6-DNA-binding complex occur at the time of HO gene activation in yeast. Genes Dev. 1991;5:2000–2013. doi: 10.1101/gad.5.11.2000. [DOI] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- White MA, Riles L, Cohen BA. A systematic screen for transcriptional regulators of the yeast cell cycle. Genetics. 2009;181:435–446. doi: 10.1534/genetics.108.098145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.