Abstract
The identification of intrinsically disordered proteins (IDPs) among the targets that fail to form satisfactory crystal structures in the Protein Structure Initiative represent a key to reducing the costs and time for determining three-dimensional structures of proteins. To help in this endeavor, several Protein Structure Initiative Centers were asked to send samples of both crystallizable proteins and proteins that failed to crystallize. The abundance of intrinsic disorder in these proteins was evaluated via computational analysis using Predictors of Natural Disordered Regions (PONDR®) and the potential cleavage sites and corresponding fragments were determined. Then, the target proteins were analyzed for intrinsic disorder by their resistance to limited proteolysis. The rates of tryptic digestion of sample target proteins were compared to those of lysozyme/myoglobin, apo-myoglobin and α-casein as standards of ordered, partially disordered and completely disordered proteins, respectively. At the next stage, the protein samples were subjected to both far-UV and near-UV circular dichroism (CD) analysis. For most of the samples, a good agreement between CD data, predictions of disorder and the rates of limited tryptic digestion was established. Further experimentation is being performed on a smaller subset of these samples in order to obtain more detailed information on the ordered/disordered nature of the proteins.
Keywords: Intrinsically disordered proteins, protein disorder prediction, Protein Structure Initiative, limited proteolysis
The Protein Structure Initiative (PSI) (or Structural Genomics Initiative, SGI) is a $764 million effort at federal, university, and industry levels to accelerate discovery in structural genomics by dramatically reducing the costs and lessening the time it takes to determine a three-dimensional (3D) protein structure. The goal of PSI, which begun in 2000, is rather ambitious – to determine the representative structures for all the common 3D folds in nature via developing high throughput pipelines for determining protein structures and running large number of proteins through these pipelines (Shapiro and Lima, 1998; Terwilliger, 2000; Williamson, 2000). If a success, such a set of common natural folds then could be used to determine (approximately) the 3D structure of any sequence by homology modeling if the correct template could be identified (Burley and Bonanno, 2002). In short, structural genomics aims to use high-throughput structure determination and computational analysis to provide 3D models of every tractable protein (Brenner, 2000). It is believed that this approach will help to discover distant evolutionary relationships invisible from sequence, which may yield novel functional insights (Brenner and Levitt, 2000).
The primary aim of target selection in PSI is structural characterization of protein families which, so far, lack a structural representative. In other words, protein targets are selected with the goal of increasing the breadth of inferences that can be made from sequence to structure and function (Terwilliger and Berendzen, 1999). To follow the progress of the initiative, PSI Structural Genomics Knowledgebase was created (http://kb.psi-structuralgenomics.org/KB/index.html). Originally, the experimental data tracking module of this Knowledgebase consisted of two components: the target progress tracking database (TargetDB: http://targetdb.rcsb.org), which provided status and tracking information on the production protein targets and their structure solution (Chen et al., 2004), and the experimental protocol tracking database (Protein Expression, Purification, and Crystallization Database (PepcDB): http://pepcdb.rcsb.org) extending the content of TargetDB with detailed experimental histories, reasons for stopping experiments, experimental protocol descriptions, and contact information collected from the PSI centers. Recently, TargetDB and PepcDB have been replaced by the TargetTrack module, which is defined now as a target registration database that provides information on the experimental progress and status of targets selected for structural determination by the PSI and other worldwide high-throughput structural biology projects (http://sbkb.org/tt/). As of May 04, 2012, from the 298,675 targets deposited by worldwide Contributing Centers in TargetTrack, 297,987 proteins were selected, 202,313 were cloned, 127,375 were expressed, of which 48,042 were shown to be soluble. From the 62,262 purified proteins, 13,440 were crystallized, of which 10,104 produced diffraction-quality crystals. In addition to those, 3,156 purified proteins produced high quality heteronuclear single quantum coherence (HSQC) data suitable for structure determination by NMR, of which 2,084 targets were assigned. So far, according to TargetTrack, 4,941 crystal and 2,036 NMR structures were solved. The work on 81,581 targets out of 298,675 targets in TargetTrack was stopped at the various production stages. In short, out of every 100 proteins that have entered the pipelines of various PSI centers, only about 3 yielded structure that was deposited to the protein databank (PDB).
Recent years evidenced an increased appreciation of intrinsically disordered proteins (IDPs, also known as intrinsically unstructured or natively unfolded proteins). These proteins fulfill crucial biological functions while lacking stable secondary and/or tertiary structure and existing instead as very dynamic structural ensembles undergoing fast conformational exchange under physiological conditions in vitro [for reviews see e.g. (Tompa, 2002; Uversky and Dunker, 2010; Uversky et al., 2000; Wright and Dyson, 1999)]. Furthermore, intrinsic disorder is commonly found in proteins associated with the pathogenesis of various human diseases, such as cancer (Iakoucheva et al., 2002), cardiovascular disease (Cheng et al., 2006), amyloidoses (Uversky, 2008), neurodegenerative diseases (Uversky, 2009a; Uversky, 2011a), genetic diseases (Midic et al., 2009a; Midic et al., 2009b), and many other maladies (Uversky et al., 2008).
It has been suggested that the functional diversity provided by disordered regions complements functions of ordered proteins (Dunker et al., 2002; Dunker et al., 2001; Iakoucheva et al., 2002; Uversky, 2011b; Vucetic et al., 2007; Xie et al., 2007a; Xie et al., 2007b). In fact, it is recognized now that a protein function arises not only from unique structures of ordered proteins (for which the ‘sequence→unique 3-D structure→specific function’ paradigm is applicable), but also from conformational ensembles of disordered proteins and even from the transitions between order and disorder in both directions. Thus, a second paradigm ‘sequence→disordered-ensemble→function’ was proposed, leading to the Protein Trinity model (Dunker and Obradovic, 2001), which suggests that the function of a protein can originate from three distinct states (ordered, molten globule and random coil) and transitions between them. This model was subsequently expanded to include a fourth state (pre-molten globule) and transitions between all four states (Uversky, 2002). Examining the functions of IDPs suggests that they are implicated in cells for signaling, regulation, and control (Dunker et al., 2002; Dunker et al., 2005; Dunker et al., 2001; Dyson and Wright, 2005; Iakoucheva et al., 2002; Uversky and Dunker, 2010; Uversky et al., 2005; Wright and Dyson, 2009), through interactions with multiple partners where high-specificity/low-affinity interactions are often requisite (Dunker and Uversky, 2008; Dunker et al., 2008a; Dunker et al., 2008b; Dunker et al., 2001; Oldfield et al., 2008). The functions attributed to IDPs are inherent to the disordered regions and involve either the region remaining disordered or undergoing a disorder-to-order transition.
Amino acid sequences encoding the disordered proteins or regions are significantly different from those that are characteristic for the ordered proteins on the basis of local amino acid composition, flexibility, hydropathy, charge, and several other factors (Radivojac et al., 2007). These sequence biases clearly put intrinsically disordered proteins into a separate class of a protein kingdom and allowed for the reliable computational prediction of IDPs or intrinsically disordered protein regions (IDPRs). The accuracies of disorder predictors have increased as larger data sets and improved machine learning techniques were employed (Ferron et al., 2006; He et al., 2009; Radivojac et al., 2007). Based on their structural properties, IDPs and IDPRs have been grouped into at least two broad classes – compact (molten globule-like) and extended (coil-like and pre-molten globule-like, also called natively unfolded proteins) (Daughdrill et al., 2005; Dunker and Obradovic, 2001; Uversky, 2002; Uversky, 2003). Since both the ability and inability of a protein to fold is encoded in its amino acid sequence, the peculiarities of primary structures of IDPs define their unique structural properties (Dunker et al., 2001; Uversky et al., 2000) and conformational behavior including their high stability against low pH and high temperature and their structural tolerance toward the unfolding by strong denaturants (Uversky, 2009b).
Some of the distinctive structural properties of IDPs and unique peculiarities of their conformational behavior were implemented for the large-scale identification of IDPs in various organisms. In fact, Cortese et al. (2005) showed that E. coli cell extracts may be enriched for IDPs using acid treatment by perchloric acid (PCA) or trichloroacetic acid (TCA) (Cortese et al., 2005). This technique stemmed from the observation that many proteins that remain soluble after acid treatment were shown to be IDPs. Another large scale IDP identification approach was based on the fact that IDPs possessed high resistance toward the aggregation induced by heat treatment (Cortese et al., 2005; Csizmok et al., 2006; Galea et al., 2006). This observation was used to enrich the E. coli and S. cerevisiae cell extracts in IDPs. Finally, a method based on the heat treatment coupled with a 2-D gel electrophoresis was elaborated to identify IDPs in cell extracts (Csizmok et al., 2006). Here, heat treated cell extracts were subjected to native and 8M urea 2-D gel electrophoresis. IDPs, due to their already unfolded state as a result of their typically low hydrophobicity and high net charge, run along or close to the diagonal of the gel. Structured globular proteins on the other hand either aggregate and precipitate upon heat treatment or unfold in 8M urea and run above the diagonal in the second dimension (Csizmok et al., 2006). Although this 2-D gel electrophoresis approach worked well for the smaller proteomes of E. coli and S. cerevisiae, the predicted higher numbers of IDPs in mammalian proteomes make this tool less practical. For this reason, Galea and coworkers used heat treatment under various conditions followed by MS identification of protein spots, resulting in an enrichment of signaling, regulatory and structural proteins, and a depletion of proteins involved in metabolic functions (Galea et al., 2006).
Importantly, because of their flexible and very dynamic nature, IDPs might represent a big challenge for protein crystallographers. Myelin basic protein (MBP) exemplifies these troublemakers (Sedzik and Kirschner, 1992). One exhaustive series of attempts to crystallize MBP for X-ray diffraction has been reported, where the authors tried 4,600 different crystallization conditions but were unable to induce crystallization of MBP (Harauz et al., 2004). Based on these observations the myelin basic protein has been suggested to belong to the category on “uncrystallizable” proteins. In the case of MBP, several additional studies suggest that this protein lacks fixed 3D structure, existing instead as in intrinsically disordered ensemble, which in turn provides the basis of its multifunctionality (Harauz et al., 2009). It can be safely assumed that many other unsuccessful crystallization attempts for numerous other proteins have not been reported, since negative results are generally assumed to be unsuitable for publication.
Since IDPs are highly abundant in nature and can represent a dramatic challenge to the crystallographic attempts, of benefit to the PSI would be to find a way to more efficiently select samples likely to yield structures and to quickly characterize those which do not possess rigid structures. We hypothesized that protein intrinsic disorder might contribute to the bottlenecks that occur at each step in the process of protein structure determination. Therefore, we have elected to study the contributions of intrinsic disorder to the last bottleneck, namely to the failure to obtain crystals from the purified, soluble proteins. To this end, we evaluated the abundance if intrinsic disorder in the PSI proteins by a variety of computational tools. At the next stage, several PSI proteins that have been successfully expressed and purified but that have so far failed to yield 3-D structures were experimentally tested for intrinsic disorder.
Materials & Methods
Materials
Myoglobin from horse skeletal muscle (M0630), apomyoglobin from horse skeletal muscle (A8673) and lysozyme form chicken egg white (L6876) were obtained from Sigma-Aldrich Company. α-Casein (100251) from bovine milk was obtained from MP Biomedicals, Inc. These proteins were supplied as lyophilized powder and were resolublized in appropriate buffers without further purification. Type IX-S trypsin from porcine pancreas (T0303) was obtained from Sigma-Aldrich Company and stored resolubilized in 1 mM HCl containing 20 mM CaCl2 (to prevent autodigestion) in 20 µl aliquots at 50 µM. Nα-Tosyl-L-lysine chloromethyl ketone hydrochloride (TLCK) (T7254) and Soybean Trypsin-Chymotrypsin Inhibitor (STCI) (T9777) were purchased from Sigma.
Samples proteins were supplied from The Berkeley Structural Genomics Center (BSGC), The New York Structural Genomics Research Consortium (NYSGRC), and Mycobacterium tuberculosis Structural Genomics Consortium (TB/MtSGC) – Los Alamos National Laboratory.
Computational Analysis
Sequences from sample proteins were analyzed using predictors of naturally disordered regions (PONDR®) to located predicted regions of disorder. Specifically, PONDR® VL-XT, VL3E, VSL2B, and VSL2P were used from www.DisProt.org. The sequences were scanned for all possible trypsin cut sites; arginine and lysine residues not immediately preceding a proline residue. Expected protease cut sites were selected as those lying in the midst of predicted disorder regions from the PONDR® VSL2P predictor.
Limited Proteolysis
Protease digestion was performed using the final trypsin concentrations of either 0.2 µM or 1 µM. Reactions were performed in duplicate in a 96-well microwell plate with time points at 0, 1, 5, 15, 30, 60, and 120 minutes. 10 µl of 1 mg/ml protein samples were aliquoted into the wells. 2.5 µl of desired trypsin concentration (to have the final protease concentration of 0.2 µM or 1 µM) was added to each well. 2.5 µl of quencher (6 mM TLCK and 60 µM STCI) was added to each reaction at indicated time points. 5 µl of mM HCl was added to the 0 time points instead of trypsin or quencher to serve as a baseline. Immediately after the addition of quencher, 5 µl of sample buffer (4 × LDS Sample Loading Buffer with 500 mM DTT in 2:1 ratio) was added to each reaction and microwell was heated at 70°C for 10 minutes in a thermocycler. The standard protocol for running reduced samples on Invitrogen’s XCell4 MidiCell electrophoresis unit using 26-well 4–12% NuPAGE® Novex bis-tris gels.
Circular Dichroism
Far-UV and near-UV CD experiments were performed on a BioLogic MOS-450 spectropolarimiter. All experiments were performed at room temperature (23°C) with sensitivity set at +/− 30 millidegrees with acquisition duration of 2 seconds over an average of 5 scans. A path-length of 0.2 mm and 1 cm was used for far-UV and near-UV, respectively. Slit widths for both 9 monochromators were set at 1 mm and 0.5 mm for far-UV and near-UV, respectively. Standards and samples were prepared in 50 mM sodium phosphate buffer, pH 8.0. Buffer blanks were subtracted from the scans and spectra were normalized to an ellipticity of zero at 250 nm and 350 nm for far-UV CD and near-UV CD, respectively.
Results and Discussion
Computational analysis of intrinsic disorder abundance in Protein Structure Initiative targets
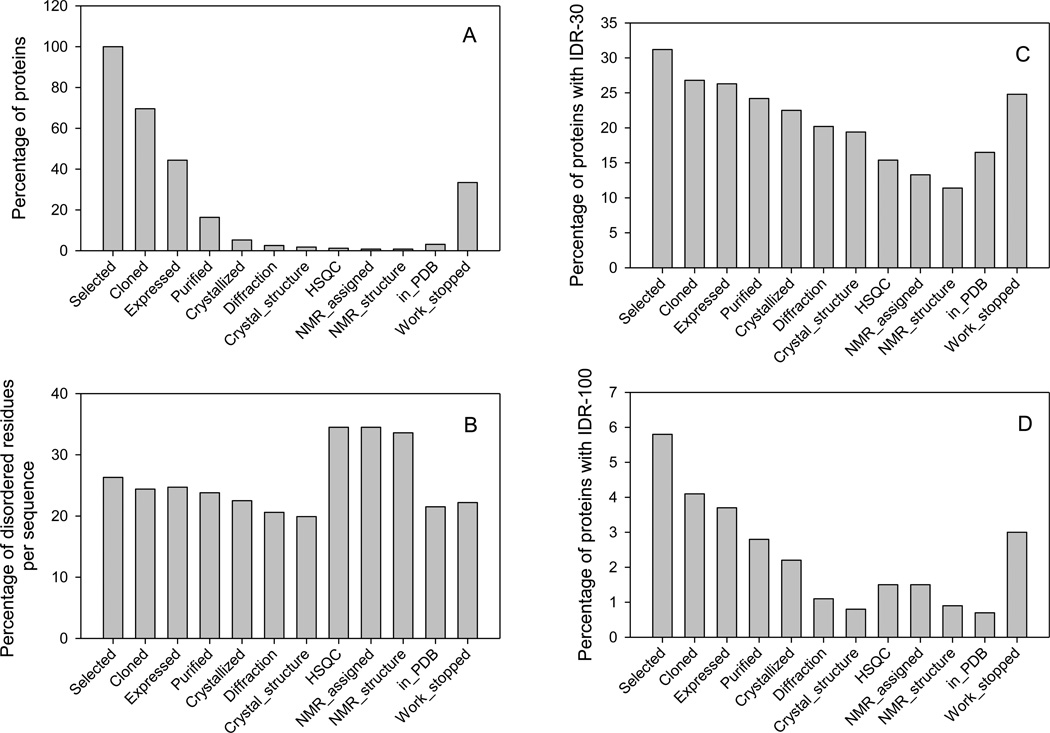
The Protein Structure Initiative (PSI) has the goals of developing high throughput pipelines for determining protein structures and then running large number of proteins through these pipelines. Figure 1A represents the distribution of target proteins from the target progress tracking database (TargetDB: http://targetdb.rcsb.org) which were successfully pushed through the different stages of these pipelines. Figure clearly shows that significant loss takes place at each stage, and that from all the targets selected so far only about 5% have been crystallized, only about 1.2% yielded satisfactory HSQC spectra, and only about 3% produced structures deposited to PDB. On the other hand, for about 33.3% target proteins work was stopped at the various production stages since these proteins were repeatedly unable to progress to the next step.
Figure 1.
Evaluation of the abundance of intrinsic disorder in proteins at various stages of the structure determination pipelines. A. The distribution of target proteins successfully passed through the different stages of these pipelines. The number of proteins at each stage is divided by the number of selected proteins. B. Abundance of predicted disordered residues in sequences of proteins at major pipeline stages. Here, at any given stage, the values calculated for each sequence were summed up and divided by the number of proteins and this stage. C. Amount of proteins containing predicted IDRs longer than 30 residues at each structure determination stage. D. Relative number of proteins with predicted long IDRs, which are at least 100 residue-long. For all plots, disorder was evaluated using the PONDR® VSL2 predictor. Sequences for this plot were obtained from the target progress tracking database (TargetDB: http://targetdb.rcsb.org).
Our hypothesis is that intrinsic disorder contributes to the bottlenecks that occur at each step in the process. In fact, disordered proteins containing large solvent-exposed hydrophobic surfaces might possess high propensity to aggregate. Furthermore, some IDPs might be characterized by low conformational stability and therefore could be degraded fast by the proteases. Finally, expression of some IDPs, which are known to be involved in signaling and regulation, might be toxic for the heterologous expression systems. Figure 1B shows that sequences of selected proteins on average contain ~26% disordered residues as predicted by PONDR® VSL2. This number decreases to 22.5% in proteins that yielded crystal structures. Proteins selected for NMR analysis have noticeably greater content of disordered residues. Proteins for which work was stopped contain ~22% disordered residues.
Since disordered regions could be evenly distributed through the protein sequence or could form long contiguous stretches, at the next step we analyzed the abundance of long predicted disordered regions in proteins at each stage of the pipeline. Figure 1C shows that the percentage of proteins with intrinsically disordered regions (IDRs) longer than 30 residues sequentially decreases mostly following the structure determination stages: although 31.2% selected proteins contain such regions, this number decreases to 19.4% and 11.1% in proteins that produced crystal or NMR structures respectively. The picture is even more convincing when length of predicted disordered regions is increased to 100 or more consecutive residues. In fact, Figure 1D shows that 5.8% selected proteins contain such long IDRs, whereas the long disordered stretches are predicted in only 0.8% and 0.9% proteins whose crystal or NMR structure was solved, respectively. Furthermore, although many of the proteins with long predicted IDRs were claimed to be crystallized, in most cases, the actually crystallized parts were domains rather than entire proteins. Once again, many proteins for which work was stopped at the various production stages contain long IDRs.
Taken together data presented in Figure 1 show that the amount of intrinsic disorder, especially long IDRs, progressively decreases while moving from the top to the bottom of the structure determination pipelines. The decrease in the amount of structural disorder is especially noticeable when the length of the predicted IDRs is taken into account (cf. Figures 1C and 1D). The facts that proteins, which are able to successfully pass to the next stage, typically contain less disorder than proteins at the previous stage and that proteins for which work was stopped typically contain noticeable amount of disorder suggest that our hypothesis is correct and intrinsic disorder likely contributes to the PSI bottlenecks. To further check this hypothesis, several crystallizable and non-crystallizable proteins obtained from the PSI Centers were analyzed by a set of computational and experimental techniques to evaluate their intrinsic disorder content.
Implemented tools for the intrinsic disorder analysis in samples from PSI centers
For this project, we have elected to study the contributions of intrinsic disorder to the protein structure determination bottleneck, namely the failure to determine 3D structures from purified, soluble proteins. To this end, a set of both crystallizable and non-crystallizable proteins was obtained from PSI Centers and analyzed by a combined approach that includes computational and experimental tools. The targets analyzed in this study were not chosen in any specific way, we simply studied proteins provided by the PSI Centers.
Each sample was analyzed computationally by several members of the PONDR® family to predict order/disorder content and then scanned for all possible trypsin cut sites. Next, limited proteolytic digestion at two trypsin concentrations was performed for each protein. The results of this analysis were compared to standards (i.e., well-characterized proteins with known disorder status, fully ordered, fully disordered, and partially disordered) in terms of rate of digestion and the presence, number, and stability of fragments. Then, far-UV and near-UV CD were measured for all the samples to investigate the overall content of the ordered secondary structure and to evaluate the rigidity of the environment of aromatic residues, respectively.
Several computational tools for intrinsic disorder prediction, PONDR® VLXT, VSL2 and VL3, CDF analysis and CH-plot, were exploited in this study. The rational for their use is briefly outlined below. PONDR® VL3 (Peng et al., 2005) is a predictor for accurate evaluation of long disordered regions. PONDR® VLXT (Dunker et al., 2001) is a general disorder predictor, which is very sensitive to potential functional sites. PONDR® VSL2 (Peng et al., 2006) is one of the most accurate disorder predictors developed so far. It is statistically better for proteins containing both structure and disorder. For these three predictors, which evaluate the per residue disorder probability, scores above 0.5 correspond to the predicted disordered regions/residues, whereas scores below 0.5 correspond to predicted ordered regions/residues. Two other computational tools, the cumulative distribution function (CDF) analysis (Dunker et al., 2000; Oldfield et al., 2005; Xue et al., 2009) and charge-hydropathy plot (CH-plot) (Uversky et al., 2000), are binary predictors, which indicate whether a given protein is ordered or disordered as whole. Ordered proteins generally lie in the upper left region of the CDF plot as a larger fraction of their residues is predicted to have lower PONDR® scores. Conversely, disordered proteins are expected to lie in the lower right corner of the CDF plot since a larger fraction of their residues is predicted to have higher disorder scores (Dunker et al., 2000; Oldfield et al., 2005; Xue et al., 2009). In CH-plot, compact proteins lie in the right-hand corner since they are generally enriched in hydrophobic residues and contain fewer charged residues. Conversely, proteins with extended disorder are characterized by low hydropathy and high net charge and therefore tend to lie in the upper left-hand region of the CH-plot (Oldfield et al., 2005; Uversky et al., 2000).
The extent of proteolytic digestion by specific proteases, such as trypsin, has been shown to correspond to flexibility in the region of the cut site and not just surface exposure (Hubbard, 1998). Depending on the amount of intrinsic disorder in a given protein, three major scenarios for the proteolytic digestion are expected. Highly disordered proteins are expected to be digested fast, typically without accumulation of stable fragments. However, some of such proteins might produce semi-stable fragments, the number and protease resistance of which are expected to be depended on the amount and stability of partially ordered structure. Highly ordered proteins are expected be digested slowly, typically with accumulation of one or several stable fragments. Partially disordered proteins (proteins with long IDRs) are expected to show the intermediate proteolytic behavior. As accessible cut sites in disordered regions are cleaved, stable fragments may emerge if enough structural stability is maintained. Therefore, by comparing the rates of digestion as determined by the disappearance of the initial protein band on SDS-PAGE and by the analysis of the digestion patterns, we can loosely characterize the degree of disorder in protein samples. Obviously, experimental mapping of intrinsically disordered regions with combined prediction and proteolysis can be readily parallelized in a high-throughput format.
There has been a long-running debate whether proteolysis occurs only in unfolded or disordered regions of proteins or whether proteolysis can occur in surface-exposed, but ordered regions of proteins. There are a several arguments supporting the former model. Particularly, if proteolysis is indeed happening at the surface-exposed and ordered sites, then it is unclear how a protease of known sequence specificity recognizes such a limited subset out of the many putative sites of proteolysis in a folded polypeptide chain. In fact, trypsin ought to cleave polypeptide at nearly every lysine-X and arginine-X bond (with the partial exception of proline at X), assuming that about 5–10% of the peptide bonds in a typical protein has to be susceptible to proteolytic attack. However, at the native (or near-native) conditions, trypsin will cut only a limited number of such bonds (or on occasion none at all) in a natively folded protein. This means that the structure and dynamics of a substrate protein play a crucial role in determining the efficiency of proteolysis (Hubbard et al., 1994; Hubbard et al., 1998). Interestingly, the analysis of the X-ray crystallographic structures of proteases with small protein inhibitors, such as BPTI, revealed that the inhibitor reactive site loop bound into the enzyme active site in the manner of a ‘perfect’ substrate. This canonical conformation is conserved throughout diverse families of small protein inhibitors of serine proteases although the overall fold and the amino acid sequence of these inhibitors are not (Bode and Huber, 1992; Laskowski and Kato, 1980).
Using this canonical conformation of the inhibitor reactive site loops as a template, Thornton and co-workers showed that limited proteolytic sites are quite different in structure from the idealized inhibitor loops, and they must therefore undergo a conformational change in order to enter the proteinase active site (Hubbard et al., 1991). Furthermore, for several proteins it has been shown that local unfolding of at least 13 residues is needed for a set of observed cut-sites to properly fit into trypsin’s active site (Hubbard et al., 1994; Hubbard et al., 1998). Hence, the position of the putative limited proteolytic site with respect to the rest of the substrate tertiary structure, and the inherent flexibility and opportunity for local unfolding must help determine its proteolytic susceptibility. Furthermore, it has been established that limited proteolytic sites are typically found within flexible solvent-exposed loop regions (as indicated by crystallographic temperature factors or B-values) (Fontana et al., 1986; Hubbard et al., 1991; Novotny and Bruccoleri, 1987), and are notably absent in the regions of regular secondary structure, especially within the β-sheets (Fontana et al., 1997a; Fontana et al., 1997b; Hubbard et al., 1994). These proteolytic sites protrude from the protein surface and are expected to be found at regions where the local packing does not inhibit the local unfolding that is deemed necessary (Hubbard et al., 1991). Fontana and co-workers (Fontana et al., 1997b) showed that apo-myoglobin is digested many orders of magnitude faster than myoglobin and the sites of digestion from several different proteases all mapped to a region of intrinsic disorder. Similarly, it has been shown that while the holoform of cytochrome c is fully resistant to proteolytic digestion and the apo-protein is digested to small peptides, the non-covalent complex of the apo-protein and heme exhibits an intermediate resistance to proteolysis, in agreement with the fact that the more folded structure of the complex makes the protein substrate more resistant to proteolysis (Spolaore et al., 2001). Thus, our view is that protease digestion is much faster in regions of intrinsic disorder and so can be used to map ordered and disordered regions. In agreement with this suggestion, earlier we have established that PONDR® indications of order and disorder are perfectly correlated with the protease digestion experiments (Iakoucheva et al., 2001a; Iakoucheva et al., 2001b): regions predicted to be ordered were generally not cut at all, while regions predicted to be disordered were rapidly cut.
Secondary structure content can be estimated from the circular dichroism spectra in the far-UV region (190–250 nm) which reveal the peculiarities of the peptide bond environment. As a result, very distinctive spectra are produced from different types of secondary structure. Extended IDPs (native coils or native pre-molten globules) are characterized by ellipticities near zero at 185 nm, large negative ellipticities in the region around 200 nm, and low negative signals near 222 nm (Daughdrill et al., 2005; Receveur-Brechot et al., 2006; Uversky and Longhi, 2010; Uversky and Dunker, 2012a; Uversky and Dunker, 2012b; Uversky and Dunker, 2012c).
Local tertiary structure may be evaluated from near-UV CD spectra (250–350 nm) which display environmental characteristics of aromatic amino acid residues (tyrosine, tryptophan, and phenylalanine) (Daughdrill et al., 2005; Receveur-Brechot et al., 2006; Uversky and Longhi, 2010; Uversky and Dunker, 2012a; Uversky and Dunker, 2012b; Uversky and Dunker, 2012c). Proteins with aromatic groups embedded into rigid tertiary structure produce specific near-UV CD spectra with a unique fingerprint distribution of peaks characteristic for each aromatic side chain type. It is expected that signals for phenylalanine, tyrosine and tryptophan generally are within the 250–270 nm, 270–290 nm, and 280–300 nm regions respectively. Since structured proteins have unique 3D structures, their aromatic groups have unique environments which create spectra that are unique. Although a lack of spectral complexity and low signal strength are indicative of a lack of tertiary structure in many IDPs, some IDPs produce intricate near-UV CD spectra implying that they retain, at least in part, some residual tertiary structure (Daughdrill et al., 2005; Receveur-Brechot et al., 2006; Uversky and Longhi, 2010; Uversky and Dunker, 2012a; Uversky and Dunker, 2012b; Uversky and Dunker, 2012c).
Simultaneous analysis of near- and far-UV CD spectra can discriminate whether the protein is in an ordered form, in an extended disordered form, or in a collapsed disordered form. That is, ordered forms give negative intensity in the ~ 205 to ~240 nm range, indicating secondary structure, and also a set of specific sharp positive or negative peaks in the ~ 250 to ~ 305 range, indicating rigidly packed aromatic side chains. In contrast, extended disorder gives very weak or even positive intensity in the ~210 to ~ 240 nm range combined with a strong negative peak in the vicinity of 200 nm, indicating the absence of regular secondary structure, and also the absence of well-defined signal in the ~ 280 to ~ 305 range, indicating absence of rigid side chain packing. Finally, collapsed disorder exhibits spectra indicating secondary structure (negative intensity in the ~ 205 to ~ 240 nm range), but with the absence of rigid aromatic chain packing (absence of sharp peaks in the ~ 250 to ~ 305 nm range).
Evaluating intrinsic disorder in standard well-characterized proteins with known disorder status
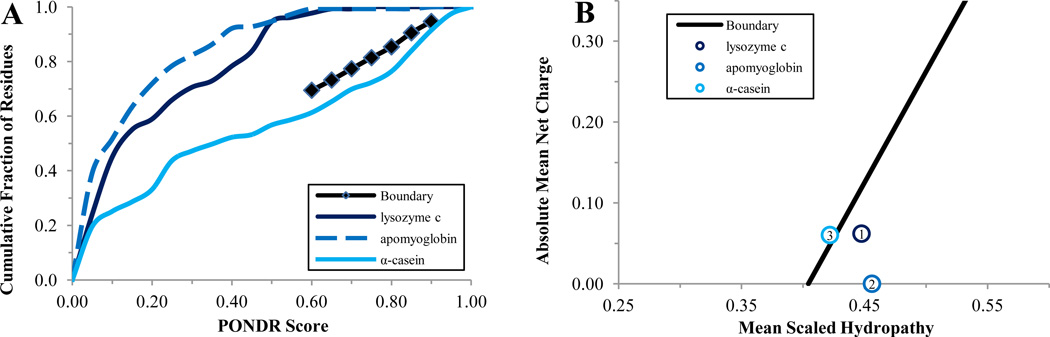
The proposed approach was first calibrated using a set of standard proteins with known disorder status. To this end, we analyzed a well-studied and easily accessible IDP (α-casein), a well-studied and easily accessible highly ordered protein (hen egg white lysozyme), and a moderately stable partially ordered protein (apo-myoglobin). Figure 2 represents the analysis of the 3 protein standards by the cumulative distribution function (CDF) and charge-hydropathy plot (CH-plot). In agreement with previous studies, α-casein was predicted to be intrinsically disordered as a whole, whereas both apomyoglobin and lysozyme were predicted to be mostly ordered. Next, we analyzed the per-residue disorder distribution in these standards. Figure 3 and Table 1 represent the results of intrinsic disorder prediction by PONDR® VSL2, VL3, and VLXT and support the notion that α-casein is essentially more disordered than apomyoglobin and lysozyme. In fact, according to PONDR® VSL2 data in Table 1, α-casein, apomyoglobin, and lysozyme were predicted to have 81.4%, 50.6% and 8.8% disordered residues with the average scores of 0.775, 0.453, and 0.275, respectively.
Figure 2.
Cumulative Distribution Function (CDF) and CH-plot of standards and example proteins: Panel A – boundary ( ), lysozyme c (
), lysozyme c ( ), apomyoglobin (
), apomyoglobin ( ), α-casein(
), α-casein( ); Panel B – boundary (
); Panel B – boundary ( ), lysozyme c (
), lysozyme c ( ), apomyoglobin(
), apomyoglobin( ), α-casein(
), α-casein( ).
).
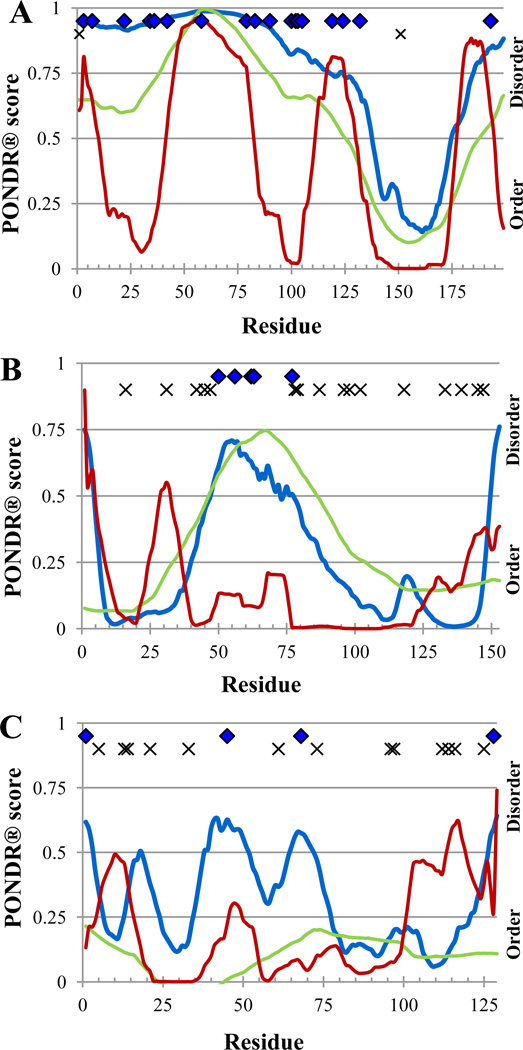
Figure 3.
PONDR® plot and trypsin digestion prediction of standards. VSL2P (blue), VL3E (green), VLXT (red), potentially accessible, disorder-based cut sites ( ), inaccessible cut sites (x)). Panel A – α-casein; Panel B – myoglobin and apo-myoglobin; Panel C – lysozyme.
), inaccessible cut sites (x)). Panel A – α-casein; Panel B – myoglobin and apo-myoglobin; Panel C – lysozyme.
Table 1.
Percent Disorder and Average PONDR Score of Standards
| Protein | % Disorder | Average PONDR Score | ||||
|---|---|---|---|---|---|---|
| VSL2 | VL3 | VLXT | VSL2 | VL3 | VLXT | |
| α-Casein | 81.4 | 68.8 | 43.2 | 0.775 | 0.579 | 0.428 |
| Apo-myoglobin | 50.6 | 26.0 | 6.5 | 0.453 | 0.317 | 0.155 |
| Lysozyme | 8.8 | 0.0 | 4.8 | 0.275 | 0.116 | 0.160 |
Earlier studies revealed that there is an excellent correlation between the PONDR® indications of order and disorder and protease cut sites in a given protein, since regions predicted to be ordered were generally uncut at all, whereas regions predicted to be disordered were rapidly cut (Iakoucheva et al., 2001a; Iakoucheva et al., 2001b). This is illustrated by the analysis of the digestion pattern of the XPA protein which showed that virtually all of the trypsin-sensitive sites were found in regions that were predicted to be disordered and virtually all of the potential cut sites which were trypsin-resistant were seen in regions that were predicted to be ordered (Iakoucheva et al., 2001a; Iakoucheva et al., 2001b). To evaluate the accessibility of the standard proteins to proteolytic attack, Figure 3 represents the probable cut sites located in disordered regions of α-casein, apomyoglobin, and lysozyme. Figure shows that almost all cut sites of α-casein are located in regions predicted to be disordered and therefore are trypsin-sensitive, whereas both apomyoglobin and lysozyme contain noticeable number of potential trypsin-resistant sites.
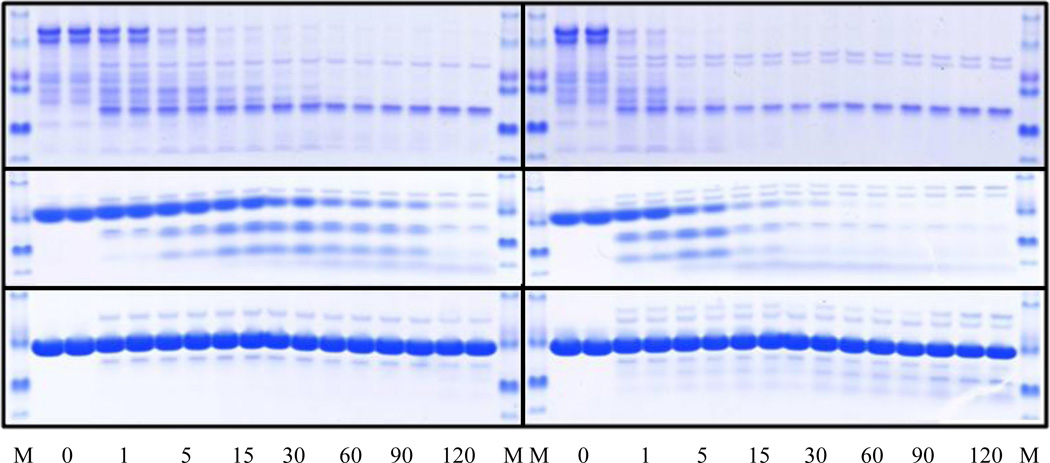
The SDS-PAGE gels provide an easy and rather accurate tool for quantitative analysis of the amount of digestion, and the overall pattern observed on the gels is highly informative. For example, if a single protein becomes cut into two pieces, it is likely that the protein has 2 domains connected by a flexible (disordered) linker. If a protein shows multiple intermediates, ultimately giving a lower molecular mass band, it likely has at least one long flexible region that is cut almost simultaneously at multiple locations. If no sizable intermediates are observed during the digestion time course, the digested protein could be completely disordered (Yang and Klee, 2002). Figure 4 illustrates the limited tryptic digestion results for a typical IDP (α-casein), a partially folded protein (apo-myoglobin) and a highly ordered protein (lysozyme). In agreement with expectations, intrinsically disordered α-casein was digested very fast, even in the presence of low trypsin concentrations (Figures 4A and B). Partially folded apomyoglobin possessed a moderate proteolytic resistance and produced sizable stable fragments (Figures 4C and D), whereas highly ordered lysozyme was not cut (Figures 4E and F).
Figure 4.
Time course of the limited trypsin digestion of standard proteins by 0.2 µM trypsin (A, C, and E) and 1 µM trypsin (B, D, and F). Plots A and B show the digestion results for α-casein. Plots C and D represent the data on the apo-myoglobin trypsinolysis. Plots E and F illustrate tryptic cleavage of lysozyme. In each plot, the first and the last lanes correspond to the molecular mass standards. Numbers reflect the time points at which the proteolysis was quenched.
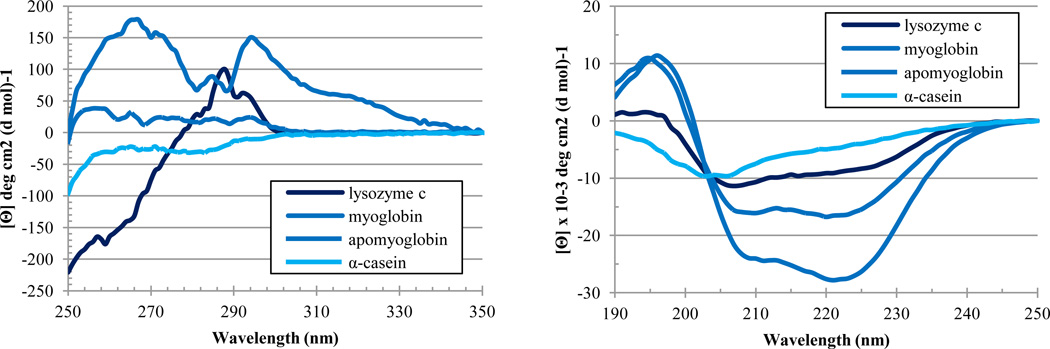
Next, structural properties of the standard proteins were analyzed using near- and far-UV CD spectroscopy. The near-UV CD spectrum of lysozyme was relatively intensive and was characterized by fine structure, possessing a broad feature-less negative peat in the vicinity of 260 nm and a series of three, relatively well-resolved positive peaks at 280, 290 and 295 nm (Figure 5A). Such near-UV CD spectrum is indicative of the well-developed tertiary structure. The near-UV CD spectrum of myoglobin was also typical for the well-folded protein, being characterized by relatively high intensity and having a pronounced fine structure, containing 4 positive peaks, a large, broad peak at 265 nm, a small peak at 285 nm, another large peak at 295 nm, and small, broad peak at 320 nm (Figure 5A). The hem removal from myoglobin is known to destabilize this protein. In the near-UV CD analysis of apo-myoglobin, this destabilization led to the dramatic decrease in the spectral intensity, being also accompanied by the noticeable simplification of the spectrum. In fact, the near-UV CD spectrum of apo-myoglobin was characterized by 4 or 5 positive peaks between 250 and 300 nm, all with relatively low amplitudes (Figure 5A). Finally, the near-UV CD spectrum of α-casein possessed low intensity and showed a broad feature-less negative peak at 280 nm, clearly indicating almost complete lack of rigid tertiary structure in this protein (Figure 5A).
Figure 5.
Spectroscopic analysis of the standard proteins. A. Near-UV CD spectra of standard proteins at 1 mg/ml. B. Far-UV CD spectra of standard proteins at 1 mg/ml.
Figure 5B represents the results of the secondary structure analysis of the standard proteins. The far-UV CD spectrum of lysozyme possessed a positive peak at 198 nm and two minima at 208 nm and 222 nm, typical for the proteins with the well-developed secondary structure (Figure 5B). Similarly, the far-UV CD spectrum of myoglobin clearly possessed features of well-folded helical protein (see Figure 5B). On the other hand, apo-myoglobin was characterized by less intensive far-UV CD spectrum, reflecting the reduction of ordered secondary structure caused by the hem removal. Finally, the far-UV CD spectrum of α-casein was characterized by features typical for the highly disordered polypeptide chain, a negative peak at 204 nm and very shallow shoulder at 222 nm (see Figure 5B).
Altogether, data retrieved for the well-characterized proteins with the different levels of intrinsic disorder clearly indicated that that the proposed approach where computational analysis is combined with the experimental tools can be used for obtaining the reliable information on the disorder status of the query protein.
Analysis of the illustrative PSI target proteins
BSGCAIR30378
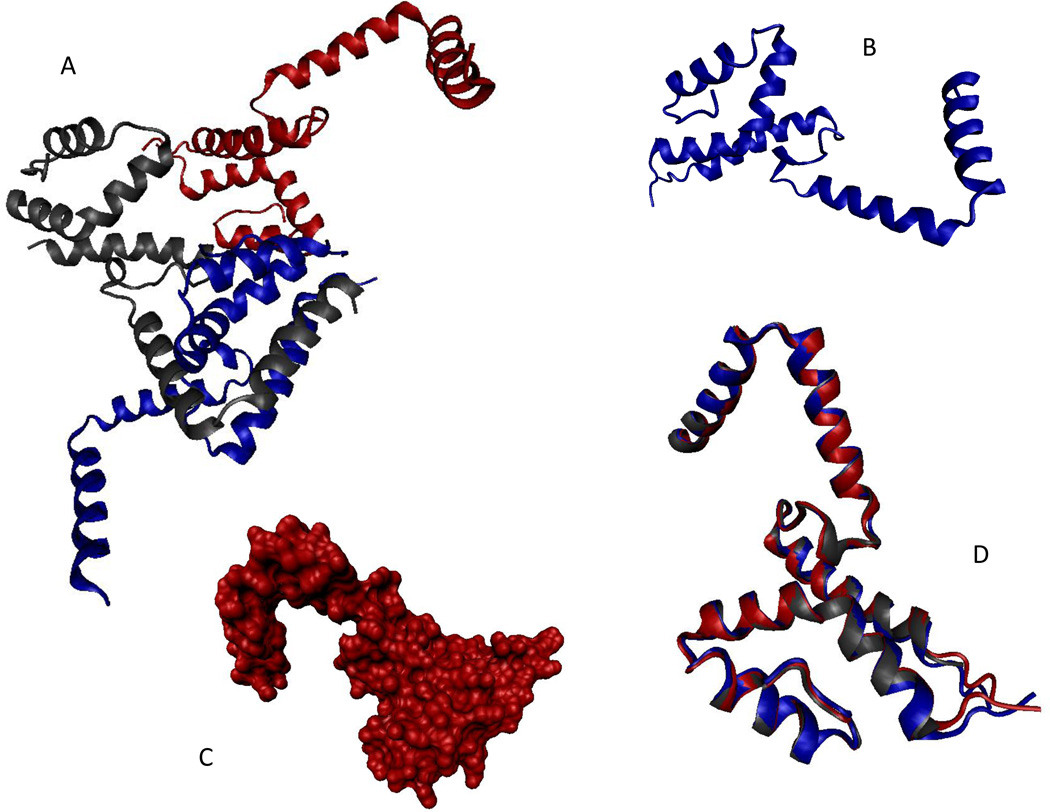
The first protein selected as an example from the set of the PSI targets was BSGCAIR30378. Since the crystal structure of this protein was successfully solved (PDB ID 2I15, see Figure 6), BSGCAIR30378 (which is the hypothetical protein MG296 homolog) was chosen as an illustration of well-ordered crystallizable proteins. Analysis of the crystal structure revealed that BSGCAIR30378 contained a potentially disordered region, fragment 71–85, disorder in which was manifested as missing electron density. Although the biological unit of BSGCAIR30378 is unknown, this protein crystallized as an oligomer. Figure 6 shows a homotrimer of BSGCAIR30378 (Figure 6A), monomers of which contain a tri-helix up-down bundle globular domain and a rather extended helical protrusion containing two helices (residues 3–17 and 21–35) without noticeable intramolecular contacts (Figure 6B). This helical protrusion is used to establish an extensive set of intermolecular contacts with neighboring BSGCAIR30378 molecules (see Figure 6A). The obscure shape of the BSGCAIR30378 monomer is further illustrated by Figure 6C representing a solvent accessible surface area of the monomer. Despite the rather unusual shape, monomers inside the BSGCAIR30378 oligomers are rather immobile, as evidenced by the perfection of their structural alignment (Figure 6D).
Figure 6.
Crystal structure of BSGCAIR30378 (PDB ID: 2I15). A. Structure of the BSGCAIR30378 trimer. B. Structure of the monomeric species (cartoon representation). C. Structure of the monomeric species (solvent accessible surface representation). D. Multiple structure alignment of the three monomeric species from the biological unit of BSGCAIR30378 (PDB ID: 2I15). The alignment was performed using the MultiProt tool (http://bioinfo3d.cs.tau.ac.il/MultiProt/) (Shatsky et al., 2004). All images were created using the VMD tool (Humphrey et al., 1996).
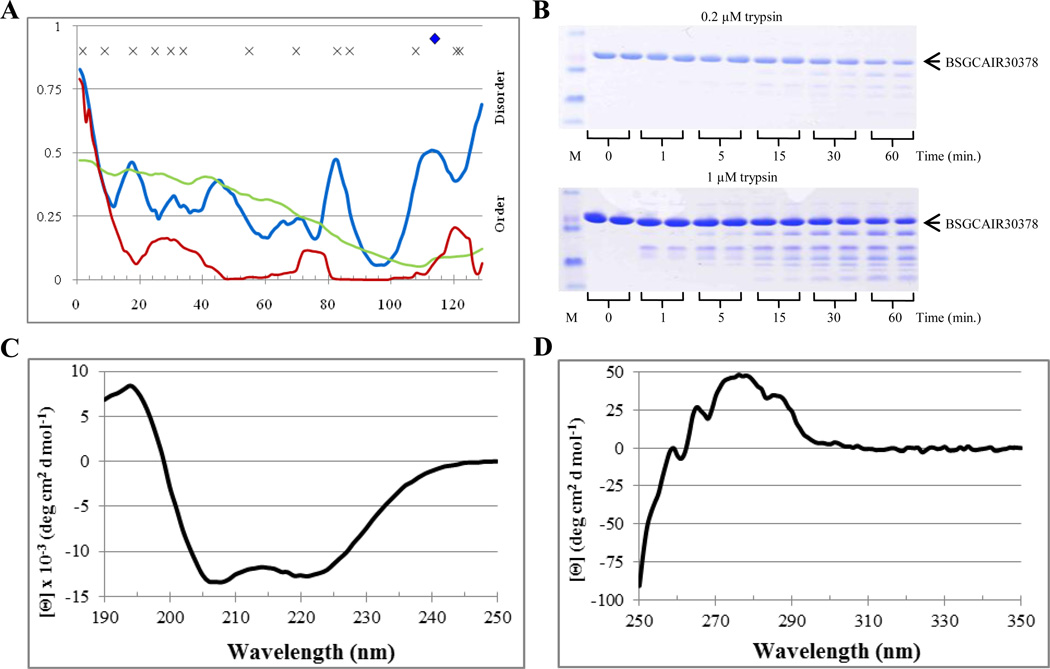
Computational analysis revealed that BSGCAIR30378 was a mostly ordered protein, which contained 10.9%, 0.0%, and 4.7% disordered residues predicted with an average scores of 0.324, 0.263, and 0.103 for PONDR VSL2, VL3, and VLXT, respectively (see Table 2). Figure 7A shows that according to these three predictors the majority of the BSGCAIR30378 residues possessed scores below 0.5. Whereas PONDR VL3 produced a smooth, featureless curve with a broad minimum around residue 110, PONDR VLXT curve stayed near zero, had three low intensity but distinctive maxima in the vicinity of residues 30, 75 and 120, and went above the score of 0.5 only at the N-terminus, possibly indicating that BSGCAIR30378 had a disordered tail. On the other hand, PONDR VSL2 curve was shifted toward the higher disorder score (~0.25), had four low intensity maxima near the residues 20, 35, 45 and 70, and two higher intensity maxima near the residues 85 and 110, and predicted disorder for both N-and C-termini. Regions of increased intrinsic disorder propensity corresponded to the loops connecting helical elements in the BSGCAIR30378 3-D structure. Interestingly, the most flexible region with the highest intrachain disorder score (near the residue 85) roughly corresponded to the region with missing electron density. Only one predicted trypsin cut site near the C-terminus of the protein was located in the putative disordered region predicted by PONDR VSL2) (Figure 7A). Based on the PONDR analysis and tryptic cut site predictions, digestion was expected to occur relatively slowly resulting in the accumulation of stable fragment(s).
Table 2.
Evaluation of the abundance of intrinsic disorder in PSI target proteins by PONDR VSL2, VL3, and VLXT together with the number of predicted disorder-based trypsin cut sites and the experimentally validated number of tryptic fragments. Targets that could be crystallized are highlighted.
| Prediction | Experimental | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Structure Genomics/Protein Structure Initiative Center |
Protein (TargetID) |
Length | MW (Da) | % Disorder (VSL2P) |
% Disorder (VL3E) |
% Disorder (VLXT) |
Average PONDR Score (VSL2P) |
Average PONDR Score (VL3E) |
Average PONDR Score (VLXT) |
Expected # Cut sites |
Other Cut Sites |
Expected # fragments |
Number of Stable Fragments |
Rate of 1 µM Digestion |
Rate of 0.2 µM000B5 Digestion |
|
| BSGC | BSGCAIR30637 | 188 | 21489.2 | 18.1% | 14.4% | 16.0% | 0.332 | 0.355 | 0.280 | 7 | 23 | 8 | 0 | ** | ** | |
| BSGC | BSGCAIR30592 | 159 | 17142.4 | 25.8% | 20.8% | 30.8% | 0.309 | 0.385 | 0.381 | 5 | 11 | 6 | 5–6 | *** | *** | |
| BSGC | BSGCAIR30556 | 354 | 40458.2 | 30.8% | 22.9% | 39.3% | 0.364 | 0.412 | 0.415 | 20 | 44 | 21 | 7–9 | ** | ** | |
| BSGC | BSGCAIR30476 | 232 | 26937.0 | 17.2% | 22.4% | 33.6% | 0.243 | 0.357 | 0.380 | 5 | 27 | 6 | 1 | * | * | |
| BSGC | BSGCAIR30468 | 283 | 31603.7 | 6.0% | 7.1% | 68.2% | 0.149 | 0.272 | 0.561 | 2 | 36 | 3 | 3 | ** | ** | |
| BSGC | BSGCAIR30347 | 231 | 25235.3 | 7.8% | 10.0% | 28.1% | 0.244 | 0.222 | 0.285 | 1 | 31 | 2 | 1 | ** | ||
| BSGC | BSGCAIR30399 | 131 | 15528.8 | 9.9% | 17.6% | 55.7% | 0.221 | 0.382 | 0.535 | 2 | 19 | 3 | 1–2 | ***** | ***** | |
| BSGC | BSGCAIR30378 | 129 | 14939.1 | 10.9% | 0.0% | 4.7% | 0.324 | 0.263 | 0.103 | 1 | 13 | 2 | 4 | * | ||
| BSGC | BSGCAIR30649 | 518 | 57556.7 | 17.4% | 3.7% | 21.6% | 0.283 | 0.138 | 0.250 | 10 | 39 | 11 | 4–5 | ** | ** | |
| BSGC | BSGCAIR30544 | 371 | 44116.7 | 16.7% | 25.6% | 20.8% | 0.275 | 0.277 | 0.233 | 11 | 47 | 12 | 4 | ***** | ***** | |
| BSGC | BSGCAIR30883 | 201 | 22978.6 | 21.4% | 12.4% | 35.8% | 0.282 | 0.329 | 0.396 | 3 | 26 | 4 | 1 | ***** | ***** | |
| BSGC | BSGCAIR30998 | 109 | 12135.4 | 81.7% | 59.6% | 36.7% | 0.773 | 0.605 | 0.394 | 12 | 4 | 13 | 1 | ***** | ***** | |
| BSGC | BSGCAIR31082 | 98 | 11534.9 | 31.6% | 0.0% | 27.6% | 0.399 | 0.152 | 0.332 | 5 | 7 | 6 | 0 | * | ||
| BSGC | BSGCAIR30932 | 108 | 12547.1 | 100.0% | 0.9% | 22.2% | 0.888 | 0.422 | 0.317 | 16 | 0 | 17 | 1 | *** | *** | |
| BSGC | BSGCAIR30686 | 329 | 36102.2 | 55.9% | 40.1% | 22.8% | 0.513 | 0.345 | 0.294 | 20 | 14 | 21 | 1 | * | ||
| BSGC | BSGCAIR30926 | 134 | 15659.7 | 100.0% | 71.6% | 50.7% | 0.878 | 0.548 | 0.467 | 20 | 1 | 21 | 2 | ** | ||
| BSGC | BSGCAIR30600 | 449 | 51131.9 | 25.4% | 34.5% | 60.4% | 0.332 | 0.429 | 0.541 | 18 | 47 | 19 | ||||
| BSGC | BSGCAIR30441 | 520 | 58181.9 | 25.8% | 9.6% | 22.9% | 0.372 | 0.283 | 0.279 | 16 | 61 | 17 | 5 | ***** | ***** | |
| NYSGXRC | 10001e | 280 | 32062.2 | 13.2% | 21.8% | 49.6% | 0.267 | 0.347 | 0.436 | 2 | 21 | 3 | 3–4 | **** | *** | |
| NYSGXRC | 10336x | 435 | 48578.3 | 15.9% | 2.3% | 21.6% | 0.274 | 0.242 | 0.287 | 7 | 34 | 8 | 1–2 | ***** | ***** | |
| NYSGXRC | 10354e | 213 | 24129.3 | 25.8% | 14.6% | 15.5% | 0.305 | 0.294 | 0.186 | 11 | 12 | 12 | 1–2 | ***** | ***** | |
| NYSGXRC | 10379l | 609 | 70570.8 | 14.6% | 10.8% | 18.9% | 0.282 | 0.225 | 0.235 | 7 | 52 | 8 | 5 | ** | ** | |
| NYSGXRC | 10379u | 618 | 70295.3 | 13.8% | 13.9% | 17.8% | 0.266 | 0.239 | 0.242 | 9 | 45 | 10 | 2 | ** | ** | |
| NYSGXRC | 10407h | 234 | 27040.2 | 25.6% | 29.5% | 34.6% | 0.357 | 0.428 | 0.415 | 8 | 19 | 9 | 1 | ***** | ***** | |
| NYSGXRC | 11004k | 140 | 16480.4 | 19.3% | 13.6% | 24.3% | 0.224 | 0.241 | 0.281 | 4 | 9 | 5 | 4 | **** | *** | |
| NYSGXRC | 11007m | 139 | 15744.9 | 15.8% | 24.5% | 41.0% | 0.260 | 0.348 | 0.394 | 1 | 13 | 2 | 1 | *** | **** | |
| NYSGXRC | 11016m | 139 | 15568.9 | 15.1% | 18.7% | 25.2% | 0.188 | 0.288 | 0.272 | 1 | 14 | 2 | 0 | * | * | |
| NYSGXRC | 9245d | 448 | 49468.6 | 5.4% | 4.5% | 10.0% | 0.137 | 0.193 | 0.232 | 3 | 39 | 4 | 0 | ** | ** | |
| NYSGXRC | 9262h | 413 | 46416.3 | 10.4% | 0.0% | 21.8% | 0.177 | 0.183 | 0.295 | 4 | 34 | 5 | 3 | **** | *** | |
| NYSGXRC | 9396a | 374 | 41800.2 | 9.6% | 0.0% | 20.2% | 0.215 | 0.148 | 0.257 | 2 | 37 | 3 | 3 | *** | ** | |
Figure 7.
Disorder analysis of BSGCAIR30378. A. PONDR® plots (VSL2P (blue), VL3 (green), VLXT (red)) and trypsin digestion prediction (potentially accessible cut sites ( ), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
In agreement with these predictions, Figure 7B shows that the rate of the BSGCAIR30378 limited digestion at both 0.2 µM and 1 µM trypsin was relatively slow, in between that of the partially and fully ordered standard, apo-myoglobin and lysozyme, respectively. Furthermore, greater than 75% of the protein remained intact after the digestion for 60 minutes at both trypsin concentrations (0.2 µM and 1 µM). At 0.2 µM trypsin, a few stable fragments had emerged after the 15 minute proteolysis, whereas at 1 µM stable fragments began to appear at the 1 minute time point. The results of computational predictions and limited trypsinolysis were further supported by the BSGCAIR30378 spectroscopic analysis. In fact, the far-UV CD spectrum of this protein was typical for a well-folded protein with pronounced α-helical structure (Figure 7C). Whereas near-UV CD spectrum confirmed the presence of relatively rigid tertiary structure in BSGCAIR30378, since this protein possessed a broad peak at 275 nm and several sharper peaks at 258 nm, 265 nm, and 286 (Figure 7D). Therefore, both computational and experimental analyses agreed that BSGCAIR30378 is a relatively ordered protein with a conformational stability between those of lysozyme and apo-myoglobins.
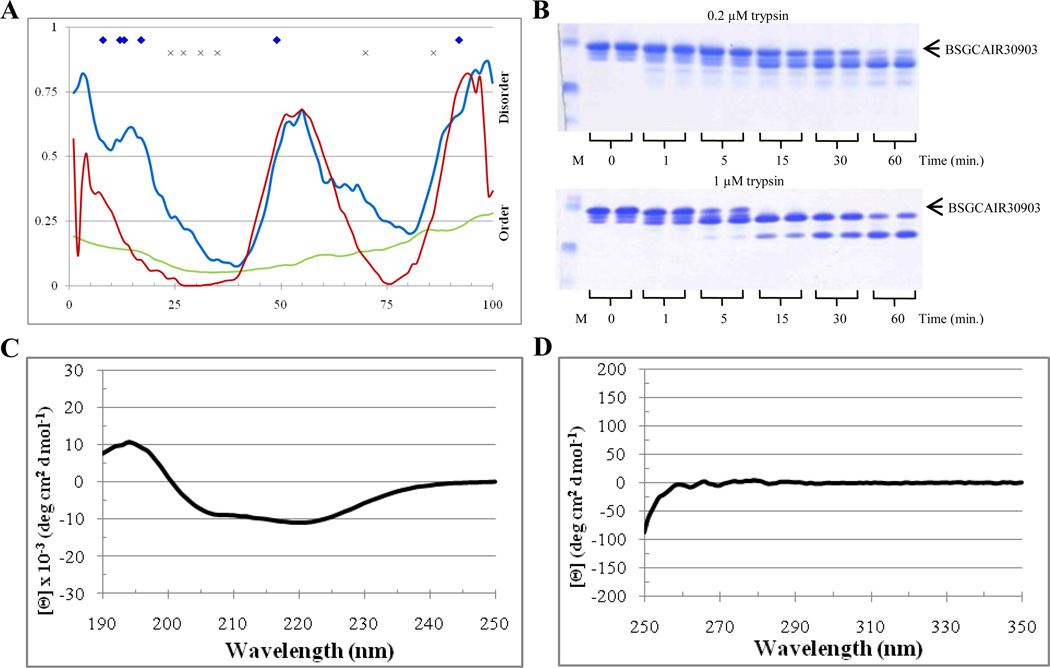
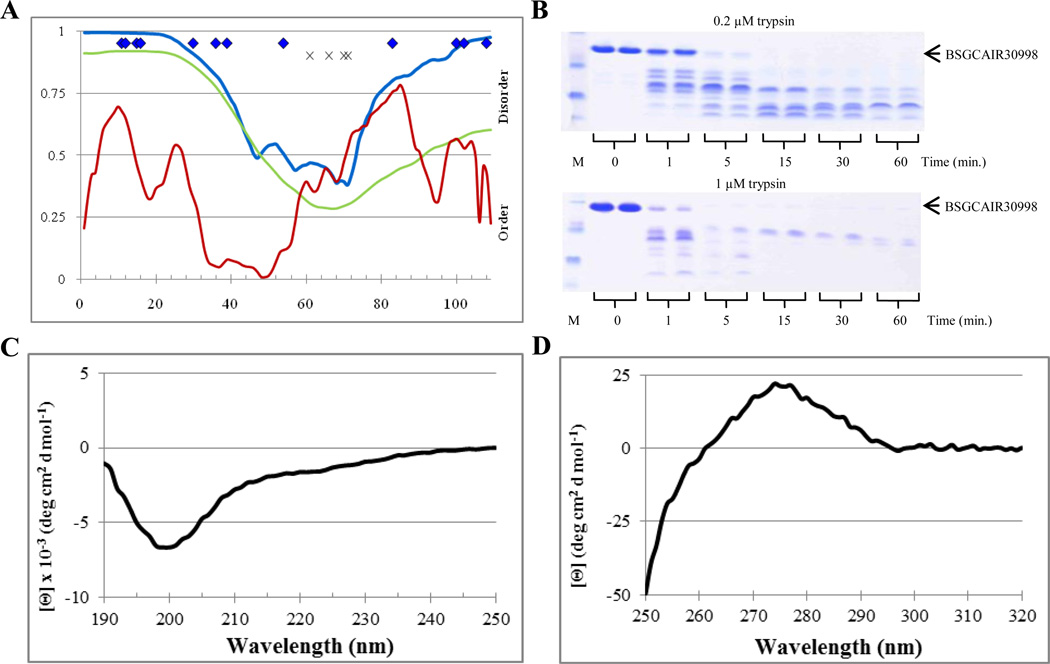
BSGCAIR30903
The second protein selected for the analysis was BSGCAIR30903, which was successfully crystallized too. In agreement with this observation, PONDR analysis indicated that BSGCAIR30903 was expected to be a mostly ordered protein. There were 42%, 0%, and 25% disordered residues predicted with the average scores of 0.425, 0.132, and 0.278 by PONDR VSL2P, VL3E, and VLXT, respectively (see Table 2). Figure 8A illustrates that although PONDR VL3 showed a very low overall disorder score, being characterized by one shallow dip, PONDR VSL2 and VL-XT both indicated two ordered regions linked by a disordered region and bounded by disordered tails. Several predicted trypsin cut sites lied in the putative disordered regions predicted by PONDR VSL2 in both termini, and one site was predicted to be located in the disordered central region of the protein while several others are located in the each of the ordered regions (Figure 8A). Based on this analysis digestion of BSGCAIR30903 was expected to result in the formation of two stable fragments that correspond to the two predicted ordered regions flanking the cut site in the disordered spike. The overall rate of digestion was expected to be relatively low.
Figure 8.
Disorder analysis of BSGCAIR30903. A. PONDR® plots (VSL2P (blue), VL3 (green), VLXT (red)) and trypsin digestion prediction (potentially accessible cut sites ( ), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
Figure 8B shows that the rate and the profile of limited trypsinolysis of BSGCAIR30903 at both 0.2 µM and 1 µM trypsin concentration were similar to those observed for the partially ordered standard, apomyoglobin. At 0.2 µM, a portion of the initial band was still visible at 60 minutes. However, no intact protein was observed after the 5 minute digestion by 1 µM trypsin. By 15 minutes at 1µM trypsin, two stable fragments emerged. The far-UV CD spectrum of BSGCAIR30903 was characteristic for a well-folded protein with the pronounced α-helical structure, possessing a positive peak at 198 nm and two pronounced minima at 208 nm and 222 nm, with the minima at 222 nm being slightly deeper (Figure 8C). The near-UV CD spectrum of this protein showed low signal, which was not surprising since BSGCAIR30903 does not contain tryptophan residues (Figure 8D). However, multiple low intensity bands are seeing in the near-UV CD spectrum of this protein suggesting that its tyrosine and phenylalanine residues might have relatively rigid environment.
NYSGXRC10336x
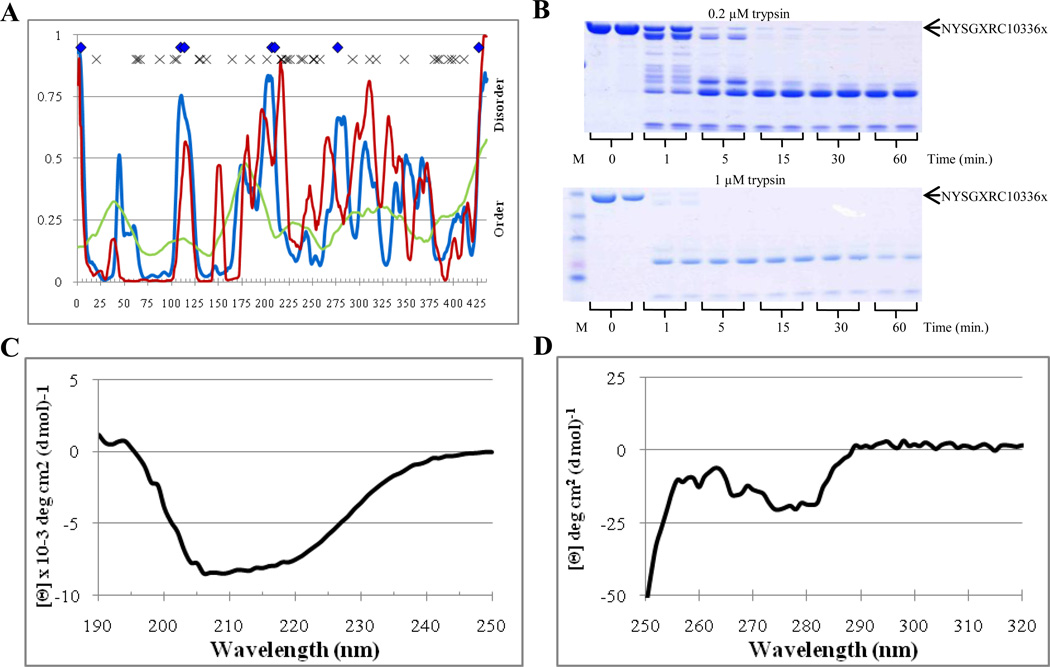
Next, we analyzed chromosomal replication initiator protein DnaA (NYSGXRC10336x), a protein which failed to crystallize. Figure 9A shows that this protein was predicted to be mostly ordered, possessing 15.9%, 2.3%, and 21.6% disordered residues predicted with the average disorder scores of 0.274, 0.242, and 0.287 by PONDR VSL2, VL3, and VLXT, respectively (see Table 2). The overall profiles of the PONDR-derived disorder propensity distributions in this protein were essentially more complex that those of BSGCAIR30378, possessing multiple peaks exceeding the 0.5 threshold (cf. Figures 7A and 9A). Seven potential trypsin cleavage sites were spread somewhat uniformly across the length of the protein in major disordered spikes predicted by PONDR VSL2 (Figure 9A). Figure 9A shows that only the first tryptic fragment (residues 1–100) was predicted to possess low intrinsic disorder content, whereas other fragments contained more disorder. Based on these observations, at least four main fragments were expected to appear at the beginning of the digestion process. These fragments were expected to have different proteolytic resistances. Furthermore, the rate of proteolysis was expected to be intermediate between that of wholly and partially disordered standards (i.e., trypsinolysis rates of α-casein and apo-myoglobin).
Figure 9.
Disorder analysis of NYSGXRC10336x. A. PONDR® plots (VSL2P (blue), VL3 (green), VLXT (red)) and trypsin digestion prediction (potentially accessible cut sites ( ), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
Figure 9B reports on the efficiency of the NYSGXRC10336x limited digestion by 0.2 µM and 1 µM trypsin. Both the rate of trypsinolysis and the proteolytic profile were closer to those of the fully disordered standard, α-casein, rather than to the partially disordered apo-myoglobin (cf. with data shown in Figure 4). At 0.2 µM, no initial protein band was visible past 5 minutes, whereas at 1 µM trypsin, no initial protein band was observed by already 1 minute of digestion. Figure 9B shows that 7 or 8 bands became visible at the 1 minute digestion at 0.2 µM giving way to 3 or 4 bands at the 5 minute digestion and 2 stable bands throughout the remaining digestion time. At 1 µM, 3 or 4 faint bands were detectable after the trypsinolysis for 1 minute, with only two stable bands being present at the remaining time points (note, the upper band might correspond to the inhibitor). The far-UV CD spectrum of NYSGXRC10336x was characterized by a positive peak at 198 nm and two minima at 208 nm and 222 nm, with the minima at 208 nm being slightly deeper (Figure 9C). The near-UV CD spectrum of this protein showed overall low intensity, possessing some fine structure and a broad negative peak at 273 nm (Figure 9D). Overall, computational and experimental data suggested that NYSGXRC10336x is a moderately stable protein with noticeable amount of intrinsic disorder, which can preclude this protein from the successful crystallization.
BSGCAIR30998
The fourth protein selected as an example was a Mycoplasma pneumoniae protein BSGCAIR30998, which also failed to crystallize. Figure 10A clearly shows that according to the PONDR analysis, BSGCAIR30998 was predicted to be a mostly disordered protein. In fact, it had 81.7%, 59.6%, and 36.7% disordered residues predicted with the average scores of 0.773, 0.605, and 0.394 by PONDR VSL2, VL3, and VL-XT, respectively (see Table 2). PONDR VL3 curve, being located mostly above the threshold 0.5, contained a pronounced dip of about 20 residues near the middle of the sequence (Figure 10A). Similarly, PONDR VSL2 analysis revealed that BSGCAIR30998 was the mostly disordered protein. On the other hand, PONDR VLXT curve contained multiple dips near the middle of the protein with the rest being mostly disordered (Figure 10A). BSGCAIR30998 was predicted to have several accessible trypsin cut sites located within the putative disordered regions at both termini and as well as four partially inaccessible trypsin cut sites located in the shallow ordered dip in the central region of the protein (Figure 10A). Based on the PONDR analysis and the tryptic cut site predictions, digestion of BSGCAIR30998 was expected to occur at a high rate and possibly to result in one stable fragment that corresponded to the removal of the two predicted disordered regions flanking the central ordered fragment. However, fast complete digestion could not be ruled out if the removal of the terminal regions would destabilize the central fragment, exposing new trypsin cut sites.
Figure 10.
Disorder analysis of BSGCAIR30998. A. PONDR® plots (VSL2P (blue), VL3 (green), VLXT (red)) and trypsin digestion prediction (potentially accessible cut sites ( ), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
), inaccessible cut sites (x)). B. SDS-PAGE analysis of limited digestion. C. Far-UV CD spectrum. D. Near-UV CD spectrum.
In agreement with these predictions, the rate and the profile of the BSGCAIR30998 limited digestion at both 0.2 µM and 1 µM trypsin were very similar to those of the fully disordered standard, α-casein. At 0.2 µM, no initial protein band was visible past 5 minutes, whereas at 1 µM trypsin, no initial protein band was observed already after 1 minute of digestion. At 0.2 µM trypsin, 5 to 6 bands became visible after the first minute of digestion, which gave way to 2 or 3 bands in throughout remaining time points. At 1 µM, 4 or 5 bands were present at 1 minute, 4 bands at 5 minutes, and only 1 band in the remaining time points. Similar to the situation with NYSGXRC10336x described above, this band might correspond to the trypsin inhibitor. This hypothesis was in agreement with the control experiments, where the corresponding concentration of trypsin inhibitor was run alone (data not shown). Both far- and near-UV CD spectra confirmed almost complete lack of ordered structure in BSGCAIR30998. The Far-UV CD spectrum resembled that of α-casein and showed a negative peak at 198 nm and very shallow shoulder at 215 nm (Figure 10C). The near-UV CD spectrum of BSGCAIR30998 possessed a small, broad, feature-less peak at 275 nm and the edge of a negative peak visible at 250 nm (Figure 10D).
Overall disorder status of analyzed PSI targets
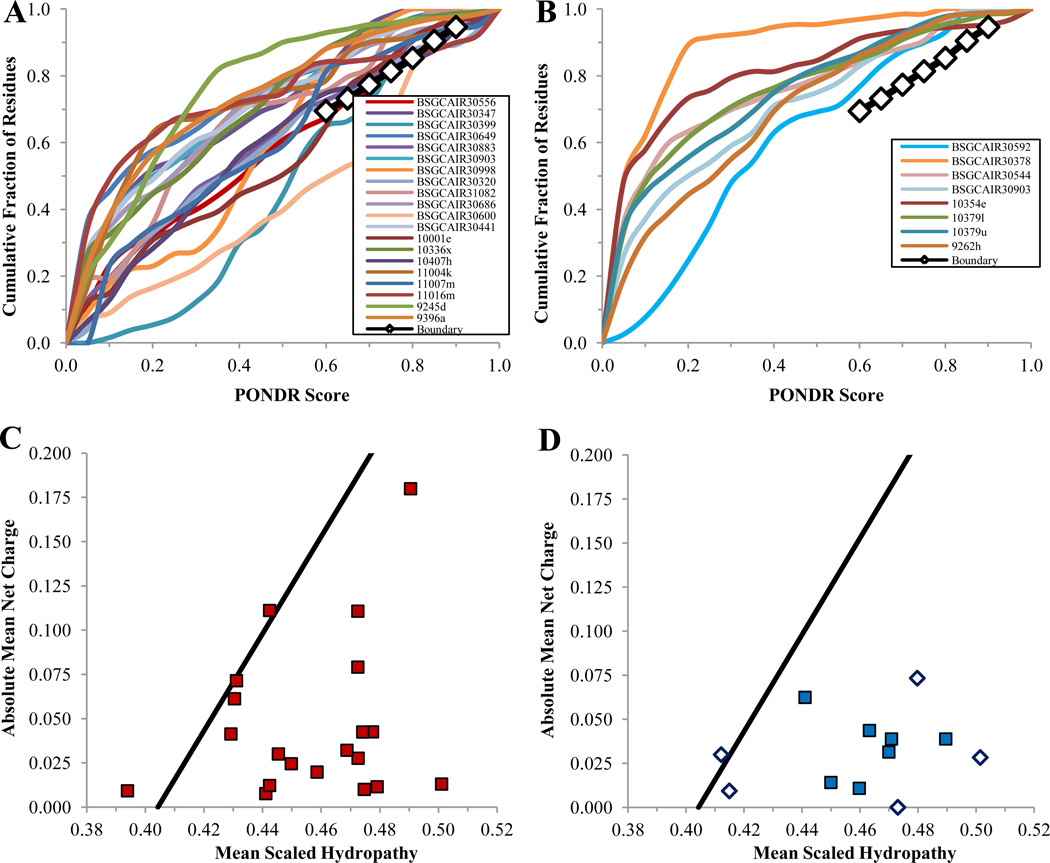
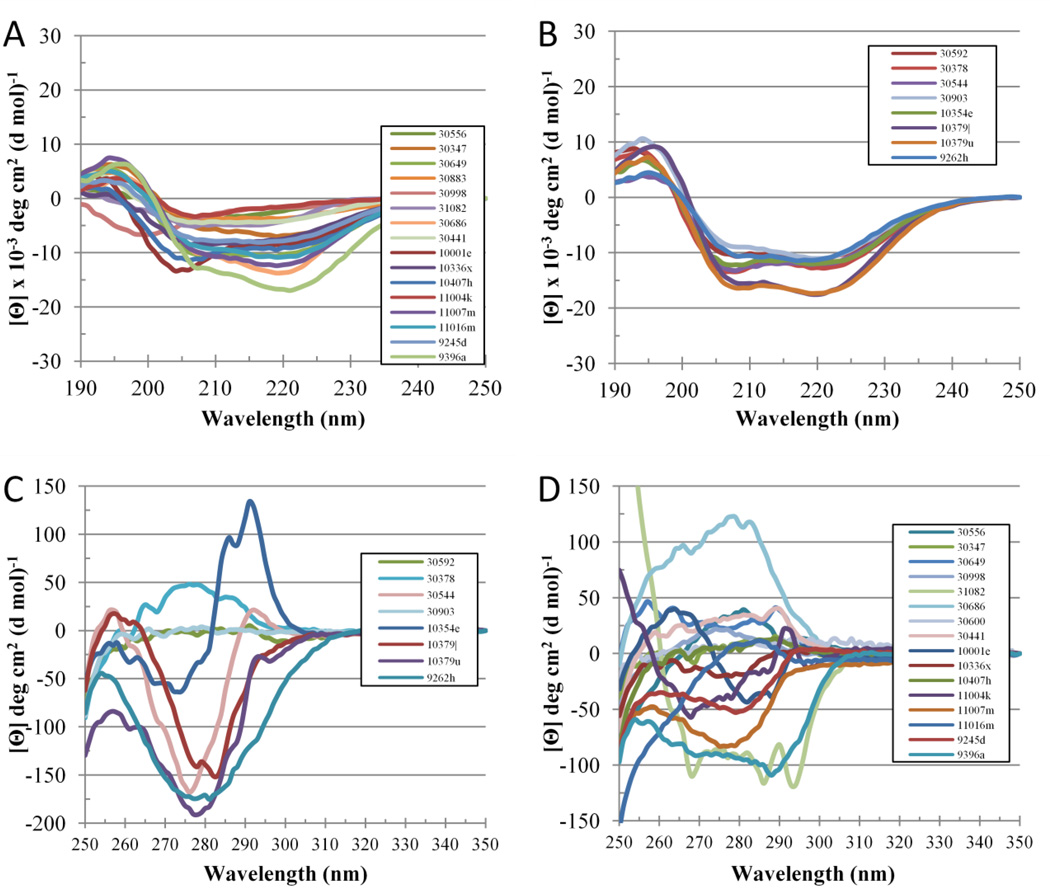
Figure 11 represents the evaluation of the overall disorder status of all non-crystallizable and crystallizable PSI targets analyzed in this study by the binary disorder identifiers, CDF and CH-plot. These data show that the majority of PSI targets were predicted as mostly ordered proteins. Binary predictors are not ideally suited for analysis of crystallizability of proteins, since it is the length of the disordered region(s) (rather than the average disorder score) that is crucial in terms of success or failure in getting crystals. For example, although the average disorder score can be low, the occurrence of a single, rather long disordered region can prevent crystallization. Therefore, as such, predictors providing score on a per residue level are more appropriate for these purposes. However, both the CDF curves and CH points of the non-crystallizable targets were in general closer to the corresponding boundaries in comparison with the curves and points corresponding to the crystallizable proteins. Table 2 represents the results of the PONDR-based prediction of the amount of disorder and the number of accessible tryptic sites in these proteins. Actual data for the limited proteolysis of the PSI target protein are tabulated here too. Finally, Figure 12 summarizes data on the spectroscopic analysis of the non-crystallizable and crystallizable targets. These data show that in agreement with the CDF- and CH-plot-based disorder evaluations, the majority of non-crystallizable proteins were predicted and shown to be more disordered and less stable than crystallizable targets from various PSI Centers.
Figure 11.
Cumulative distribution function (CDF) (plots A and B) and CH-plot analyses (plots C and D) of non-crystallizable (plots A and C) and crystallizable (plots B and D) PSI target proteins.
Figure 12.
Far- (plots A and C) and near-UV CD spectra (plots B and D) of non-crystallizable (plots A and B) and crystallizable (plots C and D) PSI targets analyzed in this study.
Overall, analyses presented in this study reveal that a large portion of expressed and purified proteins that fail to yield 3-D structures contain lengthy intrinsically disordered regions. Therefore, these data clearly suggest that the disorder evaluation in target proteins by a combination of computational and experimental tools represents a valuable approach for finding those targets which potentially will be recalcitrant to structure determination. Furthermore, this study shows that the experimental mapping of intrinsically disordered regions combined with prediction of the abundance of intrinsic disorder and abundance of disorder-based potential proteolytic sites proteolysis can be readily parallelized in a high-throughput format.
ACKNOWLEDGMENTS
This work was supported in part by the grants R01 LM007688-01A1 (to A.K.D and V.N.U.) and GM071714-01A2 (to A.K.D and V.N.U.) from the National Institute of Health, the grant EF 0849803 (to A.K.D and V.N.U.) from the National Science Foundation, and the Program of the Russian Academy of Sciences for the “Molecular and Cellular Biology” (to V.N.U.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bode W, Huber R. Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem. 1992;204:433–451. doi: 10.1111/j.1432-1033.1992.tb16654.x. [DOI] [PubMed] [Google Scholar]
- Brenner SE. Target selection for structural genomics. Nat Struct Biol. 2000;7(Suppl):967–969. doi: 10.1038/80747. [DOI] [PubMed] [Google Scholar]
- Brenner SE, Levitt M. Expectations from structural genomics. Protein Sci. 2000;9:197–200. doi: 10.1110/ps.9.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley SK, Bonanno JB. Structural genomics of proteins from conserved biochemical pathways and processes. Curr Opin Struct Biol. 2002;12:383–391. doi: 10.1016/s0959-440x(02)00330-5. [DOI] [PubMed] [Google Scholar]
- Chen L, Oughtred R, Berman HM, Westbrook J. TargetDB: a target registration database for structural genomics projects. Bioinformatics. 2004;20:2860–2862. doi: 10.1093/bioinformatics/bth300. [DOI] [PubMed] [Google Scholar]
- Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Dunker VN. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45:10448–10460. doi: 10.1021/bi060981d. [DOI] [PubMed] [Google Scholar]
- Cortese MS, Baird JP, Uversky VN, Dunker AK. Uncovering the unfoldome: enriching cell extracts for unstructured proteins by acid treatment. J Proteome Res. 2005;4:1610–1618. doi: 10.1021/pr050119c. [DOI] [PubMed] [Google Scholar]
- Csizmok VE, Szollosi E, Friedrich P, Tompa P. A novel two-dimensional electrophoresis technique for the identification of intrinsically unstructured proteins. Mol Cell Proteomics. 2006;5:265–273. doi: 10.1074/mcp.M500181-MCP200. [DOI] [PubMed] [Google Scholar]
- Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of Protein Folding. Weinheim, Germany: Wiley-VCH, Verlag GmbH & Co; 2005. pp. 271–353. [Google Scholar]
- Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4:229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008a;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008b;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- Fontana A, de Laureto PP, de Filippis V, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold. Des. 1997a;2:R17–R26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- Fontana A, Fassina G, Vita C, Dalzoppo D, Zamai M, Zambonin M. Correlation between sites of limited proteolysis and segmental mobility in thermolysin. Biochemistry. 1986;25:1847–1851. doi: 10.1021/bi00356a001. [DOI] [PubMed] [Google Scholar]
- Fontana A, Zambonin M, Polverino de Laureto P, De Filippis V, Clementi A, Scaramella E. Probing the conformational state of apomyoglobin by limited proteolysis. J. Mol. Biol. 1997b;266:223–230. doi: 10.1006/jmbi.1996.0787. [DOI] [PubMed] [Google Scholar]
- Galea CA, Pagala VR, Obenauer JC, Park CG, Slaughter CA, Kriwacki RW. Proteomic studies of the intrinsically unstructured mammalian proteome. J Proteome Res. 2006;5:2839–2848. doi: 10.1021/pr060328c. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ladizhansky V, Boggs JM. Structural polymorphism and multifunctionality of myelin basic protein. Biochemistry. 2009;48:8094–8104. doi: 10.1021/bi901005f. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ. The structural aspects of limited proteolysis of native proteins. Biochimica et biophysica acta. 1998;1382:191–206. doi: 10.1016/s0167-4838(97)00175-1. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Campbell SF, Thornton JM. Molecular recognition. Conformational analysis of limited proteolytic sites and serine proteinase protein inhibitors. J. Mol. Biol. 1991;220:507–530. doi: 10.1016/0022-2836(91)90027-4. [DOI] [PubMed] [Google Scholar]
- Hubbard SJ, Eisenmenger F, Thornton JM. Modeling studies of the change in conformation required for cleavage of limited proteolytic sites. Protein Sci. 1994;3:757–768. doi: 10.1002/pro.5560030505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SJ, Beynon RJ, Thornton JM. Assessment of conformational parameters as predictors of limited proteolytic sites in native protein structures. Protein Eng. 1998;11:349–359. doi: 10.1093/protein/11.5.349. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, Ackerman EJ. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001a;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Kimzey AL, Masselon CD, Bruce JE, Garner EC, Brown CJ, Dunker AK, Smith RD, Ackerman EJ. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 2001b;10:560–571. doi: 10.1110/ps.29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Jr, Kato I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics. 2009a;10(Suppl 1):S12. doi: 10.1186/1471-2164-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Unfoldomics of human genetic diseases: illustrative examples of ordered and intrinsically disordered members of the human diseasome. Protein Pept Lett. 2009b;16:1533–1547. doi: 10.2174/092986609789839377. [DOI] [PubMed] [Google Scholar]
- Novotny J, Bruccoleri RE. Correlation among sites of limited proteolysis, enzyme accessibility and segmental mobility. FEBS Lett. 1987;211:185–189. doi: 10.1016/0014-5793(87)81433-3. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic Disorder and Functional Proteomics. Biophys. J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- Sedzik J, Kirschner DA. Is myelin basic protein crystallizable? Neurochem Res. 1992;17:157–66. doi: 10.1007/BF00966794. [DOI] [PubMed] [Google Scholar]
- Shapiro L, Lima CD. The argonne structural genomics workshop: Lamaze class for the birth of a new science. Structure. 1998;6:265–267. doi: 10.1016/s0969-2126(98)00030-6. [DOI] [PubMed] [Google Scholar]
- Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- Spolaore B, Bermejo R, Zambonin M, Fontana A. Protein interactions leading to conformational changes monitored by limited proteolysis: apo form and fragments of horse cytochrome c. Biochemistry. 2001;40:9460–9468. doi: 10.1021/bi010582c. [DOI] [PubMed] [Google Scholar]
- Terwilliger T. Structural genomics in north america. Nat. Struct. biol. Structural Genomics Supplement. 2000:935–939. doi: 10.1038/80700. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Exploring structure space. A protein structure initiative. Genetica. 1999;106:141–147. doi: 10.1023/a:1003701500519. [DOI] [PubMed] [Google Scholar]
- Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 60:1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Amyloidogenesis of natively unfolded proteins. Curr Alzheimer Res. 2008;5:260–287. doi: 10.2174/156720508784533312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009a;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically Disordered Proteins and Their Environment: Effects of Strong Denaturants, Temperature, pH, Counter Ions, Membranes, Binding Partners, Osmolytes, and Macromolecular Crowding. Protein J. 2009b doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Flexible nets of malleable guardians: intrinsically disordered chaperones in neurodegenerative diseases. Chem Rev. 2011a;111:1134–1166. doi: 10.1021/cr100186d. [DOI] [PubMed] [Google Scholar]
- Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011b;43:1090–1103. doi: 10.1016/j.biocel.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Longhi S. Instrumental Analysis of Intrinsically Disordered Proteins: Assessing Structure and Conformation. In: Uversky VN, editor. The Wiley Series in Protein and Peptide Science. John Wiley & Sons, Inc: Hoboken, New Jersey, USA; 2010. [Google Scholar]
- Uversky VN, Dunker AK. Understanding protein non-folding. Biochim Biophys Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Experimental Tools for the Analysis of Intrinsically Disordered Protein. In: Walker J, editor. Methods in Molecular Biology. Volume II. Totowa, NJ, USA: Humana Press; 2012a. [Google Scholar]
- Uversky VN, Dunker AK. A multiparametric analysis of the intrinsically disordered proteins: Looking at intrinsic disorder through the compound eyes. Anal. Chem. 2012b doi: 10.1021/ac203096k. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Dunker AK. Experimental Tools for the Analysis of Intrinsically Disordered Protein. In: Walker J, editor. Methods in Molecular Biology. Volume I. Totowa, NJ, USA: Humana Press; 2012c. [Google Scholar]
- Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, coding sequence diversities correlated with long disordered regions. J Proteome Res. 2007;6:1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A. Creating a structural genomics consortium. Nat. Struct. biol. Structural Genomics. 2000;(Supplement):953. doi: 10.1038/80726. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007a;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res. 2007b;6:1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Oldfield CJ, Dunker AK, Uversky VN. CDF it all: consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009;583:1469–1474. doi: 10.1016/j.febslet.2009.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SA, Klee C. Study of calcineurin structure by limited proteolysis. Methods Mol Biol. 2002;172:317–334. doi: 10.1385/1-59259-183-3:317. [DOI] [PubMed] [Google Scholar]