Abstract
Nucleotide excision repair (NER) has allowed bacteria to flourish in many different niches around the globe that inflict harsh environmental damage to their genetic material. NER is remarkable because of its diverse substrate repertoire, which differs greatly in chemical composition and structure. Recent advances in structural biology and single-molecule studies have given great insight into the structure and function of NER components. This ensemble of proteins orchestrates faithful removal of toxic DNA lesions through a multistep process. The damaged nucleotide is recognized by dynamic probing of the DNA structure that is then verified and marked for dual incisions followed by excision of the damage and surrounding nucleotides. The opposite DNA strand serves as a template for repair, which is completed after resynthesis and ligation.
In global genome repair, the genome is scanned by UvrA acting in concert with UvrB. In transcription-coupled repair, RNA polymerase stalls at the damaged site and the UvrAB machinery is then recruited.
During the emergence of bacterial life some 3 billion years ago, the amount of solar radiation reaching the surface of Earth may have been two to three orders of magnitude higher than current levels. This environmental insult helped to fashion efficient DNA repair mechanisms that coemerged with bacteria and shaped the evolution of life (Canuto et al. 1982; Oró et al. 1990; Cnossen et al. 2007). UV light induces two major photoproducts in DNA: a cyclobutane pyrimidine dimer and a 6-4 photoproduct at a ratio of 3:1 that are repaired by two evolutionarily conserved processes called photoreactivation and NER. A third type of repair of UV-induced photoproducts has been found in some organisms and is covered in detail elsewhere. The removal of UV-induced cyclobutane pyrimidine dimers by NER was first described in Escherichia coli in 1964 by the pioneering work of Bill Carrier and Dick Setlow (Setlow and Carrier 1964) and independently by Dick Boyce and Paul Howard-Flanders (Boyce and Howard-Flanders 1964). During the mid-1980s, the cloning of uvr genes, the overexpression and purification of their gene products, and the subsequent reconstitution of the NER process by Dean Rupp, Aziz Sancar (Sancar et al. 1981; Sancar and Rupp 1983; Thomas et al. 1985), and Larry Grossman (Yeung et al. 1983) ushered in an exciting biochemical era of prokaryotic NER. During the past decade, the structural biology of Uvr proteins and the DNA lesions upon which they act have allowed an unprecedented understanding of the structure and function of prokaryotic NER proteins (Van Houten 1990; Theis et al. 2000; Van Houten et al. 2005; Truglio et al. 2006a; Goosen and Moolenaar 2008; Goosen 2010). Here, we review our current knowledge of prokaryotic NER, first discussing global genome repair and then transcription-coupled repair below.
OVERVIEW OF BACTERIAL NER
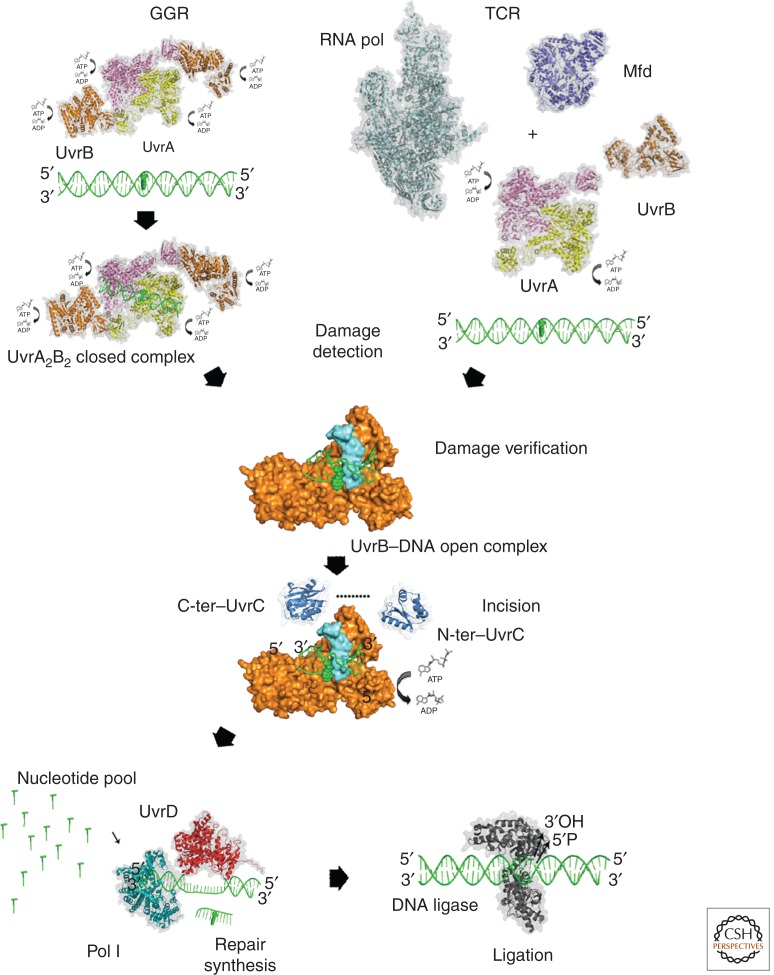
The process of NER in prokaryotes and eukaryotes is highly conserved and can be viewed in discrete steps: damage detection, damage verification, incision, excision, and DNA ligation. DNA repair is initiated in two major ways in prokaryotes (Fig. 1). First, damage can be detected by UvrA acting in concert with UvrB. Alternatively, if the damage is first encountered by an RNA polymerase (RNAP) that stalls at the damaged site, the action of a transcriptional-repair coupling factor (TRCF; also known as Mfd) is necessary to dislodge the stalled RNAP and recruit the UvrAB machinery to the damaged site. The subsequent steps in the process are the same. The damage is passed from UvrA to UvrB, which separates the two DNA strands to verify the position of the lesion, initiating the release of UvrA. UvrB forms a tight scaffold on the DNA for the arrival of UvrC, which contains two nuclease domains that cleave the phosphodiester bonds 8 nucleotides 5′ and 4–5 nucleotides 3′ to the damaged site. In some bacterial species, such as E. coli, a second nuclease, UvrC homolog (Cho), has been found that nicks 4 nucleotides farther away than the normal cleavage site on the 3′ side of the lesion (Moolenaar et al. 2002). The postincision complex is displaced by the dual action of UvrD (helicase II) and DNA polymerase I (Pol I) that together work to excise the damage-containing oligonucleotide and allow turnover of the UvrB and UvrC proteins while filling in the resulting gap using the remaining complementary strand (Caron et al. 1985; Husain et al. 1985). The final step is achieved by the action of DNA ligase, which seals the newly created repair patch (Fig. 1).
Figure 1.
Schematic representation of the prokaryotic NER pathways. In global genome repair (GGR), the genome is scanned by the heterotetrameric UvrA2–UvrB2 complex in search for damaged nucleotides causing large conformational changes. In transcription-coupled repair, the repair process is initiated by a stalled RNA polymerase on an actively transcribed gene through the interaction of the RNA polymerase with Mfd, which recruits the UvrA dimer or a UvrA2–UvrB heterotrimer to the site of the lesion. Both mechanisms converge into the same pathway and proceed with damage verification by UvrB followed by 3′ and 5′ incisions catalyzed through UvrC. The helicase activity of UvrD is required for the removal of UvrC. The incised strand is excised and repair is completed after the repair patch is synthesized by DNA polymerase I and DNA ligase seals the nick. All protein structures in this figure, with the exception of UvrB, are shown with a transparent surface and in ribbon presentation. UvrB is shown with its surface in orange for domains 1 to 3, and the β-hairpin is shown in cyan. C-ter, Carboxy terminal; N-ter, amino terminal.
One remarkable feature of NER in both prokaryotes and eukaryotes is its ability to recognize and remove a large number of DNA lesions induced by a wide variety of environmental agents that differ dramatically in their chemical and structural composition (for review, see Van Houten et al. 2005; Truglio et al. 2006a). There is good agreement that the efficiency of NER in both systems is directly correlated with the structural distortion and subsequent destabilization of the DNA helix (Liu et al. 2011). DNA undergoes rapid structural changes, and damage detection is a highly dynamic process. Pioneering work by Nick Geacintov and Suse Broyde, who have combined biochemical and structural studies with molecular dynamics, showed that the UvrABC proteins can sense the lesion-induced distortion in a highly dynamic manner (Jia et al. 2009; Liu et al. 2011). Because of its general DNA damage-detection capabilities, the UvrABC system has been found to work on DNA substrates other than photoproducts, ranging in size from abasic sites to polycyclic aromatic hydrocarbon adducts (for review, see Van Houten et al. 2005; Truglio et al. 2006a). Recent work has suggested that even protein–DNA cross-links up to 10 kDa in size, and adjacent base damage induced by oxidative damage can be efficiently processed by the UvrABC system (Minko et al. 2005; Nakano et al. 2005; Imoto et al. 2008; Salem et al. 2009).
INITIAL DAMAGE RECOGNITION: THE ROLE OF UvrA
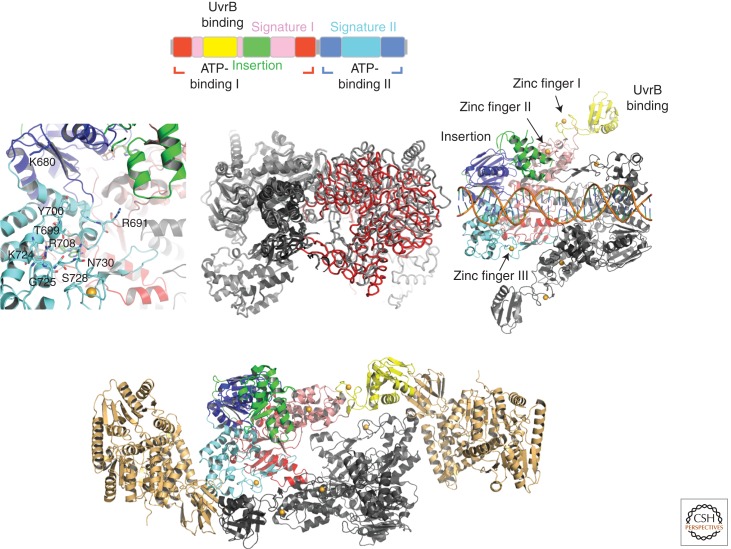
UvrA is the first of the four Uvr proteins within the NER cascade to interact with the DNA and is the initial damage sensor in the prokaryotic NER pathway. Surprisingly, UvrA’s affinity for nondamaged DNA is only two- to fivefold less than that for damaged DNA (Croteau et al. 2006). UvrA belongs to the ATP-binding cassette (ABC) superfamily of ATPases (Doolittle et al. 1986; Junop et al. 2001), which couple ATP hydrolysis to different cellular functions such as transport, ribosome biogenesis, chromosome condensation, and DNA repair (Junop et al. 2001; Hopfner and Tainer 2003; Locher 2004; Lebbink and Sixma 2005). Two nucleotide-binding domains (NBD-I and NBD-II) either from the same or different polypeptides form the ABC ATPase domain that harbors the Walker A motif and a Q loop for nucleotide binding, the Walker B motif, the D loop, the ABC signature (Leu-Ser-Gly-Gly-Gln), and a His loop (see Fig. 2) (Smith et al. 2002). Interestingly, each UvrA monomer possesses two ABC modules, and within the dimeric UvrA, four nucleotide-binding sites are potentially formed through intermolecular or intramolecular interactions (Gorbalenya and Koonin 1990). The first structure of UvrA from the thermophilic prokaryote Bacillus stearothermophilus (PDB entry 2R6F), from the Verdine group, clearly showed that all four nucleotide-binding sites are formed in an intramolecular fashion; that is, the nucleotides are located within the monomers, thus providing a direct explanation for why UvrA can also dimerize in the absence of a nucleotide (Fig. 2) (Pakotiprapha et al. 2008). All four NBDs in the structure were occupied with ADP, which was present in the crystallization buffer.
Figure 2.
(Top) Different UvrA domains are shown schematically in the linear polypeptide using the same color codes as are in the ribbon presentations for the different domains within UvrA. (Left) Enlargement of the DNA-binding region of UvrA (PDB entry 2R6F) from B. stearothermophilus. (Center) Superposition of the apo- and DNA-bound UvrA structures. For clarity, two of the monomers are shown as Cα-trace in gray and red, whereas the other two are shown in different shades of gray and in ribbon presentation. (Right) The UvrA2–DNA complex from Thermotoga maritima (PDB entry 3PIH). The color code is the same throughout the figure with the exception of the superposition, and the DNA is shown in orange for the backbone and blue-green spokes for the bases. (Bottom) Structure of the UvrA2–UvrB2 complex (PDB entry 3UWX). The two UvrB molecules are shown in gold.
Specific to its function, UvrA also possesses additional domains. Within the amino-terminal NBD, two additional domains are inserted, one of which has been identified as the UvrB-binding domain (residues 118–257) and the other as the insertion domain (residues 287–398). Furthermore, each UvrA monomer coordinates three Zn ions. The first Zn ion is located between signature domain I (part of NBD-I) and the UvrB-binding domain, the second is located between signature domain I and the insertion domain, and the third connects the helical region of signature domain II (part of NBD-II) to the dimer interface. Thus, all three Zn ions seem to have an important structural role in addition to the proposed role of the third Zn ion that is believed to interact with DNA to facilitate the recognition specificity for the damage (Navaratnam et al. 1989; Visse et al. 1993; Wang et al. 1994; Moolenaar et al. 2000; Croteau et al. 2006; Truglio et al. 2006a).
Despite the immense insights gained from the B. stearothermophilus UvrA structure and subsequent structures of UvrA from Deinococcus radiodurans (Timmins et al. 2009), it remained unclear how DNA is bound to UvrA and, even more importantly, how damage is perceived. The subsequent structural characterization of UvrA from Thermotoga maritima (PDB entry 3PIH) in complex with double-stranded DNA (dsDNA) containing damage on both strands by Nowotny’s group revealed how the DNA is bound within the UvrA dimer and, importantly, provided a first glimpse of where the damage could be located (Jaciuk et al. 2011). A comparison of the apo–UvrA structure with the DNA-bound structure clearly showed that most of the domains within the monomers can be readily superimposed, especially the two ATP-binding domains, which seem to form a rigid core. The only exceptions are the insertion domain and signature domain II. The insertion domain, however, seems to be very flexible, and the investigators cannot exclude the possibility that the difference in position is due to crystal contacts, whereas the altered position of signature domain II leads to an open dimer conformation. The relative position of the two UvrA monomers to each other thus changes upon DNA binding, which seems to be required for binding of the distorted, damage-containing DNA (Fig. 2). The DNA is bound in a cleft formed by the UvrA dimer, and interactions of the UvrA dimer with the DNA can only be observed with the DNA backbone, thus explaining how UvrA can bind to different lesions in different sequence contexts (Truglio et al. 2006a). Importantly, it was also observed that the most extensive interactions between the protein and the DNA are restricted to four consecutive nucleotides at the ends of the dsDNA fragment and signature domain II of T. maritima UvrA (residues Gly-670, Thr-679, Tyr-680, Arg-688, Lys-704, Ser-705, Ser-708, and Asn-710), thus implying that various DNA substrates with vastly varying backbone distortions could be held in place. In addition, the analysis of Bacillus caldotenax UvrA revealed that Lys-680 and Arg-691 contribute significantly to DNA binding, because a Lys-680-Ala/Arg-691-Ala UvrA variant binds a 50-bp damage-containing substrate 37-fold less tightly (Croteau et al. 2008). Interestingly, the Zn finger that is inserted into signature domain II points toward the damage within the duplex but does not form direct contacts to the DNA. A superposition with the apo–UvrA structure with bound ADP reveals that this Zn finger would sterically interfere with DNA binding if it would maintain its same position and is thus rotated away from the dimer interface. These observations are in good agreement with biochemical data that showed that deletion of the Zn finger increases the affinity for fluorescein-containing DNA but also reduces the specificity for modified DNA (Croteau et al. 2006).
The only caveat in this structural analysis is the use of a DNA duplex that contains damage on both strands and is thus not an ideal substrate for NER (Jaciuk et al. 2011). The authors observed that fluorescein-modified thymines are slightly pushed out of the double helix, leading to an increase in the stretch and shear values of the surrounding bases. The DNA shows an overall unwinding of ∼20° and bending by 15°. On the basis of this observation, they speculate that these weaker base pairs can cause a general destabilization of the double helix that is sensed by the protein and thus allows the recognition of lesions that differ greatly in structure and chemistry by an indirect readout mechanism. As a dimer, UvrA is ideally positioned to probe the DNA for the possibility of bending and unwinding. One could envision one hand (the signature II domains) in each monomer holding on to the DNA at a certain distance away from the lesion and probing for the presence of an altered nucleotide by bending the DNA, assessing the rigidity of the DNA in between. This dynamic model of damage recognition such that UvrA is constantly testing the structural mobility of the DNA is supported by biochemical and molecular dynamic modeling data in which a benzo[a]pyrene diol epoxide-N2-guanine adduct that is flanked by Cs is processed twofold more slowly compared to the same lesion flanked by Ts (Ruan et al. 2007; Cai et al. 2009). Such a dynamic recognition mechanism immediately also explains the necessity for damage verification. In this scenario, UvrA would never interact directly with the damage and an additional protein is required to ensure that the locally destabilized region is a result of the presence of a lesion.
UvrB: THE CENTRAL PLAYER IN PROKARYOTIC NER
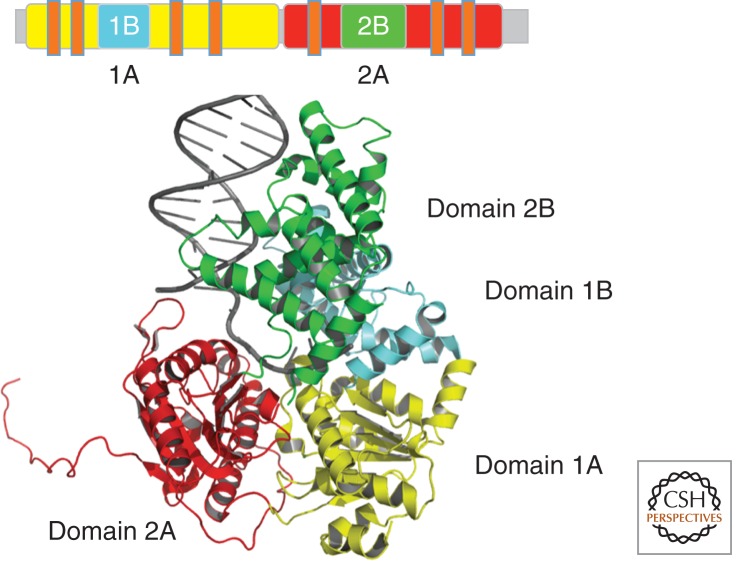
The above-described recognition mechanism calls for a second protein that verifies the presence of the damage. UvrB assumes a central position in the NER pathway because it interacts not only with UvrA but also with all other proteins in the pathway: UvrC, UvrD, and DNA Pol I. UvrB is structurally well characterized and has been solved in its apo form (PDB entries 1D9Z, 1D2M, 1C4O, and 1T5L), in complex with ATP (PDB entry 1D9X) and in complex with DNA (PDB entries 2FDC, 2NMV, and 2D7D) (Machius et al. 1999; Nakagawa et al. 1999; Theis et al. 1999; Truglio et al. 2004, 2006a; Eryilmaz et al. 2006; Waters et al. 2006). UvrB is a superfamily 2 (SF2) DEAD-box family helicase that exhibits only weak helicase and ATPase activity (Theis et al. 2000); the latter is only activated after interaction with UvrA and damaged DNA or when its autoinhibitory domain 4 is removed (Wang et al. 2006).
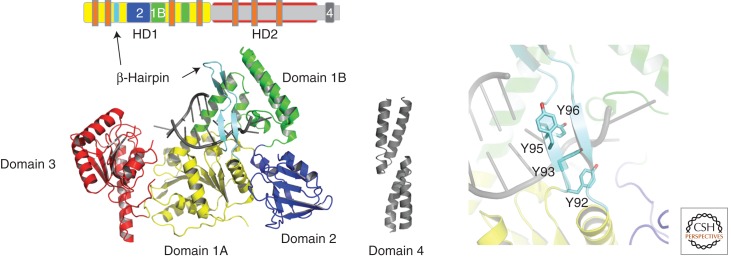
The protein contains the classical SF2 RecA-like domains (Fig. 3, domains 1A and 3) and three auxiliary domains specific to its function. Domain 1B provides additional interactions with the DNA, whereas domain 2 exclusively interacts with UvrA and domains 2 and 4 have been shown to interact with UvrC and UvrA (Truglio et al. 2006a). Domain 4 is linked to domain 3 by a flexible linker and so far has not been solved as part of the entire protein but only as a separate fragment (PDB entries 1E52 and 1Q0J) (Sohi et al. 2000; Alexandrovich et al. 2001). A critical feature with respect to the damage verification process is the β-hairpin in the first RecA-like domain that bridges domains 1A and 1B (Theis et al. 1999). Several studies identified this structural motif as well as specific residues within the β-hairpin as important determinants in the damage verification process (Moolenaar et al. 2001, 2005; Skorvaga et al. 2002, 2004). The structure of a UrvB–DNA complex (PDB entry 2FDC) further supported two previously proposed concepts: the “padlock” model (Theis et al. 2000) and the base-flipping model (Malta et al. 2006, 2008). This new structure showed that one DNA strand threads behind the β-hairpin, in which one base is flipped into a hydrophobic pocket, whereas the other remains on the surface of the protein (Truglio et al. 2006b). So far, however, it is unclear as to which strand harbors the damage. Several studies suggest that the damage is located on the outer strand and directly interacts with one of the tyrosine residues of the β-hairpin, which is located on the outer surface of this motif (Sancar et al. 1984; Zou et al. 2004). Other experiments are compatible with the suggestion that the damage is located on the inner strand (Malta et al. 2006; Waters et al. 2006), positioning it directly in front of the ploughshare of the helicase, that is, the β-hairpin. In this model, Tyr-96 could interrogate the DNA for damaged bases through stacking interactions (Van Houten and Snowden 1993; Skorvaga et al. 2002, 2004). Based on this model, it was assumed that it would be sterically impossible for the damage to be accommodated behind the β-hairpin, thereby creating the signal that the damage is present. Surprisingly, however, recent molecular modeling and molecular dynamic simulations have shown that benzo[a]pyrene-modified DNA could be accommodated on the inner strand and positioned in a pocket behind the β-hairpin (Jia et al. 2009). Furthermore, the investigators suggest that the position of the damage is highly dependent on the size, shape, and conformation of the lesion and could therefore be located behind the β-hairpin or directly in front of the β-hairpin as suggested above.
Figure 3.
(Top) The UvrB structure is shown schematically within the linear polypeptide. In addition to the colors that match the color coding of the different domains in the structure below, the location of the helicase motifs is indicated by small orange bars. (Left) Overall structure of UvrB in the presence of DNA (PDB entry 2FDC). (Center) The dimeric carboxy-terminal domain of UvrB is shown in ribbon presentation in gray. (Right) Enlargement of the region of the β-hairpin. Residues Tyr-92, Tyr-93, Tyr-95, and Tyr-96 are shown in stick presentation.
An additional important aspect regarding UvrB is its functional composition. Several studies suggested that UvrB or the preincision complex containing UvrB bound to damaged DNA forms a dimer (Hildebrand and Grossman 1999; Verhoeven et al. 2002b; Moolenaar et al. 2005; Wang et al. 2006), and the structural analysis of the carboxy-terminal fragment of UvrB further supported this hypothesis (Sohi et al. 2000; Alexandrovich et al. 2001). However, other studies indicate that UvrB is monomeric (Orren and Sancar 1989; DellaVecchia et al. 2007) and the UvrB crystal structures containing domains 1–3 provided no indication for a dimer. Very recently, the structure of Bacillus subtilis UvrB was solved in complex with single-stranded DNA and a nonhydrolyzable ATP analog, AMPPCP, at 3.25-Å resolution; for the first time, the structure reveals a dimer interface along domains 2 and 3 as well as the β-hairpin, and thus, the dimer assumes a head-to-head orientation (Webster et al. 2012). However, the relevance of this dimer interface with respect to UvrBs from other organisms requires further analysis, because the investigators also state that the location of residues 590–595 in the monomeric B. caldotenax structure would interfere with the observed dimer interface and, interestingly, the electron density of the B. subtilis structure does not extend beyond residue 590.
THE UvrA–UvrB COMPLEX
UvrA and UvrB work together as a complex to accomplish damage recognition and damage verification. Fluorescence resonance transfer studies from the Goosen group (Malta et al. 2007) have shown unambiguously that the complex formed between UvrA and UvrB contains two molecules of each protein. This stoichiometry was also confirmed using red or green Qdot-labeled UvrBs that were found to interact only in the presence of UvrA (Kad et al. 2010).
Recent single-molecule studies using a novel DNA tightrope approach and oblique angle fluorescent microscopy have provided initial insights into how the UvrAB complex scans DNA for damage. It was shown that UvrA alone forms a dimer on the DNA and performs a three-dimensional (3D) search in the presence and absence of ATP. UvrA only binds for a short time of about 7 sec to the DNA and can also jump from one DNA molecule to another (Kad et al. 2010). In the presence of UvrB, ∼17% of the complexes perform a one-dimensional search accompanied by a sixfold increase in residence time, whereas the other UvrA2–UvrB2 complexes continue a 3D search. The sliding on the DNA was complex, showing random walks and directed and paused motions. Combined, these search mechanisms could significantly increase successful damage recognition (Kad et al. 2010), and this research thus provides a first glimpse of how the NER proteins can scan an entire genome efficiently for the presence of lesions.
Currently, it is not known how the DNA is bound to the UvrA2–UvrB2 complex nor the relative orientation of the two UvrB molecules with respect to the UvrA dimer in the presence of damaged DNA. Previous studies suggested that the DNA is bent and wrapped around UvrB (Shi et al. 1992; Hsu et al. 1994; Verhoeven et al. 2001, 2002b), whereas no wrapping of UvrA was observed (Van Houten et al. 1987, 1988; Shi et al. 1992; Verhoeven et al. 2001). More recently, capillary electrophoresis coupled with laser-induced fluorescence polarization studies showed that UvrA facilitates wrapping of the DNA around UvrB (Wang et al. 2009). Interestingly, wrapping may be coupled to the helicase activity of UvrB. A β-hairpin mutant that lost its helicase activity also displayed a smaller degree of wrapping. Furthermore, wrapping was reduced when a DNA substrate was used that contained mismatches around the lesion that mimic a melted DNA structure, thus suggesting that wrapping is important for local melting of the DNA around the lesion (Wang et al. 2009). However, this process may only be required in mesophilic organisms and was not observed in thermophilic prokaryotes.
In the fluorescence studies mentioned above, the investigators also show that UvrB forms a dimer in the presence of UvrA, that is, in the UvrA2–UvrB2 complex (Malta et al. 2007). This dimeric form of the UvrA2–UvrB2 complex was observed in the absence of DNA but also in the presence of damaged DNA and, importantly, when both proteins are in the ATP-bound state. These observations are in contrast to the very recently published structure of the UvrA2–UvrB2 complex from Geobacillus stearothermophilus (Pakotiprapha et al. 2012), in which the two UvrB molecules are located ∼145 Å apart from each other and positioned ∼80 Å away from the lesion (Fig. 2). These discrepancies could highlight the importance of the bound nucleotide toward the conformation of the different complexes. Unfortunately, identification of the nucleotides bound in the UvrA2–UvrB2 structure was not possible because of the low resolution of the structure. The investigators compare two distinct UvrA conformations: an open-tray UvrA dimer, previously observed in the UvrA–DNA complex (Jaciuk et al. 2011), and a closed-groove configuration in the UvrA2–UvrB2 complex, in which the DNA-binding surface in the UvrA dimer adopts a deep and narrow channel. The transition from one form to the other can be achieved through a helical rotation of each UvrA monomer along a shared axis along the DNA-binding surface. In the closed conformation, UvrA interacts with UvrB through domain 2 of UvrB and the UvrB-interacting domain within UvrA as well as through UvrB’s domain 1B and UvrA’s signature domain II. In the open-tray conformation, these simultaneous interactions are not possible and are most likely restricted to domain 2 of UvrB and the UvrB-interacting domain of UvrA.
Based on these conformational differences, the investigators suggest a mechanism for efficient damage recognition. Initially, the UvrA2–UvrB2 complex may bind in its open-tray form to DNA, thus allowing binding to damaged and nondamaged DNA. The interaction with DNA leads to a conformational change toward the closed-groove state that is only possible in the presence of nondamaged DNA. If the closed-groove conformation can be accomplished, nondamaged DNA was encountered that would trigger the reversal to the open-tray state. If, on the other hand, damaged DNA is encountered, the transition to the closed-groove conformation would be inhibited and thus the UvrA2–UvrB2 complex would be trapped most likely in the open or partially open state. Subsequently, UvrA would dissociate, leaving UvrB bound to the DNA in the preincision complex. Both UvrB molecules would then be located at a distance of ∼80 Å from the lesion and must translocate toward the damage. This movement would be accomplished on opposite DNA strands toward the lesion, yielding the UvrB dimer in close proximity to the lesion. Loading of UvrC would interrupt the UvrB dimer interface, and concomitantly one UvrB molecule would dissociate from the DNA.
This model seems very reasonable with the data derived from the UvrA2–UvrB2 complex structure. However, the fluorescence resonance energy transfer data described above suggest that the two UvrB molecules in the UvrA2–UvrB2 complex are sufficiently close to allow energy transfer, which is a function of 1/r6, and thus propose that the UvrB molecules are within 4–9 nm of each other in the UvrA2–UvrB2 complex. Combined, these results clearly indicate that more data are required to decipher the damage-recognition process. One could easily envision a combination of all three conformations: the open tray, the closed groove, as well as the conformation in which the two UvrB molecules are in close proximity to each other. Determining at which point in the pathway each of these conformations will be assumed will require more snapshots of the UvrA2–UvrB2 complex, possibly in different nucleotide-bound states, as well as in the presence of unmodified and damaged DNA. To this end, it is interesting to note that, as mentioned above, UvrB changes UvrA’s 3D search into a sliding mechanism (Kad et al. 2010) and that binding of UvrB can overcome mutations in UvrA that greatly decrease UvrA’s overall binding affinity to allow proper loading of UvrB and subsequent incisions (Croteau et al. 2008). Together, these two observations indicate that UvrB makes significant contact with DNA within the UvrA2–UvrB2–DNA complex.
THE ENDONUCLEASES UvrC AND Cho
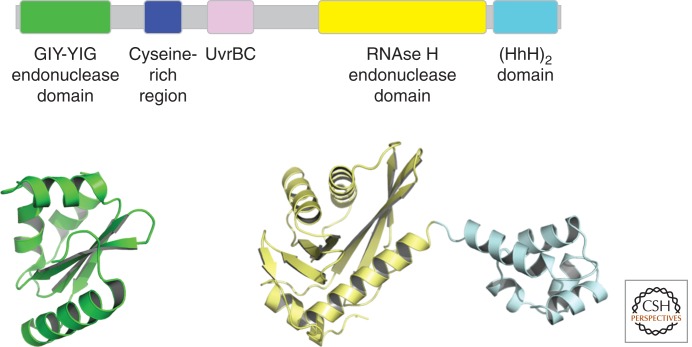
Once damage recognition has been achieved and the preincision complex has formed, NER proceeds by the recruitment of UvrC. It is currently unclear whether two UvrB molecules (Verhoeven et al. 2002b; Moolenaar et al. 2005) are present at this stage or whether just one UvrB molecule is present (Orren and Sancar 1989). UvrC interacts with UvrB’s carboxy-terminal domain to form a complex that leads to the subsequent incision of the damage-containing strand (Hsu et al. 1995). The UvrC protein is the smallest of the four Uvr proteins and contains two distinct catalytic sites that are responsible for the 3′ and 5′ incision reactions and can be inactivated independently (Lin and Sancar 1992; Verhoeven et al. 2000; Truglio et al. 2005; Karakas et al. 2007). The amino-terminal domain harbors the catalytic residues for the 3′ incision that is achieved at the fourth or fifth phosphodiester bond 3′ to the damaged site. The fold and sequence of this domain share homology with the catalytic domains of members of the GIY-YIG endonuclease superfamily (Fig. 4) (Truglio et al. 2005). The structure of this domain revealed that the surface toward the active site can be described as shallow and concave and could readily accommodate dsDNA (Truglio et al. 2005). This is in good agreement with biochemical data showing that the 3′ endonuclease site requires the presence of dsDNA (Zou and Van Houten 1999). Interestingly, however, this domain by itself does not bind to DNA and requires other regions of UvrC for catalysis. An additional feature of this domain is the presence of only one metal ion in the active site coordinated by a glutamic acid and five water molecules. On the basis of this geometry, a novel one-metal mechanism for cleavage of the phosphodiester bond was proposed (Truglio et al. 2005), in which the function of the Lewis acid is fulfilled by the divalent cation, the general acid is a metal-coordinated water molecule, and the general base is a tyrosine residue.
Figure 4.
(Top) Schematic representation of UvrC. No complete structure of UvrC has been solved so far. (Bottom) The two structures from T. maritima represent the 3′ catalytic GIY-YIG domain (PDB entry 1YCZ) and the 5′ RNase H-like catalytic domain as well as the very carboxy-terminal HhH motif (PDB entry 2NRT).
In between the two catalytic domains, UvrC harbors a UvrB-interacting region (Hsu et al. 1995) that is required for 3′ incision (Moolenaar et al. 1995, 1998) but not for 5′ incision (Moolenaar et al. 1995). The carboxy-terminal half of UvrC contains the catalytic domain responsible for 5′ incision at the eighth phosphodiester bond 5′ to the damaged site and, remarkably, despite the lack of sequence conservation, shares significant structural homology with the RNase H enzyme family, with an uncommon DDH triad instead of the common DDE triad at the active site (Karakas et al. 2007). Further studies suggested that the DDH triad should be extended to a DDKH motif. Interestingly, the surface opposite from the active site contains several positively charged conserved amino acids, and a mutational analysis showed that this surface is important to create a protein–DNA interface that influences both incision reactions (Karakas et al. 2007).
A flexible linker connects the 5′ catalytic domain to the carboxyl terminus that harbors two helix-hairpin-helix (HhH) motifs (Aravind et al. 1999). These motifs are also found in other proteins and have been shown to interact nonspecifically with DNA through contacts to the phosphate backbone of the minor groove (Shao and Grishin 2000; Singh et al. 2002). The HhH motif in UvrC is essential for 5′ incision and may also be required for 3′ incision when the lesion is located within a certain sequence context (Verhoeven et al. 2002a). On the basis of the structure of the carboxy-terminal half of UvrC containing both the catalytic domain responsible for 5′ incision and the HhH motifs, a model was proposed as to how UvrC may form a productive protein–DNA complex to accomplish the second incision reaction (Karakas et al. 2007).
An additional member of the NER pathway in some branches of prokaryotes (including E. coli, Salmonella, Mycobacterium, and Clostridium) is the endonuclease Cho (UvrC homolog), which is up-regulated during the SOS response in E. coli (Moolenaar et al. 2002; Van Houten et al. 2002). Interestingly, UvrC, in contrast, is the only one of the three UvrABC proteins that is not SOS up-regulated (Moolenaar et al. 1987). Cho shares homology with the amino-terminal half of UvrC and accordingly can catalyze the 3′ incision, albeit at a distance, which is 4 nucleotides farther away from the lesion compared to the 3′ incision performed by UvrC. It was proposed that Cho may be a backup endonuclease in some bacteria when the lesion may be too bulky or other steric blocks occur and thereby prevent 3′ incision by UvrC (Moolenaar et al. 2002; Van Houten et al. 2002). One could speculate that the bacterial species possessing Cho live in a specific niche and that this environment causes a unique type of DNA damage. As a member of the NER pathway, Cho forms a complex with UvrB but requires a different region for this interaction in comparison to UvrC, which may also explain the difference in incision because it may be positioned differently toward the DNA in the UvrB–Cho–DNA complex (Moolenaar et al. 2002).
POSTINCISION EVENTS
As mentioned above, both UvrD and Pol I are required for multiple rounds of NER (Caron et al. 1985; Husain et al. 1985). E. coli UvrD is a DNA helicase belonging to superfamily 1 (SF1) members including RecBCD, PcrA, and Rep helicases and has a broad role in DNA transactions including mismatch repair, replication, recombination, and NER (Dillingham 2011). UvrD, like other SF1 and SF2 helicases, is folded into four domains: 1A, 1B, 2A, and 2B (see Fig. 5). The ternary UvrD–DNA–ATP complexes of Wei Yang’s group provided snapshots of UvrD in several different conformations along its reaction trajectory and gave important insights into its mechanism of DNA unwinding (Lee and Yang 2006). Splicing together these fascinating structures into molecular movies revealed that ATP binding at the interface of domains 2A and 1A induces a large conformational change in the protein, causing one-base-pair unwinding. Subsequent ATP hydrolysis allows translocation of the UvrD complex in a 3′ → 5′ direction on the single-stranded DNA through the motion of the gating helix of 1B. Analysis of site-specific point mutations indicated that residues 620–623 in domain 2A containing a Tyr-621 at the tip of the separation pin are essential for helix unwinding. Additional residues in the GIG motif and the adjacent Arg-421 of 2B make important contacts with the DNA duplex. Biochemical analysis showed that UvrD is capable of unwinding at a nick, and it would appear that dissociation of DNA strands >18 bp long requires UvrD to act as a dimer to translocate on both strands (Runyon et al. 1990; Brosh and Matson 1997; Fischer et al. 2004). However, the precise stoichiometry of UvrD at the site of the UvrB–UvrC–DNA postincision complex is not yet known. It is also unknown how UvrD binds to the 5′ incision site, which is probably occluded by UvrB and/or UvrC. Bulk biochemical studies have suggested that UvrD acts to release UvrC and the damage-containing oligonucleotides, whereas Pol I is required to displace UvrB, which stays bound to the nondamaged strand (Orren et al. 1992; Manelyte et al. 2009). These bulk studies need to be confirmed using direct visualization of single molecules using the tools of atomic force microscopy (Wang et al. 2008) and fluorescence microscopy (Kad et al. 2010).
Figure 5.
Schematic representation of UvrD from E. coli. The two RecA-like domains 1A and 2A are shown in yellow and red, respectively, and the inserted domains 1B and 2B in cyan and green. The DNA is shown in gray (PDB entry 2IS2). In the sequence above, the helicase motifs are indicated by orange bars.
UvrD has been shown to interact with UvrA and UvrB separately but not with the UvrAB complex (Ahn 2000). Subsequent work using two-hybrid screens and surface plasmon resonance approaches indicated that the unstructured carboxyl terminus of UvrD is required for its interaction with the first 414 amino acids of UvrB, although deletion of this UvrD domain did not greatly sensitize E. coli to killing by UV light (Manelyte et al. 2009). This may be due to the additional interaction domain 1A of UvrD, which was shown to interact with UvrB by two-hybrid analysis. These two regions of UvrD have also been shown to interact with MutL during mismatch repair (Matson and Robertson 2006). It is interesting to note that B. subtilis PcrA, which acts like UvrD to displace UvrC, also contains an unstructured carboxy-terminal domain that is absent from Rep helicase (Manelyte et al. 2009). In addition, UvrAB has been shown to stimulate UvrD’s helicase activity at a nick, which was specific for UvrD and probably not related to UvrAB helix-destabilizing activity, because Rep helicase was not be stimulated by UvrAB (Atkinson et al. 2009).
Repair synthesis absolutely requires the action of both UvrD and Pol I (Husain et al. 1985; Orren et al. 1992), and Pol I cannot access the nick without previous removal of UvrC by the action of UvrD. Analysis of the repair synthesis reaction in the presence of DNA ligase indicated that ∼90% of the events were simple gap filling and <10% involved nick translation to repair tracts >12–13 nucleotides long (Van Houten et al. 1988). Gap filling did not require UvrD and addition of UvrD did not alter the size of the repair patch.
COUPLING REPAIR TO TRANSCRIPTION
A remarkable feature of RNAP (RNA polymerase) is that once transcription is underway, it is tightly bound to DNA and does not dissociate. Therefore, RNAP stalled at a DNA lesion on the transcribed strand represents a special problem that the bacterial cell must resolve. RNAP appears to arrest at most lesions that are repaired by NER (for review, see Dreij et al. 2008).
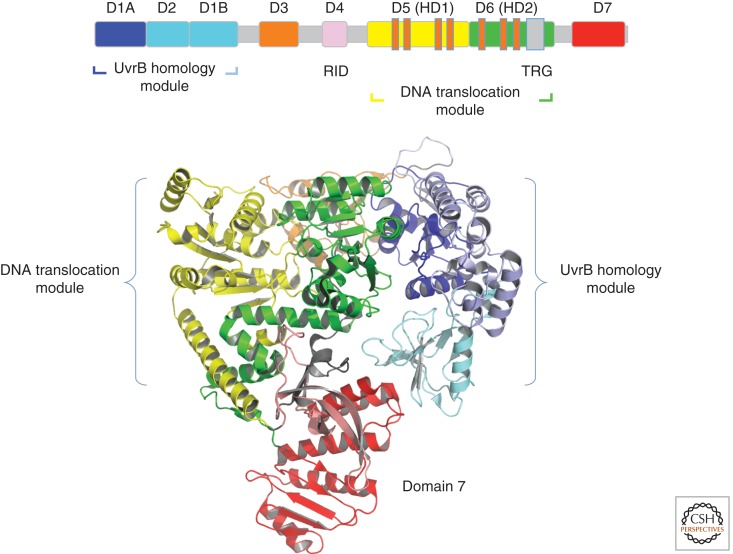
Seminal work by Chris Selby and Aziz Sancar led to the model showing that the TRCF/Mfd protein is both necessary and sufficient to displace the stalled RNAP and recruit the NER machinery to the damaged site. The complete E. coli Mfd structure was solved by Nigel Savery’s and Seth Darst’s groups and gave great insight into how Mfd functions to facilitate repair at a transcription-blocking lesion (Deaconescu et al. 2006). The monomeric E. coli Mfd (130 kDa) was found to be folded into eight domains: the amino-terminal region, comprising a three-domain UvrB homology module, BHM (D1A, D2, and D1B) (Assenmacher et al. 2006); a poorly conserved region, D3; an RNAP interaction domain, D4 (RID); a translocation module containing seven SF2 helicase motifs in domains 5 and 6; and a carboxy-terminal autoinhibitory domain 7 (Fig. 6).
Figure 6.
Mfd: The seven domains of the TRCF from E. coli, shown schematically (top) and in ribbon presentation (bottom) (PDB entry 2EYQ). The UvrB homology module is indicated in different shades of blue, and the remaining domains are displayed in orange, violet, yellow, green, and red. In the schematic presentation of the polypeptide, the helicase motifs are shown in small orange bars and the TRG is shown in gray.
The RID of Mfd interacts directly with RNAP through its β subunit. Critical contacts for this interaction were elucidated by the structural characterization of the complex containing the Mfd-RID domain bound to the RNAP-β1 domain, and the interface includes both salt bridges and hydrophobic contacts among three β strands in Mfd and two β strands in the β subunit, thereby forming an extended β sheet (Westblade et al. 2010). Once Mfd binds to stalled RNAP molecules, using its translocation module and ATP hydrolysis, Mfd pushes RNAP in the direction of transcription. The force required to move RNAP is large and is probably greater than RNAP itself, which is one of the strongest DNA translocation motors known to biology. However, this translocation is probably insufficient for RNAP dissociation at a lesion. Savery (2007) proposed that Mfd causes a collapse of the transcription bubble through helical stress-induced translocation into RNAP. In addition to the translocation domain, Mfd shares significant homology with RecG, another SF2 family member and DNA motor that is involved in replication fork regression and recombination (Rudolph et al. 2010).
Deletion of the carboxy-terminal domain or expression of carboxy-terminal domains 4–7 activates Mfd ATPase (Smith et al. 2007; Murphy et al. 2009). These studies have three important implications. First, domain 7 acts as an autoinhibitory domain that maintains Mfd in the repressed state. Second, Mfd undergoes large conformational changes when it collides with a stalled RNAP, causing its amino-terminal domain to move away from the autoinhibitory domain. This conformational switch allows the BHM of Mfd to engage UvrA’s UvrB-interacting domain, through specific contacts that should be comparable to the interactions observed between UvrA and UvrB (Deaconescu et al. 2012; Pakotiprapha et al. 2012). Finally, this conformational change derepresses the ATPase motor to allow Mdf to translocate on the DNA.
A working model first proposed by Hopfner suggested that a UvrA dimer binds one UvrB molecule and one Mfd molecule (Assenmacher et al. 2006). Recent studies suggest that UvrA’s interaction with Mfd is sufficient to overcome UvrA mutations that abolish genome-wide repair by inhibiting the ability to discriminate damaged from nondamaged DNA (Manelyte et al. 2010). Thus, it is possible that a heterotetrameric complex of two UvrAs, one UvrB, and one Mfd molecule might form at the site of a stalled RNAP, which, once displaced from the damage site, could rapidly engage the damage.
A head-on collision of RNAP with the replication machinery represents a serious train wreck of a problem for the bacterial cell. O’Donnell’s group has shown that Mfd allows a replication restart by displacing RNAP and that this replication restart did not require DnaB (Pomerantz and O’Donnell 2010; McGlynn et al. 2012). Mfd is widely conserved in bacteria, and recently Mfd was shown to depress spontaneous mutation during the stationary phase in B. subtilis (Ross et al. 2006). Mfd was also found to potentially decrease transcriptional mutagenesis by decreasing RNAP bypass of abasic or nicked sites (Smith and Savery 2008) by direct dissociation, although it had no effect on the rate of transcription through an 8-oxodG adduct or a uracil.
Campylobacter jejuni is a major food-borne pathogen, and resistance to fluoroquinolones, the major class of antibiotics used to treat enteric infections, is not uncommon. Recently, C. jejuni containing mfd mutations was shown to have a 100-fold reduction in the rate of spontaneous mutations to ciprofloxacin resistance, whereas overproduction of Mfd increased the rate of mutations (Han et al. 2008).
NusA is a protein associated with RNAP and aids in both transcriptional elongation and termination. Graham Walker has recently shown that the nusA11(ts) mutant is very sensitive to killing by nitrofurazone and 4-nitroquinolone-1-oxide but not through UV damage (Cohen and Walker 2011). Because UvrA is essential for resistance to nitrofuranzome, it has been proposed that NusA helps to recruit UvrA to lesions resulting from this chemical (Cohen and Walker 2011; Savery 2011). This intriguing model awaits direct evidence of an interaction of NusA with stalled RNAP and UvrA at the site of a nitrofuranzone lesion and/or demonstration of a loss of transcription-coupled repair for this lesion in a nusA11 mutant.
CONCLUSIONS
The tremendous interest in NER accompanied by new insights obtained through structural, biochemical, and biophysical methods, as well as the development and use of new single-molecule methods, have provided deep insights into the prokaryotic NER pathway. Nevertheless, many steps in this complex process await further characterization. For example, it is now known that the damage recognition complex is composed of two UvrA and two UvrB molecules, but exactly how these proteins interact dynamically with damaged nucleotides to achieve high specificity remains largely elusive. Further along the NER pathway, similar questions arise. Are two UvrB molecules bound at the site of a lesion? How is damage transferred from UvrA to UvrB? Because UvrC binds tightly to DNA, what keeps its dual nucleases from making spurious incisions? How does the UvrB–UvrC–DNA complex assemble and which conformational changes have to be accomplished to ensure that both incisions take place? Isolation and crystallization of new intermediates, as well as new single-molecule approaches with damaged DNA substrates and eventually with intact bacterial cells, will help to answer these important questions. It will be exciting to see how they will be addressed in the future and will thus allow the assembly of the different pieces of the puzzle to obtain a full picture of this most important DNA repair mechanism.
ACKNOWLEDGMENTS
This work is supported by a grant from the National Institutes of Health, 1R01ES019566 to B.V.H. and by the Deutsche Forschungsgemeinschaft (KI-562/2 and Forschungszentrum FZ-82) to C.K.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Ahn B 2000. A physical interaction of UvrD with nucleotide excision repair protein UvrB. Mol Cells 10: 592–597 [DOI] [PubMed] [Google Scholar]
- Alexandrovich A, Czisch M, Frenkiel TA, Kelly GP, Goosen N, Moolenaar GF, Chowdhry BZ, Sanderson MR, Lane AN 2001. Solution structure, hydrodynamics and thermodynamics of the UvrB C-terminal domain. J Biomol Struct Dyn 19: 219–236 [DOI] [PubMed] [Google Scholar]
- Aravind L, Walker DR, Koonin EV 1999. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res 27: 1223–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenmacher N, Wenig K, Lammens A, Hopfner KP 2006. Structural basis for transcription-coupled repair: The N terminus of Mfd resembles UvrB with degenerate ATPase motifs. J Mol Biol 355: 675–683 [DOI] [PubMed] [Google Scholar]
- Atkinson J, Guy CP, Cadman CJ, Moolenaar GF, Goosen N, McGlynn P 2009. Stimulation of UvrD helicase by UvrAB. J Biol Chem 284: 9612–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce RP, Howard-Flanders P 1964. Release of ultraviolet light-induced thymine dimers from DNA in E. coli K-12. Proc Natl Acad Sci 51: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM Jr, Matson SW 1997. A point mutation in Escherichia coli DNA helicase II renders the enzyme nonfunctional in two DNA repair pathways. Evidence for initiation of unwinding from a nick in vivo. J Biol Chem 272: 572–579 [DOI] [PubMed] [Google Scholar]
- Cai Y, Patel DJ, Geacintov NE, Broyde S 2009. Differential nucleotide excision repair susceptibility of bulky DNA adducts in different sequence contexts: Hierarchies of recognition signals. J Mol Biol 385: 30–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canuto VM, Levine JS, Augustsson TR, Imhoff CL 1982. UV-radiation from the young sun and oxygen and ozone levels in the prebiological paleoatmosphere. Nature 296: 816–820 [Google Scholar]
- Caron PR, Kushner SR, Grossman L 1985. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc Natl Acad Sci 82: 4925–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnossen I, Sanz-Forcada J, Favata F, Witasse O, Zegers T, Arnold NF 2007. Habitat of early life: Solar X-ray and UV radiation at Earth’s surface 4–3.5 billion years ago. J Geophys Res-Planet 112: E02008 [Google Scholar]
- Cohen SE, Walker GC 2011. New discoveries linking transcription to DNA repair and damage tolerance pathways. Transcription 2: 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, DellaVecchia MJ, Wang H, Bienstock RJ, Melton MA, Van Houten B 2006. The C-terminal zinc finger of UvrA does not bind DNA directly but regulates damage-specific DNA binding. J Biol Chem 281: 26370–26381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, DellaVecchia MJ, Perera L, Van Houten B 2008. Cooperative damage recognition by UvrA and UvrB: Identification of UvrA residues that mediate DNA binding. DNA Repair (Amst) 7: 392–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA 2006. Structural basis for bacterial transcription-coupled DNA repair. Cell 124: 507–520 [DOI] [PubMed] [Google Scholar]
- Deaconescu AM, Sevostyanova A, Artsimovitch I, Grigorieff N 2012. Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface. Proc Natl Acad Sci 109: 3353–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaVecchia MJ, Merritt WK, Peng Y, Kirby TW, DeRose EF, Mueller GA, Van Houten B, London RE 2007. NMR analysis of [methyl-13C]methionine UvrB from Bacillus caldotenax reveals UvrB–domain 4 heterodimer formation in solution. J Mol Biol 373: 282–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS 2011. Superfamily I helicases as modular components of DNA-processing machines. Biochem Soc Trans 39: 413–423 [DOI] [PubMed] [Google Scholar]
- Doolittle RF, Johnson MS, Husain I, Van Houten B, Thomas DC, Sancar A 1986. Domainal evolution of a prokaryotic DNA repair protein and its relationship to active-transport proteins. Nature 323: 451–453 [DOI] [PubMed] [Google Scholar]
- Dreij K, Burns JA, Dimitri A, Nirenstein L, Noujnykh T, Scicchitano DA 2008. DNA damage and transcription elongation: Consequences and RNA integrity. In The chemical biology of DNA damage (ed. Geacintov NE, Broyde S), pp. 399–437 Wiley-VCH, Weinheim, Germany [Google Scholar]
- Eryilmaz J, Ceschini S, Ryan J, Geddes S, Waters TR, Barrett TE 2006. Structural insights into the cryptic DNA-dependent ATPase activity of UvrB. J Mol Biol 357: 62–72 [DOI] [PubMed] [Google Scholar]
- Fischer CJ, Maluf NK, Lohman TM 2004. Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J Mol Biol 344: 1287–1309 [DOI] [PubMed] [Google Scholar]
- Goosen N 2010. Scanning the DNA for damage by the nucleotide excision repair machinery. DNA Repair (Amst) 9: 593–596 [DOI] [PubMed] [Google Scholar]
- Goosen N, Moolenaar GF 2008. Repair of UV damage in bacteria. DNA Repair (Amst) 7: 353–379 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV 1990. Superfamily of UvrA-related NTP-binding proteins. Implications for rational classification of recombination/repair systems. J Mol Biol 213: 583–591 [DOI] [PubMed] [Google Scholar]
- Han J, Sahin O, Barton YW, Zhang Q 2008. Key role of Mfd in the development of fluoroquinolone resistance in Campylobacter jejuni. PLoS Pathog 4: e1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand EL, Grossman L 1999. Oligomerization of the UvrB nucleotide excision repair protein of Escherichia coli. J Biol Chem 274: 27885–27890 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Tainer JA 2003. Rad50/SMC proteins and ABC transporters: Unifying concepts from high-resolution structures. Curr Opin Struct Biol 13: 249–255 [DOI] [PubMed] [Google Scholar]
- Hsu DS, Takahashi M, Delagoutte E, Bertrand-Burggraf E, Wang YH, Norden B, Fuchs RP, Griffith J, Sancar A 1994. Flow linear dichroism and electron microscopic analysis of protein-DNA complexes of a mutant UvrB protein that binds to but cannot kink DNA. J Mol Biol 241: 645–650 [DOI] [PubMed] [Google Scholar]
- Hsu DS, Kim ST, Sun Q, Sancar A 1995. Structure and function of the UvrB protein. J Biol Chem 270: 8319–8327 [DOI] [PubMed] [Google Scholar]
- Husain I, Van Houten B, Thomas DC, Abdel-Monem M, Sancar A 1985. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci 82: 6774–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imoto S, Bransfield LA, Croteau DL, Van Houten B, Greenberg MM 2008. DNA tandem lesion repair by strand displacement synthesis and nucleotide excision repair. Biochemistry 47: 4306–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaciuk M, Nowak E, Skowronek K, Tanska A, Nowotny M 2011. Structure of UvrA nucleotide excision repair protein in complex with modified DNA. Nat Struct Mol Biol 18: 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Kropachev K, Ding S, Van Houten B, Geacintov NE, Broyde S 2009. Exploring damage recognition models in prokaryotic nucleotide excision repair with a benzo[a]pyrene-derived lesion in UvrB. Biochemistry 48: 8948–8957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junop MS, Obmolova G, Rausch K, Hsieh P, Yang W 2001. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell 7: 1–12 [DOI] [PubMed] [Google Scholar]
- Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B 2010. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single-molecule imaging of quantum-dot-labeled proteins. Mol Cell 37: 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Truglio JJ, Croteau D, Rhau B, Wang L, Van Houten B, Kisker C 2007. Structure of the C-terminal half of UvrC reveals an RNase H endonuclease domain with an Argonaute-like catalytic triad. EMBO J 26: 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbink JH, Sixma TK 2005. Variations on the ABC. Structure 13: 498–500 [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang W 2006. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127: 1349–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Sancar A 1992. Active site of (A)BC excinuclease. I. Evidence for 5′ incision by UvrC through a catalytic site involving Asp399, Asp438, Asp466, and His538 residues. J Biol Chem 267: 17688–17692 [PubMed] [Google Scholar]
- Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, et al. 2011. Probing for DNA damage with β-hairpins: Similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 10: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher KP 2004. Structure and mechanism of ABC transporters. Curr Opin Struct Biol 14: 426–431 [DOI] [PubMed] [Google Scholar]
- Machius M, Henry L, Palnitkar M, Deisenhofer J 1999. Crystal structure of the DNA nucleotide excision repair enzyme UvrB from Thermus thermophilus. Proc Natl Acad Sci 96: 11717–11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malta E, Moolenaar GF, Goosen N 2006. Base flipping in nucleotide excision repair. J Biol Chem 281: 2184–2194 [DOI] [PubMed] [Google Scholar]
- Malta E, Moolenaar GF, Goosen N 2007. Dynamics of the UvrABC nucleotide excision repair proteins analyzed by fluorescence resonance energy transfer. Biochemistry 46: 9080–9088 [DOI] [PubMed] [Google Scholar]
- Malta E, Verhagen CP, Moolenaar GF, Filippov DV, van der Marel GA, Goosen N 2008. Functions of base flipping in E. coli nucleotide excision repair. DNA Repair (Amst) 7: 1647–1658 [DOI] [PubMed] [Google Scholar]
- Manelyte L, Guy CP, Smith RM, Dillingham MS, McGlynn P, Savery NJ 2009. The unstructured C-terminal extension of UvrD interacts with UvrB, but is dispensable for nucleotide excision repair. DNA Repair (Amst) 8: 1300–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manelyte L, Kim YI, Smith AJ, Smith RM, Savery NJ 2010. Regulation and rate enhancement during transcription-coupled DNA repair. Mol Cell 40: 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson SW, Robertson AB 2006. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res 34: 4089–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Savery NJ, Dillingham MS 2012. The conflict between DNA replication and transcription. Mol Microbiol 85: 12–20 [DOI] [PubMed] [Google Scholar]
- Minko IG, Kurtz AJ, Croteau DL, Van Houten B, Harris TM, Lloyd RS 2005. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry 44: 3000–3009 [DOI] [PubMed] [Google Scholar]
- Moolenaar GF, van Sluis CA, Backendorf C, van de Putte P 1987. Regulation of the Escherichia coli excision repair gene uvrC. Overlap between the uvrC structural gene and the region coding for a 24 kD protein. Nucleic Acids Res 15: 4273–4289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar GF, Franken KL, Dijkstra DM, Thomas-Oates JE, Visse R, van de Putte P, Goosen N 1995. The C-terminal region of the UvrB protein of Escherichia coli contains an important determinant for UvrC binding to the preincision complex but not the catalytic site for 3′-incision. J Biol Chem 270: 30508–30515 [DOI] [PubMed] [Google Scholar]
- Moolenaar GF, Bazuine M, van Knippenberg IC, Visse R, Goosen N 1998. Characterization of the Escherichia coli damage-independent UvrBC endonuclease activity. J Biol Chem 273: 34896–34903 [DOI] [PubMed] [Google Scholar]
- Moolenaar GF, Herron MF, Monaco V, van der Marel GA, van Boom JH, Visse R, Goosen N 2000. The role of ATP binding and hydrolysis by UvrB during nucleotide excision repair. J Biol Chem 275: 8044–8050 [DOI] [PubMed] [Google Scholar]
- Moolenaar GF, Höglund L, Goosen N 2001. Clue to damage recognition by UvrB: Residues in the β-hairpin structure prevent binding to non-damaged DNA. EMBO J 20: 6140–6149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar GF, van Rossum-Fikkert S, van Kesteren M, Goosen N 2002. Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair. Proc Natl Acad Sci 99: 1467–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar GF, Schut M, Goosen N 2005. Binding of the UvrB dimer to non-damaged and damaged DNA: Residues Y92 and Y93 influence the stability of both subunits. DNA Repair (Amst) 4: 699–713 [DOI] [PubMed] [Google Scholar]
- Murphy MN, Gong P, Ralto K, Manelyte L, Savery NJ, Theis K 2009. An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd. Nucleic Acids Res 37: 6042–6053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa N, Sugahara M, Masui R, Kato R, Fukuyama K, Kuramitsu S 1999. Crystal structure of Thermus thermophilus HB8 UvrB protein, a key enzyme of nucleotide excision repair. J Biochem 126: 986–990 [DOI] [PubMed] [Google Scholar]
- Nakano T, Katafuchi A, Terato H, Suzuki T, Van Houten B, Ide H 2005. Activity of nucleotide excision repair enzymes for oxanine cross-link lesions. Nucleic Acids Symp Ser (Oxf) 49: 293–294 [DOI] [PubMed] [Google Scholar]
- Navaratnam S, Myles GM, Strange RW, Sancar A 1989. Evidence from extended X-ray absorption fine structure and site-specific mutagenesis for zinc fingers in UvrA protein of Escherichia coli. J Biol Chem 264: 16067–16071 [PubMed] [Google Scholar]
- Oró J, Miller SL, Lazcano A 1990. The origin and early evolution of life on Earth. Annu Rev Earth Planet Sci 18: 317–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Sancar A 1989. The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc Natl Acad Sci 86: 5237–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Selby CP, Hearst JE, Sancar A 1992. Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J Biol Chem 267: 780–788 [PubMed] [Google Scholar]
- Pakotiprapha D, Inuzuka Y, Bowman BR, Moolenaar GF, Goosen N, Jeruzalmi D, Verdine GL 2008. Crystal structure of Bacillus stearothermophilus UvrA provides insight into ATP-modulated dimerization, UvrB interaction, and DNA binding. Mol Cell 29: 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakotiprapha D, Samuels M, Shen K, Hu JH, Jeruzalmi D 2012. Structure and mechanism of the UvrA-UvrB DNA damage sensor. Nat Struct Mol Biol 19: 291–298 [DOI] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M 2010. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 327: 590–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C, Pybus C, Pedraza-Reyes M, Sung HM, Yasbin RE, Robleto E 2006. Novel role of mfd: Effects on stationary-phase mutagenesis in Bacillus subtilis. J Bacteriol 188: 7512–7520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Liu T, Kolbanovskiy A, Liu Y, Ren J, Skorvaga M, Zou Y, Lader J, Malkani B, Amin S, et al. 2007. Sequence context- and temperature-dependent nucleotide excision repair of a benzo[a]pyrene diol epoxide-guanine DNA adduct catalyzed by thermophilic UvrABC proteins. Biochemistry 46: 7006–7015 [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Briggs GS, Lloyd RG 2010. Is RecG a general guardian of the bacterial genome? DNA Repair (Amst) 9: 210–223 [DOI] [PubMed] [Google Scholar]
- Runyon GT, Bear DG, Lohman TM 1990. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc Natl Acad Sci 87: 6383–6387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AM, Nakano T, Takuwa M, Matoba N, Tsuboi T, Terato H, Yamamoto K, Yamada M, Nohmi T, Ide H 2009. Genetic analysis of repair and damage tolerance mechanisms for DNA-protein cross-links in Escherichia coli. J Bacteriol 191: 5657–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Rupp WD 1983. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33: 249–260 [DOI] [PubMed] [Google Scholar]
- Sancar A, Kacinski BM, Mott DL, Rupp WD 1981. Identification of the uvrC gene product. Proc Natl Acad Sci 78: 5450–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A, Franklin KA, Sancar GB 1984. Escherichia coli DNA photolyase stimulates uvrABC excision nuclease in vitro. Proc Natl Acad Sci 81: 7397–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savery NJ 2007. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol 15: 326–333 [DOI] [PubMed] [Google Scholar]
- Savery N 2011. Prioritizing the repair of DNA damage that is encountered by RNA polymerase. Transcription 2: 168–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Carrier WL 1964. The disappearance of thymine dimers from DNA: An error-correcting mechanism. Proc Natl Acad Sci 51: 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao X, Grishin NV 2000. Common fold in helix-hairpin-helix proteins. Nucleic Acids Res 28: 2643–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Q, Thresher R, Sancar A, Griffith J 1992. Electron microscopic study of (A)BC excinuclease. DNA is sharply bent in the UvrB-DNA complex. J Mol Biol 226: 425–432 [DOI] [PubMed] [Google Scholar]
- Singh S, Folkers GE, Bonvin AM, Boelens R, Wechselberger R, Niztayev A, Kaptein R 2002. Solution structure and DNA-binding properties of the C-terminal domain of UvrC from E. coli. EMBO J 21: 6257–6266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skorvaga M, Theis K, Mandavilli BS, Kisker C, Van Houten B 2002. The β-hairpin motif of UvrB is essential for DNA binding, damage processing, and UvrC-mediated incisions. J Biol Chem 277: 1553–1559 [DOI] [PubMed] [Google Scholar]
- Skorvaga M, DellaVecchia MJ, Croteau DL, Theis K, Truglio JJ, Mandavilli BS, Kisker C, Van Houten B 2004. Identification of residues within UvrB that are important for efficient DNA binding and damage processing. J Biol Chem 279: 51574–51580 [DOI] [PubMed] [Google Scholar]
- Smith AJ, Savery NJ 2008. Effects of the bacterial transcription-repair coupling factor during transcription of DNA containing non-bulky lesions. DNA Repair (Amst) 7: 1670–1679 [DOI] [PubMed] [Google Scholar]
- Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF 2002. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell 10: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AJ, Szczelkun MD, Savery NJ 2007. Controlling the motor activity of a transcription-repair coupling factor: Autoinhibition and the role of RNA polymerase. Nucleic Acids Res 35: 1802–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohi M, Alexandrovich A, Moolenaar G, Visse R, Goosen N, Vernede X, Fontecilla-Camps JC, Champness J, Sanderson MR 2000. Crystal structure of Escherichia coli UvrB C-terminal domain, and a model for UvrB-UvrC interaction. FEBS Lett 465: 161–164 [DOI] [PubMed] [Google Scholar]
- Theis K, Chen PJ, Skorvaga M, Van Houten B, Kisker C 1999. Crystal structure of UvrB, a DNA helicase adapted for nucleotide excision repair. EMBO J 18: 6899–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis K, Skorvaga M, Machius M, Nakagawa N, Van Houten B, Kisker C 2000. The nucleotide excision repair protein UvrB, a helicase-like enzyme with a catch. Mutat Res 460: 277–300 [DOI] [PubMed] [Google Scholar]
- Thomas DC, Levy M, Sancar A 1985. Amplification and purification of UvrA, UvrB, and UvrC proteins of Escherichia coli. J Biol Chem 260: 9875–9883 [PubMed] [Google Scholar]
- Timmins J, Gordon E, Caria S, Leonard G, Acajjaoui S, Kuo MS, Monchois V, McSweeney S 2009. Structural and mutational analyses of Deinococcus radiodurans UvrA2 provide insight into DNA binding and damage recognition by UvrAs. Structure 17: 547–558 [DOI] [PubMed] [Google Scholar]
- Truglio JJ, Croteau DL, Skorvaga M, DellaVecchia MJ, Theis K, Mandavilli BS, Van Houten B, Kisker C 2004. Interactions between UvrA and UvrB: The role of UvrB’s domain 2 in nucleotide excision repair. EMBO J 23: 2498–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio JJ, Rhau B, Croteau DL, Wang L, Skorvaga M, Karakas E, DellaVecchia MJ, Wang H, Van Houten B, Kisker C 2005. Structural insights into the first incision reaction during nucleotide excision repair. EMBO J 24: 885–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truglio JJ, Croteau DL, Van Houten B, Kisker C 2006a. Prokaryotic nucleotide excision repair: The UvrABC system. Chem Rev 106: 233–252 [DOI] [PubMed] [Google Scholar]
- Truglio JJ, Karakas E, Rhau B, Wang H, DellaVecchia MJ, Van Houten B, Kisker C 2006b. Structural basis for DNA recognition and processing by UvrB. Nat Struct Mol Biol 13: 360–364 [DOI] [PubMed] [Google Scholar]
- Van Houten B 1990. Nucleotide excision repair in Escherichia coli. Microbiol Rev 54: 18–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B, Snowden A 1993. Mechanism of action of the Escherichia coli UvrABC nuclease: Clues to the damage recognition problem. Bioessays 15: 51–59 [DOI] [PubMed] [Google Scholar]
- Van Houten B, Gamper H, Sancar A, Hearst JE 1987. DNase I footprint of ABC excinuclease. J Biol Chem 262: 13180–13187 [PubMed] [Google Scholar]
- Van Houten B, Gamper H, Hearst JE, Sancar A 1988. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J Biol Chem 263: 16553–16560 [PubMed] [Google Scholar]
- Van Houten B, Eisen JA, Hanawalt PC 2002. A cut above: Discovery of an alternative excision repair pathway in bacteria. Proc Natl Acad Sci 99: 2581–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B, Croteau DL, DellaVecchia MJ, Wang H, Kisker C 2005. “Close-fitting sleeves”: DNA damage recognition by the UvrABC nuclease system. Mutat Res 577: 92–117 [DOI] [PubMed] [Google Scholar]
- Verhoeven EE, van Kesteren M, Moolenaar GF, Visse R, Goosen N 2000. Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J Biol Chem 275: 5120–5123 [DOI] [PubMed] [Google Scholar]
- Verhoeven EE, Wyman C, Moolenaar GF, Hoeijmakers JH, Goosen N 2001. Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J 20: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven EE, van Kesteren M, Turner JJ, van der Marel GA, van Boom JH, Moolenaar GF, Goosen N 2002a. The C-terminal region of Escherichia coli UvrC contributes to the flexibility of the UvrABC nucleotide excision repair system. Nucleic Acids Res 30: 2492–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven EE, Wyman C, Moolenaar GF, Goosen N 2002b. The presence of two UvrB subunits in the UvrAB complex ensures damage detection in both DNA strands. EMBO J 21: 4196–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visse R, de Ruijter M, Ubbink M, Brandsma JA, van de Putte P 1993. The first zinc-binding domain of UvrA is not essential for UvrABC-mediated DNA excision repair. Mutat Res 294: 263–274 [DOI] [PubMed] [Google Scholar]
- Wang J, Mueller KL, Grossman L 1994. A mutational study of the C-terminal zinc-finger motif of the Escherichia coli UvrA protein. J Biol Chem 269: 10771–10775 [PubMed] [Google Scholar]
- Wang H, DellaVecchia MJ, Skorvaga M, Croteau DL, Erie DA, Van Houten B 2006. UvrB domain 4, an autoinhibitory gate for regulation of DNA binding and ATPase activity. J Biol Chem 281: 15227–15237 [DOI] [PubMed] [Google Scholar]
- Wang H, Tessmer I, Croteau DL, Erie DA, Van Houten B 2008. Functional characterization and atomic force microscopy of a DNA repair protein conjugated to a quantum dot. Nano Lett 8: 1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lu M, Tang MS, Van Houten B, Ross JB, Weinfeld M, Le XC 2009. DNA wrapping is required for DNA damage recognition in the Escherichia coli DNA nucleotide excision repair pathway. Proc Natl Acad Sci 106: 12849–12854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters TR, Eryilmaz J, Geddes S, Barrett TE 2006. Damage detection by the UvrABC pathway: Crystal structure of UvrB bound to fluorescein-adducted DNA. FEBS Lett 580: 6423–6427 [DOI] [PubMed] [Google Scholar]
- Webster MP, Jukes R, Zamfir VS, Kay CW, Bagneris C, Barrett T 2012. Crystal structure of the UvrB dimer: Insights into the nature and functioning of the UvrAB damage engagement and UvrB-DNA complexes. Nucleic Acids Res 40: 8743–8758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westblade LF, Campbell EA, Pukhrambam C, Padovan JC, Nickels BE, Lamour V, Darst SA 2010. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res 38: 8357–8369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung AT, Mattes WB, Oh EY, Grossman L 1983. Enzymatic properties of purified Escherichia coli uvrABC proteins. Proc Natl Acad Sci 80: 6157–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Van Houten B 1999. Strand opening by the UvrA2B complex allows dynamic recognition of DNA damage. EMBO J 18: 4889–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Ma H, Minko IG, Shell SM, Yang Z, Qu Y, Xu Y, Geacintov NE, Lloyd RS 2004. DNA damage recognition of mutated forms of UvrB proteins in nucleotide excision repair. Biochemistry 43: 4196–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]