Abstract
The commensal microbiota affects many aspects of mammalian health including control of the immune system to such a extent that a “commensalocentric” view of the maintenance of overall health could be suggested. Autoimmunity is a case of mistaken identity: The immune system reacts to self-tissues and cells as if they were pathogens. Autoimmune reactions can be both advanced or blocked by the commensal microbiota, which can affect innate and adaptive arms of immune responses as well as the mechanisms of “innate–adaptive connection.” Whether specific microbial lineages affect immunity and autoimmunity (the “specific lineage hypothesis”) or multiple lineages can tip the homeostatic balance that regulates host/microbiota homeostasis toward reduced or enhanced host reactivity (the “balanced signal hypothesis”) is yet unknown. The complexity of host/microbiota interactions needs to be fully appreciated in order to find the means for prophylaxis and treatment of autoimmune disorders.
Microbes can promote autoimmunity (e.g., via molecular mimicry); they may also provide protection against autoimmunity.
IMPORTANCE OF COMMENSAL MICROBIOTA FOR MAINTENANCE OF HEALTH
The communities of nonpathogenic microorganisms populating various niches within the animal or human body are known as the “commensal microbiota.” There are three major aspects of host–commensal interactions. First, commensals affect many functions of animal physiology to the extent that axenic or germ-free (sterile) organisms are not viable under regular conditions. That is mostly due to the lack of essential nutrients (such as vitamins) normally produced by commensals and shared with the host. The rapid demise of the host in the absence of commensals without a properly supplemented diet shows its dependence on the microbiota, whereas “genome rationalization” (lack of “essential metabolic pathways”) in many microbes indicates their dependence on the host and on the other members of microbial communities. This also explains the difficulties of in vitro cultivation of the majority of mammalian microbiota members. The term “superorganism” (Lederberg 2000) describing the animal host–microbial consortium as an inseparable whole is quite fair.
The second, most important function of the microbiota is the “colonization resistance” (Stecher and Hardt 2008) that makes it hard for pathogens to take a foothold in the organs that serve as habitats for commensal microbes.
Finally, the microbiota affects development of the host. In more primitive metazoans such as hydra (Rahat and Dimentman 1982) or in highly complex animals such as a fly (Shin et al. 2011), the lack of microbiota leads to abnormalities in body development, whereas in vertebrates the lack of microbiota manifests in an underdeveloped gut and immune system. Microbes also control other functions such as vascularization of the intestine (Stappenbeck et al. 2002; Fagundes et al. 2012), and they even influence the work of the central nervous system by affecting the production of soluble mediators (Fagundes et al. 2012). The influence on immune system development, however, is mostly reversible, because reconstitution of germ-free animals with a normal microbiota restores most of the immune functions. However, species–specific host–microbe interactions seem to be evolutionarily conserved to an extent that even closely related microbiotas (such as mouse and rat microbial communities) cannot fully restore the immunological functions of the other gnotobiotic host. At the same time, the addition of a single species that went through evolutionary adaptation to the host, when added to the foreign microbiota transplant is able to restore the immune functions (Chung et al. 2012). Moreover, for at least some features of the adaptive immune response (production of IgA antibodies), the restoration is transient and leaves no memory of the event as soon as the animal returns to the germ-free state (Hapfelmeier et al. 2010). Nevertheless, some immune functions (either induced or suppressed by the microbiota) may be lost for good if the microbiota is absent during a critical period for the development of a particular cell type. One recent study has found that accumulation of invariant NKT (iNKT) cells is not observed in germ-free mice; however, a limited window of time exists when colonization of germ-free mice leads to restoration of the normal numbers of iNKT cells (Olszak et al. 2012).
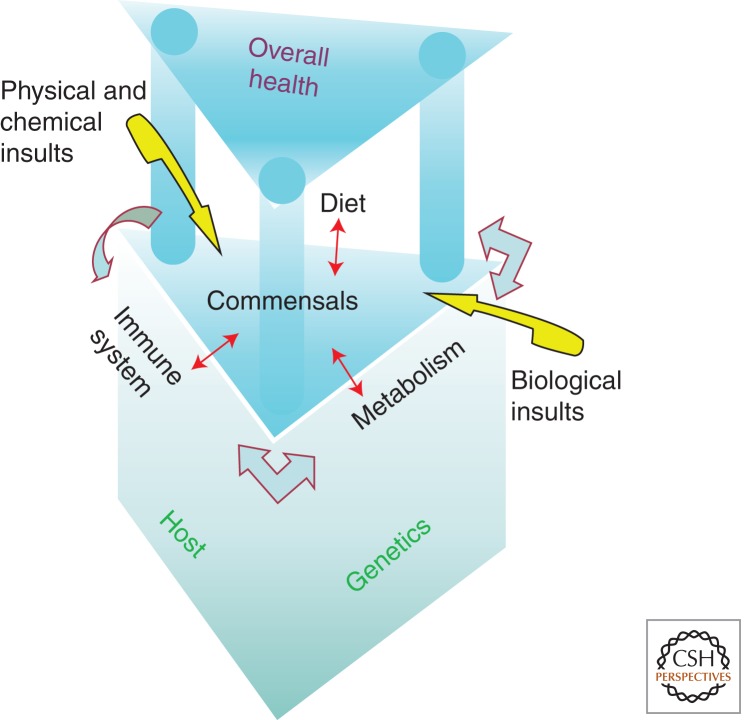
These three key features of commensal–host interactions can be predictably involved in the control of the host’s health, including development of autoimmunity. One can adopt a “commensalocentric” view of the animal or human health (Fig. 1), in which the microbiota and its interactions with ever-changing environmental cues determine the homeostatic maintenance of the higher organism. In fact, dietary influences on health were shown to be amplified by transfer of microbiota from affected animals (Ley et al. 2006; Turnbaugh et al. 2006). Commensals are actively involved in interaction with pathogens, either by mediating colonization resistance or by limiting damage to the host through mechanisms known as “tolerance to pathogens” or “tolerance to infection” (Ayres and Schneider 2012; Medzhitov et al. 2012). These complex mechanisms are able to limit damage evoked by the pathogen itself or by the immune response to the pathogen. It is quite obvious that the genetic makeup of the host must play a central role in maintenance of health and interactions with the environment.
Figure 1.
Commensalocentric view of the homeostatic maintenance of the host’s health. Commensals are directly affected by what the host consumes (diet), by the host’s metabolism, and by the immune system (red arrows). These interactions are reciprocal, and the microbiota affects all three aspects of the host’s life by either direct influences or through behavioral modifications, whereas the immune system, metabolism, and diet are also connected by mutual influences (gray arrows). It is important to recognize that the host’s genetics has fundamental influences on immune and metabolic functions, including the outcomes of interactions with the microbiota. Similarly, the environment can exert its influences on health through modification of the microbiota (yellow arrows).
ENVIRONMENT AND AUTOIMMUNITY
Environmental influences include physical (UV light, radioactivity), chemical (toxins and irritants), and biological insults. Biological insults include dietary challenges, invasion with parasites, viral infections, and colonization by microbes (both commensal and pathogenic). Although many of these are likely to work in concert, here we focus on the role of commensal microbiota.
Hard evidence from animal studies and correlative evidence from studies in humans both indicate that the microbiota is involved in autoimmunity. The term “autoimmunity” is used here to refer to adaptive (clonal) immune responses mediated by T and B lymphocytes that are directed against self-antigens. Autoimmune diseases must be distinguished from purely inflammatory diseases (such as gout, pseudogout, or Mediterranean fever) without an adaptive component and from inflammatory diseases that have an adaptive component, but it is not directed against self-antigens (such as inflammatory bowel disease [IBD]). All three categories of diseases can be collectively termed “diseases of the activated immune system” (Chervonsky 2010). They all cause damage to the host’s organs and tissues, but such damage, as well as damage evoked during normal immune responses to pathogens, is not autoimmune by our definition. The key to understanding how the microbiota could be involved in autoimmunity is in the theory formulated by Charles Janeway, which explained the principles of initiation of the adaptive immune responses (Janeway 1989). The central point of this theory is the innate–adaptive connection. According to Janeway, only the innate immune system is capable of self–nonself-discrimination due to recognition of conserved pathogen-associated molecular patterns (PAMPs) by a limited number of nonclonal pattern recognition receptors (PRRs). The conserved PAMPs are different from the virulence factors harbored by pathogens and are present in commensal microbiota as well. They are frequently termed MAMPs (microbe-associated molecular patterns), but we will continue to use the term “PAMPs,” especially because this complication was evident to Janeway himself. The adaptive immune system, on the other hand, recognizes antigens in their native form (B cells) or as a processed peptide in complex with major histocompatibility complex (MHC) molecules (T cells). The repertoires of clonal receptors are large but are not evolutionarily selected to avoid self-reactivity. Moreover, self-reactivity is a prerequisite for T cells to survive the quality control in the thymus and be allowed to egress to the periphery. Mechanisms of central tolerance (deletion and inactivation of self-reactive lymphocytes) and their active inhibition by regulatory T cells (Tregs) exist to minimize autoimmunity, but potentially autoreactive cells are always present in the host (Kim et al. 2007b, 2009). Thus, adaptive immune responses have to be activated only on a per-need basis, only when a microbial pathogen or a parasite is challenging the host. Innate sensing provides three factors that are all necessary for the “innate–adaptive connection”: activation of antigen processing and presentation by antigen-presenting cells (APCs), activation of costimulation, and secretion of cytokines that also shape the type of adaptive immunity. The innate–adaptive connection can be easily viewed as a center stage for the microbiota to exert its influence. We have previously suggested a classification for the diseases of the activated immune system based on the involvement of the innate–adaptive connection (Chervonsky 2010).
According to this classification, autoimmune diseases can be divided into two large groups: Group I is composed of diseases that require the innate–adaptive connection for their onset, and Group II, those for which this connection is not critical. Group II diseases are mostly monogenic disorders that are based on the loss of control over one of the principal mechanisms controlling adaptive immunity, such as negative selection or generation of Tregs. Loss of negative selection of T cells due to mutations in the transcription factor AIRE (Anderson et al. 2002; Mathis and Benoist 2009) leads to a multiorgan autoimmunity known as autoimmune polyendocrinopathy-candidiases-ectodermal dystrophy (APECED) (Peterson et al. 2004), whereas mutations in the transcriptional regulator of Tregs FoxP3 lead to a multiorgan autoimmunity termed Scurfy or immune dysregulation, polyendocrinopathy, enteropathy, or X-linked syndrome (IPEX) (for review, see Josefowicz et al. 2012). An important factor that separates Group I from Group II is that the commensal microbiota does not affect the latter group but may play a role in the pathogenesis of Group I diseases. Experimental evidence for this idea comes from studies of germ-free and gnotobiotic (populated with defined microbiota) animals. The general approach is to produce germ-free animals that under conventional conditions develop one sort of autoimmunity or another, and then ask whether the outcome changes when the microbiota is absent or replaced with known microbes. These studies have revealed that Group II diseases were insensitive to the presence of microbiota: sterile mice lacking AIRE, FoxP3, or AID (Table 1). The situation with Group I is very different. Here the tested models could be separated into four groups: (1) germ-free animals were as susceptible as conventionally raised animals, (2) some features of the disease were lost, but other features were still present, (3) diseases did not affect germ-free mice at all or the disease was greatly attenuated, and (4) diseases that require commensals (Table 1).
Table 1.
Importance of commensal microbiota for Group II autoimmunity revealed by experiments with germ-free and gnotobiotic animals
| A. Monogenic diseases are insensitive to commensal regulation IPEX (FoxP3 deletion in adult mice) (Chinen et al. 2010) APECED (AIRE knockout) (Gray et al. 2007) Activation-induced cytidine deaminase (Aicda) deficiency-dependent gastritis (Hase et al. 2008) |
| B. Diseases that develop independently of commensals Type 1 diabetes in mice (Wen et al. 2008) and rats (Rossini et al. 1979) |
| C. Diseases that develop independent of commensals, but commensals amplify the disease Models of rheumatoid arthritis (RA) in collagen-immunized rats (Bjork et al. 1994) RA in K/BxN mice (Wu et al. 2010) Systemic lupus erythematosus (SLE in MRL.lpr mice) (Maldonado et al. 1999) Experimental encephalomyelitis (EAE) caused by immunization with MOG peptide (Lee et al. 2011) |
| D. Diseases that require commensals EAE (T-cell receptor transgenic model) (Berer et al. 2011) Ankylosing enthesopathy in B10.BR mice (Rehakova et al. 2000; Sinkorova et al. 2008) RA in IL1R-antagonist KO (Abdollahi-Roodsaz et al. 2008) |
The experiments listed in the table are not exhaustive.
These experiments lead to the obvious conclusion that the influence of the microbiota on Group I diseases can range from having no effect to being indispensable for induction of autoimmunity. This set of experiments, however, does not take into consideration the protective properties of the microbiota. Take type 1 diabetes as an example. The disease develops in genetically susceptible germ-free mice and rats with high incidence. At the same time, it has been well established that the incidence of the disease varies significantly among rodents housed in different animal facilities (from almost zero to ∼100%) (Pozzilli et al. 1993). The variability in incidence is blamed on environmental factors, of which variability in microbiota is likely to be the major one. Importantly, the incidence of the disease found in susceptible germ-free diabetes-prone nonobese diabetic (NOD) mice is very similar to the incidence observed in conventional animal facilities with a high percentage of diabetic mice. The slopes of the incidence curves are also very similar (Wen et al. 2008). Thus, in type 1 diabetes, the microbiota can stay neutral or prevent/attenuate the disease development, but it does not normally accelerate the disease. It is assumed that the conventional microbiota is free of “specific pathogens,” that is, microbes that are known to induce diseases in experimental animals. It does not mean that they are free of “pathobionts,” or microbes that do not cause pathology under normal circumstances and thus avoid being put on the specific pathogen list. This category of commensal microorganisms could be a good candidate for playing some role in protection against autoimmunity. Bone fide pathogens can either suppress or provoke autoimmunity. Even the same pathogens can be protective or stimulatory of the same disease, like the Coxsackie B3 viruses in the mouse model of type 1 diabetes (Drescher et al. 2004). Injection of Poly:IC, which mimics viral infections, can also lead to protection of susceptible animals from (Serreze et al. 1989) or induction of type 1 diabetes in resistant animals (Moriyama et al. 2002), although it required additional antigenic stimulation. The timing, the dose of infection, or agonist administration mattered in these experiments. How that works is not known and it is very important to understand.
MECHANISMS BY WHICH MICROBES PROMOTE AUTOIMMUNITY
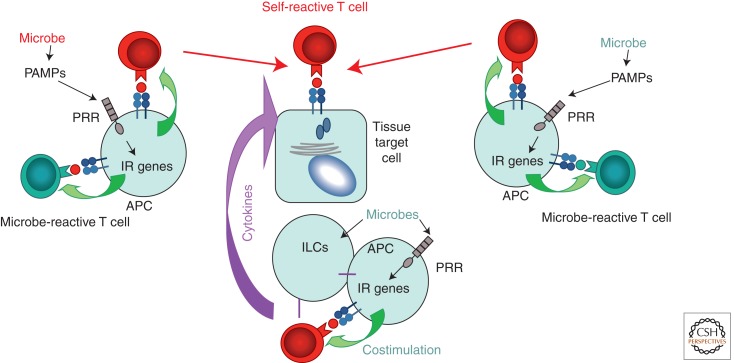
When microbes are critical for development of autoimmunity, several mechanisms can be discriminated (Fig. 2).
Figure 2.
Mechanisms by which microbes contribute to initiation or severity of autoimmunity. In all cases the end point is the destruction of tissue target cells (center) by self-reactive cells (in red) or antibodies (not shown). Microbial peptides could be identical or similar enough to self-peptides (left) to induce cross-reactivity to self-tissues—“molecular mimicry.” Innate–adaptive connection mechanisms (antigen presentation and costimulation; green arrows) are required to activate self-reactive cells. The mimicry is not necessary if antigen-presenting cells (APCs) are activated by microbes through innate receptor signaling and happen to express self-antigen when a self-reactive T cell is present. This is “bystander activation” (right). The third mechanism is “amplification of autoimmunity by cytokines” elicited by microbial activation of professional APCs and the so-called innate lymphoid cells (ILCs), which together produce or elicit cytokine production by T cells. These cytokines target self-reactive T cells and their target tissues (bottom). They can also reduce the efficiency of regulatory cells, which are responsible for active tolerance to self.
First, a mechanism known as molecular mimicry could be at the core of autoimmunity. The best example of such influence and a clear example of Group I immunity is rheumatoid fever, a disease caused by destruction of myocardium due to cross-reactivity (molecular mimicry) with streptococcal antigens (Malkiel et al. 2000). Cross-reactivity by itself is hardly a unique situation because T cells are intrinsically cross-reactive. However, activation of T cells by organ-specific antigens in the absence of infection (in other words, when the innate–adaptive connection is not activated) does not normally happen. Mice expressing an antigen of the lymphocyte choriomeningitis virus (LCMV) in the pancreas are healthy even when they are double-transgenic for a T-cell receptor (TCR) that reacts with this very viral antigen—unless, of course, the mice are infected with live LCMV (Lang et al. 2005). Then the pancreas is quickly destroyed. In both cases, the innate–adaptive connection stimulates reactivity to existing self-antigens, and provision of costimulation and inflammatory cytokines induces autoimmunity. Pathobionts that belong to commensal microbiota can act through the same mechanism. Klebsiella pneumoniae is believed to carry antigens mimicking MHC class I molecule HLA-B27 and, thus, induce ankylosing spondylitis (Rashid and Ebringer 2012).
Second, it is also possible that the microbe and the host do not share structurally similar antigens, but induction of costimulation and cytokine production by an APC that is activated by infection, and at the same time presents self-antigens, will lead to activation of autoimmunity. This is the mechanism known as “bystander activation.”
Finally, and very relevant to commensal microbiota, is the ability of specific commensal lineages to induce production of cytokines that can contribute to the pathogenesis of autoimmunity. The best-known example is the one of segmented filamentous bacteria (SFB) that stimulate generation of Th17 and likely Th1 types of T-cell responses (Gaboriau-Routhiau et al. 2009; Ivanov et al. 2009). Whereas Th17 cells are critical for defense against certain types of pathogens (Conti et al. 2009), they also contribute to autoimmunity. Th17 cells induced by SFB can affect autoimmune reactions in remote organs, such as joints (Wu et al. 2010).
In all cases, autoreactive TCRs preexist the challenge. This is true for both Group I and Group II autoimmune diseases. That Group II diseases occur in germ-free animals suggests that autoimmune T cells are activated in the absence of the innate–adaptive connection, which sounds contradictory to what had been postulated above. There are two possibilities to explain the discrepancy: (1) Germ-free conditions are only free from live microbes, but not from microbial products. These products are present in the food and bedding, and some can resist autoclaving (Kane et al. 2011). (2) Endogenous nucleic acids in the extracellular space and within cells including endogenous viruses, mitochondrial DNA, and cellular DNA from dying cells are always present. They are promptly removed by nucleases (Napirei et al. 2000), and special DNA sensors exist to reduce their influence on the adaptive immune system (Stetson et al. 2008). However, tonic signaling that keeps the adaptive immune system in an alert state must be constantly present. That is evident from experiments with ablation of Treg cells in adult animals including germ-free rodents (Chinen and Rudensky 2012). In addition, some level of expression of costimulatory molecules, such as CD80, is known to be present even in resting APCs. Thus, division of autoimmune diseases into two groups includes the understanding that background tuning of the adaptive immune responses by innate mechanisms is inevitably present in both groups.
MICROBIAL LINEAGES AND AUTOIMMUNITY
Specific lineages of commensals were found to be involved in development of autoimmunity. The importance of these lineages has been supported by experiments with germ-free animals colonized with microbes of interest. SFB contributed to inflammation in a model of arthritis (Wu et al. 2010), and a defined mixture of several bacteria (but not Lactobacillus sp.) was capable of induction of development of ankylosing enthesopathy (Sinkorova et al. 2008) in gnotobiotic mice. Importantly, the influence of a particular lineage is disease-specific. A recent report found that SPF NOD mice carrying SFB were protected from type 1 diabetes (Kriegel et al. 2011). These observations support a seemingly logical conclusion that some microbial lineages are better suited for induction or prevention of a certain disease development than others. This statement is true to some extent and resonates with the current view on the role of microbiota in affecting health in general. But it has the same limitations. Many studies single out particular bacterial lineages that strongly affect immune responses in the gut and beyond. The best-studied bacterial lineages in this respect are Bacteroides fragilis and the SFB. B. fragilis produces a polysaccharide A (PSA) that stimulates production of the anti-inflammatory cytokine IL-10 (for review, see Surana and Kasper 2012). SFB stimulates IL-17 very efficiently through a yet-unknown molecular mechanism. These results are very important, because they show that the microbiota can affect inflammation and autoimmunity and are likely to lead to an understanding of the underlying mechanisms. There are, however, several considerations that need to be discussed:
Different Microbial Lineages Can Fulfill the Same Functions. A relatively random consortium of intestinal microbiota termed Altered Schaedler’s Flora (ASF) can also induce IL-17 production, and most efficiently if IL-10 signaling is inactivated (Geuking et al. 2011). In addition, humans, who generally do not carry SFB, still mount Th17 responses. These findings indicate that other microbes can substitute for SFB.
Microbial Consortia Can Induce a Specific Type of Response. A relatively large group of Clostridia bacilli can induce Tregs in the lamina propria of the large intestine (Atarashi et al. 2011). It is currently unknown whether some members of the group do it better than others or the whole consortium is needed.
Microbes with Established Function Can Cause Opposite Effects. For example, B. fragilis can cause inflammation (Polk and Kasper 1977), and SFB can reduce organ-specific autoimmunity in the form of type 1 diabetes (Kriegel et al. 2011). However, this latter finding requires the following amendment:
Microbial Effects in Large Communities Are Hard to Ascribe. SFB can be used as an example to illustrate the point. Our unpublished results suggest that monocolonization of GF NOD mice or transfer of SPF, SFB-negative microbiota to SFB-monocolonized gnotobiotic mice does not affect diabetes development in female mice (A Chervonsky, unpubl.). These results do not agree with the previously published data (Kriegel et al. 2011) and with the new data from the same group showing that the exposure of SFB-free SPF mice to SFB from monocolonized mice leads to reduction in diabetes development (D Mathis, pers. comm.). That difference in the results requires a plausible explanation. One option is that SFB are not the perpetrators of the effect and require help from (or provide help to) other microbes (variable between the colonies) that then act to reduce autoimmunity. The other explanation would be that SFB lineages used by the two groups differ in their ability to affect the development of autoimmune diabetes. Regardless of the reason for discrepancies, the story underscores the complexity of the host–microbiota interactions.
In Vivo Experiments with Colonization of Germ-Free Mice May Show Results that Differ from Transfer Experiments in SPF Animals. Many beneficial (anti-inflammatory) bacteria that are used as probiotics cannot efficiently colonize adult SPF animals because of colonization resistance provided by the host’s microbiota. At the same time, colonization of the germ-free host is not a problem for these microbes. Whereas introduction of a commercial probiotic consortium (VSL3) into SPF NOD mice was found to reduce type 1 diabetes development, which correlated with production of IL-10 by local APCs (Calcinaro et al. 2005), we have found no effect of the consortium in germ-free NOD mice colonized by their mothers (data not shown). The discrepancy in the results could mean that other members of the microbiota are necessary for the antidiabetic effect. Or, alternatively, the reason could be more prosaic: Because the microbial mix does not colonize SPF animals, the gastric gavage of the microbes has to be applied regularly. This is hardly an innocuous procedure and can lead to accidental systemic administration of bacteria.
What Bacteria Can Do in In Vitro Experiments May Not Directly Translate into What Effects They Have In Vivo. When admixed with the enormous preexisting community, the supplemented microbes face different conditions that vary in availability of the niche space, nutrients, and quorum sensing signals. In this case, the analysis of transcriptional changes in colonizing microbiota has significant value. An example of such analysis performed in human twin pairs and gnotobiotic animals receiving lactobacteria as a probiotic revealed that although the composition of commensal consortia did not change significantly, the metabolic pathways did (McNulty et al. 2011). These results suggest that transcriptional analysis of bacteria used to affect diseases should complement in vitro observations.
Besides the Influence from Other Commensals and the Environment on the Behavior of a Specific Microbial Lineage, the Host’s Genetics Can Be a Decisive Factor. For example, ASF failed to induce Th17 cells in the Swiss Webster outbred strain, but did it in several gnotobiotic mice of other genetic backgrounds (Geuking et al. 2011).
HOW DOES THE MICROBIOTA QUENCH THE DEVELOPMENT OF AUTOIMMUNITY?
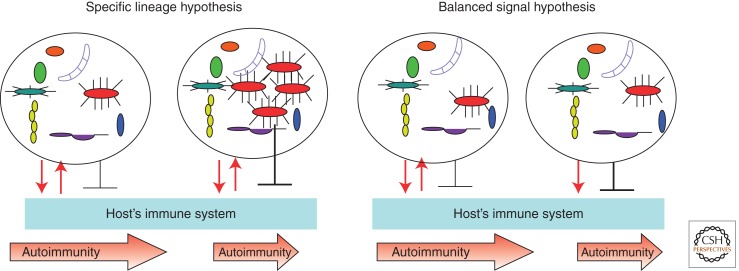
The now well-established role of unique microbial lineages in intestinal homeostasis supports the idea that individual lineages within complex communities of commensal microbes can affect the development of autoimmunity. This view can be termed the “specific lineage hypothesis” (Fig. 3). It suggests that the composition of commensals that has been acquired neonatally and challenged during the life of the individual must include specific lineages that can affect certain aspects of health including tolerance to self, and that the individuals lacking these specific lineages would not be able to maintain related functions. By the same token, animals or individuals carrying mutations that can affect microbial composition (such as mutations affecting the innate and adaptive immune systems) would experience a change in microbiota affecting the outcome of homeostatic changes including autoimmunity. A different view that could be termed the “balanced signal hypothesis” predicts that the presence of specific lineages is not required because specific functions (both proinflammatory and anti-inflammatory) could be taken care of by multiple lineages. In this case, the host’s genetics defines the interactions with the microbiota, and the outcome of these interactions affects homeostasis including autoimmunity. The “balanced signal” implies that microbes can elicit opposing effects. The microbiota does this because it faces two problems during its symbiotic coexistence with the host: It needs to reduce the host’s impact on itself (negative regulation of antimicrobial, inflammatory responses), whereas the host needs to control the microbiota by constantly sensing disturbances and preventing invasion beyond the barrier tissues (positive sensing and response).
Figure 3.
The two hypotheses explain how microbes could provide protection against autoimmunity. The “specific lineage hypothesis” predicts that in genetically predisposed animals or humans, microbiota could provide signals that induce antimicrobial effectors that are neutralized by microbial inhibitory signals (reciprocating red arrows). As a result, the microbiota stays in a homeostatic relationship with the host (leftmost panel) and the disease progresses independently of microbiota. When a specific microbial lineage is expanded (next panel), it blocks the development of autoimmunity. It does so to improve its own odds of staying in this expanded state by suppressing the host’s inflammatory and adaptive responses. Autoimmunity is quenched as a side effect. The “balanced signal hypothesis” predicts that the host’s interactions with microbiota are independent of the precise microbiota composition and that the host’s genetics plays a critical role in the conversation with microbes. Whereas a balanced host response to commensals and the commensals’ effort to reduce this response do not affect disease development (middle right panel), the inability of the host to control the microbiota properly (such as in mice lacking PRR signaling because of a knockout of the adaptor molecule MyD88) (Wen et al. 2008) (rightmost panel) results in the dominance of negative signaling provided by the microbiota and reduction of autoimmunity. Both hypotheses predict that tolerizing effects would be lost in germ-free conditions.
From an experimental point of view, the discrimination between the two hypotheses is not simple. For example, a genetic ablation of TLR2 in NOD mice led to reduction in diabetes incidence by ∼50% in SPF mice (Kim et al. 2007a; Wen et al. 2008). At the same time, germ-free TLR2-deficient mice had an incidence of type 1 diabetes that did not differ from that of germ-free NOD mice (data not shown). One could propose that TLR2-negative mice lose control over some microbes that now thrive and reduce the incidence of diabetes. Or, one could suggest that the positive (antimicrobial) signaling is lost, and that negative, anti-inflammatory signaling induced by the microbiota is prevailing. Even if some changes in microbiota composition are found in knockout animals, their causative role is not so simple to prove.
The actual mechanisms that the microbiota uses to affect autoimmunity vary. There are some general mechanisms, such as maintenance of the integrity of the barriers colonized by commensals (Rakoff-Nahoum et al. 2004), and more specific mechanisms that amplify the tolerance-inducing pathways. These mechanisms are discussed in many reviews. The point that we are making here is that in many cases in which autoimmunity develops independently of the microbiota, it is a case of mistaken identity: No pathogen is present, but an immune response develops that targets self-tissues. The effector mechanisms that are induced are no different from the mechanisms used against pathogens. There are two points to make: First, genetic glitches in the innate–adaptive connection mechanisms are likely to be responsible for autoimmunity (Chervonsky 2010), and, second, other mechanisms that are normally involved in host–pathogen interactions may apply to the case of autoimmunity. One such mechanism, or rather a complex system of responses, are known as tolerance to pathogens. It is defined as the maintenance of a host’s high fitness without affecting the levels of pathogen burden. The two goals of tolerance to pathogens are to reduce the damage to the host delivered by the pathogen, and to reduce the damage to the host delivered by the immune response. These mechanisms are not well studied yet in higher animals, but some important observations have been made that implicate commensal microbes in participating in tolerance to pathogens (Cohen-Poradosu et al. 2011; Ayres et al. 2012; Chervonsky 2012). We now need to test whether these mechanisms are applicable to autoimmunity as well.
ACKNOWLEDGMENTS
My work is supported by National Institutes of Health (NIH) grants R01 AI072627, R01 AI082418, R01 DK090422, JDRF grant 17-2011- 519, and NIH grant P30DK042086. I am grateful to Joseph Pickard for editing the manuscript.
Footnotes
Editors: Diane J. Mathis and Alexander Y. Rudensky
Additional Perspectives on Immune Tolerance available at www.cshperspectives.org
REFERENCES
- Abdollahi-Roodsaz S, Joosten LA, Koenders MI, Devesa I, Roelofs MF, Radstake TR, Heuvelmans-Jacobs M, Akira S, Nicklin MJ, Ribeiro-Dias F, et al. 2008. Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J Clin Invest 118: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, et al. 2002. Projection of an immunological self shadow within the thymus by the AIRE protein. Science 298: 1395–1401 [DOI] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Schneider DS 2012. Tolerance of infections. Annu Rev Immunol 30: 271–294 [DOI] [PubMed] [Google Scholar]
- Ayres JS, Trinidad NJ, Vance RE 2012. Lethal inflammasome activation by a multi-drug resistant pathobiont upon antibiotic disruption of the microbiota. Nat Med 18: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, Wekerle H, Krishnamoorthy G 2011. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 479: 538–541 [DOI] [PubMed] [Google Scholar]
- Bjork J, Kleinau S, Midtvedt T, Klareskog L, Smedegard G 1994. Role of the bowel flora for development of immunity to hsp 65 and arthritis in three experimental models. Scand J Immunol 40: 648–652 [DOI] [PubMed] [Google Scholar]
- Calcinaro F, Dionisi S, Marinaro M, Candeloro P, Bonato V, Marzotti S, Corneli RB, Ferretti E, Gulino A, Grasso F, et al. 2005. Oral probiotic administration induces interleukin-10 production and prevents spontaneous autoimmune diabetes in the non-obese diabetic mouse. Diabetologia 48: 1565–1575 [DOI] [PubMed] [Google Scholar]
- Chervonsky AV 2010. Influence of microbial environment on autoimmunity. Nat Immunol 11: 28–35 [DOI] [PubMed] [Google Scholar]
- Chervonsky AV 2012. Intestinal commensals: Influence on immune system and tolerance to pathogens. Curr Opin Immunol 24: 255–260 [DOI] [PubMed] [Google Scholar]
- Chinen T, Rudensky AY 2012. The effects of commensal microbiota on immune cell subsets and inflammatory responses. Immunol Rev 245: 45–55 [DOI] [PubMed] [Google Scholar]
- Chinen T, Volchkov PY, Chervonsky AV, Rudensky AY 2010. A critical role for regulatory T cell-mediated control of inflammation in the absence of commensal microbiota. J Exp Med 207: 2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, et al. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149: 1578–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Poradosu R, McLoughlin RM, Lee JC, Kasper DL 2011. Bacteroides fragilis–stimulated interleukin-10 contains expanding disease. J Infect Dis 204: 363–371 [DOI] [PubMed] [Google Scholar]
- Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, et al. 2009. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med 206: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher KM, Kono K, Bopegamage S, Carson SD, Tracy S 2004. Coxsackievirus B3 infection and type 1 diabetes development in NOD mice: Insulitis determines susceptibility of pancreatic islets to virus infection. Virology 329: 381–394 [DOI] [PubMed] [Google Scholar]
- Fagundes CT, Amaral FA, Teixeira AL, Souza DG, Teixeira MM 2012. Adapting to environmental stresses: The role of the microbiota in controlling innate immunity and behavioral responses. Immunol Rev 245: 250–264 [DOI] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, et al. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31: 677–689 [DOI] [PubMed] [Google Scholar]
- Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, McCoy KD, Macpherson AJ 2011. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity 34: 794–806 [DOI] [PubMed] [Google Scholar]
- Gray DH, Gavanescu I, Benoist C, Mathis D 2007. Danger-free autoimmune disease in AIRE-deficient mice. Proc Natl Acad Sci 104: 18193–18198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Lawson MA, Slack E, Kirundi JK, Stoel M, Heikenwalder M, Cahenzli J, Velykoredko Y, Balmer ML, Endt K, et al. 2010. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328: 1705–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hase K, Takahashi D, Ebisawa M, Kawano S, Itoh K, Ohno H 2008. Activation-induced cytidine deaminase deficiency causes organ-specific autoimmune disease. PLoS ONE 3: e3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA Jr 1989. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54: 1–13 [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY 2012. Regulatory T cells: Mechanisms of differentiation and function. Annu Rev Immunol 30: 531–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334: 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Han MS, Chung KW, Kim S, Kim E, Kim MJ, Jang E, Lee HA, Youn J, Akira S, et al. 2007a. Toll-like receptor 2 senses β-cell death and contributes to the initiation of autoimmune diabetes. Immunity 27: 321–333 [DOI] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY 2007b. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol 8: 191–197 [DOI] [PubMed] [Google Scholar]
- Kim J, Lahl K, Hori S, Loddenkemper C, Chaudhry A, deRoos P, Rudensky A, Sparwasser T 2009. Cutting edge: Depletion of Foxp3+ cells leads to induction of autoimmunity by specific ablation of regulatory T cells in genetically targeted mice. J Immunol 183: 7631–7634 [DOI] [PubMed] [Google Scholar]
- Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D 2011. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci 108: 11548–11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang KS, Recher M, Junt T, Navarini AA, Harris NL, Freigang S, Odermatt B, Conrad C, Ittner LM, Bauer S, et al. 2005. Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat Med 11: 138–145 [DOI] [PubMed] [Google Scholar]
- Lederberg J 2000. Infectious history. Science 288: 287–293 [DOI] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, Mazmanian SK 2011. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci 108: 4615–4622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI 2006. Microbial ecology: Human gut microbes associated with obesity. Nature 444: 1022–1023 [DOI] [PubMed] [Google Scholar]
- Maldonado MA, Kakkanaiah V, MacDonald GC, Chen F, Reap EA, Balish E, Farkas WR, Jennette JC, Madaio MP, Kotzin BL, et al. 1999. The role of environmental antigens in the spontaneous development of autoimmunity in MRL-lpr mice. J Immunol 162: 6322–6330 [PubMed] [Google Scholar]
- Malkiel S, Liao L, Cunningham MW, Diamond B 2000. T-Cell-dependent antibody response to the dominant epitope of streptococcal polysaccharide, N-acetyl-glucosamine, is cross-reactive with cardiac myosin. Infect Immun 68: 5803–5808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D, Benoist C 2009. AIRE. Annu Rev Immunol 27: 287–312 [DOI] [PubMed] [Google Scholar]
- McNulty NP, Yatsunenko T, Hsiao A, Faith JJ, Muegge BD, Goodman AL, Henrissat B, Oozeer R, Cools-Portier S, Gobert G, et al. 2011. The impact of a consortium of fermented milk strains on the gut microbiome of gnotobiotic mice and monozygotic twins. Sci Transl Med 3: 106ra106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Schneider DS, Soares MP 2012. Disease tolerance as a defense strategy. Science 335: 936–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H, Wen L, Abiru N, Liu E, Yu L, Miao D, Gianani R, Wong FS, Eisenbarth GS 2002. Induction and acceleration of insulitis/diabetes in mice with a viral mimic (polyinosinic-polycytidylic acid) and an insulin self-peptide. Proc Natl Acad Sci 99: 5539–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Möröy T 2000. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet 25: 177–181 [DOI] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336: 489–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson P, Pitkanen J, Sillanpaa N, Krohn K 2004. Autoimmune polyendocrinopathy candidiasis ectodermal dystrophy (APECED): A model disease to study molecular aspects of endocrine autoimmunity. Clin Exp Immunol 135: 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polk BF, Kasper DL 1977. Bacteroides fragilis subspecies in clinical isolates. Ann Intern Med 86: 569–571 [DOI] [PubMed] [Google Scholar]
- Pozzilli P, Signore A, Williams AJ, Beales PE 1993. NOD mouse colonies around the world—Recent facts and figures. Immunol Today 14: 193–196 [DOI] [PubMed] [Google Scholar]
- Rahat M, Dimentman C 1982. Cultivation of bacteria-free Hydra viridis: Missing budding factor in nonsymbiotic hydra. Science 216: 67–68 [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118: 229–241 [DOI] [PubMed] [Google Scholar]
- Rashid T, Ebringer A 2012. Autoimmunity in rheumatic diseases is induced by microbial infections via crossreactivity or molecular mimicry. Autoimmune Dis 2012: 539282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reháková Z, Capková J, Stěpánková R, Sinkora J, Louzecká A, Ivanyi P, Weinreich S 2000. Germ-free mice do not develop ankylosing enthesopathy, a spontaneous joint disease. Hum Immunol 61: 555–558 [DOI] [PubMed] [Google Scholar]
- Rossini AA, Williams RM, Mordes JP, Appel MC, Like AA 1979. Spontaneous diabetes in the gnotobiotic BB/W rat. Diabetes 28: 1031–1032 [DOI] [PubMed] [Google Scholar]
- Serreze DV, Hamaguchi K, Leiter EH 1989. Immunostimulation circumvents diabetes in NOD/Lt mice. J Autoimmun 2: 759–776 [DOI] [PubMed] [Google Scholar]
- Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334: 670–674 [DOI] [PubMed] [Google Scholar]
- Sinkorova Z, Capkova J, Niederlova J, Stepankova R, Sinkora J 2008. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2(k)) male mice. Hum Immunol 69: 845–850 [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Hooper LV, Gordon JI 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci 99: 15451–15455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Hardt WD 2008. The role of microbiota in infectious disease. Trends Microbiol 16: 107–114 [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana NK, Kasper DL 2012. The yin yang of bacterial polysaccharides: Lessons learned from B. fragilis PSA. Immunol Rev 245: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JL 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444: 1027–1031 [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. 2008. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 455: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, Mathis D 2010. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32: 815–827 [DOI] [PMC free article] [PubMed] [Google Scholar]