Abstract
The mammalian heart is the first functional organ, the first indicator of life. Its normal formation and function are essential for fetal life. Defects in heart formation lead to congenital heart defects, underscoring the finesse with which the heart is assembled. Understanding the regulatory networks controlling heart development have led to significant insights into its lineage origins and morphogenesis and illuminated important aspects of mammalian embryology, while providing insights into human congenital heart disease. The mammalian heart has very little regenerative potential, and thus, any damage to the heart is life threatening and permanent. Knowledge of the developing heart is important for effective strategies of cardiac regeneration, providing new hope for future treatments for heart disease. Although we still have an incomplete picture of the mechanisms controlling development of the mammalian heart, our current knowledge has important implications for embryology and better understanding of human heart disease.
The heart is the first organ to function in an embryo. Heart development has been well studied, and this knowledge will be critical to develop strategies for cardiac regeneration.
1. INTRODUCTION
The heart is the first organ to function in the embryo, and this function is essential for survival during fetal life. The beating heart is the most readily detectable sign of life. In humans, defects in heart formation occur in 1%–2% of the population, resulting in a broad range of congenital heart defects. Heart development is a finely tuned complicated process that requires precise differentiation and assembly of many cell types. Heart development has been well studied, and the pathways that coordinately regulate cardiac differentiation and morphogenesis are well defined (Fig. 1). The study of mouse heart development has been particularly important for understanding congenital heart disease and for developing strategies for cardiac regeneration.
Figure 1.
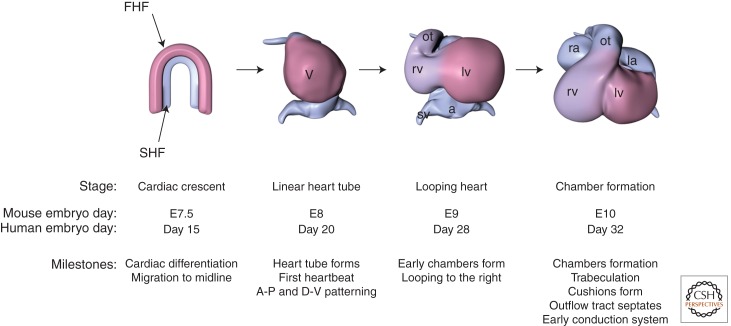
Major steps in heart development. Diagrams of developing hearts (ventral views) are shown. At the cardiac crescent stage, the two heart fields represent different cardiac precursors. The first heart field (FHF, pink) contributes to the left ventricle (lv), and the second heart field (SHF, blue) contributes to the right ventricle (rv) and later to the outflow tract (ot), sinus venosus (sv), and left and right atria (la and ra, respectively). Days of development for mouse and human are shown below. (From Bruneau 2008, with permission.)
In the early heart, cells expressing cardiac markers coalesce at the midline of the embryos to form a linear heart tube that soon begins to contract. The linear heart tube expands and begins a series of contortions. This cardiac looping shifts the heart tube toward the right of the embryo, the first overt sign of breaking left–right symmetry. Major morphogenetic events form the rudimentary chambers of the heart: the two atria and two ventricles. Septa form between the pairs of chambers to isolate each chamber, and valves soon connect the atria to the ventricles. The heart matures during the later stages of fetal life and is fully functional at birth. Morphogenesis of the mouse heart resembles that of the human, and thus has been critical for understanding human congenital heart disease.
2. CARDIAC PROGENITORS AND EARLY DIFFERENTIATION
The cells that make up the heart originate from mesoderm emerging from the primitive streak. The earliest known committed cardiac precursors express the T-box transcription factor Eomesodermin (Eomes) (Costello et al. 2011). Eomes-expressing cells contribute quite specifically to the cardiovascular system, including the heart and vasculature. Eomes is important for activating Mesp1, another transcription factor whose expression was considered to be one of the earliest markers of cardiac precursors. Identification of Mesp1 as a marker of early cardiac mesoderm was based on Cre-loxP-based genetic tracing that suggested that Mesp1-expressing cells contribute predominantly to the heart (Saga et al. 1999). In fact, the Mesp1 lineage is much broader, encompassing almost all of the trunk mesenchyme, the limbs, and the vasculature (Yoshida et al. 2008). Notably, accurate lineage contributions based on genetic tracing approaches depend on a critical combination of the fidelity of the Cre-expressing transgene and the homogeneous expression of the reporter transgene. In Mesp1- and Eomes Cre lines, a knockin approach is likely to ensure fidelity, but the reporter strain can greatly affect interpretation of the lineage contribution. In differentiated embryonic stem (ES) cells, Mesp1 is also active during early stages of cardiac differentiation, but as in vivo, Mesp1 expression predicts many mesodermal lineages (Bondue et al. 2008; David et al. 2008; Lindsley et al. 2008). Therefore, Mesp1 expression characterizes cardiac progenitors, among multiple cell types.
Classic embryological manipulations pinpointed the origins of the heart in the gastrula. Using cell transplantation and labeling of live embryos, followed by embryo culture, precursors of the heart were identified as occupying anterior mesoderm, proximal to the embryonic–extraembryonic boundary (Tam et al. 1997; Kinder et al. 1999). An important insight from these experiments was that the populations contributing to the heart are plastic, in that cells taken from a different location or developmental time point could contribute to the heart when placed in the appropriate location. This plasticity points toward the cellular environment as more important than the initial identity of the cells. These landmark experiments broadly located the cardiac precursors, but did not identify them specifically. Therefore, the cells fated specifically to the heart have not yet been identified and characterized.
Hints about the molecular identifying marks labeling cardiac precursors have been gleaned from embryos and cultured ES cells. From the molecular cues that drive cardiac differentiation in vivo, a strategy for differentiating ES cells into cardiac myocytes was devised (Kattman et al. 2006; Moretti et al. 2006; Yang et al. 2008; Kattman et al. 2011). This led to identification of Flk1 and PDGFRα as cell-surface markers that label a population of mesoderm destined to become cardiomyocytes. Flk1 was identified in these studies, but further mouse genetic labeling experiments showed Flk1+ derivatives contribute to many other mesodermal derivatives (Ema et al. 2006). PDGFRα was identified in Nkx2-5-expressing cardiac precursors (Prall et al. 2007) and provides added specificity with Flk1, to identify and isolate cardiac progenitors (Kattman et al. 2011). However, these do not mark cardiac precursors exclusively, but instead allow enrichment of cardiac progenitors.
2.1. Allocation of Progenitors
Cardiac progenitors originating from mesoderm are rapidly allocated to two major populations, referred to as heart fields (Buckingham et al. 2005; Vincent and Buckingham 2010). The first heart field (FHF) is thought to contribute to the left ventricle and parts of the atria. Adjacent to the FHF, the second heart field (SHF) contributes predominantly to the arterial pole of the heart (e.g., outflow tract and right ventricle) and also to the venous pole (e.g., sinus venosus and atria). Unlike the FHF, the SHF actively contributes cardiac precursors in early organogenesis, whereas the FHF is more rapidly incorporated into the differentiating heart. The SHF can be identified by the expression of Isl1, although its expression is much broader than just the SHF. Isl1 was associated with the SHF from Cre-mediated genetic tracing. Although Isl1 was not expressed to any significant degree in the differentiating heart, the descendants of the Isl1-expressing cells populated large segments of the heart (Cai et al. 2003). Isl1 has since been used as a marker of the SHF, but as with any genetic label, it must be interpreted carefully. Isl1 is active in cardiac neural crest (Engleka et al. 2012), which complicates analyses of mouse deletion phenotypes that use Isl1::Cre to manipulate genes. Other markers, such as Fgf10 and a specific enhancer of the Mef2c gene, also mark a portion of the SHF, a more anterior domain referred to as the anterior heart field, which gives rise to the outflow tract and right ventricle (Kelly et al. 2001; Zaffran et al. 2004; Verzi et al. 2005).
A similar conclusion was reached, with some notable added insight, by a retrospective lineage tracing approach. Meilhac and colleagues used a genetic labeling approach that relies on the random activation of a marker, localized to the nucleus lacZ, which has an insertion that disrupts its function (Meilhac et al. 2003, 2004a). Rare recombination events activate the gene in a few cells, permanently labeling the cell lineage. The modified reporter gene was inserted at a gene expressed in all cardiac myocytes during development, the α cardiac actin (Actc1) gene. During embryonic development random recombination events activate the transgene, but its expression is restricted to cardiac myocytes, thus neatly labeling clones of cells in the developing heart. This strategy was very useful to retrospectively analyze hundreds of mouse embryos, and derive conclusions based on the distribution of clones. The main conclusion was that two main cardiac progenitor populations exist. One arose very early and had common progenitors for all regions of the heart except the outflow tract, and one segregated later to contribute to the outflow tract, right ventricle, and atria, but not the left ventricle. These results are comparable to those obtained from genetic tracing experiments, except for one key distinction: prediction of an early common cardiac progenitor. This retrospective approach does not allow identification or isolation of the distinct progenitors.
The SHF has been well studied using the genetic tools available. In addition to the broad contributions to the outflow tract and right ventricle, cells labeled by the Mef2cAHF::Cre transgene are also found in a region called the dorsal mesenchymal protrusion, or DMP (Hoffmann et al. 2009). The DMP invades the posterior wall of the atria, and DMP-derived cells contribute to the primary atrial septum, which participates in separating the left and right atria from one another.
The SHF also contributes to the posterior, venous pole of the heart. Here, expression of SHF markers overlaps with other cardiac marker genes, and it is not clear if these are true progenitors (Galli et al. 2008). The cell lineage allocation is more complex: Sinus venosus progenitors are a separate population, marked by the absence of Nkx2-5 (and perhaps Isl1), and instead by the unique presence of Tbx18 (Christoffels et al. 2006; Mommersteeg et al. 2010). Thus, three distinct cardiac progenitor populations are evident, each with a distinct signature and contribution to the developing heart. The allocation of cardiac lineages is likely to be more complex, as more specific markers are identified.
2.2. Signals that Induce the Heart Progenitors
Cardiac differentiation is induced by signaling cues from adjacent tissues. In early mesoderm formation, graded levels of the TGFB-family member Nodal are important for specifying different types of mesoderm. Higher levels of Nodal favor cardiac mesoderm (Brennan et al. 2001). After specification of cardiac mesoderm, bone morphogenic protein (BMP) and Wnt signals are modulated in the early stages of cardiac differentiation. Wnt signaling initially promotes cardiogenesis, but later is inhibitory as progenitors begin to differentiate into various cardiac derivatives (Naito et al. 2006; Kwon et al. 2007). Wnt/β-catenin-induced expansion of cardiac precursors requires Isl1 down-regulation, which promotes cardiac differentiation (Kwon et al. 2009). BMP function is not clear in the earliest stages of cardiac commitment, although inhibiting BMP signaling seems important to promote the emergence of cardiac mesoderm (Yuasa et al. 2005). Later BMPs seem to be important for expansion of cardiac progenitors, as genetic deletion of the BMP receptor in Mesp1-expressing mesoderm results in major defects in heart formation (Klaus et al. 2007). In cardiac progenitors, BMPs maintain the balance between precursor expansion and differentiation (Prall et al. 2007). Notch signaling is also important in cardiac precursors: Deleting Notch1 in the SHF leads to a greater number of cardiac precursors, in part by increasing the activity of Wnt signaling and thus promoting expansion of cardiac progenitors (Kwon et al. 2009).
2.3. Transcriptional Regulation of Cardiac Differentiation
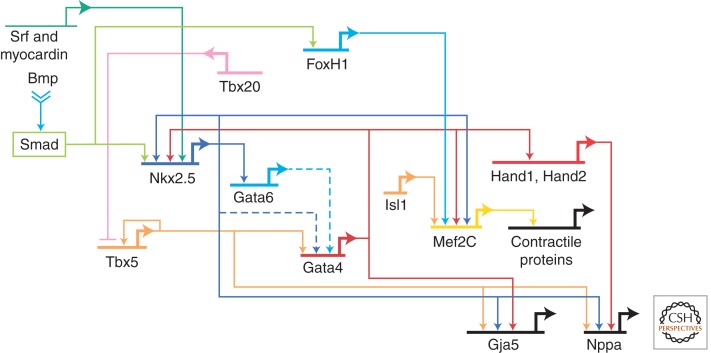
Precise regulation of tissue-specific gene transcription is essential for the correct differentiation and patterning of the heart. Transcription factors regulate aspects of heart development, including terminal differentiation, identity of cardiac chambers, establishment of patterning boundaries, and formation of transcriptional gradients (Olson 2006; Srivastava 2006). For example, the homeodomain transcription factor Nkx2-5 is critical for terminal differentiation of the myocardium; without Nkx2-5, the heart does not progress beyond a primitive arrangement of chamber primordia (Lyons et al. 1995). Paradoxically, Nkx2-5 is required for restricting the numbers of progenitors, by restricting the expression of BMPs, and thus maintaining the balance between progenitor proliferation and differentiation (Prall et al. 2007). The T-box transcription factor Tbx5 is required for the growth and differentiation of the posterior segment of the heart, the atrial and left ventricular precursors (Bruneau et al. 2001b). The GATA factors GATA4 and GATA6 redundantly regulate the onset of cardiac differentiation (Zhao et al. 2008). Similarly, serum response factor (SRF) modulates expression of multiple cardiac genes (Niu et al. 2008). Other transcription factors (e.g., bHLH Hand proteins) control aspects of chamber differentiation (Srivastava et al. 1997; Firulli et al. 1998). Several of these transcription factors functionally interact to regulate downstream gene expression. Although networks of interacting transcription factors are being defined, the full complement of their target genes is not clear, nor is how these factors are integrated at the level of chromatin to activate transcriptional programs. In chromatin localization studies of Nkx2-5, Gata4, Tbx5, and Mef2a in culture, cardiac transcription factors often bind together at genes that are coregulated during heart development, suggesting a high degree of cooperativity between cardiac transcription factors (Fig. 2) (He et al. 2011).
Figure 2.
Transcriptional networks in heart development. A representative transcriptional network is shown. (Adapted from Davidson and Erwin 2006.)
In addition to cooperative activation of gene expression, several cardiac transcription factors function to sculpt cardiac gene expression during patterning of the early developing heart. As an example, Tbx5 and Tbx2 function antagonistically in the atrioventricular (AV) canal. Tbx5 is expressed throughout the atrial and left ventricular myocardium, including the intervening AV canal. In the AV canal, a related T-box transcription factor, Tbx2, is expressed. In chamber myocardium, Tbx5 activates chamber-specific genes, such as Nppa, in cooperation with Nkx2-5. In the AV canal, Tbx2 competes for Tbx5 binding to target DNA elements, and to Nkx2-5, and represses the same genes (Habets et al. 2002). This antagonistic activity readily patterns the developing heart.
Heterozygous mutations of several transcription factor genes that are important in early heart development cause human congenital heart disease (CHD) (Bruneau 2008). In Holt-Oram syndrome, TBX5 mutations cause atrial septal defects (ASDs), ventricular septal defects (VSDs), and conduction system disease (Mori and Bruneau 2004). Mutations in NKX2-5 cause similar defects and others, such as tetralogy of Fallot or Ebstein’s anomaly of the tricuspid valve (Schott et al. 1998; Benson et al. 1999). GATA4 mutations result in a subset of defects caused by TBX5 and NKX2-5 mutations, ASDs and VSDs (Garg et al. 2003). Deficiency in another T-box gene, TBX1, is at the root of 22q11.2 microdeletion syndrome (DiGeorge syndrome); this finding came from advanced mouse genome engineering, which sought to recapitulate human chromosomal microdeletions (Lindsay et al. 2001; Merscher et al. 2001). Two very important insights came from these studies. First, developmentally important transcription factors are critically important in the etiology of CHD. Second, the dosage of these factors must be precisely regulated, as most disease-causing mutations appear to result in haploinsufficiency. This latter finding hints at mechanisms of disease owing to decreased dosage. Furthermore, most of these disease-related transcription factors interact with each other, and their interactions are stoichiometrically regulated. In crosses between Tbx5, Nkx2-5, and Gata4 mutant mice, these factors genetically interacted, and compound heterozygosity of any two genes leads to far more severe CHDs than haploinsufficiency of single genes (Moskowitz et al. 2007; Maitra et al. 2008).
2.4. Epigenetic Control of Cardiac Differentiation
Transcription factors function within the regulatory confines of chromatin, the dense arrangement of DNA around the histone octamers that forms nucleosomes (Kouzarides 2007; Li et al. 2007). With its formidable packaging, chromatin provides an important level of gene regulation, via the packing density and chemical modification of unstructured histone “tails.” Regulation of the developing heart greatly relies on epigenetic regulation by chromatin remodeling and histone modifications (Bruneau 2010; Chang and Bruneau 2012). Mapping epigenetic regulation during cardiac differentiation on a global scale revealed complex but specific temporal patterns of histone modifications coordinately regulate cardiac genes (Wamstad et al. 2012). Therefore, complex coordination of epigenetic regulators on sets of cardiac genes is likely an essential component of cardiac differentiation, with important implications for heart development.
In the heart, chromatin remodelers have been uncovered as interacting with DNA-binding transcription factors to modify their target gene expression. For example, Baf60c (Lickert et al. 2004) is one isoform of three subunits that are part of the Brg1/Brm-associated factor (BAF) complexes. These important chromatin-remodeling complexes shift nucleosomes side to side to regulate access to regulatory DNA. Baf60c is expressed very early in precardiac mesoderm at late gastrulation and prenodal plate stages and later in the heart, somites, and the developing central nervous system. RNAi-mediated knockdown of Baf60c in mouse embryos yields malformed hearts and aberrant gene expression. Baf60c functions as a bridge, creating or reinforcing molecular interactions between cardiac transcription factors (GATA4, Nkx2-5, and Tbx5) and Brg1, thus presumably bringing the BAF complex to target genes bound by GATA4, Nkx2-5, or Tbx5 (Lickert et al. 2004). Baf60c is one of a trio of required cardiogenic transcription factors that include GATA4 and Tbx5, which activate the entire cardiac program de novo in noncardiac mesoderm (Takeuchi and Bruneau 2009). Baf60c allows initial binding of GATA4 to target loci, indicating that it is essential for the cardiogenic activity of GATA4 and Tbx5.
Insights into the importance of BAF complexes come from deletion of the main BAF complex ATPase, Brg1. Deleting Brg1 in the developing endocardium showed that BAF complexes can be required for very specific functions in establishing endocardial–myocardial signaling: Loss of endocardial Brg1 derepresses a major gene involved in controlling the matrix between the endocardium and myocardium, disrupting cardiac morphogenesis (Stankunas et al. 2008). This finding illustrates the fine degree of regulation by BAF complexes, and shows that they can be just as important as repressors of gene expression, in addition to their activation function. Deleting Brg1 in the differentiating myocardium more broadly deregulates gene expression, leading to defects in chamber morphogenesis (Hang et al. 2010; Takeuchi et al. 2011). Interesting from the perspective of interactions with disease-causing transcription factors, mice heterozygous for Brg1 deletion have significant CHDs, indicating dosage dependency of BAF complexes in the heart (Takeuchi et al. 2011). Brg1 haploinsufficiency is considerably worsened by combined haploinsufficiency of Tbx5, Nkx2-5, or Tbx20 (Takeuchi et al. 2011). This indicates a strong genetic interaction between DNA-binding factors and BAF complexes, which predicts a tightly regulated stoichiometric relationship between transcription factors and chromatin-remodeling complexes. These data suggest transcription factor haploinsufficiency in CHD involves impaired recruitment of BAF complexes to target genes. This important insight highlights the key interplay between DNA-binding factors and chromatin-remodeling complexes.
The potential role of histone acetylation in cardiac development is highlighted largely by work performed on the HATs p300 and CBP and their interactions with cardiac transcriptional regulators. Mice lacking p300 have multiple defects in embryogenesis, including cardiac defects such as reduced ventricular trabeculation and impaired expression of cardiac genes (Yao et al. 1998). p300 interacts with cardiac transcription factors, such as MEF2D and GATA factors (Kakita et al. 1999; Dai and Markham 2001; Slepak et al. 2001; Kawamura et al. 2005). Thus, p300-mediated histone acetylation is likely to be a widespread mechanism for coactivating cardiac genes. Tbx5 interacts with TAZ, a WW-domain-containing transcriptional regulator, which recruits the HATs p300 and PCAF, thereby enhancing Tbx5-dependent trans-activation of the Nppa promoter (Murakami et al. 2005). This cooperativity is prevented by mutations in TBX5 in patients with Holt-Oram syndrome, in which CHDs are prevalent. Thus, defective Tbx5-mediated histone acetylation may contribute to the congenital heart defects associated with the syndrome. Tbx5 and other T-box transcription factors, as well as SRF, interact with Tip60, a MYST-family HAT, to potently activate target genes, including SRF itself (Kim et al. 2006).
Repressing transcription by epigenetic regulators is also important for normal heart development. Histone deacetylases (HDACs), which repress gene expression, have been largely characterized from mouse knockouts as important for adaptive gene regulation in the adult heart, but their roles in the developing heart are emerging. Loss of individual HDAC genes yields mice with little or no detectable embryonic cardiac defects, but mice lacking multiple HDAC genes die in utero from cardiac defects (McKinsey et al. 2001; Montgomery et al. 2007). Of potential significance to the developing heart, HDAC function is highly regulated by cellular signaling processes. Thus, in the developing heart, HDACs may have complex functions that depend on interactions with DNA-binding transcription factors and growth factor signaling pathways. Indeed, two transcription factors essential for heart development—Smyd1 (also known as mBop) and homeodomain-only protein (Hopx)—function in part by associating with HDACs (Gottlieb et al. 2002; Trivedi et al. 2010). SRF, which is important for activating several cardiac genes, interacts with HDACs through Hopx (Kook et al. 2003). The Hopx/HDAC interaction is also important for regulating the function of GATA4, by directly modulating its acetylation (Trivedi et al. 2010). Thus, through their associations with transcription factors, HDACs are important for heart development.
One of the best-studied repressive marks is the trimethylation of lysine 27 of histone H3 (H3K27me3). This mark is laid down by the polycomb repressive complex 2 (PRC2), which comprises three core subunits: Suz12, Eed, and the catalytic histone methyltransferase, Ezh2 (Surface et al. 2010). Ezh2 deletion in cardiac precursors leads to defective cardiac morphogenesis, including thinned myocardial walls and ventricular septation defects (He et al. 2012a). This is perhaps as a result of impaired proliferation, as a result of the derepression of negative regulators of the cell cycle. Interestingly, cardiac progenitor genes that would normally no longer be expressed in the heart are still on, indicating that PRC2 function is essential to shut off progenitor genes permanently once they are no longer needed in differentiating cardiomyocytes. A slightly different ablation of Ezh2, in a more restricted domain of cardiac progenitors, leads not to defects in cardiac morphogenesis, but instead to cardiac enlargement after birth (Delgado-Olguin et al. 2012). This results mainly from lack of repression of one transcription factor that functions in cardiac precursors, the homeodomain transcription factor encoded by Six1. When its expression persists, Six1 can still activate noncardiac genes, including genes normally restricted to skeletal muscle. Therefore, epigenetic repression of developmental regulators by PRC2 is an essential component of early cardiac development. Ezh2 may also function independently of histone methylation, as a direct methylator of GATA4; GATA4 methylation represses its transcription activation potential (He et al. 2012b).
2.5. Lessons Learned from Early Heart Development
Studying early heart development yielded many important lessons that are relevant to lineage specification and differentiation. Transcriptional regulation of cardiac differentiation underscored the importance of combinatorial factor interactions in robust regulation of tissue-specific gene expression. Furthermore, from modeling human CHD in the mouse, these interactions are dosage sensitive, indicating a very fine integration of transcriptional inputs (Bruneau et al. 2001b; Jerome and Papaioannou 2001; Lindsay et al. 2001; Merscher et al. 2001). Understanding how transcription factors and chromatin remodelers interface contributes to a model by which transcription factors promote or repress target gene expression in a specific cell type. The study of early gene expression and its regulators has also led to important insights into mechanisms of lineage determination and lineage allocation that are relevant to most mammalian cell types. Integrating these investigations will yield a comprehensive view of cell lineage determination and regulation of differentiation.
3. CARDIAC ORGANOGENESIS
Heart formation includes several important steps that precisely organize the various cellular subtypes into chambers and associated structures. Along the way, precise patterning of gene expression underlies many of the events that drive organogenesis (see Fig. 3 for examples).
Figure 3.
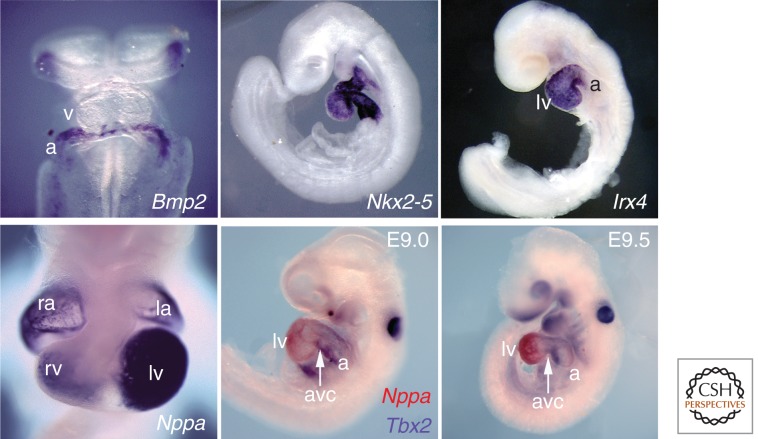
Patterning the developing heart. In situ hybridization in mouse embryos illustrates the complexity and precision of patterning. Shown are in situ hybridizations for Bmp2 (with expression in atrial precursors at E8.5), Nkx2-5 (expressed in all cardiac cells, at E9.25), Irx4 (expressed in ventricular [lv] but not atrial [a] cells at E9.25), Nppa (with a complex chamber expression pattern at E10.5), and the mutually exclusive pattern of Nppa and Tbx2 (Nppa in chamber myocardium and Tbx2 in AV canal [Venter et al. 2001]) at E9.25 and E9.5. a, atrium; avc, atrioventricular canal; lv, left ventricle; rv, right ventricle; v, ventricle. (Adapted from Bruneau 2003, 2011.)
3.1. Establishing the Heart Field
On differentiation, the bilateral regions of mesoderm that will form the heart migrate toward the midline to form a linear heart tube. Defective convergence of the two sides of the heart-forming region results in cardia bifida, in which cardiac differentiation proceeds independently in two separate areas, and a normal heart does not form. In striking cases, cardiac development proceeds some distance. Sometimes, the paired hearts form primitive chambers (Li et al. 2004). Although the cellular mechanisms required for bilateral migration and fusion are not well understood, several transcriptional regulators are known to be important for this process. Mesp1 is required early on, presumably to allow migration of cardiac precursors to their appropriate location (Saga et al. 1999). Gata4 is also essential for cardiac fusion (Kuo et al. 1997; Molkentin et al. 1997), although it is not clear whether this is a cell-autonomous requirement in early cardiomyocytes or a function in adjacent endoderm. The proprotein convertase furin also has essential functions in regulating embryonic ventral closure, and in its absence cardia bifida is a common phenotype (Roebroek et al. 1998; Constam and Robertson 2000). Furin may activate several TGF-β family members, including Nodal, Lefty, and BMPs. Perhaps related to Furin’s function, BMP2 secreted from visceral endoderm is required for embryonic patterning events, including normal migration of cardiac precursors (Madabhushi and Lacy 2011).
3.2. Chamber Formation
At the onset of heart looping, the chambers begin to form. The first evident chamber is the single ventricle that bulges out from the looping heart tube. As this chamber grows, a morphologically recognizable right ventricle begins to form, and the atria begin to grow into the laterally paired appendages visible behind the more evident ventricular chambers. One interesting hypothesis suggests how the ventricular and atrial chambers grow, whereas the rest of the heart tube remains more or less in its original tubular form. Based on patterns of gene expression, and later on examining patterns of cell proliferation, it was proposed that the chambers of the heart “balloon” out from the primitive heart tube (Christoffels et al. 2000; Moorman and Christoffels 2003). This ballooning model is attractive as it neatly explains what appeared to be an overly complex means of allocating cells to a particular structure. The model of directional growth leading to morphogenesis of structures of specific shape and cellular orientation is directly supported by the same retrospective lineage analyses that defined the early cardiac lineages: By examining smaller clones of labeled cells, the patterns of cell division that cells had followed could be understood during recent organogenesis (Meilhac et al. 2003; Meilhac et al. 2004b; Bajolle et al. 2006). For example, ventricular growth was observed as radial, explaining the large rounded shape of the primitive ventricles. Conversely, growth in the outflow tract is initially linear, corresponding to its elongation, and then is radial, which corresponds to the broadening and rotation of the outflow tract.
Lineage analyses confirmed that the chambers form as thought (Meilhac et al. 2003, 2004b), but allocating cells from the primitive heart tube to the forming chambers might be more complicated. For example, Tbx2-expressing cells, which are predominantly confined to the AV canal (Fig. 3), contribute many cells to the adjacent left ventricle (Aanhaanen et al. 2009).
Importantly, “primitive” segments of the heart are retained and result in regions of slowed conduction and minimal contraction. This is apparent during chamber morphogenesis as the AV canal forms. Patterning cues establish boundaries along the primitive heart tube, within which the AV canal forms. This region of the heart expresses a distinct complement of genes, especially those involved in conduction of impulses, such as connexins. The AV canal myocardium is important in coordinated impulse propagation: It forms the insulating conduction system tissue that directs the impulses that initiate in the atrium along a specific insulated path (Hoogaars et al. 2004; Bakker et al. 2010; Aanhaanen et al. 2011). Signals that initiate and maintain formation of the AV valves arise from the AV canal. The specialization of the conduction system and valve formation will be reviewed later in this section.
Little is understood about what regulates these growth processes, except that differential rates of proliferation may drive early chamber morphogenesis (Soufan et al. 2006; van den Berg et al. 2009; Aanhaanen et al. 2011). The Tbx2 transcription factor, which is restricted to the AV canal, is important for maintaining a lower level of proliferation in this segment of the heart (Aanhaanen et al. 2011).
3.3. Trabeculation
Formation of trabeculae, the fingerlike projections that protrude inside the developing chambers, is important in cardiac chamber morphogenesis. The trabeculae ensure the growth of the heart from a thin-walled organ to one that can pump against high pressures. A trabeculated heart can also exert force, while being easily oxygenated before development of coronary circulation. Trabeculae are highly conserved in evolution and, in more primitive hearts, such as amphibians and reptiles, are retained into adulthood. Several signaling pathways are deployed to ensure trabeculation, including neuregulin/ErbB, Notch, Ephrin/Eph, and Bmp10. Bmp10 is expressed within developing trabeculae and is essential for proliferation of trabecular myocardium (Chen et al. 2004). Nrg1, produced from the endocardium, signals to ErbB receptors in the myocardium to allow trabeculation (Gassmann et al. 1995; Meyer and Birchmeier 1995), in a pathway that appears independent of Bmp10 (Grego-Bessa et al. 2007). EphrinB2 and its receptor EphB4 are also important for trabeculation (Wang et al. 1998). Notch signaling in the endocardium adjacent to the trabeculae is also essential for trabeulation, and is implicated in many signaling pathways: Notch signaling sustains Bmp10 expression and promotes expression of Nrg1 and EphrinB2 in the endocardium (Grego-Bessa et al. 2007).
3.4. Chamber Septation
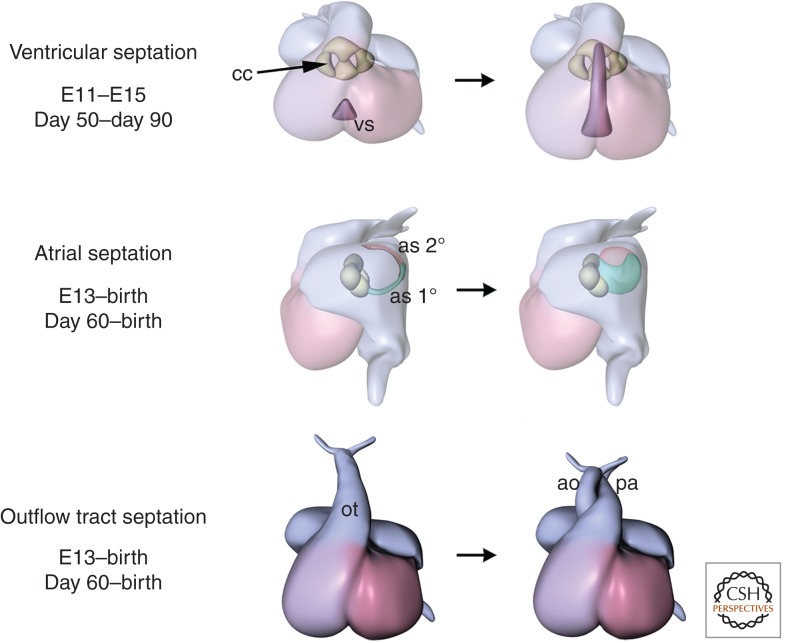
Chamber septation involves separation of the left and right sides of the atria and ventricles by growth of specific regions of myocardium (Fig. 4). Septation of the ventricles is simpler and occurs at the junction of the two heart fields. Ventricular septation defects are the second most common CHDs. However, the patterning events that dictate ventricular septation and the subsequent cellular changes that govern septal morphogenesis are not understood.
Figure 4.
Later steps in cardiac morphogenesis. Shown are ventricular septation, atrial septation, and septation of the outflow tracts. ao, aorta; as 1°, primum atrial septum; as 2°, secundum atrial septum, cc, cardiac cushions; ot, outflow tract; pa, pulmonary artery; vs, ventricular septum. Days of development for mouse and human are shown below. (From Bruneau 2008, with permission.)
Early embryological studies determined that the interventricular septum (IVS) is formed by outgrowth of two adjacent populations of cells in a small segment of the left and right ventricles. By genetic lineage analysis, the left and right components of the IVS do not mix during IVS outgrowth (Meilhac et al. 2004a; Franco et al. 2006). These observations suggest patterning of the developing ventricles that dictates the location of the IVS.
Several genes have been implicated in forming of the IVS, including Tbx5, Gata4, Nkx2-5, Sall4, and the Hand family of transcription factors. Although these genes are all important for forming the IVS, their loss results in pleiotropic effects on the heart as a whole or in defects in, but not a complete absence of, IVS formation. Therefore, the molecular determinants that pattern the location of the IVS and promote its formation are not well understood.
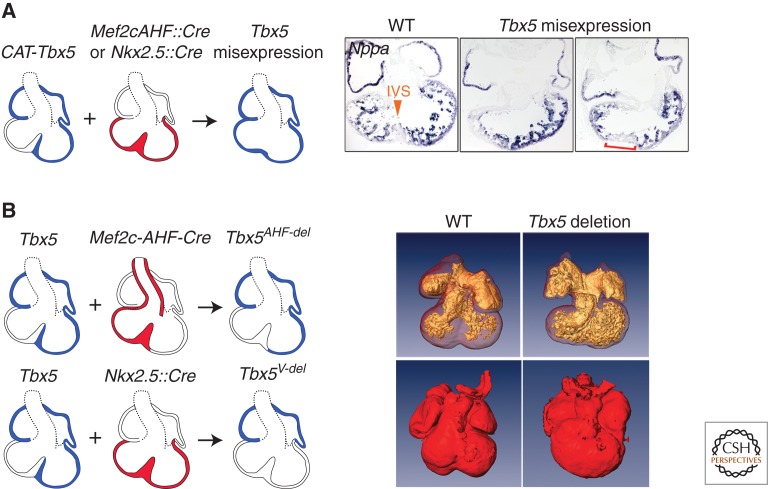
At least one gene might be directly related to patterning the myocardium guiding IVS formation. Tbx5 is expressed in a dynamic pattern that extends, in the ventricles, just up to the region where the IVS forms (Bruneau et al. 1999; Koshiba-Takeuchi et al. 2009). Genetic manipulation of Tbx5 in the chick and mouse suggested that the position of the IVS relative to the ventricles is determined by the boundary of cells that do and do not express Tbx5 (Fig. 5) (Takeuchi et al. 2003; Koshiba-Takeuchi et al. 2009). Misexpression of Tbx5 across the boundary between the left and right ventricles eliminates IVS formation (Koshiba-Takeuchi et al. 2009). In mice with Tbx5 deleted from ventricular myocardium or a segment of the ventricular myocardium that includes the cells that will contribute to the IVS, grossly normal hearts form, but fail to initiate IVS formation (Koshiba-Takeuchi et al. 2009). This suggests Tbx5 in the specific region of ventricular myocardium that contributes to the IVS is critical for IVS formation. Therefore, the patterning of Tbx5 expression is essential for IVS formation, but how this boundary functions to form an IVS is unknown.
Figure 5.
Function of a Tbx5 boundary in ventricular septation. (A) Effect of misexpression of Tbx5 on septum formation. Left: diagrammatic representation of the experimental design and resulting phenotype. Right: in situ hybridization for Nppa on sections from wild-type (WT) and Tbx5 misexpression hearts, showing absence of septum formation and expansion of expression of Nppa. (B) Phenotype resulting from deletion of Tbx5 from ventricular myocardium (V-del). Two distinct strategies are shown. Left: Optical projection tomography scans of heart from WT and Tbx5 deletion hearts. Note the lack of septation in the absence of Tbx5. (From Koshiba-Takeuchi et al. 2009, with permission.)
Tbx5 is also critical for atrial septation, via the integration of diverse cell signaling pathways. Its function in the endocardium is required for establishing signaling cascades that ensure septal formation (Nadeau et al. 2010). An earlier role in the origin of the atrial septum primum from the SHF implicates Tbx5 in regulating response of the posterior SHF to Hedgehog signaling, and provides a mechanism for the dosage-sensitive defects in atrial septation that result from Tbx5 haploinsufficiency (Xie et al. 2012). Deletion of one copy of Tbx5 in endocardium or myocardium does not reproduce the septation defects seen in Tbx5+/− mice, but deletion in the Hh-responsive cells of the SHF does, showing that Tbx5 is required very early on to establish atrial septum precursors. Tbx5 directly regulates Hh-pathway genes, and also may act in parallel to regulate Osr1 (Xie et al. 2012).
3.5. Cushions and Valves
As the heart septates, connections between the atria and ventricles are maintained, and valves ensure unidirectional blood flow. Valve formation arises from complex coordinated signaling between the myocardium and the overlying endocardium. Signals from the myocardium initiate local endothelial-to-mesenchymal transformation (EMT). EMT location depends on the initial patterning of the heart segments and thus the localized expression of EMT-inducing factors, such as Bmp2 (Ma et al. 2005). Bmp2 is also required for patterning the AV canal and outflow tract, in a feedback loop involving Tbx2, and for the expression of genes required for the establishment of the cardiac jelly, in which transformed endocardial cells migrate (Harrelson et al. 2004; Ma et al. 2005). Patterning of the AV canal also involves Notch signaling in the endocardium, which represses Bmp2 outside of its domain of expression (Luna-Zurita et al. 2010). TGFβ signaling is critical for initiating expression of the Snail-family transcription factors Snai1 and Snai2, which are essential for conferring invasive properties to cells undergoing EMT (Tao et al. 2011). Finally, a fourth pathway involving myocardial calcineurin/NFAT signaling represses VEGF expression in the valve-forming region of the AV canal (Chang et al. 2004). In the endocardium, Gata4 activity is required to activate EMT-inducing factors, including ErbB3 (Rivera-Feliciano et al. 2006). Notch and BMP are essential for endocardial EMT, with BMP signaling functioning upstream of Notch (Timmerman et al. 2004; Luna-Zurita et al. 2010).
Septation of the outflow tract (OFT) and establishment of the pulmonary and aortic valves follows a similar yet distinct set of signaling pathways as AV valve formation. For example, although VEGF signaling is required for complete EMT in the AV canal and OFT, this role is more important in the OFT than for AVC endocardial cushion formation (Stankunas et al. 2010). The OFT elongates from the SHF, where intricate signaling events are coordinated. Within the forming OFT, cells of the cardiac neural crest migrate in, establishing themselves as an important source of cells for the cushions of the distal outflow tract. The proximal cushions of the OFT are derived from the underlying endothelium. A fine interplay between signals secreted from the mesoderm of the SHF to the adjacent neural crest and endocardial cells is essential for their survival and differentiation (Park et al. 2006, 2008). Fgf8 signals, under control of the T-box transcription factor Tbx1 (Hu et al. 2004), are essential for an autocrine signaling loop that activates production of other growth factors, including BMPs, which subsequently signal to the adjacent neural crest and endothelial cells (Stottmann et al. 2004; Park et al. 2008). The role of Tbx1 in initiating the signaling is particularly interesting, as heterozygous loss of human TBX1 underlies 22q11 microdeletion syndrome or DiGeorge syndrome (Jerome and Papaioannou 2001; Lindsay et al. 2001; Merscher et al. 2001); the cardiovascular manifestations of this syndrome were thought to be owing to a defect in neural crest formation, but Tbx1 is also expressed in branchial arch and SHF mesoderm (Hu et al. 2004; Xu et al. 2004). Instead of a cell-autonomous function of Tbx1 within neural crest cells, it activates autocrine and paracrine signals that influence OFT development and cardiac morphogenesis.
After cushion formation is initiated in the OFT and AV canal, subsequent steps in valve formation involve elongation and maturation of the valve tissue (Hinton et al. 2006). Similar signaling is deployed. For example, calcineurin/NFAT signaling in the endocardium is deployed again, following its initial function in the myocardium (Chang et al. 2004). Although less well understood than the early stages of valvulogenesis, maturation of valves involves profound changes in gene expression and cellular identity, with the loss of early markers such as Twist1 and Tbx20, and the acquisition of a tendonlike phenotype, with expression of Sox9 and Scleraxis (Lincoln et al. 2007; Levay et al. 2008).
3.6. Cell-Type Specialization and Formation of the Conduction System
During heart formation, various cell types specialize. Myocardial cells, in particular, specialize into atrial, ventricular, pacemaker, AV node, and His-Purkinje cells, the latter three cell types being the major constituents of the cardiac conduction system. Patterning and intrinsic cellular programs are responsible for this diversity of cell-type specialization.
The factors leading to establishment of atrial and ventricular cells, corresponding to the left and right chambers, are unclear. The determinants of atrial versus ventricular identity are also unclear, although candidates have been proposed (Pereira et al. 1999; Xavier-Neto et al. 1999; Bruneau et al. 2000, 2001a). Diversification into left and right ventricular myocytes is likely owing to antero-posterior patterning of the heart tube and specific expression of transcription factors, such as Tbx5 and Hand1 (Bruneau et al. 1999; McFadden et al. 2004). The left and right atria are patterned based on left-right signaling cascades that initiate early on in establishing the embryonic body plan (Shiratori and Hamada 2006; Galli et al. 2008). In particular, left-sided expression of the Pitx2 transcription factor establishes left and right atrial identity, in part by suppressing in the left atrium the initiation of pacemaker tissue differentiation, which is critical for normal initiation and conduction of the heartbeat (Mommersteeg et al. 2007; Wang et al. 2010).
Within the atrial and ventricular tissues, cell-type specialization forms the fibers that conduct impulses coordinately for each heartbeat. The patterning of the sinoatrial node, where impulses initiate, is dictated by the localized expression of Tbx3, which partly represses a working myocardium phenotype, while promoting the expression of ion channels that are essential for the spontaneous depolarization that is a key feature of sinoatrial node pacemaker cells (Hoogaars et al. 2007; Frank et al. 2012). Tbx3 is also important for forming the atrioventricular conduction system, which includes the atrioventricular node, where impulses slow to ensure coordination between atrial and ventricular contraction. Tbx3 expression outlines the atrioventricular conduction system (Hoogaars et al. 2004), partly overlapping the expression of Tbx2 (Aanhaanen et al. 2009). Tbx3 is important for forming the atrioventricular node (Frank et al. 2012), but Tbx2 specifies the insulating tissues that ensure that impulses pass from the atria to the ventricles only via the atrioventricular node (Aanhaanen et al. 2011). Without this important electrical insulation, accessory pathways develop that lead to lethal arrhythmias (Aanhaanen et al. 2011). The distal conduction system, the fast conducting fibers of the His-Purkinje system, is patterned by the concerted dosage-sensitive function of Tbx5 and Nkx2-5, which act upstream of a third transcription factor, Id2 (Moskowitz et al. 2004, 2007). It is partly established by the function of the Irx3 transcription factor, which acts predominantly to maintain a balance between gap junction proteins to ensure electrical isolation of the fibers from the rest of the myocardium to ensure fast impulse propagation (Zhang et al. 2011). Finally, the patterning of cardiac repolarization coordinates the resetting of currents so that the next heartbeat can spread; this is accomplished by a transmural gradient of the Irx5 transcription factor, which establishes an inverse gradient of ion channels (Costantini et al. 2005).
4. HEART REPAIR AND MAINTENANCE
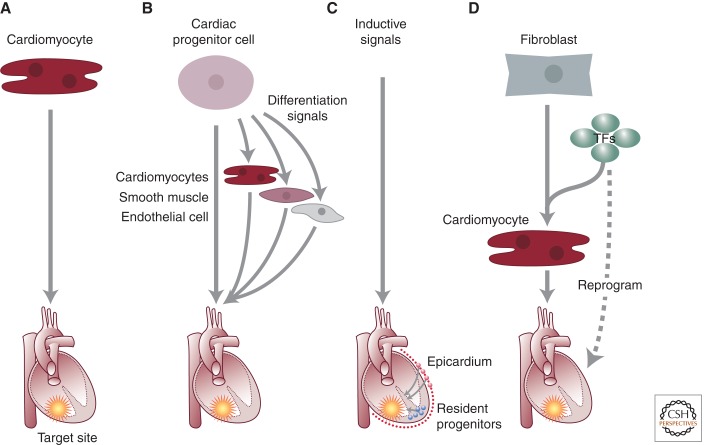
The mammalian heart cannot regenerate after injury. Because the hearts of other vertebrates, such as fish, regenerate (Poss et al. 2002; Poss 2010), mammalian hearts might have a hidden regenerative potential. Indeed, mammalian hearts have a short-lived capacity for regeneration, which disappears a few days after birth (Porrello et al. 2011). This temporary regenerative capacity seems related to proliferative potential of existing cardiomyocytes (Porrello et al. 2011), which exit the cell cycle almost permanently shortly thereafter, although other experimental approaches have led to suggestions of a role for c-kit+ progenitors in neonatal cardiac regeneration (Jesty et al. 2012). The knowledge acquired from understanding specification and allocation of cardiac lineages may provide strategies for cardiac regeneration (Fig. 6). Regeneration in zebrafish is based on proliferation of cardiomyocytes (Jopling et al. 2010; Kikuchi et al. 2010), and parallel mechanisms might exist between the adult zebrafish and the neonatal mammalian heart. The turnover of cardiac myocytes in the uninjured human heart has been calculated to be ∼50% over a lifetime, indicating very little if any ongoing proliferation (Bergmann et al. 2009) (although this estimate has been questioned and is unresolved [Anversa et al. 2012]). After injury, such as after a myocardial infarction, there also does not seem to be significant myocardial proliferation. Several approaches have been suggested to increase cardiac proliferation, and although some success has been reported (Kuhn et al. 2007; Bersell et al. 2009), the numbers of proliferating adult cardiomyocytes are far from those needed to restore an injured heart.
Figure 6.
Strategies for cardiac regeneration. Various strategies that have been suggested are shown, including (A) implantation of in vitro-generated cardiomyocytes, (B) differentiation and implantation of cardiac progenitors, (C) mobilization of endogenous precursors by inductive signals, or (D) direct reprogramming. (From Alexander and Bruneau 2010.)
4.1. Endogenous Stem Cells: Evidence for and against
Endogenous cardiac stem cells in the adult heart might be an excellent starting point for devising strategies to promote cardiac regeneration. Several studies claimed to isolate cardiac progenitors of various types from the mouse heart, using a variety of surface markers (Laflamme and Murry 2005, 2011; Leri et al. 2005). For the most part, cells with cardiogenic properties are difficult to identify in the endogenous heart, and thus their identity and lineage origin are not clear. When reintroduced into a damaged heart, these cells seldom become new cardiac myocytes. Thus, under these conditions, they do not possess cardiogenic properties. For example, c-kit-expressing cells were thought to be resident cardiac stem cells (Beltrami et al. 2003). In the neonatal heart, these cells can function as bona fide cardiac precursors (Jesty et al. 2012). However, their ability to contribute to regenerating adult heart was not confirmed (Tallini et al. 2009; Zaruba et al. 2010; Jesty et al. 2012). In vitro, when differentiated from ES cells, the c-kit+ population varies in its potential for cardiogenesis (Wu et al. 2006; Yang et al. 2008; Zaruba et al. 2010). Isl1-expressing progenitors have been detected postnatally, but their numbers are very small and all but gone in adulthood (Laugwitz et al. 2005, 2008). Finally, a cardiac mesenchymal stem cell-like population was identified in the perivascular niche of the adult mouse heart (Chong et al. 2011). These cells are distinct from c-kit+ cells and express PDGFRα; careful genetic tracing determined that they are not bone marrow derived, and are from a mesodermal lineage, likely originating from the epicardium. Whether these cells can participate in endogenous heart repair is not clear.
The epicardium has been suggested broadly as a source of cells that can regenerate myocardium after injury. Wilm’s tumor-1 (WT1)-expressing cells, carefully pulse-chase labeled using the Cre-loxP system, have been shown to contribute to some extent to producing new cardiomyocytes after injury and priming with Thymosin β4 (TB4) (Smart et al. 2011). In these experiments, following myocardial infarction and TB4 treatment, subsets of WT1+ cells expressed Isl1 and Nkx2-5, and lineage-marked cells contributed new cTnT+, functionally integrated cardiomyocytes. The number of new cardiomyocytes that appear to be epicardially derived is very small, but it is intriguing that a population of cells that would normally not contribute new myocardium could be coaxed into doing so under the right conditions.
The best evidence for endogenous renewal of cardiac myocytes and, by extrapolation, the existence of cardiac stem cells comes from a genetic experiment in the mouse. Cardiac myocytes were genetically labeled with cardiac-specific inducible Cre recombinase transgenes (Myh6::MerCreMer [Sohal et al. 2001]) during a short time, and the persistence of the label was followed over the lifetime of the mouse (Hsieh et al. 2007). If the number of labeled cells does not change, the originally labeled cells would still be present, and few or no new cardiomyocytes would have been generated. If the number of labeled cells decreases, new cells that did not receive the original genetic tag would have been generated. Although the origin of these cells cannot be ascertained, they would necessarily have arisen later than the early pulse of genetic recombination. During normal aging, there was little change in the number of labeled cells. However, after injuring the heart by imposing a myocardial infarction, the heart had fewer labeled cells. Thus, in response to injury, over time, cardiac myocytes were renewed from sources other than the genetically labeled cells.
4.2. Strategies for Enhancing Tissue Repair/Regeneration
The knowledge accumulated in studies of the developing heart has become useful in developing strategies to induce the formation of new cardiomyocytes (Fig. 6). Two distinct approaches based on pathways that regulate cardiac differentiation have been deployed to attempt to regenerate injured myocardium.
One approach has been to differentiate ES cells, or induced pluripotent stem (iPS) cells, into cardiomyocytes that could then be implanted into an injured heart. Based on the endogenous signaling cues that are important for inducing cardiac cells in vivo, protocols have been derived that achieve varying degrees of cardiac differentiation from ES and iPS cells (Laflamme et al. 2007; Yang et al. 2008; Kattman et al. 2011). When implanted in an injured mouse or rat heart, however, human ES cell-derived cardiomyocytes cannot achieve long-term contribution to the myocardium, or do not couple effectively (Laflamme et al. 2007; Laflamme and Murry 2011). The lack of long-term engraftment has been considered to be an important hindrance to any potential for use of pluripotent cell-derived cardiomyocytes. It may be that the use of small, rapidly beating rodent hearts is the main source of failure in these sets of experiments. Use of a guinea-pig infarct model has shown that, in fact, human ES cell-derived cardiomyocytes can very effectively incorporate into injured myocardium, couple to their endogenous neighbors, and survive long term (Shiba et al. 2012). Importantly, in this experimental setting, the implantation of in vitro-generated human cardiomyocytes does not lead to arrhythmogenic events, and in fact, is antiarryhthmic.
An attractive scenario in regenerative medicine is to induce one cell type to become another to replace lost or damaged cells. This is known as direct reprogramming (Graf and Enver 2009). In the heart, loss of cardiomyocytes after infarct is accompanied by an overproliferation of fibroblasts, which contribute to formation of scar tissue that impairs the proper contraction of the heart. A desirable strategy would be to convert the excess of fibroblasts into functional cardiomyocytes, which would both replenish the heart with functional units and reduce the amount of scar tissue. Recent studies have shown great promise (Ieda et al. 2010; Qian et al. 2012; Song et al. 2012). In cell culture, introducing three transcription factors (Gata4, Mef2c, and Tbx5 [GMT]), or these three factors plus Hand2 (GHMT), induced cardiac fibroblasts to radically change phenotypes to resemble cardiomyocytes (Ieda et al. 2010; Song et al. 2012). These factors induced fibroblasts to express a full battery of cardiac genes, form sarcomeres, develop cardiomyocyte-like electrical activity, and in a few cases even elicit beating activity. This approach also worked in dermal fibroblasts, indicating that the potential of the introduced factors was not limited to only one cell type. Thus, a limited set of transcription factors could impose an entire cardiac program on a noncardiac cell. This approach has also been extended to human ES cells, to determine if initial success with driving mesoderm to become cardiomyocytes (Takeuchi and Bruneau 2009) could be applied to pluripotent cell in culture. The combination of GATA4, TBX5, NKX2-5, and BAF60c was indeed effective in inducing cardiomyocytes from human ES cells, but at low efficiency (Dixon et al. 2011).
Would a directed differentiation approach be effective in the endogenous heart? This has turned out to be promising (Qian et al. 2012; Song et al. 2012). Injection of viruses overexpressing GMT or GHMT into a mouse heart immediately after a myocardial infarction led to expression of the added factors only in the highly proliferating fibroblasts as ascertained by Cre-loxP-based cell marking. Overexpressing GMT or GHMT in vivo led to generation of new functional cardiomyocytes. A large percentage (∼25%) of infected fibroblasts changed their phenotype to become remarkably similar to endogenous cardiomyocytes: They had defined sarcomeres, were rod shaped and binucleated, expressed gap junction proteins and connected to their neighbors, and contracted. Therefore, these factors converted endogenous fibroblasts into functional cardiomyocytes. The increased efficiency in vivo contrasts with the initial foray in cell culture, and predicts that the environment in vivo is more favorable for directed differentiation than culture conditions. The de novo production of cardiomyocytes significantly affected the heart. Cardiac function was partially restored and infarct scar size was greatly reduced. The reduction in scar size cannot be accounted for solely by the production of new myocytes, so there must be some paracrine effects on fibroblast proliferation that contributes as well. This study suggests functional myocardium can be reliably regenerated in vivo by direct reprogramming of endogenous cardiac fibroblasts (Qian et al. 2012; Song et al. 2012). Combined with additional factors, specific types of cardiomyocytes might be generated, such as biological pacemakers, by the inclusion of Tbx3 (Bakker et al. 2012).
4.3. Outlook to the Future: Epigenetic Reprogramming?
The outlook for cardiac regeneration is encouraging. Nevertheless, significant challenges remain, and it is not clear if strategies that work in a small heart, such as mouse, will work in the much larger hearts of humans. Furthermore, safe delivery of factors that repair the heart will be needed. Direct reprogramming is an exciting and promising approach; lessons learned from reprogramming of somatic cells to induced pluripotent cells may be instructive to develop small-molecule-based strategies of cardiac reprogramming (Xu et al. 2008; Lin et al. 2009; Yamanaka and Blau 2010; Zhu et al. 2010; Robinton and Daley 2012). Reprogramming strategies are likely to benefit from epigenetic manipulation of the genome, to broadly affect or enhance direct conversion of one cell type to another. Significant knowledge of the epigenetic blueprint of the heart will be necessary for this approach to become practical.
5. CONCLUSIONS
The developing heart is a paradigm of organogenesis, and its study has yielded important insights into progenitor and stem cell biology, lineage commitment, and morphogenesis. It illustrates how little we still understand about the formation of this complex and important organ. The study of heart development has also provided key insights into the basis of CHD, which is still prevalent and devastating for the children suffering from them, and their families. Finally, the knowledge obtained from deciphering the cues that form the heart will be critical to developing strategies for cardiac regeneration.
ACKNOWLEDGMENTS
I thank Gary Howard for editorial assistance. Work in my laboratory is supported by grants from the NHLBI, CIRM, and AHA.
Footnotes
Editors: Patrick P.L. Tam, W. James Nelson, and Janet Rossant
Additional Perspectives on Mammalian Development available at www.cshperspectives.org
REFERENCES
- Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, et al. 2009. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res 104: 1267–1274 [DOI] [PubMed] [Google Scholar]
- Aanhaanen WT, Boukens BJ, Sizarov A, Wakker V, de Gier-de Vries C, van Ginneken AC, Moorman AF, Coronel R, Christoffels VM 2011. Defective Tbx2-dependent patterning of the atrioventricular canal myocardium causes accessory pathway formation in mice. J Clin Invest 121: 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JM, Bruneau BG 2010. Lessons for cardiac regeneration and repair through development. Trends Mol Med 16: 426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anversa P, Leri A, Kajstura J 2012. Biased DNA segregation during stem cell division. Circ Res 110: 1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajolle F, Zaffran S, Kelly RG, Hadchouel J, Bonnet D, Brown NA, Buckingham ME 2006. Rotation of the myocardial wall of the outflow tract is implicated in the normal positioning of the great arteries. Circ Res 98: 421–428 [DOI] [PubMed] [Google Scholar]
- Bakker ML, Moorman AF, Christoffels VM 2010. The atrioventricular node: Origin, development, and genetic program. Trends Cardiovasc Med 20: 164–171 [DOI] [PubMed] [Google Scholar]
- Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, et al. 2012. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res 94: 439–449 [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, et al. 2003. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114: 763–776 [DOI] [PubMed] [Google Scholar]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, et al. 1999. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest 104: 1567–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, et al. 2009. Evidence for cardiomyocyte renewal in humans. Science 324: 98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersell K, Arab S, Haring B, Kuhn B 2009. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell 138: 257–270 [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C 2008. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell 3: 69–84 [DOI] [PubMed] [Google Scholar]
- Brennan J, Lu CC, Norris DP, Rodriguez TA, Beddington RS, Robertson EJ 2001. Nodal signalling in the epiblast patterns the early mouse embryo. Nature 411: 965–969 [DOI] [PubMed] [Google Scholar]
- Bruneau BG 2003. The developing heart and congenital heart defects: A make or break situation. Clin Genet 63: 252–261 [DOI] [PubMed] [Google Scholar]
- Bruneau BG 2008. The developmental genetics of congenital heart disease. Nature 451: 943–948 [DOI] [PubMed] [Google Scholar]
- Bruneau BG 2010. Chromatin remodeling in heart development. Curr Opin Genet Dev 20: 505–511 [DOI] [PubMed] [Google Scholar]
- Bruneau BG 2011. Atrial natriuretic factor in the developing heart: A signpost for cardiac morphogenesis. Can J Physiol Pharmacol 89: 533–537 [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Logan M, Davis N, Levi T, Tabin CJ, Seidman JG, Seidman CE 1999. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol 211: 100–108 [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Tanaka M, Schott JJ, Izumo S, Cepko CL, Seidman JG, Seidman CE 2000. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev Biol 217: 266–277 [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Fatkin D, Xavier-Neto J, Georgakopoulos D, Maguire CT, Berul CI, Kass DA, Kuroski-de Bold ML, de Bold AJ, et al. 2001a. Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol Cell Biol 21: 1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner D, Gessler M, Nemer M, Seidman CE, et al. 2001b. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106: 709–721 [DOI] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, Zaffran S 2005. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 6: 826–835 [DOI] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S 2003. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell 5: 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CP, Bruneau BG 2012. Epigenetics and cardiovascular development. Annu Rev Physiol 74: 41–68 [DOI] [PubMed] [Google Scholar]
- Chang CP, Neilson JR, Bayle JH, Gestwicki JE, Kuo A, Stankunas K, Graef IA, Crabtree GR 2004. A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118: 649–663 [DOI] [PubMed] [Google Scholar]
- Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, et al. 2004. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development 131: 2219–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JJ, Chandrakanthan V, Xaymardan M, Asli NS, Li J, Ahmed I, Heffernan C, Menon MK, Scarlett CJ, Rashidianfar A, et al. 2011. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 9: 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, et al. 2000. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol 223: 266–278 [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Mommersteeg MT, Trowe MO, Prall OW, de Gier-de Vries C, Soufan AT, Bussen M, Schuster-Gossler K, Harvey RP, Moorman AF, et al. 2006. Formation of the venous pole of the heart from an Nkx2-5-negative precursor population requires Tbx18. Circ Res 98: 1555–1563 [DOI] [PubMed] [Google Scholar]
- Constam DB, Robertson EJ 2000. Tissue-specific requirements for the proprotein convertase furin/SPC1 during embryonic turning and heart looping. Development 127: 245–254 [DOI] [PubMed] [Google Scholar]
- Costantini D, Arruda EP, Agarwal P, Kim K-H, Zhu Y, Zhu W, Lebel M, Cheng C, Park CY, Pierce SA, et al. 2005. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell 123: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, Arnold SJ 2011. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol 13: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YS, Markham BE 2001. p300 functions as a coactivator of transcription factor gata-4. J Biol Chem 276: 37178–37185 [DOI] [PubMed] [Google Scholar]
- David R, Brenner C, Stieber J, Schwarz F, Brunner S, Vollmer M, Mentele E, Muller-Hocker J, Kitajima S, Lickert H, et al. 2008. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1-mediated blockade of Wnt-signalling. Nat Cell Biol 10: 338–345 [DOI] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH 2006. Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800 [DOI] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou DC, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG 2012. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet 44: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JE, Dick E, Rajamohan D, Shakesheff KM, Denning C 2011. Directed differentiation of human embryonic stem cells to interrogate the cardiac gene regulatory network. Mol Ther 19: 1695–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M, Takahashi S, Rossant J 2006. Deletion of the selection cassette, but not cis-acting elements, in targeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal progenitors. Blood 107: 111–117 [DOI] [PubMed] [Google Scholar]
- Engleka KA, Manderfield LJ, Brust RD, Li L, Cohen A, Dymecki SM, Epstein JA 2012. Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res 7: 922–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firulli AB, McFadden DG, Lin Q, Srivastava D, Olson EN 1998. Heart and extra-embryonic mesodermal defects in mouse embryos lacking the bHLH transcription factor Hand1. Nat Genet 18: 266–270 [DOI] [PubMed] [Google Scholar]
- Franco D, Meilhac SM, Christoffels VM, Kispert A, Buckingham M, Kelly RG 2006. Left and right ventricular contributions to the formation of the interventricular septum in the mouse heart. Dev Biol 2: 366–375 [DOI] [PubMed] [Google Scholar]
- Frank DU, Carter KL, Thomas KR, Burr RM, Bakker ML, Coetzee WA, Tristani-Firouzi M, Bamshad MJ, Christoffels VM, Moon AM 2012. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc Natl Acad Sci 109: E154–E163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli D, Dominguez JN, Zaffran S, Munk A, Brown NA, Buckingham ME 2008. Atrial myocardium derives from the posterior region of the second heart field, which acquires left-right identity as Pitx2c is expressed. Development 135: 1157–1167 [DOI] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. 2003. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424: 443–447 [DOI] [PubMed] [Google Scholar]
- Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G 1995. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378: 390–394 [DOI] [PubMed] [Google Scholar]
- Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, et al. 2002. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet 31: 25–32 [DOI] [PubMed] [Google Scholar]
- Graf T, Enver T 2009. Forcing cells to change lineages. Nature 462: 587–594 [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, et al. 2007. Notch signaling is essential for ventricular chamber development. Dev Cell 12: 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM 2002. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: Implications for cardiac chamber formation. Genes Dev 16: 1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, Cheng H-L, Shang C, Ashley E, Zhou B, Chang C-P 2010. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466: 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE 2004. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 131: 5041–5052 [DOI] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT 2011. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci 108: 5632–5637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, et al. 2012a. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res 110: 406–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT 2012b. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev 26: 37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton RB Jr, Lincoln J, Deutsch GH, Osinska H, Manning PB, Benson DW, Yutzey KE 2006. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res 98: 1431–1438 [DOI] [PubMed] [Google Scholar]
- Hoffmann AD, Peterson MA, Friedland-Little JM, Anderson SA, Moskowitz IP 2009. Sonic hedgehog is required in pulmonary endoderm for atrial septation. Development 136: 1761–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Tessari A, Moorman AF, De Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM 2004. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res 62: 489–499 [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, et al. 2007. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev 21: 1098–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT 2007. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 13: 970–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T, Yamagishi H, Maeda J, McAnally J, Yamagishi C, Srivastava D 2004. Tbx1 regulates fibroblast growth factors in the anterior heart field through a reinforcing autoregulatory loop involving forkhead transcription factors. Development 131: 5491–5502 [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D 2010. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerome LA, Papaioannou VE 2001. Di George syndrome phenotype in mice mutant for the T-box gene. Tbx1 Nat Genet 27: 286–291 [DOI] [PubMed] [Google Scholar]
- Jesty SA, Steffey MA, Lee FK, Breitbach M, Hesse M, Reining S, Lee JC, Doran RMAYN, Fleischmann BK, et al. 2012. c-kit+ precursors support postinfarcation myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci 109: 13380–13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC 2010. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464: 606–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakita T, Hasegawa K, Morimoto T, Kaburagi S, Wada H, Sasayama S 1999. p300 protein as a coactivator of GATA-5 in the transcription of cardiac-restricted atrial natriuretic factor gene. J Biol Chem 274: 34096–34102 [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM 2006. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell 11: 723–732 [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G 2011. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8: 228–240 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Ono K, Morimoto T, Wada H, Hirai M, Hidaka K, Morisaki T, Heike T, Nakahata T, Kita T, et al. 2005. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. J Biol Chem 280: 19682–19688 [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME 2001. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell 1: 435–440 [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD 2010. Primary contribution to zebrafish heart regeneration by gata4+ cardiomyocytes. Nature 464: 601–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Merlo X, Wilson C, Lough J 2006. Co-activation of atrial natriuretic factor promoter by Tip60 and serum response factor. J Biol Chem 281: 15082–15089 [DOI] [PubMed] [Google Scholar]
- Kinder SJ, Tsang TE, Quinlan GA, Hadjantonakis AK, Nagy A, Tam PP 1999. The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development 126: 4691–4701 [DOI] [PubMed] [Google Scholar]
- Klaus A, Saga Y, Taketo MM, Tzahor E, Birchmeier W 2007. Distinct roles of Wnt/β-catenin and Bmp signaling during early cardiogenesis. Proc Natl Acad Sci 104: 18531–18536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kook H, Lepore JJ, Gitler AD, Lu MM, Wing-Man Yung W, Mackay J, Zhou R, Ferrari V, Gruber P, Epstein JA 2003. Cardiac hypertrophy and histone deacetylase-dependent transcriptional repression mediated by the atypical homeodomain protein Hop. J Clin Invest 112: 863–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba-Takeuchi K, Mori AD, Kaynak B, Cebra-Thomas J, Sukonnik T, Georges RO, Latham S, Beck L, Henkelman RM, Black BL, et al. 2009. Reptilian heart development and the molecular basis of cardiac chamber evolution. Nature 461: 95–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, Keating MT 2007. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med 13: 962–969 [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev 11: 1048–1060 [DOI] [PubMed] [Google Scholar]
- Kwon C, Arnold J, Hsiao EC, Taketo MM, Conklin BR, Srivastava D 2007. Canonical Wnt signaling is a positive regulator of mammalian cardiac progenitors. Proc Natl Acad Sci 104: 10894–10899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D 2009. A regulatory pathway involving Notch1/β-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol 11: 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE 2005. Regenerating the heart. Nat Biotechnol 23: 845–856 [DOI] [PubMed] [Google Scholar]
- Laflamme MA, Murry CE 2011. Heart regeneration. Nature 473: 326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, et al. 2007. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol 25: 1015–1024 [DOI] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, et al. 2005. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433: 647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugwitz KL, Moretti A, Caron L, Nakano A, Chien KR 2008. Islet1 cardiovascular progenitors: A single source for heart lineages? Development 135: 193–205 [DOI] [PubMed] [Google Scholar]
- Leri A, Kajstura J, Anversa P 2005. Cardiac stem cells and mechanisms of myocardial regeneration. Physiol Rev 85: 1373–1416 [DOI] [PubMed] [Google Scholar]
- Levay AK, Peacock JD, Lu Y, Koch M, Hinton RB Jr, Kadler KE, Lincoln J 2008. Scleraxis is required for cell lineage differentiation and extracellular matrix remodeling during murine heart valve formation in vivo. Circ Res 103: 948–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zhou D, Lu MM, Morrisey EE 2004. Advanced cardiac morphogenesis does not require heart tube fusion. Science 305: 1619–1622 [DOI] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432: 107–112 [DOI] [PubMed] [Google Scholar]
- Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, et al. 2009. A chemical platform for improved induction of human iPSCs. Nat Meth 6: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln J, Kist R, Scherer G, Yutzey KE 2007. Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev Biol 305: 120–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, et al. 2001. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 410: 97–101 [DOI] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Murphy TL, Langer EM, Cai M, Mashayekhi M, Wang W, Niwa N, Nerbonne JM, Kyba M, et al. 2008. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell 3: 55–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL 2010. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest 120: 3493–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP 1995. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev 9: 1654–1666 [DOI] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF 2005. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 132: 5601–5611 [DOI] [PubMed] [Google Scholar]
- Madabhushi M, Lacy E 2011. Anterior visceral endoderm directs ventral morphogenesis and placement of head and heart via BMP2 expression. Dev Cell 21: 907–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, Garg V 2008. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol 326: 368–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN 2004. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development 132: 189–201 [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN 2001. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev 11: 497–504 [DOI] [PubMed] [Google Scholar]
- Meilhac SM, Kelly RG, Rocancourt D, Eloy-Trinquet S, Nicolas JF, Buckingham ME 2003. A retrospective clonal analysis of the myocardium reveals two phases of clonal growth in the developing mouse heart. Development 130: 3877–3889 [DOI] [PubMed] [Google Scholar]
- Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME 2004a. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell 6: 685–698 [DOI] [PubMed] [Google Scholar]
- Meilhac SM, Esner M, Kerszberg M, Moss JE, Buckingham ME 2004b. Oriented clonal cell growth in the developing mouse myocardium underlies cardiac morphogenesis. J Cell Biol 164: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merscher S, Funke B, Epstein JA, Heyer J, Puech A, Lu MM, Xavier RJ, Demay MB, Russell RG, Factor S, et al. 2001. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104: 619–629 [DOI] [PubMed] [Google Scholar]
- Meyer D, Birchmeier C 1995. Multiple essential functions of neuregulin in development. Nature 378: 386–390 [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 11: 1061–1072 [DOI] [PubMed] [Google Scholar]
- Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, et al. 2007. Molecular pathway for the localized formation of the sinoatrial node. Circ Res 100: 354–362 [DOI] [PubMed] [Google Scholar]
- Mommersteeg MT, Dominguez JN, Wiese C, Norden J, de Gier-de Vries C, Burch JB, Kispert A, Brown NA, Moorman AF, Christoffels VM 2010. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc Res 87: 92–101 [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, Hill JA, Richardson JA, Olson EN 2007. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 21: 1790–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman AF, Christoffels VM 2003. Cardiac chamber formation: Development, genes, and evolution. Physiol Rev 83: 1223–1267 [DOI] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. 2006. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127: 1151–1165 [DOI] [PubMed] [Google Scholar]
- Mori AD, Bruneau BG 2004. TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol 19: 211–215 [DOI] [PubMed] [Google Scholar]
- Moskowitz IPG, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, Roden D, Berul CI, Seidman CE, Seidman JG 2004. The T-Box transcription factor TBX5 is required for the patterning and maturation of the murine cardiac conduction system. Development 131: 4107–4116 [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Kim JB, Moore ML, Wolf CM, Peterson MA, Shendure J, Nobrega MA, Yokota Y, Berul C, Izumo S, et al. 2007. A molecular pathway including id2, tbx5, and nkx2-5 required for cardiac conduction system development. Cell 129: 1365–1376 [DOI] [PubMed] [Google Scholar]
- Murakami M, Nakagawa M, Olson EN, Nakagawa O 2005. A WW domain protein TAZ is a critical coactivator for TBX5, a transcription factor implicated in Holt-Oram syndrome. Proc Natl Acad Sci 102: 18034–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau M, Georges RO, Laforest B, Yamak A, Lefebvre C, Beauregard J, Paradis P, Bruneau BG, Andelfinger G, Nemer M 2010. An endocardial pathway involving Tbx5, Gata4, and Nos3 required for atrial septum formation. Proc Natl Acad Sci 107: 19356–19361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I 2006. Developmental stage-specific biphasic roles of Wnt/β-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci 103: 19812–19817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z, Iyer D, Conway SJ, Martin JF, Ivey K, Srivastava D, Nordheim A, Schwartz RJ 2008. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc Natl Acad Sci 105: 17824–17829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN 2006. Gene regulatory networks in the evolution and development of the heart. Science 313: 1922–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM 2006. Required, tissue-specific roles for Fgf8 in outflow tract formation and remodeling. Development 133: 2419–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM 2008. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development 135: 3599–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira FA, Qiu Y, Zhou G, Tsai MJ, Tsai SY 1999. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev 13: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA 2011. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD 2010. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat Rev Genet 11: 710–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT 2002. Heart regeneration in zebrafish. Science 298: 2188–2190 [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Sollaway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, et al. 2007. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls second heart field progenitor specification and proliferation. Cell 128: 947–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D 2012. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485: 593–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Feliciano J, Lee KH, Kong SW, Rajagopal S, Ma Q, Springer Z, Izumo S, Tabin CJ, Pu WT 2006. Development of heart valves requires Gata4 expression in endothelial-derived cells. Development 133: 3607–3618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinton DA, Daley GQ 2012. The promise of induced pluripotent stem cells in research and therapy. Nature 481: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebroek AJ, Umans L, Pauli IG, Robertson EJ, van Leuven F, Van de Ven WJ, Constam DB 1998. Failure of ventral closure and axial rotation in embryos lacking the proprotein convertase Furin. Development 125: 4863–4876 [DOI] [PubMed] [Google Scholar]
- Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T 1999. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126: 3437–3447 [DOI] [PubMed] [Google Scholar]
- Schott J-J, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron B, Seidman CE, Seidman JG 1998. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281: 108–111 [DOI] [PubMed] [Google Scholar]
- Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H, et al. 2012. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 489: 322–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H, Hamada H 2006. The left-right axis in the mouse: From origin to morphology. Development 133: 2095–2104 [DOI] [PubMed] [Google Scholar]
- Slepak TI, Webster KA, Zang J, Prentice H, O’Dowd A, Hicks MN, Bishopric NH 2001. Control of cardiac-specific transcription by p300 through myocyte enhancer factor-2D. J Biol Chem 276: 7575–7585 [DOI] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, et al. 2011. De novo cardiomyocytes from within the activated adult heart after injury. Nature 474: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD 2001. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible cre protein. Circ Res 89: 20–25 [DOI] [PubMed] [Google Scholar]
- Song K, Nam Y, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, et al. 2012. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485: 599–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufan AT, van den Berg G, Ruijter JM, de Boer PA, van den Hoff MJ, Moorman AF 2006. Regionalized sequence of myocardial cell growth and proliferation characterizes early chamber formation. Circ Res 99: 545–552 [DOI] [PubMed] [Google Scholar]
- Srivastava D 2006. Making or breaking the heart: From lineage determination to morphogenesis. Cell 126: 1037–1048 [DOI] [PubMed] [Google Scholar]
- Srivastava D, Thomas T, Lin Q, Kirby ML, Brown D, Olson EN 1997. Regulation of cardiac mesodermal and neural crest development by the bHLH transcription factor, dHAND. Nat Genet 16: 154–160 [DOI] [PubMed] [Google Scholar]
- Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, et al. 2008. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell 14: 298–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K, Ma GK, Kuhnert FJ, Kuo CJ, Chang CP 2010. VEGF signaling has distinct spatiotemporal roles during heart valve development. Dev Biol 347: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Choi M, Mishina Y, Meyers EN, Klingensmith J 2004. BMP receptor IA is required in mammalian neural crest cells for development of the cardiac outflow tract and ventricular myocardium. Development 131: 2205–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA 2010. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell 7: 288–298 [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459: 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Ohgi M, Koshiba-Takeuchi K, Shiratori H, Sakaki I, Ogura K, Saijoh Y, Ogura T 2003. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development 130: 5953–5964 [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguin P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, et al. 2011. Chromatin remodeling complex dosage modulates transcription factor function in heart development. Nat Commun 2: 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallini YN, Greene KS, Craven M, Spealman A, Breitbach M, Smith J, Fisher PJ, Steffey M, Hesse M, Doran RM, et al. 2009. c-kit expression identifies cardiovascular precursors in the neonatal heart. Proc Natl Acad Sci 106: 1808–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, Parameswaran M, Kinder SJ, Weinberger RP 1997. The allocation of epiblast cells to the embryonic heart and other mesodermal lineages: The role of ingression and tissue movement during gastrulation. Development 124: 1631–1642 [DOI] [PubMed] [Google Scholar]
- Tao G, Levay AK, Gridley T, Lincoln J 2011. Mmp15 is a direct target of Snai1 during endothelial to mesenchymal transformation and endocardial cushion development. Dev Biol 359: 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, et al. 2004. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 18: 99–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA 2010. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell 19: 450–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg G, Abu-Issa R, de Boer BA, Hutson MR, de Boer PA, Soufan AT, Ruijter JM, Kirby ML, van den Hoff MJ, Moorman AF 2009. A caudal proliferating growth center contributes to both poles of the forming heart tube. Circ Res 104: 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. 2001. The sequence of the human genome. Science 291: 1304–1351 [DOI] [PubMed] [Google Scholar]
- Verzi MP, McCulley DJ, De Val S, Dodou E, Black BL 2005. The right ventricle, outflow tract, and ventricular septum comprise a restricted expression domain within the secondary/anterior heart field. Dev Biol 287: 437–449 [DOI] [PubMed] [Google Scholar]
- Vincent SD, Buckingham ME 2010. How to make a heart: The origin and regulation of cardiac progenitor cells. Curr Top Dev Biol 90: 1–41 [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151: 206–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ 1998. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 93: 741–753 [DOI] [PubMed] [Google Scholar]
- Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF 2010. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci 107: 9753–9758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH 2006. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 127: 1137–1150 [DOI] [PubMed] [Google Scholar]
- Xavier-Neto J, Neville CM, Shapiro MD, Houghton L, Wang GF, Nikovits W, Stockdale FE, Rosenthal N 1999. A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development 126: 2677–2687 [DOI] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, Friedland-Little JM, Zhang K, Moskowitz IP 2012. Tbx5-Hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell 23: 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Morishima M, Wylie JN, Schwartz RJ, Bruneau BG, Lindsay EA, Baldini A 2004. Tbx1 has a dual role in the morphogenesis of the cardiac outflow tract. Development 3217–3227 [DOI] [PubMed] [Google Scholar]
- Xu Y, Shi Y, Ding S 2008. A chemical approach to stem-cell biology and regenerative medicine. Nature 453: 338–344 [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM 2010. Nuclear reprogramming to a pluripotent state by three approaches. Nature 465: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, et al. 2008. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453: 524–528 [DOI] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch’ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator 300. Cell 93: 361–372 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Vivatbutsiri P, Morriss-Kay G, Saga Y, Iseki S 2008. Cell lineage in mammalian craniofacial mesenchyme. Mech Dev 125: 797–808 [DOI] [PubMed] [Google Scholar]
- Yuasa S, Itabashi Y, Koshimizu U, Tanaka T, Sugimura K, Kinoshita M, Hattori F, Fukami S, Shimazaki T, Ogawa S, et al. 2005. Transient inhibition of BMP signaling by Noggin induces cardiomyocyte differentiation of mouse embryonic stem cells. Nat Biotechnol 23: 607–611 [DOI] [PubMed] [Google Scholar]
- Zaffran S, Kelly RG, Meilhac SM, Buckingham ME, Brown NA 2004. Right ventricular myocardium derives from the anterior heart field. Circ Res 95: 261–268 [DOI] [PubMed] [Google Scholar]
- Zaruba MM, Soonpaa M, Reuter S, Field LJ 2010. Cardiomyogenic potential of C-kit+-expressing cells derived from neonatal and adult mouse hearts. Circulation 121: 1992–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SS, Kim KH, Rosen A, Smyth JW, Sakuma R, Delgado-Olguin P, Davis M, Chi NC, Puviindran V, Gaborit N, et al. 2011. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His-Purkinje network. Proc Natl Acad Sci 108: 13576–13581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Watt AJ, Battle MA, Li J, Bondow BJ, Duncan SA 2008. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol 317: 614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S 2010. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7: 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]